Abstract
Various studies demonstrated that bone morphogenetic proteins (BMPs) and their antagonists contribute to the development of cancers. Chordin‐like 2 (CHRDL2) is a member of BMP antagonists. However, the role and its relative mechanism of CHRDL2 in osteosarcoma remains unclear. In the present study, we demonstrated that the expression of CHRDL2 was significantly upregulated in osteosarcoma tissues and cell lines compared with adjacent tissues and human normal osteoblast. Inhibition of CHRDL2 decreased the proliferation and colony formation of osteosarcoma cells in vitro, as well as the migration and invasion. CHRDL2 overexpression induced the opposite effects. CHRDL2 can bind with BMP‐9, thus decreasing BMP‐9 expression and the combination to its receptor protein kinase ALK1. It was predicted that BMP‐9 regulates PI3K/AKT pathways using gene set enrichment analysis. Inhibition of CHRDL2 decreased the activation of PI3K/AKT pathway, while overexpression of CHRDL2 upregulated the activation. Increasing the expression of BMP‐9 reversed the effects of CHRDL2 overexpression on the activation of PI3K/AKT pathway, as well as the proliferation and metastasis of osteosarcoma cells. Take together, our present study revealed that CHRDL2 upregulated in osteosarcoma tissues and cell lines, and promoted osteosarcoma cell proliferation and metastasis through the BMP‐9/PI3K/AKT pathway. CHRDL2 maybe an oncogene in osteosarcoma, as well as novel biomarker for the diagnosis of osteosarcoma.
Keywords: BMP‐9/PI3K/AKT, CHRDL2, metastasis, osteosarcoma, proliferation
Abbreviations
- AKT
AKT serine/threonine kinase
- BMP
bone morphogenetic protein
- BMP‐9
bone morphogenetic protein 9
- CHRDL2
chordin like 2
- GSEA
gene set enrichment analysis
- PI3K
phosphatidylinositol 3‐kinase
1. INTRODUCTION
Osteosarcoma is the most commonly primary bone tumor type with genetically unstable and highly malignant (Yang et al., 2016). Osteosarcoma mainly affects children and adolescents and which is the second leading cause of tumor‐related death of children and adolescents (Zhao et al., 2018). In the United States, about 900 osteosarcoma patients are diagnosed each year. The biological characteristics of osteosarcoma most generally manifested a highly rate of transfer to lungs (Morrow & Khanna, 2015). Given the introduction of surgery and neoadjuvant chemotherapy as a primary therapy for osteosarcoma, the 5‐year survival rate of patients without metastasis has rapidly increased to nearly 80%, but since the 1970s, there have been no reports of major survival improvements (Thanapprapasr et al., 2017). Therefore, uncovering the molecular mechanism of the development of osteosarcoma may help the therapy of osteosarcoma.
Bone morphogenic proteins (BMPs), a vital secreted cytokines, belongs to the TGF‐β family of proteins, and the abnormal expression of them can be observed in various cancers including osteosarcoma (Wu et al., 2016). However, the role of BMPs in cancer was contradictory. Martinez et al. (2017) found that bone morphogenic protein 4 promoted tumor progression in bladder cancer. However, Hu et al. (2016) showed that bone morphogenic protein 6 inhibited the metastasis of breast cancer cells via inhibiting the expression of matrix metallopeptidase 1. Lv et al. (2013) demonstrated that BMP‐9 had potential to inhibit the proliferation and migration of osteosarcoma cells. There are many factors influence the expression and activity of BMPs, such as BMP antagonists, a class secreted protein which can block the interaction between BMPs and their homologous cell surface receptors (Bach et al., 2018). Chordin‐like 2 (CHRDL2), an antagonist of BMPs, can prevent the interaction between BMPs and their cognate cell surface receptor (Fujisawa et al., 2009). It has been reported that the overexpression of CHRDL2 predict the poor prognosis of colorectal cancer (Sun et al., 2017). However, the role of CHRDL2 in the development of osteosarcoma still remained unclear.
In the current study, we explored the role and its molecular mechanism of CHRDL2 in osteosarcoma. We demonstrated that CHRDL2 played as onco‐gene in osteosarcoma, and promoted the proliferation and metastasis via BMP‐9/PI3K/AKT pathway. CHRDL2 may be a novel biomarker for the diagnosis of osteosarcoma, as well as potential target for therapy.
2. MATERIALS AND METHODS
2.1. Clinical samples
Human osteosarcoma tissues and corresponding adjacent tissues were obtained from maternal and child health hospital of Guiyang. All the specimens were obtained with informed consent for patients and were approved by the Ethics Committee of maternal and child health hospital of Guiyang. The study was performed in accordance with the principles embodied in the Declaration of Helsinki.
2.2. Real‐time quantitative polymerase chain reaction (PCR)
Total RNA in the tissues and cells was extracted using TRIZOL reagent (Yeasen) according to the protocol provided by manufacturer's. The complementary DNA used to examine targeted genes were synthesized using the PrimeScriptTM RT reagent kit (Yeasen). The following primers used in our study were showed as follow: CHRDL2 forward primer, 5’‐CTGGCACCCCTACTTGGAG‐3’, and CHRDL2 reverse primer, 5’‐GCGGTAACAACTCACATGGG‐3’; β‐actin forward primer, 5’‐CATGTACGTTGCTATCCAGGC‐3’, and β‐actin reverse primer, 5’‐CTCCTTAATGTCACGCACGAT‐3’. The response condition was presented as the following: One initial cycle for 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 31 s at 60°C. β‐Actin acted as the control and tested the relative expression of CHRDL2. Relative expression of the targeted genes was calculated via analysis.
2.3. Immunohistochemical staining (IHC)
Osteosarcoma and adjacent tissues were fixed, dehydrated, and embedded in paraffin (Servicebio), and then cut into 2‐µm specimens. Specimens were deparaffinized and rehydrated with graded xylene and alcohols, respectively. After restoration by sodium citrate and blocking using H2O2 and bovine serum albumin (Servicebio), the specimens were incubated with primary anti‐CHRDL2 antibody (Cat No. ab198786; Abcam) and anti‐BMP‐9 antibody (Cat No: ab30588; Abcam) for 16 h at 4°C. Subsequently, the sections were immunohistochemically stained with horseradish peroxidase‐conjugated goat anti‐rabbit antibodies (Servicebio) for 2 h at room temperature. After incubation with the Cell and Tissue Staining HRP‐DAB Kit (Servicebio), an orthophoto light microscope was used to collect images (magnification ×200 and ×400).
2.4. Cell culture and cell transfection
Human osteosarcoma cell lines (MG63, U2OS, Saos‐2, and 143B) and human normal osteoblast (hFOB) were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines including MG63, U2OS, Saos‐2, and 143B were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) at 37°C in a humidified atmosphere incubator containing 5% CO2, while hFOB was cultured at 34°C (other conditions were same). The lentiviruses for BMP‐9 overexpression (Lv‐BMP‐9), CHRDL2 overexpression (Lv‐CHRDL2) and the corresponding negative control were obtained from Sangon. Lentiviral of shRNA against CHRDL2 (sh‐CHRDL2) and scramble control (sh‐scramble) was obtained from Genechem. The sequence of sh‐CHRDL2 was AAGTCAGGAAGCAAGACTT. To obtaining stably transfected cell line, osteosarcoma cells were treated with 0.5 μg/ml puromycin (Selleck) for 2 weeks after 48 h transfection.
2.5. Western bolt
Cells were collected and lysed using a RIPA buffer (Boster Biological Technology) containing protease inhibitor cocktail and PMSF (Boster Biological Technology). After centrifugation (12,500 rpm/15 min), separate proteins were collected from cellular debris. Bicinchoninic acid method was used to determine the protein concentration. Proteins (30 μg) were separated under 90 V by voltage via the 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto the polyvinylidene difluoride membrane (Millipore). After blocking the membranes with TBST containing 5% skim milk for 2 h at room temperature, the membranes were incubated with primary anti‐CHRDL2, anti‐BMP‐9, BMP‐7 (Cat No: ab269586; Abcam), BMP‐4 (Cat No: ab124715; Abcam), BMP‐2 (Cat No: ab276041; Abcam), ALK1 (Cat No: 14745‐1‐AP; Proteintech), anti‐PI3K (Cat No: 60225‐1‐Ig; Proteintech), anti‐p‐PI3K (Cat No: 17366; Cell Signaling Technology), anti‐AKT (Cat No: 10176‐2‐AP; Proteintech), anti‐p‐AKT (Cat No: 66444‐1‐Ig; Proteintech), anti‐MMP‐9 (Cat No: 10375‐2‐AP; Proteintech) antibodies (all 1:1000 dilution) overnight at 4°C. After washing with TBST three times, the membranes were incubated with the secondary antibody (dilution 1:3000) for 2 h at room temperature. Finally, the signals were detected by the Photoshop Image Analysis software CS3 (Adobe Systems).
2.6. Cell counting kit‐8 (CCK‐8) assay
Osteosarcoma cells (3 × 103cells/well) were placed into 96‐well plates and cultured at 37°C in a humidified atmosphere incubator containing 5% CO2. Then, in 0 h (cell adherence), 24, 48, and 72 h, a total of 10 µl of CCK8 reagent was added into each well maintaining 2 h and was measured by a spectrophotometer (Bio‐Rad Laboratories Inc.) at a wavelength of 450 nm.
2.7. Colony formation
Osteosarcoma cells were seeded onto six‐well plates (1 × 103 cells/well; dish diameter = 6 cm) and maintained for 14 days. Then, the colonies were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min. After washing residual crystal violet solution, the colonies (>10 mm2) were counted.
2.8. Wound healing assay
Osteosarcoma cells were seeded on the six‐well plate and cultured at 37°C. While the convergence degree of cells was >95%, a 20 μl sterilized tip was used to create a wound. After washing the floating cells, the medium was replaced with the free‐FBS DMEM medium. Cells were then photographed in 0 and 24 h. The percentage of the wound healing at 24 h compared with 0 h was measured using Image Pro Plus software.
2.9. Transwell assay
Total 100 μl of matrigel (Millipore) was in each transwell chamber (Millipore) before use. After 2 h, 1 × l04 osteosarcoma cells were placed in the upper chambers, while DMEM medium contained 10% FBS was added in the lower chambers. At 24 h, chambers were fixed and stained with 0.1% crystal violet solution for 10 min. After removing cells which retained in the upper chambers, five random fields of each chamber were photographed under an inverted light microscope (Olympus).
2.10. Immunoprecipitation
Total protein in cells was dissolved using mild RIPA solution (Boster) and then collected via centrifuge at 4°C. Sixty microliters protein was added into 1.5 ml centrifuge tubes, and then incubated with 3 µg antibodies against CHRDL2, ALK1, and IgG at 4°C overnight, respectively. For each sample, 30 μl Protein A/G Plus‐Agarose (MCE) was injected to form immunocomplex. After 6 h. the agarose beads were collected and washed by 1 ml phosphate‐buffered saline for five times to remove free antibodies and proteins. Then, agarose beads were mixed with ×2 loading buffer and boiled at 100°C for 5 min. Finally, the proteins were used to perform western blot to detect the expression of CHRDL2, ALK1, BMP‐2, BMP‐4, BMP‐7, and BMP‐9.
2.11. Gene set enrichment analysis (GSEA)
To predicting the pathways regulated by CHRDL2 in osteosarcoma, GSEA was performed. Gene expressed‐profile GSE99671 and GSE87624 were downloaded from Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/gds/). After annotation of probe and standardization of data, the osteosarcoma tissues in these two profiles were divided into high and low expression group according the medium expression of CHRDL2. Then, GSEA software (UC San Diego and Broad Institute, https://www.gsea-msigdb.org/gsea/index.jsp) was used to analyze the pathways enriched in high CHRDL2 expression group tissues. p < .05 was set as cut‐off to consider pathways significantly enriched in.
2.12. Statistical analysis
All statistical analyses in our present study were performed by GraphPad Prism 6.0 tool. Statistical significance of differences between two groups using Student's t test, while analysis of multigroups was performed by one‐way analysis of variance and LSD‐t test. p < .05 was considered statistically significant.
3. RESULTS
3.1. CHRDL2 was upregulated in osteosarcoma tissues and cell lines
The expression of CHRDL2 in osteosarcoma (N = 25) and corresponding adjacent normal tissues (N = 25) was detected using quantitative reverse transcription PCR (qRT‐PCR) method. Results showed that the messenger RNA (mRNA) level of CHRDL2 was increased in osteosarcoma tissues compared with adjacent tissues (p < .05, Figure 1a). Similarly, results of IHC demonstrated that the protein level of CHRDL2 was upregulated in osteosarcoma tissues compared with adjacent tissues (Figure 1b). Then, qRT‐PCR and western blot were used to detect the expression of CHRDL2 in osteosarcoma cell lines and osteoblast. Results showed that both the mRNA and protein level of CHRDL2 was increased in MG63, Saos‐2, U2OS, and 143B osteosarcoma cells compared with osteoblast hFOB (all p < .05, Figure 1c,d).
Figure 1.
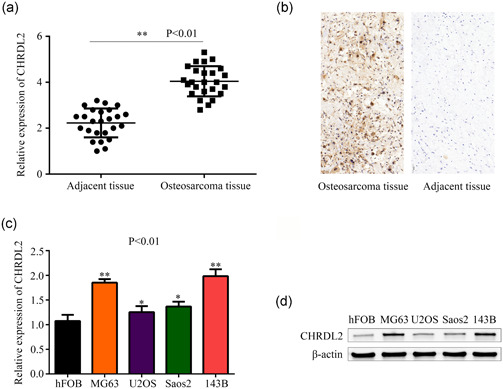
CHRDL2 was highly expressed in osteosarcoma tissues and cell lines. (a) qRT‐PCR was used to detected the expression of CHRDL2 in osteosarcoma tissues (n = 25) and adjacent tissues (n = 25). (b) Immunohistochemistry was used to detect the expression of CHRDL2 in osteosarcoma tissues and adjacent tissues (magnification ×200. (c) qRT‐PCR was used to detected the mRNA levels of CHRDL2 in normal osteoblast hFOB cells and osteosarcoma cell lines MG63, U2OS, Saos‐2, and 143B. (d) Western blot was used to detected the protein levels of CHRDL2 in normal osteoblast hFOB cells and osteosarcoma cell lines MG63, U2OS, Saos‐2, and 143B. *p < .05; **p < .01. CHRDL2, chordin‐like 2; mRNA, messenger RNA; qRT‐PCR, quantitative reverse transcription polymerase chain reaction
3.2. Inhibition of CHRDL2 suppressed the proliferation and metastasis of osteosarcoma cells
Lentiviral of shRNA against CHRDL2 (sh‐CHRDL2) was used to construct stably lowly expressed CHRDL2 cell lines. Results of qRT‐PCR and western blot showed that sh‐CHRDL2 significantly decreased the expression of CHRDL2 in MG63 and 143B cells (all p < .05, Figure 2a,b). CCK‐8 assay was performed to detect the effect of CHRDL2 inhibition on the proliferation of MG63 and 143B cells. The results indicated that inhibition of CHRDL2 obviously decreased the proliferation of MG63 and 143B cells (all p < .05, Figure 2c). Similarly, colony formation assays demonstrated that inhibition of CHRDL2 decreased the colony number of MG63 and 143B cells (all p < .05, Figure 2d). Furthermore, would healing assay and transwell assay revealed that suppression of CHRDL2 significantly reduced the migration and invasion of MG63 and 143B cells (all p < .05, Figure 2e,f).
Figure 2.
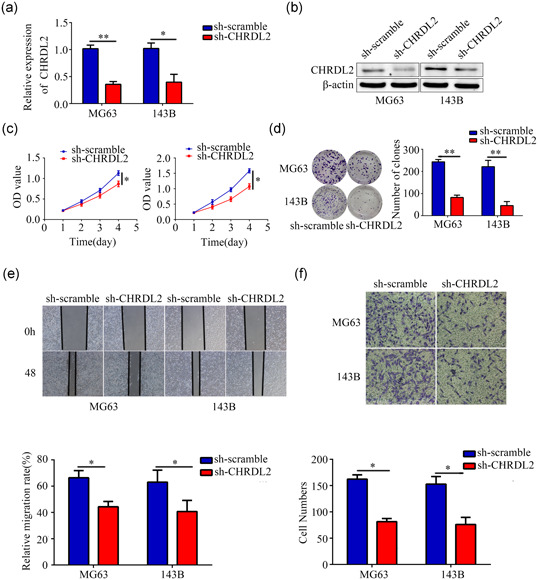
Inhibition of CHRDL2 decreased osteosarcoma cell proliferation and mobility. (a) The efficiency of targeted CHRDL2 short hairpin RNA (sh‐CHRDL2) was detected by PCR. (b) The efficiency of sh‐CHRDL2 was detected by western blot. (c) CCK8 assays were performed to detect the effects of CHRDL2 inhibition on osteosarcoma cell proliferation. (d) Colony formation assays were performed the he effect of CHRDL2 inhibition on osteosarcoma cell colony formation. (e) Wound healing assay was used to detect the effects of CHRDL2 inhibition on osteosarcoma cell migration. (f) Transwell assay was used to detect the effects of CHRDL2 inhibition on osteosarcoma cell invasion. *p < .05; **p < .01. CCK‐8, cell counting kit‐8; CHRDL2, chordin‐like 2; PCR, qRT‐PCR, polymerase chain reaction
3.3. Overexpression of CHRDL2 promoted the proliferation and metastasis of osteosarcoma cells
CHRDL2 overexpression lentiviral (Lv‐CHRDL2) was used to construct stably highly expressed CHRDL2 cell lines. Results of qRT‐PCR and western blot showed that the expression of CHRDL2 in Lv‐CHRDL2 group was significantly increased in Saos2 and U2OS cells (all p < .05, Figure 3a,b). CCK‐8 and colony formation assays showed that overexpression of CHRDL2 significantly increased the proliferation and colony formation of Saos2 and U2OS cells (all p < .05, Figure 3c,d). Wound healing assay and transwell assays indicated that the migration and invasion of Saos2 and U2OS cells was higher in Lv‐CHRDL2 compared control group (all p < .05, Figure 3e,f).
Figure 3.
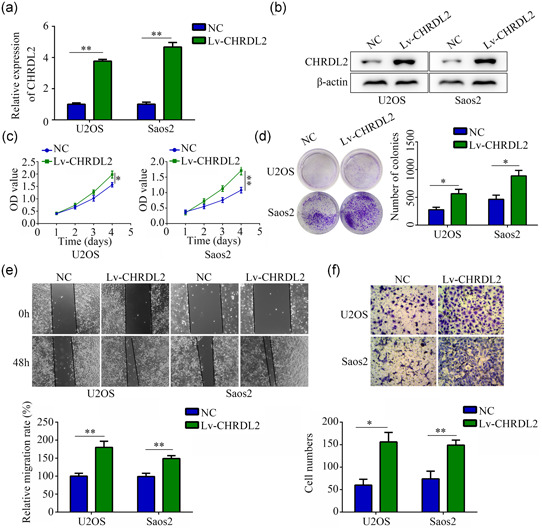
Overexpression of CHRDL2 promoted osteosarcoma cell proliferation and mobility. (a) The efficiency of CHRDL2 overexpressed lentivirus (lv‐CHRDL2) was detected by PCR. (b) The efficiency of lv‐CHRDL2 was detected by western blot. (c) CCK8 assays were performed to detect the effects of CHRDL2 overexpression on osteosarcoma cell proliferation. (d) Colony formation assays were performed to detect the effects of CHRDL2 overexpression on osteosarcoma cell colony formation. (e) Wound healing assay was used to detect the effects of CHRDL2 overexpression on osteosarcoma cell migration. (f) Transwell assay was used to detect the effects of CHRDL2 overexpression on osteosarcoma cell invasion. *p < .05; **p < .01. CCK‐8, cell counting kit‐8; CHRDL2, chordin‐like 2; PCR, qRT‐PCR, polymerase chain reaction
3.4. CHRDL2 blocked BMP‐9 and regulated PI3K/AKT pathway
CHRDL2 was a antagonist of BMPs, which can bind with BMPs and blocked them to interact with corresponding cognate cell surface receptor (Fujisawa et al, 2009). Therefore, we then determined whether CHRDL2 had potential to block BMPs in osteosarcoma. Results of immunoprecipitation revealed that CHRDL2 bind with BMP‐9, but no BMP‐7, BMP‐4, and BMP‐2 (Figure 4a). Inhibition of CHRDL2 increased the expression of BMP‐9, while overexpression of CHRDL2 decreased its expression (Figure 4b,c). Moreover, we found that inhibition of CHRDL2 increased the combination of BMP‐9 to its receptor ALK1 (Figure 4d). In addition, we found that BMP‐9 was lowly expressed in osteosarcoma tissues while CHRDL2 was overexpressed (Figure 4e). As the previous study indicated that BMP‐9 can bind with ALK1 and inhibit PI3K/AKT pathway (Song et al., 2019), we then determine whether CHRDL2 had a relationship with PI3K/AKT pathway. GSEA performed in gene expression of GSE99671 and GSE87624 both showed that CHRDL2 had potential to regulate PI3K/AKT pathway (Figure 4f). Similarly, we found that inhibition of CHRDL2 significantly decreased the expression p‐PI3K, p‐AKT, and MMP‐9, while overexpression of CHRDL2 induced the opposite effects (all p < .05, Figure 4g,h).
Figure 4.
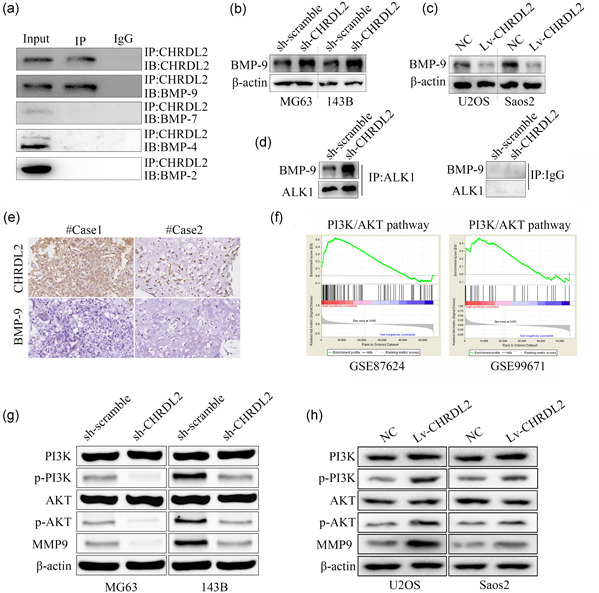
CHRDL2 blocked BMP‐9 and regulated PI3K/AKT pathway. (a) Immunoprecipitation demonstrated the binding between BMP‐9, BMP‐7, BMP‐4, BMP‐2, and CHRDL2. (b,c) Western blot was used to detect the expression of BMP‐9 while CHRDL2 was overexpressed or inhibited. (d) Immunoprecipitation was performed to detect the combination between BMP‐9 and ALK1 while CHRDL2 was inhibited. (e) Immumohistochemical staining was used to detect the expression relationship between CHRDL2 and BMP‐9 in osteosarcoma tissues (magnification ×400). (f) GSEA analysis showed CHRDL2 had potential to regulate PI3K/AKT pathway according to the gene expression profile GSE87624 and GSE99671. (g) Western blot was used to detect the expression of PI3K, p‐PI3K, AKT, p‐AKT, and MMP‐9 while CHRDL2 was inhibited. (h) Western blot was used to detect the expression of PI3K, p‐PI3K, AKT, p‐AKT, and MMP‐9 while CHRDL2 was overexpressed. BMP‐9, bone morphogenetic protein 9; CHRDL2, chordin‐like 2; GSEA, gene set enrichment analysis
3.5. Overexpression of BMP‐9 reversed the effects of CHRDL2 in osteosarcoma
To determine whether overexpression of BMP‐9 can block the effects of CHRDL2 in osteosarcoma, we then overexpression of BMP‐9 in CHRDL2 overexpressed cells. Results showed that overexpression of BMP‐9 blocked the stimulative effects of CHRDL2 on the expression of p‐PI3K, p‐AKT, and MMP‐9 (all p < .05, Figure 5a). Similarly, CCK‐8 and colony formation assay showed that overexpression of BMP‐9 reversed the stimulative effects of CHRDL2 overexpression on the proliferation and colony formation of U2OS and Saos2 cells (all p < .05, Figure 5b,c). Furthermore, wound healing assay and transwell assay showed that overexpression of BMP‐9 in U2OS and Saos2cells decreased the stimulative effects of CHRDL2 overexpression on the migration and invasion (all p < .05, Figure 5d,e).
Figure 5.
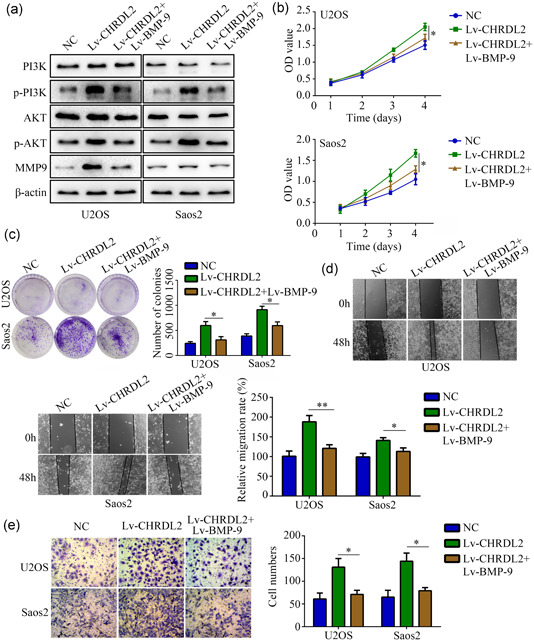
Overexpressed BMP‐9 reversed the effects of CHRDL2 overexpression on regulating PI3K/AKT pathway, proliferation and mobility. (a) Western blot was used to detect the expression of PI3K, p‐PI3K, AKT, p‐AKT, and MMP‐9 while BMP‐9 was overexpressed in CHRDL2 overexpression cells. (b) CCK‐8 was used to detect the proliferation while BMP‐9 was overexpressed in CHRDL2 overexpression cells. (c) Colony formation assay was used to detect the colony formation while BMP‐9 was overexpressed in CHRDL2 overexpression cells. (d,e) Wound healing assay and transwell assay were used to determine the mobility while BMP‐9 was overexpressed in CHRDL2 overexpression cells. *p < .05; **p < .01. BMP‐9, bone morphogenetic protein 9; CHRDL2, chordin‐like 2; GSEA, gene set enrichment analysis
4. DISCUSSION
BMPs and their antagonists are multifunctional cytokines which involved in various biology function of cells including growth, differentiation, and mobility (Chen et al., 2020). Dysregulation of BMPs and their antagonists has been proved to be one of the obvious biomarkers in osteosarcoma, and detection of the expression of BMPs and their antagonists in osteosarcoma has been demonstrated in several research (Gobbi et al., 2002; Nguyen et al., 2014). In the present study, we determined the role and molecular mechanism of CHRDL2, a antagonist of BMPs, in osteosarcoma. We found that CHRDL2 was upregulated in osteosarcoma tissues and cells compared with adjacent tissues and human normal osteoblast. Similarly, we demonstrated that suppression of CHRDL2 inhibited the proliferation, colony formation and mobility of osteosarcoma, while overexpression of CHRDL2 had opposite effects. This is the first evidence we provided that CHRDL2 may play as oncogene in osteosarcoma.
The effects of BMPs were contradictory in osteosarcoma. For example, BMP‐12 had positive effects to promote the proliferation of osteosarcoma cells through increasing the expression of alkaline phosphatase (Furuya et al., 1999). Increasing the expression of BMP‐7 indicated the enhanced proliferation and migration of osteosarcoma cells (Jiang et al., 2017). However, BMP‐4 which regulated by LIM homeobox 9 inhibited the development of osteosarcoma (Li et al., 2019). BMP‐2 had potential to inhibit the stemness of osteosarcoma cells (Wang et al., 2011). Overexpression of BMP‐9 decreased the proliferation and migration of osteosarcoma via decreasing the activity of PI3K/AKT pathway (Li et al., 2012). BMP antagonists had opposite effect with the BMPs. In the present study, we found that CHRDL2 bind with BMP‐9 and decreased its expression. Therefore, we speculated that CHRDL2 promoted osteosarcoma cells proliferation and metastasis via blocking BMP‐9.
PI3K/AKT pathway is vital pathway regulating a series of biology function of cells, including proliferation, differentiation, and migration (Jiang et al., 2020). In osteosarcoma, the change in expression levels and activity of proteins involved in the PI3K/AKT pathway were most common. Dysregulated PI3K/AKT pathway promoted osteosarcoma cells proliferation, metastasis, and drug resistance (Zhang et al., 2015). Various onco‐genes had been demonstrated to regulate the PI3K/AKT in osteosarcoma (Cohen‐Solal et al., 2015, Meng et al., 2020). Present study indicated that BMP‐9 combined to receptor ALK1, which had potential to inhibit the activity of PI3K/AKT pathway. We then determine the relationship between CHRDL2, BMP‐9, and PI3K/AKT pathway in the present study. We found that inhibition of CHRDL2 increased the combination of CHRDL2 to ALK1. Through GSEA, CHRDL2 was predicted to regulate PI3K/AKT pathway. Suppression of CHRDL2 decreased the activity of PI3K/AKT pathway, while overexpression of CHRDL2 increased the activity. Overexpression of BMP‐9 reversed the effect of CHRDL2 on regulating the activity of PI3K/AKT, as well as proliferation and metastasis of osteosarcoma.
In conclusion, our present study demonstrated that CHRDL2 was upregulated in osteosarcoma, which promoting the proliferation and metastasis via regulating BMP‐9/PI3K/AKT pathway. CHRDL2 may be a novel biomarker for diagnosis of biomarker, as well clinical targets for therapy.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
This study was supported by Qian Kehe SY (Grant No.: [2013]3042 for Houping Chen); the Qian Kehe talent perform (Grant No.: [2018]5706 for Hui Chen; [2018]15801 for Yingjie Nie); and Doctoral funding from Guizhou Provincial People's Hospital (Grant No.: GZSYBS[2015]10 for Yingjie Nie).
Chen H, Pan R, Li H, et al. CHRDL2 promotes osteosarcoma cell proliferation and metastasis through the BMP‐9/PI3K/AKT pathway. Cell Biol Int. 2021;45:623–632. 10.1002/cbin.11507
Houping Chen, Runsang Pan, and Hao Li contributed equally to this study.
Contributor Information
Hui Chen, Email: gzsrmyychenhui@163.com.
Xiangyan Zhang, Email: zxy35762@126.com.
Yingjie Nie, Email: nienyj@hotmail.com.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- Bach, D. H. , Park, H. J. , & Lee, S. K. (2018). The dual role of bone morphogenetic proteins in cancer. Molecular Therapy Oncolytics, 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Xu, H. , Yao, Y. , Xu, T. , Yuan, M. , Zhang, X. , Lv, Z. , & Wu, M. (2020). BMP signaling in the development and regeneration of cranium bones and maintenance of calvarial stem cells. Frontiers in Cell and Developmental Biology, 8, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen‐Solal, K. A. , Boregowda, R. K. , & Lasfar, A. (2015). RUNX2 and the PI3K/AKT axis reciprocal activation as a driving force for tumor progression. Molecular Cancer, 14, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa, T. , Huang, Y. , Sebald, W. , & Zhang, J. L. (2009). The binding of von Willebrand factor type C domains of chordin family proteins to BMP‐2 and Tsg is mediated by their SD1 subdomain. Biochemical and Biophysical Research Communications, 385, 215–219. [DOI] [PubMed] [Google Scholar]
- Furuya, K. , Nifuji, A. , Rosen, V. , & Noda, M. (1999). Effects of GDF7/BMP12 on proliferation and alkaline phosphatase expression in rat osteoblastic osteosarcoma ROS 17/2.8 cells. Journal of Cellular Biochemistry, 72, 177–180. [DOI] [PubMed] [Google Scholar]
- Gobbi, G. , Sangiorgi, L. , Lenzi, L. , Casadei, R. , Canaider, S. , Strippoli, P. , Lucarelli, E. , Ghedini, I. , Donati, D. , Fabbri, N. , Warzecha, J. , Yeoung, C. , Helman, L. , Picci, P. , & Carinci, P. (2002). Seven BMPs and all their receptors are simultaneously expressed in osteosarcoma cells. International Journal of Oncology, 20, 143–147. [PubMed] [Google Scholar]
- Hu, F. , Zhang, Y. , Li, M. , Zhao, L. , Chen, J. , Yang, S. , & Zhang, X. (2016). BMP‐6 inhibits the metastasis of MDA‐MB‐231 breast cancer cells by regulating MMP‐1 expression. Oncology Reports, 35, 1823–1830. [DOI] [PubMed] [Google Scholar]
- Jiang, N. , Dai, Q. , Su, X. , Fu, J. , Feng, X. , & Peng, J. (2020). Role of PI3K/AKT pathway in cancer: The framework of malignant behavior. Molecular Biology Reports, 47, 4587–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Z. H. , Peng, J. , Yang, H. L. , Fu, X. L. , Wang, J. Z. , Liu, L. , Jiang, J. N. , Tan, Y. F. , & Ge, Z. J. (2017). Upregulation and biological function of transmembrane protein 119 in osteosarcoma. Experimental & Molecular Medicine, 49, e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Yang, Y. , Jiang, S. , Ni, B. , Chen, K. , & Jiang, L. (2012). Adenovirus‐mediated overexpression of BMP‐9 inhibits human osteosarcoma cell growth and migration through downregulation of the PI3K/AKT pathway. International Journal of Oncology, 41, 1809–1819. [DOI] [PubMed] [Google Scholar]
- Li, S. Q. , Tu, C. , Wan, L. , Chen, R. Q. , Duan, Z. X. , Ren, X. L. , & Li, Z. H. (2019). FGF‐induced LHX9 regulates the progression and metastasis of osteosarcoma via FRS2/TGF‐beta/beta‐catenin pathway. Cell Division, 14, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, Z. , Yang, D. , Li, J. , Hu, M. , Luo, M. , Zhan, X. , Song, P. , Liu, C. , Bai, H. , Li, B. , Yang, Y. , Chen, Y. , Shi, Q. , & Weng, Y. (2013). Bone morphogenetic protein 9 overexpression reduces osteosarcoma cell migration and invasion. Molecules and Cells, 36, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, V. G. , Rubio, C. , Martinez‐Fernandez, M. , Segovia, C. , Lopez‐Calderon, F. , Garin, M. I. , Teijeira, A. , Munera‐Maravilla, E. , Varas, A. , Sacedón, R. , Guerrero, F. , Villacampa, F. , de la Rosa, F. , Castellano, D. , López‐Collazo, E. , Paramio, JM , Vicente, Á. , & Dueñas, M. (2017). BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clincal Cancer Research, 23, 7388–7399. [DOI] [PubMed] [Google Scholar]
- Meng, C. Y. , Zhao, Z. Q. , Bai, R. , Zhao, W. , Wang, Y. X. , Xue, H. Q. , Sun, L. , Sun, C. , Feng, W. , & Guo, S. B. (2020). MicroRNA22 mediates the cisplatin resistance of osteosarcoma cells by inhibiting autophagy via the PI3K/Akt/mTOR pathway. Oncology Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, J. J. , & Khanna, C. (2015). Osteosarcoma genetics and epigenetics: emerging biology and candidate therapies. Critical Reviews in Oncogenesis, 20, 173–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, A. , Scott, M. A. , Dry, S. M. , & James, A. W. (2014). Roles of bone morphogenetic protein signaling in osteosarcoma. International Orthopaedics, 38, 2313–2322. [DOI] [PubMed] [Google Scholar]
- Song, B. , Li, X. F. , Yao, Y. , Xu, Q. Q. , Meng, X. M. , Huang, C. , & Li, J. (2019). BMP9 inhibits the proliferation and migration of fibroblast‐like synoviocytes in rheumatoid arthritis via the PI3K/AKT signaling pathway. International Immunopharmacology, 74, 105685. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Liu, X. , Gao, H. , Zhang, L. , Ji, Q. , Wang, Z. , Zhou, L. , Wang, Y. , Sui, H. , Fan, Z. , & Li, Q. (2017). Overexpression of colorectal cancer oncogene CHRDL2 predicts a poor prognosis. Oncotarget, 8, 11489–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanapprapasr, K. , Nartthanarung, A. , Thanapprapasr, D. , & Jinawath, A. (2017). pFAK‐Y397 overexpression as both a prognostic and a predictive biomarker for patients with metastatic osteosarcoma. PLOS One, 12, e0182989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Park, P. , Zhang, H. , La Marca, F. , Claeson, A. , Valdivia, J. , & Lin, C. Y. (2011). BMP‐2 inhibits the tumorigenicity of cancer stem cells in human osteosarcoma OS99‐1 cell line. Cancer Biology & Therapy, 11, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. , Chen, G. , & Li, Y. P. (2016). TGF‐beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Research, 4, 16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Li, X. , Yang, Y. , He, Z. , Qu, X. , & Zhang, Y. (2016). Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death & Disease, 7, e2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Yu, X. H. , Yan, Y. G. , Wang, C. , & Wang, W. J. (2015). PI3K/Akt signaling in osteosarcoma. Clinica Chimica Acta, 444, 182–192. [DOI] [PubMed] [Google Scholar]
- Zhao, G. , Gao, Z. , Zhang, Q. , Tang, X. , Lv, Y. , Zhang, Z. , Zhang, Y. , Tan, Q. , Peng, D. , Jiang, D. , & Guo, Q. N. (2018). TSSC3 promotes autophagy via inactivating the Src‐mediated PI3K/Akt/mTOR pathway to suppress tumorigenesis and metastasis in osteosarcoma, and predicts a favorable prognosis. Journal of Experimental & Clinical Cancer Research, 37, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


