Abstract
To combat antimicrobial resistance (AMR), policymakers need an overview of evolution and trends of AMR in relevant animal reservoirs, and livestock is monitored by susceptibility testing of sentinel organisms such as commensal E. coli. Such monitoring data are often vast and complex and generates a need for outcome indicators that summarize AMR for multiple antimicrobial classes. Model‐based clustering is a data‐driven approach that can help to objectively summarize AMR in animal reservoirs. In this study, a model‐based cluster analysis was carried out on a dataset of minimum inhibitory concentrations (MIC), recoded to binary variables, for 10 antimicrobials of commensal E. coli isolates (N = 12,986) derived from four animal species (broilers, pigs, veal calves and dairy cows) in Dutch AMR monitoring, 2007–2018. This analysis revealed four clusters in commensal E. coli in livestock containing 201 unique resistance combinations. The prevalence of these combinations and clusters differs between animal species. Our results indicate that to monitor different animal populations, more than one indicator for multidrug resistance seems necessary. We show how these clusters summarize multidrug resistance and have potential as monitoring outcome indicators to benchmark and prioritize AMR problems in livestock.
Keywords: antimicrobial drug resistance, cluster analysis, E. coli, epidemiological monitoring, multidrug resistance, policy making
Impacts.
This study delivers clusters that differentiate levels of multidrug resistance and show differences in antimicrobial resistance (AMR) between livestock reservoirs. Risk managers can use these clusters as outcome indicators of AMR monitoring in livestock for public health hazards, to assess the need for or effects of interventions
It is the first study with a data‐driven approach to summarize resistance phenotypes over antimicrobial classes in bacterial isolates from animals
This analysis resulted in a deeper understanding in the patterns in co‐occurrence of resistance to more than one antimicrobial for commensal E. coli isolates from animals
1. INTRODUCTION
Antimicrobial resistance (AMR) threatens the core of modern medicine, since effective antimicrobials are prerequisites not only for curing infectious diseases, but are also part of routine surgery procedures to prevent bacterial infections (World Health Organization, 2015). Action plans have been developed by international healthcare organizations and governments to restrict AMR in both human and veterinary medicine and avoid possible transmission of AMR from animal or environmental reservoirs to healthcare settings (World Health Organization, 2015; O'Neill, 2016). One crucial aspect of these action plans is monitoring of AMR in relevant reservoirs (O'Neill, 2016).
In food animals in the EU, AMR monitoring is performed following EU legislation (European Commission, 2013), by annually performing standardized antimicrobial susceptibility testing of a fixed number of indicator organisms like commensal Escherichia coli, and food borne pathogens such as Salmonella and Campylobacter species from animals and food thereof. Such monitoring programmes result in complex data, which are only interpretable by experts. However, policymakers need a clear overview of development of AMR in reservoirs to further develop, adjust and validate implemented policies timely. Policy informing organs like European Food Safety Authority (EFSA) have expressed the need for outcome indicators of AMR in tested microorganisms such as commensal E. coli (European Food Safety Authority, 2017). These outcome indicators should summarize AMR for multiple antimicrobial classes in a bacterial population, to allow an overall assessment of AMR in samples from relevant reservoirs, such as food animals.
Candidates for such AMR indicators have been tested on datasets of commensal indicator E. coli from food animals by EFSA, European Centre for Disease Prevention and Control (ECDC) and European Medicines Agency (EMA) (European Food Safety Authority, 2017). Suggestions were for example the pan‐susceptible proportion and multidrug resistance proportion (resistant to three or more antimicrobial classes) (European Food Safety Authority, 2017). In this joint scientific opinion, it was concluded that the pan‐susceptible proportion is the most suitable summary indicator, because a high negative correlation was found with overall AMU. Resistance to three or more antimicrobials was suggested as secondary outcome indicator when very few isolates in an animal population were fully susceptible, and the proportion of fluoroquinolone‐resistant isolates and the prevalence of ESBL‐producing E. coli were suggested as other secondary indicators. However, as mentioned in that joint scientific opinion, the pan‐susceptible proportion or resistance to three or more antimicrobials may not be specific enough to adjust AMU policy, especially for sector‐specific measures (European Food Safety Authority, 2017). Other suggestions in literature have been to weigh antimicrobial classes, ranked by their relevance for public health as performed by a panel of experts in Havelaar et al.(2017), or to weigh observed AMR for antimicrobial classes by antimicrobial use (AMU) of that same class in an animal population; examples are Dorado‐Garcia et al. (2016), Laxminarayan and Klugman (2011). The downside of applying weights to such calculations is that subjective choices have to be made, which may bias the results.
Currently, objective arguments for suitable AMR monitoring outcome indicators are lacking. Few studies have succeeded in reducing complexity of AMR monitoring data over antimicrobial classes in understandable output parameters. To develop specific and applicable outcome indicators of AMR monitoring, we performed a model‐based cluster analysis on a dataset of minimum inhibitory concentrations (MIC) for 10 antimicrobials of commensal E. coli isolates derived from four animal species (broilers, pigs, veal calves and dairy cows) in Dutch AMR monitoring, 2007–2018. Here, we show how model‐based clustering can be used as a data‐driven method to summarize resistance patterns, resulting in four clusters that have potential as monitoring outcome indicators to follow AMR trends and effects of AMU (‐interventions) in livestock.
2. METHODS
The data used for this analysis were MIC of 12,986 bacterial isolates, all being randomly isolated commensal indicator E. coli isolates from faecal or caecal samples of livestock as prescribed by EU legislation(3): 3,602 from broiler chickens, 2,958 from dairy cows, 3,491 from slaughter pigs and 2,935 from veal calves. All isolates were collected in the Dutch national monitoring programme for AMR in livestock, from 2007 to 2018. Details of data collection and antimicrobial susceptibility testing in this monitoring programme were described extensively by Hesp et al. (2019).
We used as definition for antimicrobial resistance: non‐wild type susceptibility, based on epidemiological cut‐off (ECOFF) values as defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (European Committee on Antimicrobial Susceptibility Testing, 2019). The MIC of all 12,986 isolates were recoded to binary variables (0 for susceptible, 1 for resistant) using the EUCAST ECOFF values (in parenthesis), for ten different antibiotics: gentamicin (GEN, 2 mg/L), ceftazidime (TAZ, 0.5 mg/L), cefotaxime (FOT, 0.25 mg/L), chloramphenicol (CHL, 16 mg/L), trimethoprim (TMP, 2 mg/L), sulfamethoxazole (SMX, 64 mg/L), ampicillin (AMP, 8 mg/L), tetracycline (TET, 8 mg/L), nalidixic acid (NAL, 16 mg/L) and ciprofloxacin (CIP, 0.064 mg/L). These antimicrobials were included in the analysis because they were continuously tested in the susceptibility panel from 2007 to 2018. They represent the following antimicrobial classes: aminoglycosides (GEN); 3rd generation cephalosporins (FOT/TAZ); amphenicols (CHL); folate pathway inhibitors (TMP/SUL); aminopenicillins (AMP); tetracyclines (TET); and (fluoro) quinolones (NAL/CIP).
After recoding the MIC to binary variables, the data were explored with multivariate analyses, using the 12,986 isolates as (statistical) units with their resistance for the 10 antimicrobials as binary outcome variables (accordingly, 10 variables). Dimension reduction techniques were explored to describe the multidrug resistance patterns. Principal component analysis and multiple correspondence analysis were considered but rejected, because these methods summarize pairwise correlations and associations (i.e., joint resistance patterns) only, thus largely neglecting multiple resistance of higher order, and the interpretation of their output is complicated. Instead, we chose for model‐based cluster analysis (Vermunt, 2002) also known as latent class analysis, which derives clusters from data based on a statistical model, without the need to choose heuristically a similarity coefficient as in hierarchical clustering. For binary variables, the model has four key assumptions: (a) each unit belongs to one of K clusters (although the posterior membership is a probability, i.e., fuzzy), (b) the resistance probability of each outcome variable (probability that the outcome is 1) depends on the cluster, (c) for each cluster, the joint probability of the outcome variables is the product of the individual resistance probabilities (i.e., the outcome variables within a cluster are independent) and (d) the overall joint distribution is a mixture of the joint probabilities of the clusters with mixing proportions equal to the relative cluster sizes. The Flexmix package (Leisch, 2004) in R (R Core Team, 2017) was used to fit this model for K = 10, with 1,000 random restarts in the stepFlexmix function. The most likely number of clusters was chosen by the model based on the lowest information criterion, specifically the integrated completed likelihood criterion (ICL), because ICL gives more parsimonious solutions than Akaike's information criterion (AIC) and Bayesian information criterion (BIC) (Biernacki et al., 2000). Finally, isolates are assigned to the cluster on the basis of maximum posterior membership probability according to the model described in Appendix S1. Other resistant isolates than in this dataset can be predicted similarly (Appendix S1). The assumption that the outcome variables are independent within clusters may appear counterintuitive, because resistances from the same antimicrobial class are likely to co‐occur. However, such associations are used constructively in the model by placing isolates with such co‐occurrences in a separate cluster with high probabilities of occurrence for the antimicrobials of that specific class. The high probabilities ensure that there is high co‐occurrence, even under independence (as products of high probabilities are still high). Co‐occurrence is thus not contradictory with independence within clusters. Co‐occurrence is modelled simply by high probabilities of occurrence in a cluster. Note that if the outcome variables would be independent in the full data set, the method would not subdivide the isolates in clusters (the result would be one cluster). In other words, model‐based cluster analysis uses the associations between the outcome variables (i.e., the antimicrobials) to subdivide the isolates into clusters. The composition of the clusters was further investigated, by analysing the occurrence of combinations of resistance phenotypes within the clusters and how they differed between animal species. The clusters from this analysis were compared with the outcome indicators suggested by ECDC, EFSA and EMA to show how the clusters relate to those indicators, and to investigate the potential of model‐based clustering to quantitatively summarize monitoring outcomes.
3. RESULTS
Model‐based clustering showed that four clusters best described the data (illustration in Figure S1). The composition of the four clusters, that is the mean probability of resistance per antimicrobial per cluster, is presented in Table 1 and a graphical representation can be found in Figure 1. Out of the 1,024 possible combinations of resistance within individual isolates, 201 unique combinations were found. An overview of the overall frequency from high to low of the 201 resistance combinations is presented in supplementary material (Table S1) with the cluster they were assigned to and their posterior membership probabilities. The posterior probabilities in Table S1 of all resistant combinations to belong to cluster 1–4 are model generated and are calculated according to Appendix S1. Tables [Link], [Link], [Link], [Link] present resistance combinations per cluster, per species. A graphical representation of the yearly proportions of isolates in the clusters over time for the different animal species can be found in Figure 2.
TABLE 1.
Resistance probability per cluster (rows) of a commensal E. coli isolate against an antimicrobial (columns). Isolates (N = 12,986) are from broilers, dairy cows, slaughter pigs and veal calves in the Netherlands, 2007–2018
| GEN a | TAZ | FOT | CHL | TMP | SMX | AMP | TET | NAL | CIP | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 (n = 7,566) | 0.01 | 0 | 0 | 0 | 0.01 | 0.01 | 0.05 | 0.16 | 0 | 0 |
| Cluster 2 (n = 698) | 0.08 | 0.02 | 0.02 | 0.06 | 0.05 | 0.12 | 0.33 | 0.18 | 0.96 | 0.99 |
| Cluster 3 (n = 3,289) | 0.04 | 0.04 | 0.04 | 0.26 | 0.77 | 0.94 | 0.66 | 0.81 | 0 | 0 |
| Cluster 4 (n = 1,433) | 0.16 | 0.12 | 0.13 | 0.41 | 0.90 | 0.98 | 0.88 | 0.80 | 0.97 | 0.99 |
| Overall proportion of resistance b | 0.04 | 0.02 | 0.03 | 0.12 | 0.30 | 0.36 | 0.31 | 0.39 | 0.16 | 0.16 |
GEN, gentamicin; TAZ, ceftazidime; FOT, cefotaxime; CHL, chloramphenicol; TMP, trimethoprim; SMX, sulfamethoxazole; AMP, ampicillin; TET, tetracycline; NAL, nalidixic acid; CIP, ciprofloxacin.
Overall proportion of resistance in the full data, that is the fraction, out of all isolates (N = 12,986), resistant against an antimicrobial.
FIGURE 1.
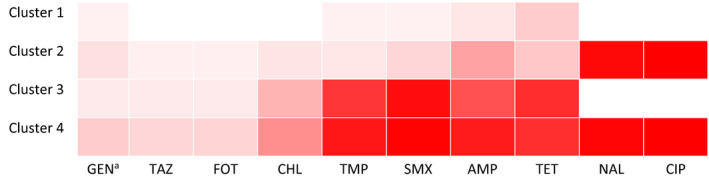
Heatmap showing the resistance probability for the ten tested antimicrobialsa per cluster in the four clusters from model‐based clustering, in commensal E. coli isolates (N = 12,986) from broilers, dairy cows, slaughter pigs and veal calves from the Netherlands, 2007–2018. aGEN, gentamicin; TAZ, ceftazidime; FOT, cefotaxime; CHL, chloramphenicol; TMP, trimethoprim; SMX, sulfamethoxazole; TET, AMP, ampicillin; NAL, nalidixic acid; CIP, ciprofloxacin
FIGURE 2.
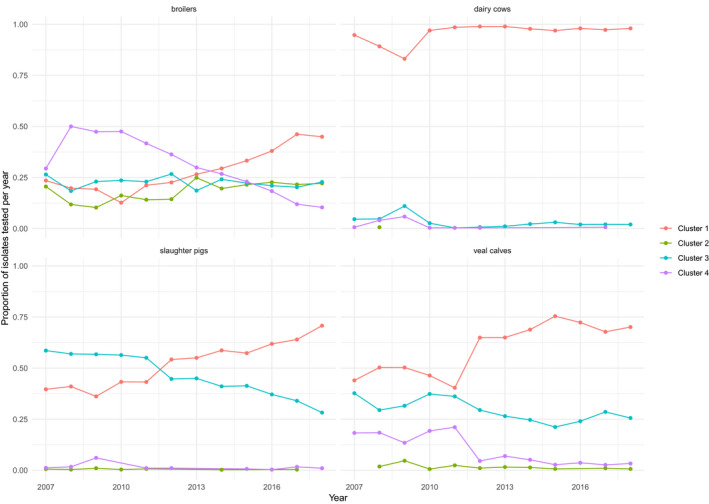
Proportion of isolates in the four clusters (1–4) derived from model‐based clustering of multidrug resistance in commensal E. coli isolates (N = 12,986) of broilers, dairy cows, slaughter pigs and veal calves from the Netherlands, 2007–2018
Cluster 1 (n = 7,566 isolates) is the cluster which is mostly pan‐susceptible, besides (mostly) single resistance against TET, AMP, AMP TET, SMX, TMP, GEN, GEN TET and CHL with low probability (<0.16) (Table 1; Table S2). The single resistance phenotype with only TET resistance is almost exclusively present in pigs (Table S2). Cluster 2 (n = 698) is mostly susceptible, but carrying only CIP and NAL resistance, sometimes in combination with other resistance traits, of which AMP and TET have the highest resistance probabilities (0.33 and 0.18, respectively). Cluster 2 is almost exclusively present in broilers and not in other animal species (Table 2; Table S3). Cluster 3 (n = 3,289) mostly consists of multidrug‐resistant isolates, with high probability of resistance against SMX, TET, TMP and AMP (proportions of 0.94, 0.81, 0.77 and 0.66 of the isolates respectively, Table 1), and against CHL with moderate probability (0.26) but with low probability (<0.05) against 3rd generation cephalosporins FOT and TAZ (Table 1; Table S4). Cluster 4 (n = 1,433) contains the most multidrug‐resistant isolates: almost all (0.99) are resistant against CIP NAL (Table 1, S5), and to SMX, TET, TMP, AMP and CHL with even higher probability than in cluster 3, and with some probability (0.12 and 0.13, respectively) to 3rd generation cephalosporins (TAZ, FOT). To summarize the clusters:
TABLE 2.
Distribution of the clusters per animal species (with the overall distribution in the last row)
| N a | Cluster | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Broilers | 3,602 | 0.28 b | 0.18 | 0.22 | 0.31 |
| Dairy cows | 2,958 | 0.96 | 0.00 | 0.03 | 0.01 |
| Slaughter pigs | 3,491 | 0.53 | 0.00 | 0.46 | 0.01 |
| Veal calves | 2,935 | 0.62 | 0.01 | 0.28 | 0.08 |
| Relative cluster size | 12,986 | 0.58 c | 0.05 | 0.25 | 0.11 |
Number of isolates tested per animal species, from 2007 to 2018.
Proportion, out of all isolates tested for this animal species, that belong to this cluster.
Relative cluster size, that is the proportion of all 12,986 isolates that belong to this cluster.
Cluster 1: Isolates that are mostly susceptible against all tested antimicrobials.
Cluster 2: Isolates that are mostly susceptible against all tested antimicrobials, except the (fluoro) quinolones.
Cluster 3: Multidrug‐resistant isolates that are (fluoro)quinolone susceptible.
Cluster 4: Multidrug‐resistant isolates that are also (fluoro)quinolone resistant.
Note that the clusters are numbered in such a way that the resistance increases in probability with cluster number for each antimicrobial, except for (fluoro)quinolones (CIP and NAL). Clusters 1 and 3 are (fluoro) quinolone susceptible, whereas clusters 2 and 4 are highly (fluoro)quinolone resistant.
Table 2 shows how the isolates from the different animal species are divided over the four clusters and Table 3 presents the comparison between the four clusters and the outcome indicators defined by ECDC, EFSA and EMA. All isolates with pan‐susceptibility (the primary outcome indicator defined by ECDC, EFSA and EMA) from the total of isolates tested (n = 12,986) belong to cluster 1 (bottom of Table 3; Table S2). Isolates belonging to the secondary outcome indicator (as defined by ECDC, EFSA and EMA) multidrug‐resistant (>=3) are divided over three clusters: 2, 3 and 4. The highest proportion of multidrug resistance is found in cluster 3 (0.21), followed by cluster 4 (0.11), and lastly 0.05 of the multidrug‐resistant isolates belongs to cluster 2 (Table 3).
TABLE 3.
A‐B Relation between the four clusters from model‐based clustering and outcome indicators as proposed by ECDC, EFSA and EMA(4): pan‐susceptibility (Pan‐S), resistant to three or more classes (>=3), and ciprofloxacin resistance (CIP‐R), shown as proportions of commensal E. coli isolates (N = 12,986) of broilers, dairy cows, slaughter pigs and veal calves from the Netherlands, 2007–2018, shown for the dataset overall (Table 3A) and stratified per animal species (Table 3B)
| Table 3 | ||||||
|---|---|---|---|---|---|---|
| Animal species | Indicator | Proportion per animal species e | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
| Table 3A | ||||||
| Total (n = 12,986) | Indicator | Overall proportion a | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
| Pan‐S b | 0.46 | 0.46 | 0.00 | 0.00 | 0.00 | |
| >=3 c | 0.33 | 0.00 | 0.01 | 0.21 | 0.11 | |
| CIP‐R d | 0.16 | 0.00 | 0.05 | 0.00 | 0.11 | |
| Table 3B | ||||||
| Broilers (n = 3,602) | Pan‐S f | 0.20 | 0.20 | 0.00 | 0.00 | 0.00 |
| >=3 g | 0.55 | 0.00 | 0.05 | 0.19 | 0.31 | |
| CIP‐R h | 0.49 | 0.00 | 0.18 | 0.00 | 0.31 | |
| Dairy cows (n = 2,958) | Pan‐S | 0.95 | 0.95 | 0.00 | 0.00 | 0.00 |
| >=3 | 0.02 | 0.00 | 0.00 | 0.02 | 0.01 | |
| CIP‐R | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | |
| Slaughter pigs (n = 3,491) | Pan‐S | 0.33 | 0.33 | 0.00 | 0.00 | 0.00 |
| >=3 | 0.38 | 0.00 | 0.00 | 0.36 | 0.01 | |
| CIP‐R | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | |
| Veal calves (n = 2,935) | Pan‐S | 0.46 | 0.46 | 0.00 | 0.00 | 0.00 |
| >=3 | 0.33 | 0.00 | 0.01 | 0.24 | 0.08 | |
| CIP‐R | 0.09 | 0.00 | 0.01 | 0.00 | 0.08 | |
Overall proportion of isolates from the total number of isolates (N = 12,986), belonging to this outcome indicator by ECDC, EFSA and EMA (4). Note: these indicators are not mutually exclusive.
Proportion of pan‐susceptible isolates per cluster.
Proportion of isolates resistant to three or more classes per cluster.
Proportion of ciprofloxacin resistant isolates per cluster.
Total proportion per animal species belonging to this outcome indicator.
Proportion of pan‐susceptible isolates per animal species per cluster.
Proportion of isolates resistant to three or more classes per animal species per cluster.
Proportion of ciprofloxacin resistant isolates per animal species per cluster.
4. DISCUSSION
The purpose of this study was to use a data‐driven method, model‐based clustering, to summarize AMR in bacterial isolates over antimicrobial classes, in order to develop suitable outcome indicators of AMR monitoring based on objective arguments. Model‐based clustering delivered four clusters as a dimension reduction of the complex data of 12,986 isolates tested with a panel of 10 antimicrobials of eight antimicrobial classes. These four identified clusters have potential to serve as AMR monitoring outcome indicators. We show a proof‐of‐principle of how model‐based clustering can be used to develop data‐driven summary indicators to assess AMR monitoring outcome. In addition, this analysis resulted in a deeper understanding of the patterns in co‐occurrence of resistance to more than one antimicrobial per isolate, as expressed phenotypically in commensal E. coli. These patterns were identified by 201 unique resistance combinations in this dataset, divided over the four clusters (Tables [Link], [Link], [Link], [Link], [Link]). The prevalence of these combinations is different for the animal populations tested. It goes beyond the extent of this study to investigate all multidrug resistance patterns individually. Here, we discuss our main results and remarkable findings: the method creates clusters that differentiate levels of multidrug resistance, with or without resistance to (fluoro)quinolones and 3rd generation cephalosporins.
Quinolone resistance splits the clusters: clusters 1 and 3 without CIP NAL and cluster 2 and 4 with CIP NAL resistance (Table 1; Figure 1). Furthermore, 3rd generation cephalosporin resistance also differs between clusters: clusters 3 and 4 contain isolates with resistance against FOT TAZ, but clusters 1 and 2 contain hardly any. Hence, this cluster analysis divides multidrug‐resistant isolates over three different categories: relatively susceptible but with the fluoro(quinolones) CIP NAL (cluster 2), multidrug resistant mostly without resistance to these critically important antimicrobials for human medicine (World Health Organization, 2019) (cluster 3), and multidrug resistance including resistance to critically important antimicrobials (cluster 4). Most isolates were assigned to a cluster with high certainty, but not all, as can be seen from a few examples of less certain posterior memberships in Table S1.
We conclude from this analysis that the isolates of the four animal species are differently distributed over the resistance clusters, Table 2. As an example, almost all isolates of dairy cows are in cluster 1, whereas broiler isolates are distributed over all clusters. This is in line with AMU in dairy cows, which has for many years been much lower than in broilers (SDa, 2018). Regarding the broilers, most isolates belong to the multidrug‐resistant cluster 4 (0.31), corresponding with the relatively high level of resistance against critically important antimicrobials in these animals. In pigs, the proportion of isolates in the multidrug‐resistant cluster 3 (without resistance against critically important antimicrobials) is high (0.46) and, in contrast, is low in cluster 4 (0.01). Veal calves have multidrug‐resistant isolates mainly belonging to cluster 3 (0.28) and to a lesser extend to cluster 4 (0.08).
We illustrate the variation in time trends of the four clusters in different animal reservoirs in Figure 2. Interestingly, the susceptible cluster 1 increases over time in all animal species. And in pigs, multidrug‐resistant cluster 3 decreases over time. In broilers, the highly multidrug‐resistant cluster 4 decreases over time. These findings are in line with the overall reduction in AMU in all animal species in the Netherlands since 2009 (Dorado‐Garcia et al., 2016; SDa, 2018; Veldman et al., 2019), and more specifically a stop of 3rd generations cephalosporin use in broilers (SDa, 2018; Mevius and Heederik, 2014).
The four clusters we found can be used as indicators to benchmark AMR: over time, over several countries or between animal sectors, either as a reflection of AMU or to assess the overall AMR situation. For benchmarking, it is crucial to create transparency by robust metrics, preferably developed by quantitative methods (Bos et al., 2015). These clusters lead to transparency of AMR present in different reservoirs and this method is flexible for policymakers to make choices. Suggestions for benchmarking methods are for example to set an AMR benchmark threshold for the proportion of isolates in ‘susceptible’ cluster 1. Also, the cluster 3 versus cluster 4 proportion could be of interest to benchmark over different reservoirs (Table 2; Figure 2).
Our results indicate that more than one indicator is needed to describe multidrug resistance, as shown in the comparison between the four clusters from this analysis and the indicators proposed by EFSA, ECDC and EMA (European Food Safety Authority, 2017), Table 3. For example, the proportion of multidrug resistant isolates in slaughter pigs versus veal calves (0.38 and 0.33, respectively, Table 3). In pigs, almost all of these isolates belong to the non‐critical multidrug‐resistant cluster 3, but in contrast a higher proportion of the multidrug‐resistant isolates from veal calves belong to the more critical multidrug resistant cluster 4 compared to slaughter pigs (0.08 versus 0.01). Ciprofloxacin resistance, a separate outcome indicator for EFSA, is represented in clusters 2 and 4, differentiating ciprofloxacin resistance as part of multidrug resistance (cluster 4) or mostly without other resistances (cluster 2). Cluster 2 with the phenotype containing just CIP NAL resistance is almost exclusively present in broilers (Table 2, Table 3 and Table S3). (Fluoro) quinolone resistance seems to persist in broiler flocks, as also described by other studies (Roth et al., 2019; Chantziaras et al., 2018; Taylor et al., 2016; Vieira et al., 2011).
Model‐based clustering summarizes the data without loss of relevant information. As ECDC, EFSA and EMA mentioned in their recent report, for a more detailed analysis of causes for AMR in the agricultural sector, an in‐depth breakdown to the level of individual drug–microbe combinations by animal species and production sector is needed (European Food Safety Authority, 2017). Furthermore, Buyle et al.(2013) mentioned an unavoidable loss of information to occur when indicators are used to summarize large datasets. Using model‐based clustering may tackle both of these problems, providing a solution to balance between reducing complexity and loss of information. The clusters reduce complexity, but can be broken down by composition to look up specific information (see the examples of the resistance combinations in Supplementary Tables). Another advantage over the EFSA indicators is that these clusters are mutually exclusive, while the EFSA indicators are not mutually exclusive and therefore cannot give an overview of the whole dataset. This is illustrated in Table 3.
In comparison with the work of Havelaar et al. (2017), Dorado‐Garcia et al. (2016) and Laxminarayan and Klugman (2011) another important advantage of model‐based clustering is that it avoids the making of arbitrary choices, that is on what basis groups are made, because this method is data‐driven. Although the antimicrobial susceptibility panel included in the analysis will influence results, this probabilistic approach avoids weighing or prioritizing with lack of quantitative arguments. And in the model no heuristic choices such as similarity coefficients have to be made, as explained in Methods.
So far, few studies have investigated multidrug resistance patterns with a quantitative approach to summarize the data and reduce complexity. Most studies on clusters in AMR data describe hierarchical clustering of genetic data from the bacterial genome from which their genetic relatedness can be inferred. Kappell et al. (2015) applied a principal component analysis to both genetic and phenotypic AMR data of multidrug‐resistant strains, but describe only ordination to visualize patterns, not quantify them (i.e., leading to an output that can be interpreted, for ranking and prioritizing specific AMR patterns). Multivariate analyses to quantify multidrug resistance in either genetic or phenotypic AMR data have hardly been performed. Now that whole genome sequencing is becoming available for routine diagnostics and surveillance activities, methods to reduce complexity in genetic AMR data are needed. We used phenotypic data, but this approach could also be interesting to apply on genetic data.
This study concerns ecological data: commensal E. coli is a sentinel organism from samples of healthy animals at slaughter, and multidrug resistance in a commensal organism from healthy animals is not directly a public health threat. However, the patterns we found reflect either direct selection or co‐selection of AMR by AMU, or other driving mechanisms in the animal populations. These patterns can consequently be subjected to further investigating the biological mechanisms behind. Besides AMU, other drivers could play a role, for example disinfectants (biocides, i.e., quaternary ammonium compounds) (Van Gompel et al., 2019), although these positive associations were not confirmed in two other studies (Maertens et al., 2019, 2020).
The cluster output of this analysis is, apart from the variability in the data itself, dependent of the selected panel of antimicrobials. Two antimicrobials of the critically important classes 3rd generation cephalosporins and (fluoro) quinolones were included as variables in the analysis. Both FOT and TAZ are included in the EUVSEC susceptibility testing panel because that increases the sensitivity for ESBL screening, and both CIP and NAL are included in that panel to monitor different types of quinolone resistance (European Food Safety Authority 2012). In this analysis, these two classes partly determine the way the data is divided into these four clusters. For those classes, the isolates resistant to one of these antimicrobials are nearly always resistant to both antimicrobials (because of cross‐resistance), therefore the model considers them to be a cluster. We checked for the influence of modelling only one antimicrobial per antimicrobial class (without TAZ and NAL, which are additional in the classes of 3rd generation cephalosporins and quinolones to FOT and CIP, respectively). This resulted in a solution of only two clusters: one with all almost completely susceptible isolates, and the other being all the multidrug‐resistant isolates (data not shown). Apparently, in this data, once an isolate is resistant to one antimicrobial, it has a high probability of being resistant to multiple antimicrobials. We considered the two‐cluster solution for eight antimicrobials less informative as it models less data and is, by design, ignorant of the importance of resistance to those two antimicrobial classes.
As a result of the characteristics of this data, the four clusters in our results have an intrinsic focus on (fluoro)quinolones and 3rd generation cephalosporins. This could be of practical use, since these antimicrobial classes are of specific interest to policy. Cluster membership varies by data which depends on the level of AMR and AMU pressure. Therefore, this clustering method should be re‐evaluated after analysing a more diverse international dataset. Next to that, it could be interesting to model cluster membership probabilities for a diversity of data, for example to estimate the probability of an isolate with resistance to one, two or three or more antimicrobial classes to belong to cluster 1–4.
In this analysis, we used a large dataset. Model‐based clustering detects associations between resistance for different antimicrobials and this is more likely to succeed in larger than in smaller data sets. However, small data sets can use the results of larger ones such as presented in this paper. For example, the four clusters found in our study can be used as new predefined categories to summarize resistance patterns in smaller datasets.
These clusters may be interesting benchmark indicators for EU member states, that monitor with the susceptibility panel as prescribed by EU legislation. This analysis was performed on Dutch data, so the question is whether the clusters would also be applicable for data from other countries. It could be dependent of specific AMR patterns, which may vary between ecologies of microbes, different animal sectors, and between regions and countries. To further develop this method, this analysis should be repeated for several other countries, such as the monitoring data yearly reported by all EU member states to EFSA.
In this study, isolates from all animal species were included in one analysis instead of analysing all animal species separately, this enables comparing the cluster outcomes for benchmarking purposes. However, the effect of this methodology on the cluster outcome should be further investigated. In addition, it could be interesting to use this model to investigate associations within one animal species over different European countries. Countries often have differences in food animal producing sectors, and other clusters may be found with different input data. However, in principle the method should perform the same: summarizing resistance data in a more easily understandable output than achieved so far.
In conclusion, model‐based clustering identifies clusters that summarize resistance over antimicrobials or antimicrobial classes. The four clusters we found have potential to be used by policymakers as monitoring outcome indicators, as we showed for Dutch AMR monitoring data from livestock, 2007–2018. The composition of the clusters was determined by the co‐occurrence of resistance to more than one antimicrobial per isolate, and these reflect selection and co‐selection patterns by AMU or other determinants. This study concerns ecological data from a commensal microorganism from Dutch livestock reservoirs, but this analytical method has potential value to identify clusters as outcome indicators for data from other microorganisms (e.g., foodborne pathogens such as Salmonella, Campylobacter), or data from other reservoirs.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplementary Material
Table S1
Table S2
Table S3
Table S4
Table S5
ACKNOWLEDGEMENTS
We thankfully acknowledge the technicians in the National Reference Laboratory at Wageningen Bioveterinary Research (WBVR), Joop Testerink, Marga Japing, Yvon Geurts and Alieda van Essen for determining the MIC. For help in data handling we would like to thank Alieda van Essen at WBVR.
Hesp A, ter Braak C, van der Goot J, Veldman K, van Schaik G, Mevius D. Antimicrobial resistance clusters in commensal Escherichia coli from livestock. Zoonoses Public Health. 2021;68:194–202. 10.1111/zph.12805
REFERENCES
- Biernacki, C. , Celeux, G. , & Govaert, G. (2000). Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Transactions on Pattern Analysis and Machine Intelligence, 22(7), 719–725. 10.1109/34.865189 [DOI] [Google Scholar]
- Bos, M. E. H. , Mevius, D. J. , Wagenaar, J. A. , van Geijlswijk, I. M. , Mouton, J. W. , & Heederik, D. J. J. & Netherlands Veterinary Medicines Authority (SDa) (2015). Antimicrobial prescription patterns of veterinarians: Introduction of a benchmarking approach. Journal of Antimicrobial Chemotherapy, 70(8), 2423–2425. 10.1093/jac/dkv104 [DOI] [PubMed] [Google Scholar]
- Buyle, F. , Metz‐Gercek, S. , Mechtler, R. , Kern, W. , Robays, H. , Vogelaers, D. , & Struelens, M. (2013). Development and validation of potential structure indicators for evaluating antimicrobial stewardship programs in European hospitals. European Journal of Clinical Microbiology & Infectious Diseases., 32, 1161–1170. 10.1007/s10096-013-1862-4 [DOI] [PubMed] [Google Scholar]
- Chantziaras, I. , Smet, A. , Filippitzi, M. E. , Damiaans, B. , Haesebrouck, F. , Boyen, F. , & Dewulf, J. (2018). The effect of a commercial competitive exclusion product on the selection of enrofloxacin resistance in commensal E. coli in broilers. Avian Pathology, 47(5), 443–454. 10.1080/03079457.2018.1486027 [DOI] [PubMed] [Google Scholar]
- Dorado‐Garcia, A. , Mevius, D. J. , Jacobs, J. J. , Van Geijlswijk, I. M. , Mouton, J. W. , Wagenaar, J. A. , & Heederik, D. J. (2016). Quantitative assessment of antimicrobial resistance in livestock during the course of a nationwide antimicrobial use reduction in the Netherlands. Journal of Antimicrobial Chemotherapy, 71(12), 3607–3619. 10.1093/jac/dkw308 [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (2019). Antimicrobial wild type distributions of microorganisms. Retrieved from http://www.eucast.org/mic_distributions_and_ecoffs/.
- European Food Safety Authority (2012). Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in Salmonella, Campylobacter and indicator Escherichia coli and Enterococcus spp. bacteria transmitted through food.Retrieved from. https://www.efsa.europa.eu/en/efsajournal/pub/2742.
- European Food Safety Authority (2017). ECDC, EFSA and EMA Joint Scientific Opinion on a list of outcome indicators as regards surveillance of antimicrobial resistance and antimicrobial consumption in humans and food‐producing animals. EFSA Journal, 15(10), e05017. 10.2903/j.efsa.2017.5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermunt J. K. editors. Hagenaars, A. McCutcheon, applied latent class analysis. : Cambridge University Press. 2002. pp. 89–106. [Google Scholar]
- Havelaar, A. H. , Graveland, H. , van de Kassteele, J. , Zomer, T. P. , Veldman, K. , & Bouwknegt, M. (2017). A summary index for antimicrobial resistance in food animals in the Netherlands. BMC Veterinary Research, 13(1), 305. 10.1186/s12917-017-1216-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesp, A. , Veldman, K. , van der Goot, J. , Mevius, D. , & van Schaik, G. (2019). Monitoring antimicrobial resistance trends in commensal Escherichia coli from livestock, the Netherlands, 1998 to 2016. Eurosurveillance, 24(25), 1800438. 10.2807/1560-7917.ES.2019.24.25.1800438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappell, A. D. , DeNies, M. S. , Ahuja, N. H. , Ledeboer, N. A. , Newton, R. J. , & Hristova, K. R. (2015). Detection of multi‐drug resistant Escherichia coli in the urban waterways of Milwaukee, WI. Frontiers in Microbiology, 6, 336. 10.3389/fmicb.2015.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayan, R. , & Klugman, K. P. (2011). Communicating trends in resistance using a drug resistance index. British Medical Journal Open, 1(2), e000135. 10.1136/bmjopen-2011-000135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisch, F. (2004). FlexMix: A General framework for finite mixture models and latent class regression in R. Journal of Statistical Software, 011(i08), 23274. Retrieved from https://EconPapers.repec.org/RePEc:jss:jstsof:v:011:i08. [Google Scholar]
- Maertens, H. , De Reu, K. , Meyer, E. , Van Coillie, E. , & Dewulf, J. (2019). Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Veterinary Research, 15(1), 310. 10.1186/s12917-019-2044-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens, H. , Van Coillie, E. , Millet, S. , Van Weyenberg, S. , Sleeckx, N. , Meyer, E. , Zoons, J. , Dewulf, J. , & De Reu, K. (2020). Repeated disinfectant use in broiler houses and pig nursery units does not affect disinfectant and antibiotic susceptibility in Escherichia coli field isolates. BMC Veterinary Research, 16(1), 140. 10.1186/s12917-020-02342-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevius, D. , & Heederik, D. (2014). Reduction of antibiotic use in animals “let’s go Dutch”. Journal Für Verbraucherschutz Und Lebensmittelsicherheit, 9(2), 177–181. 10.1007/s00003-014-0874-z [DOI] [Google Scholar]
- O’Neill, J. (2016). Tackling drug‐resistant infections globally: Final report and recommendations. The review on antimicrobial resistance. Chaired by Jim O'Neill., (2016). Retrieved from https://amr‐review.org/Publications.html
- R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Retrieved from https://www.R‐project.org/. [Google Scholar]
- Roth, N. , Käsbohrer, A. , Mayrhofer, S. , Zitz, U. , Hofacre, C. , & Domig, K. J. (2019). The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poultry Science, 98(4), 1791–1804. 10.3382/ps/pey539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SDa (2018). Usage of Antibiotics in Agricultural Livestock in the Netherlands in 2018. Retrieved from https://cdn.i‐pulse.nl/autoriteitdiergeneesmiddelen/userfiles/Publications/2018‐def‐rapport1.pdf.
- European Commission (2013). Commission implementing decision on the monitoring and reporting of antibmicrobial resistance in zoonotic and commensal bacteria, 2013/652/EU Stat (2013).
- Taylor, N. M. , Wales, A. D. , Ridley, A. M. , & Davies, R. H. (2016). Farm level risk factors for fluoroquinolone resistance in E. coli and thermophilic Campylobacter spp. on poultry farms. Avian Pathology, 45(5), 559–568. 10.1080/03079457.2016.1185510 [DOI] [PubMed] [Google Scholar]
- Van Gompel, L. , Luiken, R. E. C. , Sarrazin, S. , Munk, P. , Knudsen, B. E. , Hansen, R. B. , & Smit, L. A. M. (2019). The antimicrobial resistome in relation to antimicrobial use and biosecurity in pig farming, a metagenome‐wide association study in nine European countries. Journal of Antimicrobial Chemotherapy, 74(4), 865–876. 10.1093/jac/dky518 [DOI] [PubMed] [Google Scholar]
- Veldman, K. T. , Mevius, D. J. , Wit, B. , Van Pelt, W. Franz, E. , & Heederik, D. (2019). Monitoring of antimicrobial resistance and antibiotic usage in animals in The Netherlands in 2018. Retrieved from https://www.wur.nl/upload_mm/a/7/9/89640bbc‐53a2‐40f0‐ba4a‐a9a34a7bf416_Nethmap%20Maran%202019.pdf.
- Vieira, A. R. , Collignon, P. , Aarestrup, F. M. , McEwen, S. A. , Hendriksen, R. S. , Hald, T. , & Wegener, H. C. (2011). Association between antimicrobial resistance in escherichia coli isolates from food animals and blood stream isolates from humans in Europe: An ecological study. Foodborne Pathogens and Disease, 8(12), 1295–1301. 10.1089/fpd.2011.0950 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2015). Global action plan on antimicrobial resistance. Retrieved from https://www.who.int/antimicrobial‐resistance/publications/global‐action‐plan/en/. [DOI] [PubMed]
- World Health Organization (2019). Critically important antimicrobials for human medicine, 6th revision. Retrieved from https://www.who.int/foodsafety/publications/antimicrobials‐sixth/en/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1
Table S2
Table S3
Table S4
Table S5


