Abstract
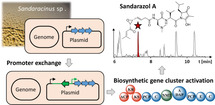
Herein, we describe a new plasmid found in Sandaracinus sp. MSr10575 named pSa001 spanning 209.7 kbp that harbors a cryptic secondary metabolite biosynthesis gene cluster (BGC). Activation of this BGC by homologous‐recombination‐mediated exchange of the native promoter sequence against a vanillate inducible system led to the production and subsequent isolation and structure elucidation of novel secondary metabolites, the sandarazols A–G. The sandarazols contain intriguing structural features and very reactive functional groups such as an α‐chlorinated ketone, an epoxyketone, and a (2R)‐2‐amino‐3‐(N,N‐dimethylamino)‐propionic acid building block. In‐depth investigation of the underlying biosynthetic machinery led to a concise biosynthetic model for the new compound family, including several uncommon biosynthetic steps. The chlorinated congener sandarazol C shows an IC50 value of 0.5 μm against HCT 116 cells and a MIC of 14 μm against Mycobacterium smegmatis, which points at the sandarazols’ potential function as defensive secondary metabolites or toxins.
Keywords: biosynthesis, horizontal gene transfer, megaplasmid, myxobacteria, secondary metabolites
Genetic activation artificially induced a plasmid‐encoded natural product biosynthetic gene cluster and activated the complex biosynthesis of a previously unknown cytotoxin named sandarazol. Its biosynthesis is unique in featuring the genetic blueprint for the formation of intriguing structural features such as an α‐chlorinated ketone, an epoxyketone, and a rare (2R)‐2‐amino‐3‐(N,N‐dimethylamino)‐propionic acid building block.
Introduction
Bacterial antibiotic resistance genes cause antimicrobial resistance (AMR) in pathogens thereby leading to enormous challenges in the treatment of infectious diseases in the clinic. [1] AMR genes are often encoded on plasmids, horizontally transferrable elements employed by bacteria to survive antibiotics treatment and effectively spread AMR. [2] In addition to that, AMR genes co‐localized with antibiotic biosynthesis gene clusters (BGCs) are important for self‐resistance during antibiotic production in bacteria capable of respective biosyntheses. Similarly to plasmid‐mediated exchange of antibiotic resistance, bacteria are able to gain an advantage over competing microorganisms by exchanging secondary metabolite BGCs in horizontal gene transfer events. [3] The mechanism for plasmid‐mediated exchange of secondary metabolite BGCs is known for plasmid‐encoded toxins, with the anthrax‐causing Bacillus anthracis as the most prominent example. [4] The Bacillus plasmid is shared between different bacilli and thereby transmits the anthrax toxin virulence factor. Exchange mechanisms of such plasmids that encode toxins enable the respective bacteria to acquire the ability for chemical warfare. As bacteria are competing with other microbes for nutrients in their ecological niches, the encoded chemical warfare molecules optimized by evolution are attractive targets in the search for novel bioactive natural products. For actinobacteria it has already been shown that defensive secondary metabolites can be encoded as multimodular BGCs on autonomously replicating plasmids. A BGC responsible for production of the highly cytotoxic mycolactones, for example, is also encoded on a large plasmid. [5] Mycobacterium ulcerans produces this cytotoxin and releases it during the infection process of human skin, while the bacterium feeds on skin cells. Streptomycetes also host plasmid‐borne antibiotics BGCs as exemplified by the esmeraldins, a series of phenazine antibiotics. [6] With respect to the total output of natural products and natural product diversity, bacteria have emerged as major players as they are prolific producers of biologically active secondary metabolites. [7] Besides the well‐described bacterial phyla such as actinobacteria, firmicutes, and cyanobacteria that are responsible for the majority of identified biologically active natural products to date, Gram‐negative proteobacteria such as myxobacteria have also shown great promise for the discovery of novel bioactive secondary metabolites.[ 8 , 9 , 10 ] Especially novel and phylogenetically distant bacterial genera among the myxobacteria, such as the recently identified genus Sandaracinus studied herein, hold promise for finding interesting novel natural products chemistry. [11] In recent years, availability of cheap and reliable DNA sequencing technologies along with the development of BGC prediction tools such as antiSMASH depict the theoretical genetically encoded bacterial secondary metabolome as to date largely underexploited.[ 12 , 13 ] However, many BGCs that can be identified in silico are “cryptic” under laboratory conditions. In this case, the production of the corresponding secondary metabolite is completely repressed or remains below the detection limit of modern analytical tools such as liquid chromatography (LC)‐coupled mass spectrometry (MS) instrumentation. Activating such cryptic clusters by genetic tools holds promise for discovering new chemistry, but currently remains a non‐automatable, labor‐intensive process. [14] Therefore, a strong focus on rational prioritization of BGCs to be activated by heterologous expression or gene cluster overexpression using heterologous promoters remains crucial to focus discovery efforts on BGCs encoding for bioactive natural products. [15] Methods employed for BGC prioritization so far include expected target‐guided mining for potential self‐resistance genes and exploring biosynthetic gene cluster complexity for modular BGC architectures as well as the presence of certain tailoring enzymes such as halogenases or epoxidases. [16] Still, the presence of a BGC on a plasmid, as an example for a BGC that is likely to be a mobile genetic element, has not yet been used as a means for BGC prioritization. As part of our ongoing efforts to isolate taxonomically diverse myxobacteria we isolated a novel myxobacterial strain belonging to the Sandaracinus clade. Surprisingly, we found that the strain's genome contains a circular 209.7 kbp plasmid called pSa001 that features a large polyketide synthase (PKS) non‐ribosomal peptide synthetase (NRPS) hybrid BGC in its sequence. Herein, we show that activation of this BGC by genetic engineering of this strain leads to production of several derivatives of the sandarazols, a previously unknown family of natural products. These compounds turned out to be potent and chemically novel toxins biosynthesized by a series of intriguing biosynthetic steps.
Results and Discussion
Biosynthetic Gene Cluster Identification and Activation
Myxobacteria are Gram‐negative δ‐proteobacteria with a rich and diverse secondary metabolism.[ 9 , 10 ] A survey of known and unknown natural products in 2300 myxobacteria showed structural novelty to be clearly correlated to phylogenetic distance and thus heavily reliant on in‐depth investigation of novel myxobacterial genera. [11] We thus chose to dive into the secondary metabolome of a myxobacterial strain called MSr10575 that was recently isolated in‐house. The strain belongs to the Sandaracinus genus, a rare myxobacterial genus little studied for secondary metabolism with the type strain Sandaracinus amylolyticus NOSO‐4T. [17] This strain was the only member in the Sorangineae clade until MSr10575 was isolated and provided the indiacens A and B, two prenylated indole secondary metabolites. [18] We chose to sequence the genome of strain MSr10575 using single‐molecule real‐time sequencing technology. [19] We subsequently annotated the BGCs in its sequence by antiSMASH to obtain an overview about the strain's secondary metabolite production potential. [12] Sequencing coverage analysis and genome assembly revealed that the genome of MSr10575 consists not only of a bacterial chromosome of 10.75 Mbp but also of an autonomously replicating plasmid of 209.7 kbp, which we named pSa001 (see SI). As sequencing coverage per base on the pSa001 plasmid is twice as large as observed for the bacterial chromosome, we assume a median value of two pSa001 copies per MSr10575 cell (see SI). The overall GC content of the plasmid is 70 % and the codon usage bias favors GC‐rich codons over AT‐rich codons for the same amino acid at a rate of 3.6 to 1. As these values do not differ significantly from the corresponding parameters for the MSr10575 chromosome, the plasmid seems well adapted to its host strain (see SI). The presence of this plasmid is distinctive for this myxobacterial strain as autonomously replicating plasmids are rarely observed in myxobacteria. The only other example of a characterized myxobacterial plasmid is pMF1 from Myxococcus fulvus. [20] In comparison to pMF1, pSa001 is not only significantly larger, but also encodes a large type 1 in‐trans acyl transferase (trans‐AT) PKS–NRPS hybrid BGC, which was named sandarazol (szo) BGC (see SI). Besides the szo BGC, the plasmid contains five ORFs encoding putative transposases and one ORF encoding a putative integrase indicating its ability to integrate into (or transfer parts of its sequence into) foreign bacterial genomes after conjugation. It might thus serve as a BGC shuttle vector for horizontal gene transfer of the BGC. Cultivation of wild‐type MSr10575 followed by extraction and liquid chromatography–tandem mass spectrometry (LC–MS2) analysis and GNPS based spectral networking of the bacterial metabolome did not reveal a family of secondary metabolites matching the BGC architecture on the plasmid in expected secondary metabolite size and fragmentation pattern (see SI). [21] We therefore assumed the plasmid borne szo BGC to be “cryptic” as seen in many other cases such as the pyxidicyclines or taromycin.[ 22 , 23 ] All the coding regions in the szo cluster spanning from szoA to szoO are encoded on the same DNA strand and the intergenic regions seemed too small to contain important elements other than ribosome binding sites. We therefore reasoned that the szo cluster is encoded as a single transcriptional unit even though it spans 44.5 kbp. Activating the szo BGC with a single promoter exchange in line with the experiments described in promoter exchange BGC activation in myxobacteria seemed therefore a reasonable strategy to unlock the corresponding natural products.[ 22 , 24 ] As we set out to create a plasmid for szo cluster overexpression by single crossover promoter exchange, we chose a plasmid with a pBelobac borne replication machinery, as this would allow the entire 209.7 kbp pSa001 to be extracted and transferred into E. coli as replicative plasmid for further investigations into the plasmid's replication mechanism. [25] While extracting the pBeloBacSa001 into E. coli is possible, retransformation of this plasmid into a plasmid‐cured version of MSr10575 needs further optimization. To investigate the secondary metabolite products of the BGC, we chose BGC activation in the native host MSr10575 first, as it was assumed MSr10575 would be best suited to produce the corresponding small‐molecule product. We activated the szo cluster on the plasmid by promoter exchange against the vanillate promoter and repressor cassette, since this tool achieved overexpression of BGCs in several other myxobacteria including Myxococcus xanthus and Pyxidicoccus fallax (see SI).[ 22 , 26 ] Compared to other promoters, the vanillate promoter and repressor system shows tight control of BGC expression as well as strong secondary metabolite production upon BGC induction in myxobacteria. [27]
To exchange the promoter of the szo cluster, the first 2 kbp of the szo BGC were PCR‐amplified, fused to a vanillate promoter and repressor cassette, and ligated into the commercial pBeloBac11. This plasmid featuring the pBeloBac replication machinery and a kanamycin resistance in its backbone was named pBeloBac‐TransAT (see SI). Promoter exchange of the szo cluster's native promoter against the vanillate cassette is achieved by homologous recombination of pBeloBac‐TransAT with the pSa001 plasmid after electroporation (Figure 1). MSr10575 clones harboring the recombined fused plasmid called pBeloBacSa001 were selected on 25 μg mL−1 kanamycin and genotypically verified by PCR (see SI). To obtain a comprehensive overview about the metabolic differences between MSr10575 wild type and MSr10575:pBeloBacSa001, analytical‐scale fermentation cultures were prepared in triplicates, extracted, and subjected to UHPLC–qTOF analysis. All detected LC–MS features in the triplicate analyses were grouped into 2D retention time–exact mass windows (so‐called buckets), and all MSr10575 wild‐type‐derived liquid chromatography–mass spectrometry (LC–MS) features were subtracted from the detected LC–MS features using principal component analysis (PCA).[ 28 , 29 ] These features were then used for selective acquisition of LC–MS2 data covering exclusively the mutant‐derived LC–MS features that are subsequently used for spectral networking in GNPS. [21] We thus obtained spectral networks comprising all LC–MS features linked to activation of the sandarazol BGC (see Figure 1 and SI).
Figure 1.
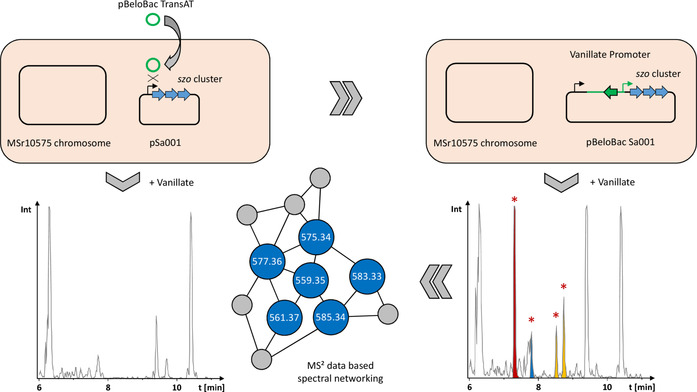
Schematic overview of the activation of the plasmid‐borne szo BGC in wild‐type Sandaracinus sp. MSr10575 by homologous recombination, the corresponding changes in the extracts’ LC–MS chromatograms, and LC–MS2‐based spectral networking for identification of the produced sandarazols.
Isolation and Structure Elucidation
The novel secondary metabolites observed in this PCA‐based analysis of LC–MS features were named the sandarazols. Besides sandarazol A (1; 575.344 Da [M+H]+; C30H47N4O7, Δ=0.64 ppm), we were able to identify and characterize seven structural variants from appearance of their characteristic MS signals, approximately half of which showed the characteristic isotope pattern for chlorine‐containing compounds such as sandarazol C (3; 583.3256 Da [M+H]+; C29H48ClN4O6, Δ=0.07 ppm). [30] The sandarazols A, B, C, and F (1, 2, 3, and 6) were isolated from vanillate‐induced large‐scale cultures of MSr10575:pBeloBac Sa001 using a sequence of different techniques such as liquid/liquid extraction, countercurrent partition chromatography (CPC), and HPLC under N2 (see SI, Figure 2). It is worth noting that not even trace amounts of the sandarazols A–G (1–7) can be detected in the MSr10575 wild‐type extracts indicating the szo cluster to be most likely fully cryptic in the wild‐type strain under the laboratory conditions applied in our study. The structures of sandarazol D (4), E (5), and G (7) were assigned by comparison of their MS2 spectra to the other derivatives (see SI). As the compounds are sensitive to oxygen and/or strongly acidic or basic pH values, compound isolation and purification was performed in ammonium‐formate‐buffered eluents and under constant N2 stream wherever possible (Figure 2).
Figure 2.
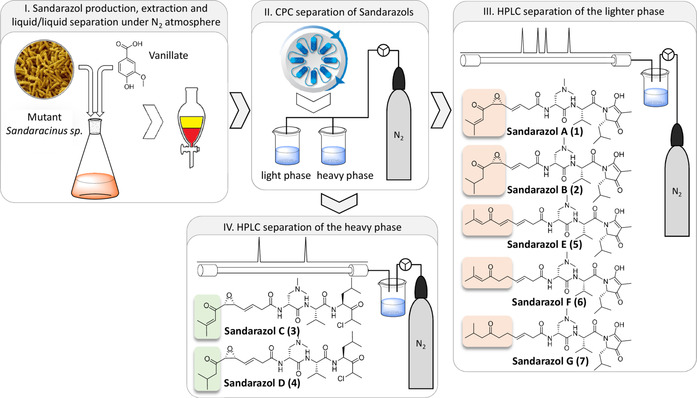
Workflow and isolation scheme for preparation of the sandarazols from Sandaracinus sp. MSr10575:pBeloBacSa001 including structure formulas for all structurally elucidated sandarazol derivatives.
1H NMR, 13C NMR and HSQC‐dept. spectra of 1 reveal two α‐protons based on their characteristic chemical shift at δ 1H=4.78 and 4.45 ppm (Figure 3). COSY and HMBC correlations of the first methine group at δ 1H=4.78 ppm to one methylene group, two methyl groups with a downfield δ 13C chemical shift of 45.2 ppm and a quaternary carbon with a characteristic amide shift, show that this α‐proton is part of 2‐amino‐3‐(N,N‐dimethyl amino)‐propionic acid (Me2Dap). The second α‐proton at δ 1H=4.45 ppm exhibits COSY and HMBC correlations to one methine group, two methyl groups, and another amide function. The second amino acid of the molecule was determined to be a valine. HMBC correlations of the valine α‐proton to the Me2Dap carboxylic acid function suggests their connection via the valine N‐terminus. Further HMBC correlations of the Me2Dap α‐proton to another amide function reveal N‐terminal elongation by the polyketide part of the molecule. Characteristic chemical shifts of two protons at δ 1H=3.47 ppm and 3.46 ppm with correlations to two vinylic double bond protons at one side and one epoxide on the other side, suggest 1 to contain an epoxyketone, as well as unsaturation in β,γ position relative to the amide bond. Typical J H–H coupling values of 15.4 Hz for the vinylic protons at the double bond indicate E configuration. [31] 1D and 2D NMR spectra reveal a trisubstituted vinyl group is conjugated to the ketone side of the epoxyketone. The C‐terminal end of valine shows correlations to another methine group with characteristic chemical shifts close to an α‐proton shift, suggesting further elongation of the molecule by another amino acid. This amino acid is determined to be leucine coupled to a propionate unit from 1D and 2D NMR data. The combination of a low‐field proton shift and high‐field carbon shift of an additional methyl group at δ 1H=1.60 and δ 13C=5.7 ppm in this part of the molecule reveals heterocyclization of a propionate‐extended valine cyclized to a tetramic acid, which also serves as connection between the valine and leucine building blocks (Figures 2 and 3).
Figure 3.
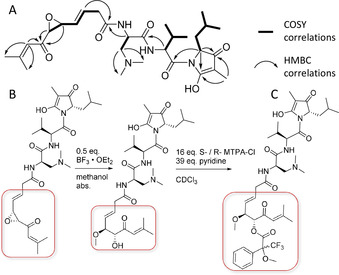
A) NMR correlations important for structure elucidation. B) Reaction sequence to open the epoxide and form a methanol adduct. C) Mosher's esterification of 1 used to elucidate the epoxide's configuration.
In contrast to 1, HSQC spectra of 2 reveal a missing double bond in the terminal moiety of the polyketide part of the molecule (Figure 2). Sandarazol F differs in the polyketide part of the molecule, as HSQC spectra reveal the two methines in the epoxide motif to be replaced by two double bond protons. In 3, however, the polyketide part equates the one of sandarazol A, but 1D and HSQC spectra show a missing tetramic acid moiety. The methyl group at δ 13C=6.0 in 1 is shifted to δ 13C=20.7 ppm in 3. In line with the hrMS spectra of 3, chlorination of the molecule is observed at the α‐position of the elongated leucine, confirmed by the characteristic chemical shifts and COSY as well as HMBC correlations of the surrounding methyl and methine groups.
All sandarazol derivatives contain five stereocenters, two of which are linked by the epoxide. The chlorinated derivatives 3 and 4 feature an additional stereocenter at the chlorinated carbon atom, which epimerizes fast at room temperature (see SI). Its original stereoconfiguration can therefore not be determined as epimerization occurs already under fermentation conditions. The stereocenters contained in the l‐valine (l‐Val), l‐leucine (l‐Leu), and (2R)‐2‐amino‐3‐(N,N‐dimethyl amino)‐propionic acid (d‐Me2Dap) building blocks were confirmed by Marfey's analysis using commercially available standards (see SI). [32]
The stereocenters in the polyketide part of the molecule are determined by the orientation of the epoxide. Direct configurative assignment was not possible, as established derivatization methods like the Mosher's method rely on reaction of functional groups like alcohols. [33] To tackle this issue, the epoxide was transformed into an alcohol by Lewis‐acid‐catalyzed addition of methanol to the epoxide (Figure 3). The resulting secondary alcohol inherits its stereochemistry from the epoxide due to retention of the epoxide's stereochemistry in the SN2‐based ring opening reaction (Figure 3). Subsequent Mosher's esterification of the secondary alcohol revealed the alcohol to be in R‐configuration and thus the epoxide in sandarazols to be R,R‐configured. With full structure elucidation, the sandarazols’ absolute configuration, as well as the szo biosynthetic gene cluster at hand we were able to devise a biosynthetic model for the sandarazols.
Biosynthesis of the Sandarazols
The sandarazol biosynthesis pathway is based on a type 1‐trans‐AT PKS–NRPS hybrid BGC spanning 44.5 kbp and 15 open reading frames (ORFs, szoA to szoO). The megasynthase genes are encoded on the ORFs szoE to szoH. The in‐trans acting AT and the very unusual in‐trans acting DH domains are encoded on szoC and szoD. Tailoring enzymes such as the epoxidase (SzoP), the halogenase (SzoI), the short‐chain reductase (SzoA), the β‐branching cassette (SzoK to SzoO), and the biosynthetic machinery to supply the (2S)‐2,3‐diamino‐propionic acid precursor (SzoB and SzoJ) are encoded on the remaining tailoring enzymes in the BGC (Figure 4).
Figure 4.
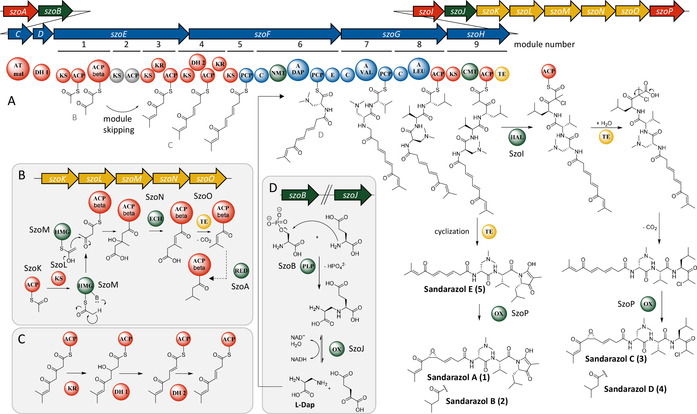
A) Scheme of the proposed sandarazol BGC and sandarazol biosynthesis by its megasynthase including the corresponding tailoring reactions. B) β‐branching reaction cascade to form the branched‐chain tail group. C) Reaction cascade that leads to the formation of two isomerized double bonds. D) Reaction sequence that supplies the amino acid l‐Dap to module 6 of the assembly line (arrows: genes; circles: domains; A: adenylation; ACP/PCP: acyl/peptidyl carrier protein; AT: acyl transferase; C: condensation; CMT/NMT: C/N methyl transferase; DH: dehydratase; E: epimerization; ECH: enoyl‐CoA hydratase; HAL: halogenase; HMG: hydroxymethylglutaryl‐CoA synthase; KR: ketoreductase; KS: ketosynthase; PLP: pyridoxal‐phosphate‐dependent enzyme; RED: reductase; TE: thioesterase; OX: oxidation).
Sandarazol biosynthesis starts with a trans‐AT PKS starter module on SzoE that loads an acetate unit onto the first acyl carrier protein (ACP). The peculiarity of this first module is the presence of a β‐branching ACP, which acts at the site where the β‐branching cassette SzoK to SzoO adds a β‐methyl branch to the first PKS extension (Figure 4 B). [34] In contrast to standard β‐branching cassettes like PyxK to PyxO in the pyxipyrrolone BGC consisting of an ACP, a ketosynthase (KS), a hydroxymethylglutaryl‐CoA synthase (HMG), and two enoyl‐CoA dehydratases (ECH), the latter ECH domain is replaced with SzoO, a thioesterase (TE). [35] This enzyme is able to replace the decarboxylation function of the second ECH as described in the curacin biosynthesis. [36] While most branched‐chain‐end moieties in polyketide synthase are biosynthesized by incorporation of an isovaleryl or an isoamyl starter unit as, for example, in fulvuthiacene biosynthesis, bongkrekic acid biosynthesis contains a similar branched‐chain starter unit biosynthesis based on a β‐branching cassette.[ 29 , 37 ] As the szo cluster does not encode enoyl reductase (ER) domains, the standalone short‐chain reductase protein szoA is likely responsible for the reduction steps leading to the production of the saturated tail group seen in the sandarazol variants 2, 4, and 7. The enzyme is also likely to be responsible for the other α,β enoyl reduction reaction next to the ketone in 6 and 7 (Figures 2 and 4). Module 2 is most likely either skipped or its ACP solely takes part in a transacylation reaction to forward the polyketide chain to module 3, as we see no polyketide extension in this module even though both the KS and the ACP in this module seem functional based on sequence alignment analysis. The following modules three to five incorporate three units of malonyl‐CoA by successive decarboxylative Claisen condensation to attach six carbon atoms to the growing polyketide chain. In this process, we observe consecutive formation of two double bonds through β‐ketoreduction by a ketoreductase (KR) and subsequent dehydration most likely performed by the in‐trans‐acting dehydratase SzoD in modules four and five. In‐trans‐acting dehydratases have already been described in trans‐AT PKS pathways such as the CurE and CorN proteins from curacin and corallopyronin, respectively.[ 38 , 39 ] The KR domain located in module 3 is most likely responsible for β‐ketoreduction in module 4, as module 4 does not contain a KR domain and no β‐ketoreduction is observed in module 3. Both double bonds formed in modules 4 and 5 are isomerized from an α,β‐double bond to a β,γ‐double bond by the double‐bond‐shifting DH2 domain encoded on szoF. [40] Similarly to the double‐bond‐shifting domains in corallopyronins, the double‐bond‐shifting DH2 on SzoF contains the first DH domain consensus motif but lacks the second DxxxQ consensus motif, a characteristic that is conserved in double‐bond‐shifting DH domains (see SI).[ 39 , 41 ] The subsequent NRPS module on szoF contains an adenylation (A) domain with a specificity code close to serine specificity according to NRPS predictor 2. This domain is assumed to accept (2S)‐2,3‐diamino‐propionic acid (l‐Dap) that is then incorporated into the nascent chain (Figure 4). [42] Biosynthesis of l‐Dap is presumed in analogy to the described pathway in Staphylococcus aureus. [43] The proteins carrying out the corresponding biosynthetic steps in MSr10575 are the SbnA homolog SzoB and the SbnB homolog SzoJ. SbnA is a pyridoxal phosphate (PLP)‐dependent aminotransferase‐type enzyme that connects the amino group of glutamate to replace the phosphate moiety of an O‐phosphoserine molecule from the bacterium's primary metabolism (Figure 4 D). [44] SbnB is an NAD‐dependent oxidase that releases α‐ketoglutarate from this intermediate and forms l‐Dap. A SAM‐dependent N‐methyl transferase domain in module 6 subsequently transfers two methyl groups to the free amino group of the attached l‐Dap building block to form l‐Me2Dap, which is subsequently epimerized to d‐Me2Dap by the following epimerase domain (Figure 4). In contrast to most N‐methyl transferase domains built into NRPS systems, which transfer a methyl group onto the α‐nitrogen atom that will later form the peptide bond, this methylation reaction consecutively transfers two methyl groups to the nitrogen atom in 3‐position of l‐Dap. [45] The following two modules incorporate l‐Val and l‐Leu according to NRPS textbook logic. [46] Incorporation of all three amino acids was proven by feeding of stable‐isotope‐labeled precursors (see SI). Module 9 is a PKS module that incorporates another malonyl‐CoA unit by decarboxylative Claisen condensation and adds an α‐methyl branch via a SAM‐dependent C‐methyl transferase domain to the molecule (Figure 4).
We did not find any non‐chlorinated, non‐cyclized sandarazols in the culture broth and cyclization of an amide nitrogen with a carboxylic acid function to form the tetramic acid heterocycle is very unlikely to occur spontaneously. We therefore suspect that chlorination of sandarazols by the halogenase SzoJ occurs on the assembly line. This reaction is likely to govern whether sandarazol is released from the megasynthase as a chlorinated open‐chain molecule prone to decarboxylation as seen in 3 or as a non‐chlorinated cyclized product like 1 (Figure 4). Halogenation by SzoJ occurs via the accepted FAD‐dependent halogenation mechanism similar to synthetic α‐chlorination of β‐keto acids. [47] This chlorinated ACP‐bound intermediate is released from the assembly line by the TE and quickly loses its terminal carboxylic acid moiety by decarboxylation, a reaction occurring spontaneously in α‐chlorinated β‐keto acids. If chlorination does not occur, the molecule is subsequently cleaved off the assembly line by a TE domain with a much slower rate, thus cyclizing the product to form a 5‐membered ring system as seen, for example, in 1 and 2 (Figure 4). To finalize sandarazol biosynthesis, epoxidation needs to occur next to the ketone moiety in the polyketide part of the molecule. Catalysis of such a reaction is likely performed by FAD‐dependent monooxygenase SzoP that putatively performs a reaction similar to the one performed by the FAD‐dependent epoxidizing styrene monooxygenase. [48] We assume that sandarazols are finally exported into the surrounding medium by an ABC exporter of broad range specificity encoded somewhere else in the MSr10575 genome, as no specific exporter system is encoded within or in close proximity to the sandarazol BGC or elsewhere on the pSa001 plasmid.
Biological Activity of the Sandarazols
Due to the low stability of the sandarazols in aerobic environments, antimicrobial and anti‐proliferative activities were only determined for 1 and 3 as representatives for one chlorinated and one cyclized member of the sandarazol compound family, respectively. Under the specified assay conditions, the non‐chlorinated 1 displays only very limited biological activity, while the chlorinated 3 shows prominent cytotoxicity as well as antibiotic activity against Mycobacterium smegmatis and a variety of other indicator bacteria for Gram‐positive pathogens (Table 1).
Table 1.
Antimicrobial and cytotoxic activities of sandarazol A (1) and C (3) as minimum inhibitory concentrations (MIC) and inhibitory concentrations at 50 % inhibition (IC50).
|
|
Microbial strain |
MIC 1 [μm] |
MIC 3 [μm] |
|
|---|---|---|---|---|
|
|
C. albicans |
>110 |
110 |
|
|
|
P. anomala |
>110 |
110 |
|
|
|
C. freundii |
>110 |
>110 |
|
|
|
A. baumanii |
>110 |
>110 |
|
|
|
S. aureus |
110 |
55 |
|
|
|
B. subtilis |
>110 |
55 |
|
|
|
E. coli |
>110 |
>110 |
|
|
|
P. aeruginosa |
>110 |
55 |
|
|
|
M. smegmatis |
>110 |
14 |
|
|
Cell line |
IC50 1 [μm] |
IC50 3 [μm] |
|---|---|---|---|
|
|
Human colon cancer HCT116 |
>64 |
0.5 |
We observe that the biological activity of the sandarazols mainly stems from the chlorinated sandarazols such as 3, which shows both the best antimicrobial and anti‐proliferative activity. Its cytotoxicity of 0.5 μm against HCT116 cells might make 3 a promising candidate for compound optimization and mode‐of‐action studies. Such approaches directed towards improvements of the sandarazols’ stability against oxygen would probably increase the observable IC50 values, as the values shown here cannot be determined under anaerobic conditions that stabilize the sandarazols. The antimicrobial and anti‐proliferative activities of 3 indicate that those secondary metabolites enhance the bacterium's capability to defend itself against competitors like other bacteria or eukaryotic microorganisms. The position of the szo BGC on the replicative plasmid pSa001 indicates that this bacterial self‐defense strategy can be transferred between bacteria via conjugation.
Conclusion and Outlook
Herein, we identified pSa001 as the second myxobacterial autonomously replicating plasmid known to date. It was found in the newly isolated Sandaracinus sp. MSr10575, the second myxobacterial isolate belonging to the Sandaracinus clade. The 209.7 kbp pSa001 plasmid itself encodes the szo biosynthetic gene cluster as well as five transposase‐ and one integrase‐type ORFs. Therefore, the plasmid's content including the szo BGC may be transferred between bacterial species, not only by conjugative transfer of the plasmid as an autonomously replicating unit, but also by transposition or integration into the acceptor strain's genome. We chose to investigate and characterize the pSa001‐borne szo BGC that turned out to be cryptic under laboratory conditions. Previous examples of such plasmid‐borne secondary metabolite pathways have shown to be responsible for esmeraldin and mycolactone production, both of which show significant biological activity.[ 5 , 6 ] The biosynthetic origin of these two compounds led to the theory that the BGC encoded on pSa001 also encodes the biosynthetic machinery for toxin production. After activation of the BGC located on the plasmid pSa001 by promoter exchange, we observed the production of the sandarazols, a series of type1‐trans‐AT PKS–NRPS hybrid secondary metabolites. The chemical structure of the sandarazols features a permanent positive charge at physiological conditions at the tertiary amine, a Michael acceptor system, an epoxyketone as well as an α‐chlorinated ketone. These compounds are therefore prone to display a variety of different reactivities that might well translate into diverse biological activities. More precisely, the sandarazols show promising anti‐bacterial and anti‐proliferative activities. Following in‐depth investigation of the szo BGC, we developed a concise biosynthetic model, which explains the sandarazol biosynthesis on its megasynthase protein. Furthermore, we shed light on a variety of uncommon biosynthetic steps such as the first description of incorporation of Me2Dap into natural products, polyketide β‐branching, isomerization of two consecutive double bonds by a single shifting DH‐domain, and α‐chlorination‐governed release of the sandarazols from the assembly line, all of which warrant further investigations of biosynthetic details. The discovery of the sandarazols does not only highlight the capability of myxobacteria to biosynthesize diverse and biologically active secondary metabolites, it also emphasizes the promise of activating biosynthetic gene clusters by inducible heterologous promoters. Vanillate‐inducible promoter systems have proven once more to be able to unlock a larger fraction of the cryptic secondary metabolome of myxobacteria in promoter exchange experiments. [8] The sandarazols combine biological activity with both their intriguing chemical structure and biosynthesis. Thus, natural products encoded on other bacterial plasmids might feature similar intriguing properties. The discovery of sandarazols therefore highlights plasmid‐encoded BGCs as valuable targets in the search for novel biologically active natural products in the future. As for myxobacteria, encoding toxin BGCs on plasmids that are transferrable between themselves may provide myxobacteria with a competitive edge in their ecological niche. This transfer of toxin production machinery would make particular sense, as myxobacteria do not live individually but rather as part of a swarm which shows wolf‐pack‐like predatory behavior. [49] Thus, distribution of the genetic means for toxin production among its kind should be advantageous for these microorganisms in their collective predation and survival strategies.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
The authors want to thank S. Schmidt for antimicrobial profiling of the sandarazols and A. Amann for the cytotoxicity assays. Furthermore, the authors want to thank N. Zaburannyi for assembly of the MSr10575 genome and D. Krug for careful revision of the manuscript. Open access funding enabled and organized by Projekt DEAL.
F. Panter, C. D. Bader, R. Müller, Angew. Chem. Int. Ed. 2021, 60, 8081.
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.1101/2020.10.06.323741).
References
- 1. Naylor N. R., Atun R., Zhu N., Kulasabanathan K., Silva S., Chatterjee A., Knight G. M., Robotham J. V., Antimicrob. Resist. Infect. Control 2018, 7, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenciani A., Morroni G., Pollini S., Tiberi E., Mingoia M., Varaldo P. E., Rossolini G. M., Giovanetti E., J. Antimicrob. Chemother. 2016, 71, 307. [DOI] [PubMed] [Google Scholar]
- 3. Soucy S. M., Huang J., Gogarten J. P., Nat. Rev. Genet. 2015, 16, 472. [DOI] [PubMed] [Google Scholar]
- 4. Luna V. A., King D. S., Peak K. K., Reeves F., Heberlein-Larson L., Veguilla W., Heller L., Duncan K. E., Cannons A. C., Amuso P., et al., J. Clin. Microbiol. 2006, 44, 2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stinear T. P., Mve-Obiang A., Small P. L., Frigui W., Pryor M. J., Brosch R., Jenkin G. A., Johnson P. D., Davies J. K., Lee R. E., et al., Proc. Natl. Acad. Sci. USA 2004, 101, 1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rui Z., Ye M., Wang S., Fujikawa K., Akerele B., Aung M., Floss H. G., Zhang W., Yu T.-W., Chem. Biol. 2012, 19, 1116. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a. Cragg G. M., Newman D. J., Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3670; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Newman D. J., Cragg G. M., J. Nat. Prod. 2016, 79, 629. [DOI] [PubMed] [Google Scholar]
- 8. Wenzel S. C., Müller R., Mol. Biosyst. 2009, 5, 567. [DOI] [PubMed] [Google Scholar]
- 9. Bader C. D., Panter F., Müller R., Biotechnol. Adv. 2020, 39, 107480. [DOI] [PubMed] [Google Scholar]
- 10. Herrmann J., Fayad A. A., Müller R., Nat. Prod. Rep. 2017, 34, 135. [DOI] [PubMed] [Google Scholar]
- 11. Hoffmann T., Krug D., Bozkurt N., Duddela S., Jansen R., Garcia R., Gerth K., Steinmetz H., Müller R., Nat. Commun. 2018, 9, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blin K., Wolf T., Chevrette M. G., Lu X., Schwalen C. J., Kautsar S. A., Suarez Duran H. G., de Los Santos E. L. C., Kim H. U., Nave M., et al., Nucleic Acids Res. 2017, 45, W36–W41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cimermancic P., Medema M. H., Claesen J., Kurita K., Wieland Brown L. C., Mavrommatis K., Pati A., Godfrey P. A., Koehrsen M., Clardy J., et al., Cell 2014, 158, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ross A. C., Gulland L. E. S., Dorrestein P. C., Moore B. S., ACS Synth. Biol. 2015, 4, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mao D., Okada B. K., Wu Y., Xu F., Seyedsayamdost M. R., Curr. Opin. Microbiol. 2018, 45, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hug J. J., Bader C. D., Remškar M., Cirnski K., Müller R., Antibiotics 2018, 7, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohr K. I., Garcia R. O., Gerth K., Irschik H., Müller R., Int. J. Syst. Evol. Microbiol. 2012, 62, 1191. [DOI] [PubMed] [Google Scholar]
- 18. Steinmetz H., Mohr K. I., Zander W., Jansen R., Gerth K., Müller R., J. Nat. Prod. 2012, 75, 1803. [DOI] [PubMed] [Google Scholar]
- 19. Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., et al., Science 2009, 323, 133. [DOI] [PubMed] [Google Scholar]
- 20. Feng J., Chen X. J., Sun X., Wang N., Li Y. Z., Plasmid 2012, 68, 105. [DOI] [PubMed] [Google Scholar]
- 21. Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., Nguyen D. D., Watrous J., Kapono C. A., Luzzatto-Knaan T., et al., Nat. Biotechnol. 2016, 34, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panter F., Krug D., Baumann S., Müller R., Chem. Sci. 2018, 9, 4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamanaka K., Reynolds K. A., Kersten R. D., Ryan K. S., Gonzalez D. J., Nizet V., Dorrestein P. C., Moore B. S., Proc. Natl. Acad. Sci. USA 2014, 111, 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cortina N. S., Krug D., Plaza A., Revermann O., Müller R., Angew. Chem. Int. Ed. 2012, 51, 811; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 836. [Google Scholar]
- 25. Shizuya H., Birren B., Kim U. J., Mancino V., Slepak T., Tachiiri Y., Simon M., Proc. Natl. Acad. Sci. USA 1992, 89, 8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hug J. J., Panter F., Krug D., Müller R., J. Ind. Microbiol. Biotechnol. 2019, 46, 319. [DOI] [PubMed] [Google Scholar]
- 27. Iniesta A. A., García-Heras F., Abellón-Ruiz J., Gallego-García A., Elías-Arnanz M., J. Bacteriol. 2012, 194, 5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffmann T., Krug D., Hüttel S., Müller R., Anal. Chem. 2014, 86, 10780. [DOI] [PubMed] [Google Scholar]
- 29. Panter F., Krug D., Müller R., ACS Chem. Biol. 2019, 14, 88. [DOI] [PubMed] [Google Scholar]
- 30. Meusel M., Hufsky F., Panter F., Krug D., Müller R., Böcker S., Anal. Chem. 2016, 88, 7556. [DOI] [PubMed] [Google Scholar]
- 31. Duddeck H. in Houben-Weyl Methods of organic chemistry, Vol. E 21 a, Thieme, Stuttgart, 1995. [Google Scholar]
- 32.
- 32a. Harada K.-i., Fujii K., Hayashi K., Suzuki M., Ikai Y., Oka H., Tetrahedron Lett. 1996, 37, 3001; [Google Scholar]
- 32b. Marfey P., Carlsberg Res. Commun. 1984, 49, 591. [Google Scholar]
- 33. Hoye T. R., Jeffrey C. S., Shao F., Nat. Protoc. 2007, 2, 2451. [DOI] [PubMed] [Google Scholar]
- 34. Maloney F. P., Gerwick L., Gerwick W. H., Sherman D. H., Smith J. L., Proc. Natl. Acad. Sci. USA 2016, 113, 10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kjaerulff L., Raju R., Panter F., Scheid U., Garcia R., Herrmann J., Müller R., Angew. Chem. Int. Ed. 2017, 56, 9614; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 9743. [Google Scholar]
- 36. Gehret J. J., Gu L. C., Gerwick W. H., Wipf P., Sherman D. H., Smith J. L., J. Biol. Chem. 2011, 286, 14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moebius N., Ross C., Scherlach K., Rohm B., Roth M., Hertweck C., Chem. Biol. 2012, 19, 1164. [DOI] [PubMed] [Google Scholar]
- 38. Akey D. L., Razelun J. R., Tehranisa J., Sherman D. H., Gerwick W. H., Smith J. L., Structure 2010, 18, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pogorevc D., Panter F., Schillinger C., Jansen R., Wenzel S. C., Müller R., Metab. Eng. 2019, 55, 201. [DOI] [PubMed] [Google Scholar]
- 40. Gay D. C., Spear P. J., Keatinge-Clay A. T., ACS Chem. Biol. 2014, 9, 2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kusebauch B., Busch B., Scherlach K., Roth M., Hertweck C., Angew. Chem. Int. Ed. 2010, 49, 1460; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 1502. [Google Scholar]
- 42. Röttig M., Medema M. H., Blin K., Weber T., Rausch C., Kohlbacher O., Nucleic Acids Res. 2011, 39, W362–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beasley F. C., Cheung J., Heinrichs D. E., BMC Microbiol. 2011, 11, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kobylarz M. J., Grigg J. C., Takayama S.-i. J., Rai D. K., Heinrichs D. E., Murphy M. E. P., Chem. Biol. 2014, 21, 379. [DOI] [PubMed] [Google Scholar]
- 45. Hornbogen T., Riechers S.-P., Prinz B., Schultchen J., Lang C., Schmidt S., Mügge C., Turkanovic S., Süssmuth R. D., Tauberger E., et al., ChemBioChem 2007, 8, 1048. [DOI] [PubMed] [Google Scholar]
- 46. Walsh C. T., Nat. Prod. Rep. 2016, 33, 127. [DOI] [PubMed] [Google Scholar]
- 47.
- 47a. Yeh E., Blasiak L. C., Koglin A., Drennan C. L., Walsh C. T., Biochemistry 2007, 46, 1284; [DOI] [PubMed] [Google Scholar]
- 47b. Guan X., An D., Liu G., Zhang H., Gao J., Zhou T., Zhang G., Zhang S., Tetrahedron Lett. 2018, 59, 2418. [Google Scholar]
- 48.
- 48a. Kantz A., Gassner G. T., Biochemistry 2011, 50, 523; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48b. Morrison E., Kantz A., Gassner G. T., Sazinsky M. H., Biochemistry 2013, 52, 6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muñoz-Dorado J., Marcos-Torres F. J., García-Bravo E., Moraleda-Muñoz A., Pérez J., Front. Microbiol. 2016, 7, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


