Abstract
The plant‐associated microbial community (microbiome) has an important role in plant–plant communications. Plants decipher their complex habitat situations by sensing the environmental stimuli and molecular patterns and associated with microbes, herbivores and dangers. Perception of these cues generates inter/intracellular signals that induce modifications of plant metabolism and physiology. Signals can also be transferred between plants via different mechanisms, which we classify as wired‐ and wireless communications. Wired communications involve direct signal transfers between plants mediated by mycorrhizal hyphae and parasitic plant stems. Wireless communications involve plant volatile emissions and root exudates elicited by microbes/insects, which enable inter‐plant signalling without physical contact. These producer‐plant signals induce microbiome adaptation in receiver plants via facilitative or competitive mechanisms. Receiver plants eavesdrop to anticipate responses to improve fitness against stresses. An emerging body of information in plant–plant communication can be leveraged to improve integrated crop management under field conditions.
Keywords: dodder, herbivore‐induced plant volatiles, holobiont, microbe‐induced plant volatiles, microbiome adaptation, mycorrhiza
Short abstract
In nature, plants need to communicate with neighbouring plants to anticipate upcoming challenges for survival in complex and competitive ecosystems. Inter‐plants signals can be transferred via volatile compounds and root exudates, or through fungal hyphae and parasitic plant organs.
1. INTRODUCTION
Plants adapt to stress via sensor elicitation, signalling cascade activation, gene expression and phenotype modification (Glazebrook, 2005; Jung, Tschaplinski, Wang, Glazebrook, & Greenberg, 2009). Signal processing time is critical for success or failure in stress responses. Plants acquired the ability to anticipate and respond to imminent dangers, which conferred ecological competence in highly dynamic ecosystems. This system is known as defence priming (Conrath, Beckers, Langenbach, & Jaskiewicz, 2015; Jung et al., 2009).
Plants can acquire early warning information through their microbiome and plant–plant signals (Gilbert & Johnson, 2017; Vahabi et al., 2018; Yi, Heil, Adame‐Alvarez, Ballhorn, & Ryu, 2009). The plant can exploit the unique capabilities of its symbionts including the mycorrhizal network, dodder (Cuscuta spp.) and endophytic fungi, which directly prime plant defence responses or transfer inter‐plant signals (da Trindade et al., 2019; Hettenhausen et al., 2017; Vahabi et al., 2018). The plant microbiome and macrobiome can modify plant‐derived inter‐plant signals such as volatiles and root exudates (Figure 1; Sharifi, Lee, & Ryu, 2018; Song, Sim, Kim, & Ryu, 2016). Herbivore‐induced plant volatiles (HIPVs) and microbe‐induced plant volatiles (MIPVs) are good examples of inter‐plant signals (Heil & Bueno, 2014; Sharifi et al., 2018).
FIGURE 1.
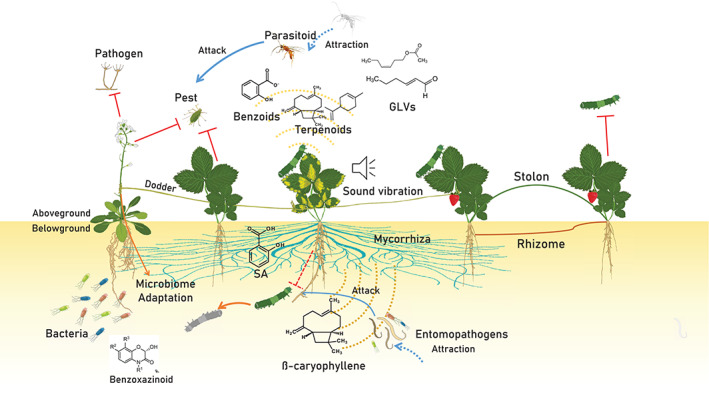
Wired and wireless phytobiome communication. Clonal plants (right) communicate via physical connections (e.g. stolons and rhizomes) or VOCs. Plants also communicate via dodder and mycorrhiza (left). Receiver plants can act as nodes to transfer defence signals against pests and pathogens to neighbouring conspecific and heterospecific plants. Volatiles and root exudates received by neighbouring plants modulate receiver plant defence systems, attract parasitoids and entemopathogens and induce plant microbiome remodelling to protect plants against imminent stress conditions [Colour figure can be viewed at wileyonlinelibrary.com]
BOX 1. Definitions: holobiont, holobiome, microbiome.
Recent reports proposed that multicellular organisms (e.g. animals and plants) and their associated unicellular organisms (e.g. microbes) could be considered as super‐organisms, or holobionts (in ancient Greek, holos means whole and biont means unit of life) (Gilbert, 2019; Suarez & Stencel, 2020). However, the definition and concept of a holobiont is still debated. We consider a holobiont as an ecological unit (assembly) of a group of organisms that gather together based on their evolutionary capability to achieve a common purpose, which is the survival of the holobiont.
The holobiome includes all living organisms, their genetic materials and their primary and secondary metabolites, as well as the molecules produced within a particular habitat (Berg et al., 2020; Sharifi & Ryu, 2017).
The microbiome includes the microbial community living in a particular habitat and their metabolites, mobile genetic elements and relic DNA (Berg et al., 2020). The microbiome helps the holobiont survive during biotic and abiotic stresses. The presence and abundance of specific microbial species in the microbiome change during successive phases of plant ontogeny and during biotic/abiotic stresses (Carrión et al., 2019; Cotton et al., 2019; Edwards et al., 2018; Gu et al., 2016).
Here, we review plant–plant communications that improve plant defence against pathogenic microbes. From multiple layers of plant–plant communications, we distinguished two distinct types: wired and wireless communications. Wired communication involves one plant sending a signal to another plant though direct contact via microbial structures and hyper‐parasitic plant organs. This can be considered as an information highway mediating plant–plant communication. Wireless communication involves signal transfer across the space separating two plants. We investigated how wired and wireless communications affect plant defence responses. We determined that these signal transduction pathways proceeded via the following three steps: signal input (extracellular signal perception generates an endogenous signal cascade); transferring signal (direct connection from signal producer to receiver through mycorrhizal network and parasitic plants, and indirect signal translocation via plant volatile compounds and exudates) and signal output (receiver plant responses to biotic and abiotic stresses).
2. WIRED COMMUNICATION
2.1. Wired connections between clonal plants via rhizomes and stolons
Some plants produce genetically identical but independent plants (clones) through specific organs such as rhizome and stolon (Bittebiere, Benot, & Mony, 2020; Vannier et al., 2019). These clonal plants are two functionally independent plants that share vascular connections and physiological integration. Clones have similar microbiomes, and represent unique models in plant–plant interaction studies (Vannier et al., 2019). Clones communicate and enhance each other's survival during biotic/abiotic stress (Karban et al., 2014; Qian, Li, Han, & Sun, 2010; Semchenko, John, & Hutchings, 2007).
Physical connections via stolon and rhizome can mitigate abiotic stress by facilitating the transfer/exchange of nutrients and metabolites, or by modulating resource use efficiency (Figure 1; Roiloa, Antelo, & Retuerto, 2014). Defence signals can be transferred via phloem from older to younger ramets to induce systemic resistance in young ramets. Older ramets do not receive defence signals from younger clones due to the direction of source‐sink gradient flow (Gómez & Stuefer, 2006). In Trifolium repens, a defence signal against Mamestra brassicae larvae can be transferred to younger ramets within 35–51 hr depending on genotypic variation. Induced but undamaged ramets lose their resistance after 28 days (Gómez, Van Dijk, & Stuefer, 2010). Besides direct plant–plant links via plant parts, higher parasitic plants are also an important link in wired communications that enhance plant fitness.
2.2. Wired connections between independent plants via dodder‐mediated interspecific signalling
Dodders (Cuscuta spp.) are plant holoparasites that acquire water and nutrients from host plants via the haustorium, which physically connects the parasite to its host. Dodder species have broad host range, and can interconnect several plant species or clusters of the same species (Figure 1) to generate a common dodder network. The common dodder network can be considered as an inter‐plant highway that translocates large numbers of proteins, RNA, metabolites and plant viruses over a distance of at least 100 cm (Hettenhausen et al., 2017; Zhuang et al., 2018). The common dodder network can translocate more than 1,500 proteins between soybean and Arabidopsis, and some of these proteins can localize in dodder seeds. Approximately 15–30% of dodder proteins have host origin, including transcription factors and R proteins that may function in signal transaction. Dodder proteins can transfer to host plant cells.
Plants can anticipate future threats by receiving neighbouring plant signals transferred through the common dodder network. Although there are few reports on the role of the common dodder network in inter‐plant signalling, the results indicate that these transferred signals are important in biotic/abiotic stress responses. When a host of the dodder plant is under abiotic stress such as high salinity, dodder transfers salinity stress signals through a cluster of plants at a rate of 1.2 cm per min, which prime salt tolerance in neighbouring receiver plants. This receiver plant priming changes the transcriptome, proline levels and stomatal conduction, so that the receiver plant stress response becomes similar to that of the donor plant (Li, Zhang, et al., 2020).
Plants infected with different herbivorous insects transfer relatively long‐distance signals to conspecific and heterospecific neighbours via the common dodder network (Hettenhausen et al., 2017; Zhuang et al., 2018). Myzus persica infestation reduces the contents of salicylic acid (SA) and jasmonic acid (JA) in dodder (Cuscuta australis) by up to 58 and 41%, respectively. Aphids can modulate hormonal signalling by injecting effector proteins into dodder plants (Rodriguez & Bos, 2013). Aphid‐infested dodder induce JA but not SA in soybean hosts, and subsequent phloem sap feeding by M. persica and chewing by Spodoptera litura causes 41 and 20% less damage, respectively, in dodder‐infected plants than in control plants. Dodder transfers signals from insect‐damaged soybean to conspecific or heterospecific plants, such as tomato and Arabidopsis. Gene expression and RNA‐seq analyses reveal intense transcriptome modification in receiver plants. An unknown signal can translocate between common dodder network–connected Arabidopsis plants at a rate of 1 cm/min. A wave of signal transduction propagated between connected Arabidopsis plants, as intracellular WRKY 40 and WRKY53 transcription factors reached maximum expression at 45 and 90 min after donor plant damage in the second and fourth plants in the cluster, respectively (Zhuang et al., 2018).
2.3. Wired connections between plants via fungal hyphae of the mycorrhizal or endophytic fungal network
Fungi can act as messengers to transfer information between independent plants. Plant‐mycorrhiza symbiosis is an ancient system that arose more than 450 million years ago (Waters, Gutjahr, Bennett, & Nelson, 2017). This symbiosis had a crucial role in land colonization by plants. Different fungal phyla have evolved different types of plant‐mycorrhiza symbiosis (Tedersoo, Bahram, & Zobel, 2020). Some Brassicaceae and Proteaceae plants do not participate in mycorrhizal relationships, and are considered to be non‐mycorrhizal symbiotic plants (Tedersoo et al., 2020). These plants can interfere in the mycorrhizal establishment of other plants (Cipollini, Rigsby, & Barto, 2012). For example, Alliaria petiolata produces glucosinolate‐derived exudate that suppresses the mycorrhizae of neighbouring plants (Cipollini et al., 2012).
Mycorrhizae have important roles in inter‐plant nutrient transfer (especially carbon, phosphorus and nitrogen), allelochemicals and signal molecules. Mycorrhizae are common in soil, as 1 g of soil contains 10–100 m of mycorrhizal fungi (Gilbert & Johnson, 2017). These fungi have broad host ranges and can connect several plants from different taxa. Closely related mycorrhiza fungi can perform anastomosis to generate a network (Figure 1), which is called a common mycorrhiza network (Deja‐Sikora, Mercy, Baum, & Hrynkiewicz, 2019). Some plant species exploit the common mycorrhiza network, especially EcM, to transfer nutrients and signals and promote the establishment and survival of conspecific seedlings (Bennett, Cahill, & van der Heijden, 2016; Hortal et al., 2017).
Plants exploit mycorrhizae fungi for long‐distance transfer of plant signals and allelochemicals. Tagetes tenuifolia secretes the lipophilic allelopathic compound thiophene and the hydrophilic allelopathic compound imazamox, which can diffuse more than 12 cm via the common mycorrhiza network (Barto et al., 2011). Mycorrhizae fungi can translocate defence signals to neighbouring unstressed plants, similar to dodder. Tomato plants infected with the nectrotrophic pathogenic fungus Alternaria solani transfer a signal to non‐infected neighbours at 18 hr post‐inoculation via mycorrhizae hypha. In the receiver plants, the relative expression levels of defence‐related genes such as phenylalanine ammonia‐lyase (PAL), lipoxygenase, polyphenol oxidase and pathogenesis‐related proteins (PR1, PR2 and PR4) were the same as those in the donor plants (Song et al., 2010). Alaux, Naveau, Declerck, and Cranenbrouck (2020) also set up a sophisticated in vitro experiment to confirm the role of mycorrhizae in plant–plant signal transduction. The results showed that a Phytophthora infestans infected tomato plant could transfer a signal to healthy neighbours via mycorrhizae hypha to activate the transient expression of JA related genes. Signal transduction via the common mycorrhiza network also has been detected for herbivore‐damaged plants (Babikova et al., 2013; Song et al., 2014).
Fungal signal transduction is not exclusive to mycorrhizae. Other mutual and pathogenic fungi with long‐term endophytic relationships with plants have the potential to transfer signals between plants. Some soil fungi can serve as a bridge between host plants. For example, Piriformospora indica transfers signals from infected Arabidopsis to neighbouring Arabidopsis plants. Alternaria brassicae activates JA pathways in infected focal plants. By contrast, endophytic fungi convert a specific JA signal to generate an ABA signal in neighbouring (receiver) plants (Vahabi et al., 2018).
Fungi serve as vectors for the transfer of plant viruses among plants. Although plants and fungi are not phylogenetically related, they can be infected by phylogenetically related viruses (Roossinck, 2019). Some plant viruses propagate in the fungal cytosol and transfer between host plants based on the host range of both fungi and viruses. A virus from the saprophytic endophyte Penicillium aurantiogriseum replicates in the host plant (Nerva, Varese, Falk, & Turina, 2017). Cucumber mosaic virus is a broad‐range virus that can survive in the pathogenic fungus Rhizoctonia solani, which has a broad host range. R. solani transfers this virus to a neighbouring host (Andika et al., 2017). There is evidence that plant virus enters fungal spores and achieves long‐distance dissemination via wind dispersal of the fungal spores. Cryphonectria transfer Cryphonectria hypovirus 1 to tobacco, which subsequently propagates and spreads systemically throughout the plant with the help of Tobacco mosaic virus (TMV) movement protein. TMV also enters fungi and propagates with the help of Cryphonectria hypovirus 1, which can spread to other hosts via fungal spores (Bian et al., 2020).
3. WIRELESS COMMUNICATION: SIGNAL INPUT‐TRANSFER‐OUTPUT MODEL
Plants normally release volatile and non‐volatile chemicals that can be exploited by other plants as a source of signal molecules (infochemicals). Here, we discuss how these plant infochemicals can be induced by microbes, insects and environmental stimuli. Infochemicals diffuse via air and soil pores to reach neighbour plants. Receiver plants can amplify the released infochemical signals to expand the effective infochemical signalling zone. Infochemicals perceived by the receiver plants can act as kin recognition signals, growth inhibitors (allelopathy), growth stimulators and defence signals (Karban, Shiojiri, Ishizaki, Wetzel, & Evans, 2013; Kong et al., 2018; Sharifi et al., 2018; Sharifi & Ryu, 2018b). Infochemicals can directly change the transcriptome and physiology of receiver plants to prime for imminent threats, or they may indirectly change the plant microbiome in neighbouring plants through a facilitative or competitive mechanism (Carvalhais et al., 2015; Li, Zhao, & Kong, 2020; Mannaa et al., 2020; Vannier, Bittebiere, Mony, & Vandenkoornhuyse, 2020).
3.1. Wireless signal input
3.1.1. Constitutive release of infochemicals as information sources
Plants continuously release a profile of volatile organic compounds (VOCs) and non‐VOCs to the root and atmosphere. These chemical profiles, concentrations and release time courses function as signals of plant presence and health status. Constitutive release of specific chemical signatures can be exploited by neighbour organisms to determine the presence and identity of neighbouring plants. Receiver plants recognize the degree of phylogenetical relationship of neighbour plants to discriminate kin and non‐kin plants (Biedrzycki, Jilany, Dudley, & Bais, 2010; Yang, Li, Xu, & Kong, 2018). In most cases, signals from healthy plants can affect neighbouring plants by modifying biomass allocation, lateral root formation and chlorophyll and phenolic compound biosynthesis, or directly inhibiting seed germination (Delory, Delaplace, Fauconnier, & du Jardin, 2016; Takigahira & Yamawo, 2019). The sagebrush volatile compounds methyl jasmonate, camphor, 1,8‐cineol, α‐thujone and nerol inhibit the germination of Nicotiana attenuate seeds (Jassbi, Zamanizadehnajari, & Baldwin, 2010). Exposing potato plants to healthy onions changes the profile of potato volatiles such that they repel aphids and attract ladybird beetles, which are aphid predators (Ninkovic et al., 2013; Vucetic et al., 2014). (E)‐nerolidol and (3E,7E)‐4,8,12‐trimethyl‐1,3,7,11‐tridecatetraene were the primary effective volatiles released by exposed potato plants (Vucetic et al., 2014).
Some plant infochemicals are released primarily by healthy plants and can be considered as a marker of plant health status. Isoprene emission levels are linked to plant health and photosynthetic efficiency. The reduction of isoprene emission after pest and disease attacks is considered as a marker for plant stress (Copolovici, Kannaste, Pazouki, & Niinemets, 2012; Jiang, Ye, Veromann, & Niinemets, 2016; Toome et al., 2010). Isoprene alleviates plant oxidative stress (van Doorn et al., 2020).
3.1.2. Microbes and insects induce the emission of specific plant volatiles
Microbes and insects modulate plant volatile emission dynamics by decreasing some VOCs and increasing others, or by inducing de novo VOC synthesis. Plant volatiles are produced by several metabolic pathways, including plastidic methylerythritol phosphate and cytosolic mevalonic acid pathways (terpenoid compounds), shikimic acid pathways (benzoid and phenylpropanoid compounds) and oxylipin pathways [green leaf volatiles (GLVs)] (Bouwmeester, Schuurink, Bleeker, & Schiestl, 2019). The emitted VOC profile is related to the plant genotype, organ and type of biotic/abiotic trigger. Different comparative triggers (e.g. chewing vs. sucking/piercing, biotroph vs. necrotroph, host vs. non‐host pathogens and saprophytic beneficial vs. parasitic pathogenic microbes) elicit distinct bouquets of VOCs, including the quantity and quality of each compound and its emission time course (Castelyn, Appelgryn, Mafa, Pretorius, & Visser, 2014; Klimm, Weinhold, & Volf, 2020; Qawasmeh, Raman, & Wheatley, 2015; Quintana‐Rodriguez et al., 2015; Sharifi et al., 2018).
VOC profiles are conventionally defined based on what triggers their emission. They include herbivore‐induced plant volatiles (HIPVs), oviposition‐induced plant volatiles, microbe‐induced plant volatiles (MIPVs) and stress‐induced plant volatiles (Kessler & Heil, 2011; Sharifi et al., 2018). These categories normally contain all of the above‐mentioned VOC groups, but the quantity/quality and emission time course for each compound carries specific information. Green leaf volatiles (GLVs) are categorized as HIPVs, especially for chewing insects, but GLVs also are emitted from microbial pathogen‐infected plants (Ameye et al., 2018). Rust disease disrupts the epidermis and induces the release of high amounts of GLVs (Jiang et al., 2016). By contrast, chewing insects feeding on maize root did not elicit the emission of GLVs, and maize root did not respond to GLVs (van Doan et al., 2020).
Plants emit VOCs in response to signalling between plants and invaders. Plant VOC profiles were altered by pathogen‐associated molecular patterns (PAMPs), herbivore‐associated molecular patterns (HAMPs), damage‐associated molecular patterns (DAMPs), effector proteins and microbial volatile compounds (Figure 2; Ameye et al., 2018; Bouwmeester et al., 2019; Rybakova et al., 2017; Sharifi et al., 2018; Wu et al., 2017). These elicitors activate defence‐related hormones (e.g. JA, SA and their cross‐talk), which in turn activate metabolic pathways that produce the main VOC groups.
FIGURE 2.
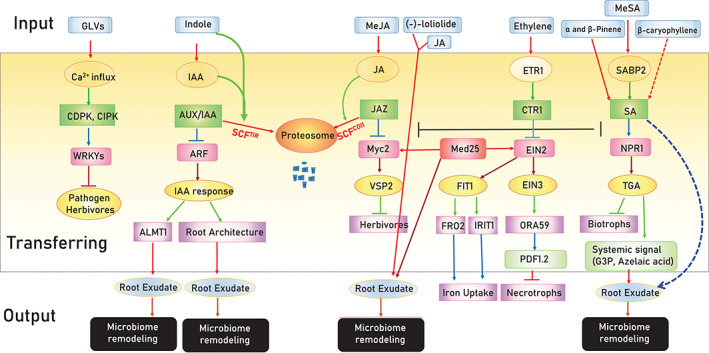
Signals from neighbouring plants modulate signalling pathways in receiver plants and induce microbiome remodelling. Signals can be sensed by receiver proteins (e.g. ETR1 sensor for ethylene) or converted to active signals [e.g. SABP2 for salicylic acid (SA)]. Signals are transmitted through well‐characterized downstream pathways that may cross‐talk with each other. These signalling pathways regulate defence mechanisms against different groups of attackers and induce plant microbiome remodelling by changing root exudation, thereby adapting the plant holobiome to respond to imminent threats. ALMT1, aluminium‐activated malate transporter; AUX1/LAX, auxin resistant 1/LIKE AUX1; CDPK, Ca2+‐dependent protein kinases; CIPK, calcineurin B like proteins (CBL)‐interacting protein kinase; CTR1, constitutive triple response 1; EIN2, ethylene insensitive 2; ETR1, ethylene response 1; FIT1, FE‐deficiency‐inducing transcription factor 1; FRO2, ferric reductase oxidase 2; G3P, glycerol‐3‐phosphate; IAA, indole‐3‐acetic acid; IRT1, iron‐regulated transporter 1; JAZ, Jasmonate Zim domain protein; Med25, mediator 25; MeJA, methyl jasmonate; MeSA, methyl salicylate; NPR, non‐expresser of PR genes; ORA59, octadecanoid‐responsive Arabidopsis 59; SABP2, SA‐binding proteins 2; SCFCOI1, Skp1‐Cul1‐F‐box protein coronatine insensitive 1 [Colour figure can be viewed at wileyonlinelibrary.com]
HAMPs such as volicitin, caeliferins and β‐glucosidase modify the volatile profiles in several plants (Alborn et al., 1997; Alborn et al., 2007; Hopke, Donath, Blechert, & Boland, 1994). These compounds can induce or suppress specific groups of volatiles to attract or repel parasitoids to host plants. Well‐adapted maize caterpillars (Spodoptera frugiperda) suppress HIPVs in maize, but not in cotton (De Lange et al., 2020).
In some plant pathogens, PAMPs (e.g. flg22, laminarin and glucan) and effector proteins (e.g. 2b) can modify plant VOCs (Chalal et al., 2015; Leitner et al., 2008; Sobhy, Bruce, & Turlings, 2018; Tu et al., 2017; Tungadi et al., 2017). PAMPs, HAMPs and effector proteins are perceived by pattern recognition receptors and R proteins in plants, and subsequently activate basal and effector‐triggered plant immune responses (Bonaventure, VanDoorn, & Baldwin, 2011; Glazebrook, 2005). Insertion of single R protein and its position in the genome can significantly change the emission of volatiles (Figure 2; Lazebnik, Tibboel, Dicke, & van Loon, 2017).
Signalling pathways (e.g. SA‐ and JA‐dependent pathways) leading to systemic resistance in inoculated and neighbouring plants have important roles in volatile biosynthesis (Orlovskis & Reymond, 2020; Wenig et al., 2019). ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) and AvrRpm1 are essential factors in systemic acquired resistance and important regulators of VOCs synthesis in Arabidopsis (Bichlmeier, 2017). Monoterpenes such as α‐ and β‐pinene also induce systemic resistance through EDS1, SA INDUCTION–DEFICIENT 2 (SID2) and NONEXPRESSOR OF PR GENES 1 (NPR1) proteins (Figure 3; Bichlmeier, 2017). Thus, any biological and chemical modulator of plant resistance can change the VOC profile or prime VOC release in response to stress. Pseudomonas protegens strain CHA0 did not change β‐caryophyllene emission or expression of the β‐caryophyllene synthase gene, but primed them in response to maize beetle Diabrotica balteata (Chiriboga et al., 2018). Bacterial pathogens Pseudomonas syringae directly induce the emission of 1‐undecanol and (Z)‐3‐hexenol volatiles in common bean, which repel spider mite (Tetranychus urticae) (Karamanoli, Kokalas, Koveos, Junker, & Farré‐Armengol, 2020). Thus, signal cross‐talk during simultaneous plant infestation with herbivores and pathogens (Eberl, Hammerbacher, Gershenzon, & Unsicker, 2018; Lazebnik et al., 2017; Peñaflor & Bento, 2019) or co‐infestation with two pests (Kroes, Weldegergis, Cappai, Dicke, & van Loon, 2017; Zhang et al., 2013) can modulate VOC emissions and attract pests and their parasitoids.
FIGURE 3.
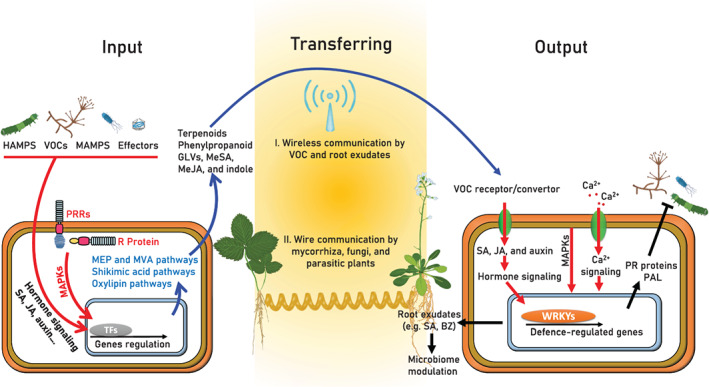
Illustration of the signal input‐transfer‐output model in plant–plant communication. Molecular patterns, volatiles and effector proteins of pests and pathogens elicit plant signalling pathways that promote volatiles emission and root exudation. Plant signals can be delivered to neighbouring plants through the atmosphere or soil (wireless communication), or transferred through mycorrhiza, fungi and odder (wired communication). Signals can be converted to their active form by receiver plant proteins. Signal perception by neighbour plants activates signalling pathways and phosphorylation cascades, which subsequently induce the expression of defence‐related proteins and metabolites. Signal perception also changes the root exudate and recruits beneficial microbes. BZ, benzoxazinoid; GLVs, green leaf volatiles; HAMPs, herbivore‐associated molecular patterns; JA, jasmonic acid; MAMPs, microbe‐associated molecular patterns; MAPKs, mitogen‐activated protein kinases; MEP, methylerythritol phosphate; MeSA, methyl salicylate; MVA, mevalonic acid; SA, salicylic acid; TFs, transcription factors; VOCs, volatile organic compounds [Colour figure can be viewed at wileyonlinelibrary.com]
3.1.3. Root exudates as inter‐plant signals
Plants release large amounts of root exudates into the soil, where they act as carbon and nitrogen sources or cues for rhizosphere organisms. The profile of root exudates can be affected by biotic and abiotic stimuli. Soil dwelling organisms and roots of neighbouring plants use some of these root exudates as a source of information (Bais, 2006; Biedrzycki et al., 2010; Carvalhais et al., 2015; Khashi u Rahman, Zhou, & Wu, 2019; Sharifi & Ryu, 2017). Plants employ root exudates to discriminate between kin and non‐kin neighbour plants. Rice use allantoin as a cue to recognize kin cultivars and respond to it by shifting root biomass allocation and increasing grain yield (Yang et al., 2018). An allantoin mutant of Arabidopsis was reported to be susceptible to P. syringae and Pectobacterium carotovorum. Exogenous application of allantoin also induces JA‐responsive genes, such as MYC2 (Table 1; Takagi et al., 2016). Root exudates such as (−)‐loliolide, jasmonic acid and salicylic acid act as inter‐plant signals and plant defence inducers (Cheol Song et al., 2016; Kong et al., 2018; Li, Zhao, & Kong, 2020). (−)‐loliolide from barnyardgrass (Echinochloa crus‐galli) root exudates induces the biosynthesis of the rice allelochemicals, momilactone B and tricin (Li, Zhao, & Kong, 2020), which can reduce the disease severity of several rice pathogens, such as Piricularia oryzae, R. solani and Fusarium oxysporum (Table 1; Kong et al., 2010; Zhao et al., 2018).
TABLE 1.
Effect of plant–plant communication via volatile organic compounds and root exudates on the suppression of plant pathogens
| Donor plant | Receiver plant | Signal molecule | Target pathogen | References |
|---|---|---|---|---|
| Volatile organic compounds | ||||
| Lima bean | Lima bean | Volatile organic compound, nonanal |
P. syringae pv. syringae PR‐2 |
Yi et al. (2009) |
| Tomato | Tomato |
(Z)‐3‐hexenyl propionate (Z)‐3‐hexenyl butyrate |
P. syringae pv. tomato stomatal closure | Lopez‐Gresa et al. (2018) |
| Maize | Maize | (Z)‐3‐hexen‐1‐yl acetate, (Z)‐3‐hexenal, linalool and β‐caryophyllene | Fusarium spp. | Piesik et al. (2011) |
| Barley and wheat | Barley and wheat | GLV, (Z)‐b‐ocimene and linalool | Fusarium spp. | Piesik et al. (2013) |
| Wheat | Wheat | GLVs, fatty acid derivatives, benzenoids and terpenoid | Puccinia triticina | Castelyn et al. (2014) |
| Lima bean | Lima bean | Limonene, linalool, nonanal, methyl salicylate and methyl jasmonate | Colletortichum lindemuthianum | Quintana‐Rodriguez et al. (2015) |
| Tobacco | Tobacco | Methyl salicylate | Tobacco mosaic virus | Shulaev, Silverman, and Raskin (1997) |
| Tea (Camellia sinensis) | Tea | (E)‐nerolidol | Colletotrichum fructicola | Chen et al. (2020) |
| A. thaliana | A. thaliana | α‐ and β‐pinene | P. syringae pv. tomato | Riedlmeier et al. (2017) |
| Maize | Maize |
GLVs, terpenes Shikimic acid pathway derivatives |
Pantoea ananatis | Delaney et al. (2015) |
| Root exudates | ||||
| Poncirus trifoliata | P. trifoliata | Salicylic acid | Xanthomonas axonopodis pv. citri | Zhang, Zou, Liu, and Wu (2019) |
| Maize | Pepper | 2,4‐Dihydroxy‐7‐methoxy‐2H‐1,4‐benzoxazin‐3(4H)‐one, 6‐methoxy‐2‐benzoxazolinone, benzothiazole and 2‐(methylthio)‐benzothiazole | P. capsici | Yang et al. (2014) |
| Tobacco | Tobacco | Salicylic acid | R. solanacearum and P. syringae pv. tabaci | Cheol Song, Sim, Kim, and Ryu (2016) |
| A. thaliana | A. thaliana | Root‐derived signal(s) | P. syringae pv. tomato | Orlovskis and Reymond (2020) |
| E. crus‐galli | Rice a | (−)‐loliolide |
P. oryzae F. oxysporum R. solani |
Kong, Xu, Zhang, and Zhang (2010), Li, Zhao, and Kong (2020) and Zhao, Cheng, Guo, Duan, and Che (2018) |
| Rice | Rice a | Allantoin |
P. syringae P. carotovorum |
Takagi et al. (2016) and Yang et al. (2018) |
This information was obtained from several reports, some of which reported these chemicals as inter‐plant signals and others as their effects on plant pathogens.
3.2. Wireless signal transfer
Plant infochemicals can diffuse around the elicited plant (producer), and are amplified by receiver plants to expand their effective zone. Infochemicals can diffuse distances of a few centimetres to several metres according to their chemical structure and vectors such as mycorrhiza and dodder. Wheat infected by Fusarium culmorum transfers signals to neighbour plants located 1–3 m distant. However, there is a negative correlation between distance and response intensity in the receiver plant. Several studies show that receiver plants also propagate signals and act as nodes in inter‐plant signalling (Chen, Yang, et al., 2019; Piesik et al., 2013; Wenig et al., 2019). Thus, neighbouring plants amplify signals in clusters of receiver plants. However, the regulation of induced signalling pathways involved in infochemical synthesis in receiver plants requires time, which results in a wave of infochemical synthesis and metabolic responses in receiver plants. Falik et al. (2012) observed that osmotic stress in Pisum sativum activated stomatal closure in the focal plant and the next three plants in the cluster within 1 hr, whereas more distant plants responded after 1 hr. In this study, focal plant stomata remained closed for 24 hr, whereas receiver plants opened their stomatal apertures because they did not directly encounter osmotic stress. Receiver plants did prepare for imminent osmotic stress, with the ability to return to a naive state if the stress was resolved. Weed plants use stress signals from P. sativum neighbours to improve their acclimation (Falik et al., 2012).
Wenig et al. (2019) showed that monoterpenes such as α‐ and β‐pinene act as immunity‐inducing signals between plants (Figure 3). Systemic acquired resistance (SAR) is an SA‐mediated induced resistance in systemic plant tissues that is effective against a broad range of plant pathogens (Wenig et al., 2019). LEGUME LECTIN‐LIKE PROTEIN1 (LLP1) is a predicted lectin that acts in the recognition of SAR signals, which also regulate intra‐ and inter‐plant monoterpene synthesis. In the non‐vascular marine alga Pyropia haitanensis, the 1‐octone‐3‐ol plant–plant defence signal can self‐stimulate in receiver plants and activate SA hormones (Chen, Yang, et al., 2019).
3.3. Wireless signal output: Receiver plant response to biotic/abiotic stresses
The transferred signal is perceived by neighbouring (receiver) plants and augments biotic/abiotic stress resistance responses. VOC‐mediated plant stress responses have been demonstrated in numerous studies, although the ethylene receptor ETR1 is the only plant VOC receptor identified to date (Chang, Kwok, Bleecker, & Meyerowitz, 1993). Future research may lead to the discovery of additional receptors for plant VOCs.
Recent studies identified different mechanisms whereby receiver plants perceive VOCs from neighbour plants. VOCs can be absorbed by a wax layer on the epidermal cell, which traps VOCs and slowly releases them to attract or repel herbivores and their parasitoids and entomopathogens (Camacho‐Coronel, Molina‐Torres, & Heil, 2020; Lin et al., 2016).
Some trapped volatiles such as methyl salicylate (MeSA), MeJA and indole can be converted to the active plant hormones SA, JA and indole‐3‐acetic acid (IAA), respectively (Figures 2 and 3; Bailly et al., 2014; Rivas‐San Vicente & Plasencia, 2011). Some enzymes metabolize trapped volatiles such as (Z)‐3‐hexenol to the more active derivative (Z)‐3‐hexenylvicianoside (Sugimoto et al., 2014). Some GLVs induce plasma membrane potential depolarization in receiver plants, thereby activating reactive oxygen species (ROS) and calcium signalling (Figure 2; Zebelo, Matsui, Ozawa, & Maffei, 2012).
The perception of VOCs modifies the transcriptome, proteome and metabolome in receiver plants (Kwon et al., 2010; van Dam & Bouwmeester, 2016; Zhang et al., 2007). In some cases, VOCs do not significantly change gene expression profiles and metabolic activity, but prime the plant to respond more rapidly and robustly to upcoming threats (Paschold, Halitschke, & Baldwin, 2006; Quintana‐Rodriguez et al., 2015). Plant volatile (Z)‐3‐hexenyl acetate directly induces JA‐ and abscisic acid–related gene expression, whereas indole primes these genes in maize against Spodoptera littoralis (Hu, Ye, & Erb, 2019). Several studies report that primed plants activate defence‐related pathways based on the attacker identity rather than the inducer (Moreira, Nell, Katsanis, Rasmann, & Mooney, 2018; Sharifi & Ryu, 2017). For example, VOCs from plants infested with general or specialized herbivores activate similar defence pathways and VOC emission profiles in healthy neighbours. By contrast, primed receiver plants mount a specific set of defence mechanisms based on the type of attacker (Moreira et al., 2018).
Infochemicals from neighbour plants can activate master regulatory systems involved in plant innate immunity, including leucine‐rich repeat‐receptor‐like kinase, mitogen‐activated protein kinases, WRKY transcription factors and systemic acquired resistance (Figures 2 and 3; Dombrowski et al., 2019; Dombrowski & Martin, 2018; Lee, Kim, Lee, Ahn, & Ryu, 2020; Mirabella et al., 2015; Wenig et al., 2019; Ye, Glauser, Lou, Erb, & Hu, 2019). d‐Lactic acid secreted by the microalga Chlorella fusca primed defence in Arabidopsis thaliana against P. syringae pv. tomato DC3000 by increasing the expression of WRKY transcription factors and cysteine‐rich receptor‐like kinases, and induced both SA‐ and JA‐dependent pathways (Lee et al., 2020).
In inter‐plant communication, VOCs modulate receiver plant physiology and directly or indirectly affect other plant holobiome members. VOCs captured by receiver plant wax display fungicidal and bactericidal activity for several days (Camacho‐Coronel et al., 2020). GLVs and terpenoid volatiles have strong fungicidal and bactericidal activity in vitro and in planta (Huang et al., 2012; Pontin, Bottini, Burba, & Piccoli, 2015; Quintana‐Rodriguez et al., 2015). VOCs can alter parasitoid attraction and entomopathogenic fungi performance in both donor and receiver plants (Desurmont, Xu, & Turlings, 2016; Lin et al., 2016; Xu et al., 2017). VOC emission in aboveground and below‐ground parts may attract or repel herbivores and plant pathogenic nematodes (Ali, Alborn, & Stelinski, 2011; D'Alessandro et al., 2014; Rasmann et al., 2005).
Root exudates act as critical triggers to activate resistance in neighbouring plants by diffusing through the soil to neighbouring roots. Root exudates such as SA transfer the SAR signal to neighbouring plants and synchronize their microbiomes (Kong, Song, Sim, & Ryu, 2020; Orlovskis & Reymond, 2020; Song et al., 2016). Plants exploit microbiome adaptation to facilitate conspecific survival according to kin selection theory, or to compete for heterospecificity. Airborne signals from wound‐damaged plants regulated the ALMT1 transporter in receiver Arabidopsis plants to release malic acid into the rhizosphere (Figure 3). Malic acid recruits B. subtilis to colonize Arabidopsis roots and induce systemic resistance to different stresses (Rudrappa, Czymmek, Pare, & Bais, 2008; Sweeney, Lakshmanan, & Bais, 2017).
A (−)‐loliolide root exudate at a physiological concentration of 5 nmol g−1 soil induces the release of the benzoxazinoid compound 2,4‐dihydroxy‐7‐methoxy‐1,4‐benzoxazin‐3‐one (DIMBOA) exudate from wheat roots (Kong et al., 2018). DIMBOA is a putative allelochemical with several other roles in the rhizosphere. DIMBOA regulates the root metabolome and exudation, which have important roles in shaping the root microbiome (Cotton et al., 2019; Kudjordjie, Sapkota, Steffensen, Fomsgaard, & Nicolaisen, 2019). DIMBOA‐treated plants recruit specific bacterial families and species such as Pseudomonas putida, thereby increasing plant resistance to several stresses (Neal & Ton, 2013). P. fluorescens increases DIMBOA and primed resistance against the fungal pathogen Setosphaeria turcica in maize (Zhou, Ma, Lu, Zhu, & Yan, 2020). The populations of bacterial plant pathogens such as Xanthomonadaceae and Agrobacterium tumefaciens decreased in benzoxazinoid‐treated plants (Cotton et al., 2019; Kudjordjie et al., 2019).
Any change in cumarin, sesquiterpenes and diadzein by airborne signals and root exudates will change plant microbiomes (Chen et al., 2019; Okutani et al., 2020; Stringlis et al., 2018). Activation of defence hormones (e.g. JA and SA) by airborne signals (e.g. MeSA, 3‐pentanol, or effectors of aphid/whitefly pest) induces microbiome adaptation in plants (Lee, Lee, & Ryu, 2012; Mannaa et al., 2020; Song, Choi, & Ryu, 2015; Yang et al., 2011). Microbiome adaptation in these examples reduces disease severity caused by several plant pathogens and pests, probably by recruiting beneficial bacteria such as B. subtilis (Lee et al., 2012; Song et al., 2015). The rhizosphere microbiome also modulates root metabolism and exudation by azelaic acid as a potential signal molecule (Figure 3; Korenblum et al., 2020). Activation of the two JA pathway branches differentially shape the root microbiome. The Arabidopsis mutants myc2 and med25 alter root exudate (Figure 3). Similar changes in some categories of root exudate were observed in mutants of both branches, but some root exudates were differentially synthesized. Clostridiales were abundant but declined in mutants of both branches. Bacillus, Lysinibacillus and Streptomyces populations increased in the med25 mutant, whereas the Enterobacteriaceae population increased in the myc2 mutant (Carvalhais et al., 2015). Med25 has an important role in regulating density recognition in Arabidopsis and changing root architecture by increasing root response to auxin (Munoz‐Parra et al., 2017).
3.4. Acoustic and electric signals
Plants utilize acoustic and electric signals as internal and inter‐plant signals. A sound vibration signal can be generated by a herbivore walking on the plant, breaking trichomes, chewing the plant and even by water stress (Caicedo‐Lopez, Contreras‐Medina, Guevara‐Gonzaleza, Perez‐Matzumotob, & Ruiz‐Ruedab, 2020; Kollasch et al., 2020). Pest species might be discriminated based on the vibration frequency they produce (Kollasch et al., 2020).
Perception of a sound vibration signal modifies plant epigenetics, transcriptome, proteome and metabolome (Ghosh et al., 2016; Jung, Kim, Jung, Jeong, & Ryu, 2020). Sound vibration signal perception modulates defence hormones such as SA, leading to activation of MAPKs, MYBs and transcription factors (Body et al., 2019; Ghosh et al., 2016). This upregulation of key enzymes and secondary metabolites, including catalase and PAL, increases the biosynthesis of phenols, alkaloids, terpenes and oxylipin‐derivative VOCs (Body et al., 2019; Ghosh et al., 2016; Kollasch et al., 2020). These metabolites enhance plant resistance or act as signals in plant–plant communication. Jung et al. (2020) reported that sound vibration induces resistance against the root pathogen Ralstonia solanacearum by modulating cytokinin signalling, increasing aliphatic glucosinolate biosynthesis through epigenetic DNA methylation by H3K27me3, and improving cell wall reinforcement by downregulating miR397b suppression of lignin accumulation–related transcripts. Vibration sensing is an evolutionarily ancient system that arose before vascular plants emerged, as microalga also have mechanosensory proteins that respond to vibration (Paika, Jin, Sim, & Jeon, 2018).
Electric signals, primarily Ca2+ signalling, have important roles in intra‐ and inter‐plant signalling (Choi et al., 2017; de Toledo et al., 2019; Simmi, Dallagnol, Ferreira, Pereira, & Souza, 2020). The oomycete pathogen Pythium aphanidermatum exploits electric signals to target host roots (Van West et al., 2002). Both acoustic and electric signals have high transmission speed and good potential for use in precision agriculture (Choi et al., 2017; Kollasch et al., 2020).
3.5. Field applications
Although these previous studies have increased our understanding of the signals and mechanisms involved in plant–plant communication, the ultimate goal is to transfer this knowledge to the agricultural field. Field applications can leverage the intrinsic potential of inter‐plant signalling by intercropping or rotating crops according to lab results, or by eliciting plant–plant communication with biological and chemical elicitors.
Plant defence inducers can be applied to induce volatile emission and trigger plant immunity against insect pests and microbial pathogens. MIPVs and HIPVs contain VOCs that inhibit pathogen growth and prime resistance in neighbouring plants (Quintana‐Rodriguez et al., 2015; Sharifi et al., 2018). Inoculating some plant rows in a field with non‐pathogenic strains of plant pathogens, microbe‐associated molecular patterns (MAMPs), or herbivore‐associated molecular patterns (HAMPs) can induce VOC release and reduce disease severity in all plants in the field. Grapevine inoculation with a sulfated laminarin MAMP increases the emission of terpenes such as (E,E)‐α‐farnesene, β‐caryophyllene and trans‐β‐ocimene, and subsequently increases resistance to downy mildew disease. VOCs release and disease resistance are significantly positively correlated (Chalal et al., 2015), and the inter‐plant signalling activity of these compounds on neighbouring plants has been reported elsewhere (Lazazzara et al., 2018; Quintana‐Rodriguez et al., 2015). The flg22 MAMP significantly induces the oxylipin volatiles nonanal, heptanal and hendecanal (Tu et al., 2017). Nonanal is an inter‐plant signal that induces systemic resistance against plant pathogens (Yi et al., 2009).
Inter‐plant infochemicals can alter volatile emission profiles and trigger immune responses (Ameye et al., 2015). (Z)‐3‐hexenyl acetate GLV primes JA‐dependent signalling against Fusarium graminearum in wheat (Ameye et al., 2015). Indole primes the expression of JA‐dependent genes and increases JA synthesis against fall armyworm (S. frugiperda) in rice (Ye et al., 2019). Some infochemicals may induce disease susceptibility depending on the pathosystem. Green leaf volatiles (GLVs) induce maize susceptibility against Colletotrichum graminicola by suppressing SA‐dependent pathways (Gorman et al., 2020). Plant growth–promoting and endophytic bacteria can induce plant volatile synthesis to attract parasitoids and entomopathogens (Bell, Naranjo‐Guevara, Santos, Meadow, & Bento, 2020; Disi, Mohammad, Lawrence, Kloepper, & Fadamiro, 2019; Maggini et al., 2020). By contrast, there has been one report showing that endophytic microbes do not have a significant effect on plant VOC profiles and the behaviour of herbivores and their parasitoids (Moisan et al., 2020).
Plant–plant communication can be enhanced by introducing fungal networks into the soil between plants and by triggering fungal spore germination and root colonization with strigolactones and beneficial bacteria. Mycorrhiza and endophytic fungi such as P. indica can transfer infochemical signals between plant species (Song et al., 2014; Vahabi et al., 2018). Soil inoculation with these fungi or promoting their populations by conservative and organic agriculture can improve inter‐plant signalling and plant priming for imminent challenges. Soil disturbance in intense tillage systems negatively affects mycorrhizae communities (Wang, Li, Li, Zhao, & Liao, 2020). Mycorrhiza colonization is controlled by strigolactones and phosphorus availability (Lopez‐Raez, Shirasu, & Foo, 2017; Waters et al., 2017). Thus, reduced application of phosphorus fertilizers and increased application of phosphate‐solubilizing bacteria and mycorrhiza can improve plant colonization by mycorrhiza. Beneficial microbes can also promote root colonization by mycorrhiza even in non‐mycorrhiza plants (Poveda, Hermosa, Monte, & Nicolás, 2019).
Plant–plant communication can be maximized by managed intercropping to locate aboveground and below‐ground plant parts in close proximity to chemicals released by neighbour plants, which can directly inhibit the germination and growth of pathogenic fungi and bacteria or repel herbivores from fields (Lazazzara et al., 2018; Quintana‐Rodriguez et al., 2015; Yang et al., 2014; Zhou, Cen, Tian, Wang, & Zhang, 2019). Volatiles from resistant cultivars contain volatiles that can directly inhibit pathogen growth or induce systemic resistance in neighbour susceptible cultivars (Lazazzara et al., 2018; Quintana‐Rodriguez et al., 2015). Inter‐plant signals can suppress plant pathogens directly or indirectly through microbiome adaptation. Intercropping of aerobic rice and watermelon reduces disease severity of F. oxysporum in watermelon (Ren et al., 2008). Rice root exudates reduce pathogen spore germination up to 41% and alter the root microbiome community structure in favour of Actinomycetes. Similarly, corn can act as a biological wall between pepper rows to inhibit Phytophthora capsici growth and promote the root microbiome (Yang et al., 2014). DIMBOA is a density‐dependent allelochemical that suppresses plant pathogens in densely cropped maize rows. Intercropped plants can emit infochemicals that alter the transcriptomes of neighbouring plants to cope with pathogens. RNA‐seq results suggest that tall fescue root exudate containing putrescine and cyclohexane‐1,2‐diol stimulates the expression of genes related to defence hormones and pathogenesis‐related proteins in tomato, and reduces stem rot disease (Zhou et al., 2019).
Plant debris such as decaying organic matter or sloughed off root cells functions as a modulator and infochemical for the next plant generation and rhizosphere microbiome. Infochemicals can remain in the ecosystem after plant death or harvest and act as signals for the next crop generation. Infochemicals have different chemical stabilities under different conditions. Plant debris in minimum‐tillage and no‐tillage systems may enhance slow‐release of infochemicals. Plant debris can affect the next crop by modifying the soil microbiome community and activity, by increasing soil fertility, or by acting as an infochemical source (Veen et al., 2019; Wang, Wu, et al., 2020).
Plant debris from root and aerial parts contains information about plant identity, life history and memory of biotic/abiotic stresses. Medicago truncatula growth and endophytic fungi are affected by neighbouring plants and by plants from the previous season. Thus, both intercropping and crop rotation affect the ecological performance of alfalfa as the holobiome (Vannier et al., 2020).
Rotation in hydroponic systems also affects the next crop's performance. Vicia faba plants infested by Acyrthosiphon pisum release soluble chemicals that increase the attractiveness of the next plant parasitoid Aphidius ervi. Similarly, lima bean infested by T. urticae increase the next season plant's attraction of the predatory mite Phytoseiulus persimilis (Delory et al., 2016; Guerrieri, Dong, & Bouwmeester, 2019).
Large‐scale formulation and application of inter‐plant infochemicals is a promising approach in integrated crop management. These compounds can be considered as synthetic pesticide alternatives and can reduce their application dose by combining synthetic pesticides and pest lure volatiles to attract and kill pest (Martel, Alford, & Dickens, 2007). However, infochemicals are highly reactive compounds with short half‐lives under natural conditions. Plant should be treated with these compounds at the proper time under optimum conditions to avoid neutral or negative effects on plant growth and defence. Micro‐ and nano‐encapsulation of infochemicals in natural and synthetic polymers for slow‐ or controlled‐release improves their effects on plant health and volatile emission (Oliveira, Varanda, & Félix, 2016; Wang, Liu, Zhan, & Liu, 2019). Plant virus particles can be used to deliver infochemicals into the rhizosphere (Chariou et al., 2019). The formulation technologies and field applications of infochemicals is reviewed elsewhere (Sharifi & Ryu, 2018a, 2020).
4. CONCLUDING REMARKS
Plants have adapted to growth in complex and competitive ecosystems during their evolutionary history. They need to compete with interspecific and conspecific plants for light, water and nutrients, and communicate with neighbouring plants to anticipate upcoming biotic/abiotic challenges (Ballaré & Austin, 2019; Effah, Holopainen, & McCormick, 2019; Hodge, Fitter, & Robinson, 2013; Hortal et al., 2017). However, only some of the competition and communication mechanisms rely on the plant genome. Plant microbiota have pivotal roles in nutrient solubility and uptake, especially nitrogen, phosphorus and iron (Adesemoye, Torbert, & Kloepper, 2008; Sharifi, Ahmadzadeh, Sharifi‐Tehrani, & Talebi‐Jahromi, 2010). Microbiota also improve water use efficiency and osmotic stress response (Fan et al., 2015; Sharifi & Ryu, 2018c). The plant holobiome leverages the collection of its member genes to optimize performance and survival. Plants have spatiotemporal layers of defence consisting of rhizosphere microbes, endophytes, pattern‐triggered immunity, effector‐triggered immunity and recruited natural enemies; each of these can efficiently suppress specific groups of attackers (Carrión et al., 2019; Sharifi & Ryu, 2017). Because of these advantages conferred by microbiota, plants donate 10–30% of their carbon and nitrogen to the rhizosphere to organize their microbiota.
Information and signal transferring systems play important roles in plant growth and survival. Plants decipher their complex habitat situations by perceiving physical and chemical cues and signals, either directly from neighbouring plants or indirectly from their symbionts such as mycorrhiza, endophytic fungi and dodder. Previous research has characterized the signal types and signal mediators involved in plant interactions with other members of the ecosystem and revealed that deaf and mute mutants of plants have reduced ecological competence. However, plants may have lost some of their communication abilities during domestication and plant breeding programs. Therefore, plant breeders and genetic engineers should have a holistic view of plants as members of the holobiome. Otherwise, small changes in genes related to inter‐plant and inter‐kingdom signals may substantially affect plant performance, a phenomenon called the butterfly effect.
Agricultural practices also affect plant–plant communication. As we mentioned above, no‐tillage and minimum tillage systems preserve common mycorrhizal networks and endophytic fungi as mediators of wired plant–plant communication. Moreover, Information can transfer between plants during intercropping or from crop to crop in the next season. Thus, recent advances in our understanding of plant–plant communication can help manage agricultural practices so that they utilize the abilities of plants to exploit ecological interactions, and survive in competitive environments.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
We appreciate Ms Meeae Hwang and Hanhee Na for drawing Figure 1. This research was supported by grants from the BioNano Health‐Guard Research Center funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea as Global Frontier Project (Grant No. HGUARD_2013M3A6B2078954), the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF‐2020M3E9A1111636), Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea [as part of the (multiministerial) Genome Technology to Business Translation Program 918017‐4] and the KRIBB Initiative Program, South Korea.
Sharifi R, Ryu C‐M. Social networking in crop plants: Wired and wireless cross‐plant communications. Plant Cell Environ. 2021;44:1095–1110. 10.1111/pce.13966
Funding information Korea Research Institute of Bioscience and Biotechnology, Grant/Award Number: KGM4572013; Ministry of Agriculture, Food and Rural Affairs, Grant/Award Number: Genome Technology to Business Translation Program; Ministry of Science, ICT and Future Planning, Grant/Award Number: HGUARD_2013M3A6B2078954; the National Research Foundation of Korea, Grant/Award Number: NRF‐2020M3E9A1111636
REFERENCES
- Adesemoye, A. O. , Torbert, H. A. , & Kloepper, J. W. (2008). Enhanced plant nutrient use efficiency with PGPR and AMF in an integrated nutrient management system. Canadian Journal of Microbiology, 54, 876–886. [DOI] [PubMed] [Google Scholar]
- Alaux, P.‐L. , Naveau, F. , Declerck, S. , & Cranenbrouck, S. (2020). Common mycorrhizal network induced JA/ET genes expression in healthy potato plants connected to potato plants infected by Phytophthora infestans . Frontiers in Plant Science, 11, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alborn, H. , Turlings, T. , Jones, T. H. , Stenhagen, G. , Loughrin, J. , & Tumlinson, J. (1997). An elicitor of plant volatiles from beet armyworm oral secretion. Science, 276, 945–949. [Google Scholar]
- Alborn, H. T. , Hansen, T. V. , Jones, T. H. , Bennett, D. C. , Tumlinson, J. H. , Schmelz, E. A. , & Teal, P. E. (2007). Disulfooxy fatty acids from the American bird grasshopper Schistocerca americana, elicitors of plant volatiles. Proceedings of the National Academy of Sciences, 104, 12976–12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, J. G. , Alborn, H. T. , & Stelinski, L. L. (2011). Constitutive and induced subterranean plant volatiles attract both entomopathogenic and plant parasitic nematodes. Journal of Ecology, 99, 26–35. [Google Scholar]
- Ameye, M. , Allmann, S. , Verwaeren, J. , Smagghe, G. , Haesaert, G. , Schuurink, R. C. , & Audenaert, K. (2018). Green leaf volatile production by plants: A meta‐analysis. New Phytologist, 220, 666–683. [DOI] [PubMed] [Google Scholar]
- Ameye, M. , Audenaert, K. , De Zutter, N. , Steppe, K. , Van Meulebroek, L. , Vanhaecke, L. , … Smagghe, G. (2015). Priming of wheat with the green leaf volatile Z‐3‐hexenyl acetate enhances defense against Fusarium graminearum but boosts deoxynivalenol production. Plant Physiology, 167, 1671–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andika, I. B. , Wei, S. , Cao, C. , Salaipeth, L. , Kondo, H. , & Sun, L. (2017). Phytopathogenic fungus hosts a plant virus: A naturally occurring cross‐kingdom viral infection. Proceedings of the National Academy of Sciences, 114, 12267–12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babikova, Z. , Gilbert, L. , Bruce, T. J. , Birkett, M. , Caulfield, J. C. , Woodcock, C. , … Johnson, D. (2013). Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecology Letters, 16, 835–843. [DOI] [PubMed] [Google Scholar]
- Bailly, A. , Groenhagen, U. , Schulz, S. , Geisler, M. , Eberl, L. , & Weisskopf, L. (2014). The inter‐kingdom volatile signal indole promotes root development by interfering with auxin signalling. The Plant Journal, 80, 758–771. [DOI] [PubMed] [Google Scholar]
- Bais, H. P. (2006). The role of root exudates in rhizosphere interations with plants and other organisms. Annual Reveiw Plant Biology, 57, 233–266. [DOI] [PubMed] [Google Scholar]
- Ballaré, C. L. , & Austin, A. T. (2019). Recalculating growth and defense strategies under competition: Key roles of photoreceptors and jasmonates. Journal of Experimental Botany, 70, 3425–3436. [DOI] [PubMed] [Google Scholar]
- Barto, E. K. , Hilker, M. , Muller, F. , Mohney, B. K. , Weidenhamer, J. D. , & Rillig, M. C. (2011). The fungal fast lane: Common mycorrhizal networks extend bioactive zones of allelochemicals in soils. PLoS One, 6, e27195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, K. , Naranjo‐Guevara, N. , Santos, R. C. , Meadow, R. , & Bento, J. M. S. (2020). Predatory earwigs are attracted by herbivore‐induced plant volatiles linked with plant growth‐promoting rhizobacteria. Insects of Jianfengling, 11, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J. A. , Cahill, J. F. , & van der Heijden, M. (2016). Fungal effects on plant–plant interactions contribute to grassland plant abundances: Evidence from the field. Journal of Ecology, 104, 755–764. [Google Scholar]
- Berg, G. , Rybakova, D. , Fischer, D. , Cernava, T. , Vergès, M.‐C. C. , Charles, T. , … Corral, G. H. (2020). Microbiome definition re‐visited: Old concepts and new challenges. Microbiome, 8, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, R. , Andika, I. B. , Pang, T. , Lian, Z. , Wei, S. , Niu, E. , … Sun, L. (2020). Facilitative and synergistic interactions between fungal and plant viruses. Proceedings of the National Academy of Sciences, 117, 3779–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichlmeier, M. (2017). Identification of systemic acquired resistance‐related volatile organic compounds and their role in plant immunity. (PhD thesis). Technische Universität München, Munich, Germany. [Google Scholar]
- Biedrzycki, M. L. , Jilany, T. A. , Dudley, S. A. , & Bais, H. P. (2010). Root exudates mediate kin recognition in plants. Communicative & Integrative Biology, 3, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittebiere, A.‐K. , Benot, M.‐L. , & Mony, C. (2020). Clonality as a key but overlooked driver of biotic interactions in plants. Perspectives in Plant Ecology, Evolution and Systematics, 43, 125510. [Google Scholar]
- Body, M. J. A. , Neer, W. C. , Vore, C. , Lin, C.‐H. , Vu, D. , Schultz, J. C. , … Appel, H. M. (2019). Caterpillar chewing vibrations cause changes in plant hormones and volatile emissions in Arabidopsis thaliana . Frontiers in Plant Science, 10, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure, G. , VanDoorn, A. , & Baldwin, I. T. (2011). Herbivore‐associated elicitors: FAC signaling and metabolism. Trends in Plant Science, 16, 294–299. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, H. , Schuurink, R. C. , Bleeker, P. M. , & Schiestl, F. (2019). The role of volatiles in plant communication. Plant Journal, 100, 892–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo‐Lopez, L. H. , Contreras‐Medina, L. M. , Guevara‐Gonzaleza, R. G. , Perez‐Matzumotob, A. E. , & Ruiz‐Ruedab, A. (2020). Effects of hydric stress on vibrational frequency patterns of Capsicum annuum plants. Plant Signaling Behavior, 15(7), e1770489–e1770482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho‐Coronel, X. , Molina‐Torres, J. , & Heil, M. (2020). Sequestration of exogenous volatiles by plant cuticular waxes as a mechanism of passive associational resistance: A proof of concept. Frontiers in Plant Science, 11, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión, V. J. , Perez‐Jaramillo, J. , Cordovez, V. , Tracanna, V. , de Hollander, M. , Ruiz‐Buck, D. , … Raaijmakers, J. (2019). Pathogen‐induced activation of disease‐suppressive functions in the endophytic root microbiome. Science, 366, 606–612. [DOI] [PubMed] [Google Scholar]
- Carvalhais, L. C. , Dennis, P. G. , Badri, D. V. , Kidd, B. N. , Vivanco, J. M. , & Schenk, P. M. (2015). Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Molecular Plant Microbe Interactions, 28, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Castelyn, H. D. , Appelgryn, J. J. , Mafa, M. S. , Pretorius, Z. A. , & Visser, B. (2014). Volatiles emitted by leaf rust infected wheat induce a defence response in exposed uninfected wheat seedlings. Australasian Plant Pathology, 44, 245–254. [Google Scholar]
- Chalal, M. , Winkler, J.B. , Gourrat, K. , Trouvelot, S. , Adrian, M. , Schnitzler, J.‐P. , Jamois, F. & Daire, X. (2015) Sesquiterpene volatile organic compounds (VOCs) are markers of elicitation by sulfated laminarine in grapevine. Frontiers in Plant Science, 6, 350. doi: 3 10.3389/fpls.2015.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. , Kwok, S. F. , Bleecker, A. B. , & Meyerowitz, E. M. (1993). Arabidopsis ethylene‐response gene ETR1: Similarity of product to two‐component regulators. Science, 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Chariou, P. L. , Dogan, A. B. , Welsh, A. G. , Saidel, G. M. , Baskaran, H. , & Steinmetz, N. F. (2019). Soil mobility of synthetic and virus‐based model nanopesticides. Nature Nanotechnology, 14, 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Yang, R. , Chen, J. , Luo, Q. , Cui, X. , Yan, X. , & Gerwick, W. H. (2019). 1‐Octen‐3‐ol, a self‐stimulating oxylipin messenger, can prime and induce defense of marine alga. BMC Plant Biology, 19, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Jiang, T. , Liu, Y. X. , Liu, H. , Zhao, T. , Liu, Z. , … Wang, G. (2019). Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Science China. Life Sciences, 62, 947–958. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Zhang, L. , Cai, X. , Li, X. , Bian, L. , Luo, Z. , … Xin, Z. (2020). (E)‐Nerolidol is a volatile signal that induces defenses against insects and pathogens in tea plants. Horticulture Research, 7, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheol Song, G. , Sim, H.‐J. , Kim, S.‐G. , & Ryu, C.‐M. (2016). Root‐mediated signal transmission of systemic acquired resistance against above‐ground and below‐ground pathogens. Annals of Botany, 118, 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiriboga, X. , Guo, H. , Campos‐Herrera, R. , Röder, G. , Imperiali, N. , Keel, C. , … Turlings, T. C. (2018). Root‐colonizing bacteria enhance the levels of (E)‐β‐caryophyllene produced by maize roots in response to rootworm feeding. Oecologia, 187, 459–468. [DOI] [PubMed] [Google Scholar]
- Choi, W.‐G. , Miller, G. , Wallace, I. , Harper, J. , Mittler, R. , & Gilroy, S. (2017). Orchestrating rapid long‐distance signaling in plants with Ca2+, ROS and electrical signals. The Plant Journal, 90, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollini, D. , Rigsby, C. M. , & Barto, E. K. (2012). Microbes as targets and mediators of allelopathy in plants. Journal of Chemical Ecology, 38, 714–727. [DOI] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G. J. , Langenbach, C. J. , & Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annual Review of Phytopathology, 53, 97–119. [DOI] [PubMed] [Google Scholar]
- Copolovici, L. , Kannaste, A. , Pazouki, L. , & Niinemets, U. (2012). Emissions of green leaf volatiles and terpenoids from Solanum lycopersicum are quantitatively related to the severity of cold and heat shock treatments. Journal of Plant Physiology, 169, 664–672. [DOI] [PubMed] [Google Scholar]
- Cotton, T. E. A. , Petriacq, P. , Cameron, D. D. , Meselmani, M. A. , Schwarzenbacher, R. , Rolfe, S. A. , & Ton, J. (2019). Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME Journal, 13, 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Trindade, R. , Almeida, L. , Xavier, L. , Lins, A. L. , Andrade, E. H. , Maia, J. G. , … da Silva, J. K. (2019). Arbuscular mycorrhizal fungi colonization promotes changes in the volatile compounds and enzymatic activity of lipoxygenase and phenylalanine ammonia lyase in Piper nigrum L. ‘Bragantina’. Plants (Basel), 8(11), 442. 10.3390/plants8110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro, M. , Erb, M. , Ton, J. , Brandenburg, A. , Karlen, D. , Zopfi, J. , & Turlings, T. C. (2014). Volatiles produced by soil‐borne endophytic bacteria increase plant pathogen resistance and affect tritrophic interactions. Plant, Cell & Environment, 37, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lange, E. S. , Laplanche, D. , Guo, H. , Xu, W. , Vlimant, M. , Erb, M. , … Turlings, T. C. (2020). Spodoptera frugiperda caterpillars suppress herbivore‐induced volatile emissions in maize. Journal of Chemical Ecology, 46(3), 344–360. https://link.springer.com/article/10.1007/s10886-020-01153-x. [DOI] [PubMed] [Google Scholar]
- de Toledo, G. R. A. , Parise, A. G. , Simmi, F. Z. , Costa, A. V. L. , Senko, L. G. S. , Debono, M.‐W. , & Souza, G. M. (2019). Plant electrome: The electrical dimension of plant life. Theoretical and Experimental Plant Physiology, 31, 21–46. [Google Scholar]
- Deja‐Sikora, E. , Mercy, L. , Baum, C. , & Hrynkiewicz, K. (2019). The contribution of endomycorrhiza to the performance of potato virus y‐infected solanaceous plants: Disease alleviation or exacerbation? Frontiers in Microbiology, 10, 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, K. J. , Breza‐Boruta, B. , Lemańczyk, G. , Bocianowski, J. , Wrzesińska, D. , Kalka, I. , & Piesik, D. (2015). Maize VOC induction after infection by the bacterial pathogen, Pantoea ananatis, alters neighbouring plant VOC emissions. Journal of Plant Diseases and Protection, 122, 125–132. [Google Scholar]
- Delory, B. M. , Delaplace, P. , Fauconnier, M.‐L. , & du Jardin, P. (2016). Root‐emitted volatile organic compounds: Can they mediate belowground plant–plant interactions? Plant and Soil, 402, 1–26. [Google Scholar]
- Desurmont, G. A. , Xu, H. , & Turlings, T. C. (2016). Powdery mildew suppresses herbivore‐induced plant volatiles and interferes with parasitoid attraction in Brassica rapa . Plant, Cell & Environment, 39, 1920–1927. [DOI] [PubMed] [Google Scholar]
- Disi, J. O. , Mohammad, H. K. , Lawrence, K. , Kloepper, J. , & Fadamiro, H. (2019). A soil bacterium can shape belowground interactions between maize, herbivores and entomopathogenic nematodes. Plant and Soil, 437, 83–92. [Google Scholar]
- Dombrowski, J. E. , Kronmiller, B. A. , Hollenbeck, V. G. , Rhodes, A. C. , Henning, J. A. , & Martin, R. C. (2019). Transcriptome analysis of the model grass Lolium temulentum exposed to green leaf volatiles. BMC Plant Biology, 19, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski, J. E. , & Martin, R. C. (2018). Activation of MAP kinases by green leaf volatiles in grasses. BMC Research Notes, 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl, F. , Hammerbacher, A. , Gershenzon, J. , & Unsicker, S. B. (2018). Leaf rust infection reduces herbivore‐induced volatile emission in black poplar and attracts a generalist herbivore. New Phytologist, 220, 760–772. [DOI] [PubMed] [Google Scholar]
- Edwards, J. A. , Santos‐Medellın, C. M. , Liechty, Z. S. , Nguyen, B. , Lurie, E. , & Eason, S. (2018). Compositional shifts in root‐associated bacterial and archaeal microbiota track the plant life cycle in field‐grown rice. PLoS Biology, 16(2), e2003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effah, E. , Holopainen, J. K. , & McCormick, A. C. (2019). Potential roles of volatile organic compounds in plant competition. Perspectives in Plant Ecology, Evolution and Systematics, 38, 58–63. [Google Scholar]
- Falik, O. , Mordoch, Y. , Ben‐Natan, D. , Vanunu, M. , Goldstein, O. , & Novoplansky, A. (2012). Plant responsiveness to root–root communication of stress cues. Annals of Botany, 110, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X. , Hu, H. , Huang, G. , Huang, F. , Li, Y. , & Palta, J. (2015). Soil inoculation with Burkholderia sp. LD‐11 has positive effect on water‐use efficiency in inbred lines of maize. Plant and Soil, 390, 337–349. [Google Scholar]
- Ghosh, R. , Mishra, R. C. , Choi, B. , Kwon, Y. S. , Won, B. D. , Park, S. C. J. , … Bae, H. (2016). Exposure to sound vibrations lead to transcriptomic, proteomic and hormonal changes in arabidopsis. Scientific Reports, 6, 33370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, L. , & Johnson, D. (2017). Plant–plant communication through common mycorrhizal networks. In Advances in Botanical Research (Vol. 82, pp. 83–97). London, UK: Elsevier. [Google Scholar]
- Gilbert, S. F. (2019). Evolutionary transitions revisited: Holobiont evo‐devo. Journal of Experimental Zoology, 332, 307–314. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Reveiw of Phytopathology, 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Gómez, S. , & Stuefer, J. F. (2006). Members only: Induced systemic resistance to herbivory in a clonal plant network. Oecologia, 147, 461–468. [DOI] [PubMed] [Google Scholar]
- Gómez, S. , Van Dijk, W. , & Stuefer, J. F. (2010). Timing of induced resistance in a clonal plant network. Plant Biology, 12, 512–517. [DOI] [PubMed] [Google Scholar]
- Gorman, Z. , Christensen, S. A. , Yan, Y. , He, Y. , Borrego, E. , & Kolomiets, M. V. (2020). Green leaf volatiles and jasmonic acid enhance susceptibility to anthracnose diseases caused by Colletotrichum graminicola in maize. Molecular Plant Pathology, 21, 702–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. , Wei, Z. , Wang, X. , Friman, V.‐P. , Huang, J. , Wang, X. , … Jousset, A. (2016). Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biology and Fertility of Soils, 52, 997–1005. [Google Scholar]
- Guerrieri, A. , Dong, L. , & Bouwmeester, H. J. (2019). Role and exploitation of underground chemical signaling in plants. Pest Management Science, 75, 2455–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, M. , & Bueno, J. C. S. (2014). Herbivore‐induced volatiles as rapid signals in systemic plant responses. Plant Signaling & Behavior, 2, 191–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen, C. , Li, J. , Zhuang, H. , Sun, H. , Xu, Y. , Qi, J. , … Wu, J. (2017). Stem parasitic plant Cuscuta australis (dodder) transfers herbivory‐induced signals among plants. Proceedings of the National Academy of Sciences, 114, E6703–E6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, A. , Fitter, A. H. , & Robinson, D. (2013). Microbial mediation of plant competition and community structure. Functional Ecology, 27, 865–875. [Google Scholar]
- Hopke, J. , Donath, J. , Blechert, S. , & Boland, W. (1994). Herbivore‐induced volatiles: The emission of acyclic homoterpenes from leaves of Phaseolus lunatus and Zea mays can be triggered by a β‐glucosidase and jasmonic acid. FEBS Letters, 352, 146–150. [DOI] [PubMed] [Google Scholar]
- Hortal, S. , Lozano, Y. M. , Bastida, F. , Armas, C. , Moreno, J. L. , Garcia, C. , & Pugnaire, F. I. (2017). Plant–plant competition outcomes are modulated by plant effects on the soil bacterial community. Scientific Reports, 7, 17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L. , Ye, M. , & Erb, M. (2019). Integration of two herbivore‐induced plant volatiles results in synergistic effects on plant defence and resistance. Plant, Cell & Environment, 42, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M. , Sanchez‐Moreiras, A. M. , Abel, C. , Sohrabi, R. , Lee, S. , Gershenzon, J. , & Tholl, D. (2012). The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)‐beta‐caryophyllene, is a defense against a bacterial pathogen. New Phytologist, 193, 997–1008. [DOI] [PubMed] [Google Scholar]
- Jassbi, A. R. , Zamanizadehnajari, S. , & Baldwin, I. T. (2010). Phytotoxic volatiles in the roots and shoots of Artemisia tridentata as detected by headspace solid‐phase microextraction and gas chromatographic‐mass spectrometry analysis. Journal of Chemical Ecology, 36, 1398–1407. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Ye, J. , Veromann, L.‐L. , & Niinemets, Ü. (2016). Scaling of photosynthesis and constitutive and induced volatile emissions with severity of leaf infection by rust fungus (Melampsora larici‐populina) in Populus balsamifera var. suaveolens . Tree Physiology, 36(7), 856–872. [DOI] [PubMed] [Google Scholar]
- Jung, H. W. , Tschaplinski, T. J. , Wang, L. , Glazebrook, J. , & Greenberg, J. T. (2009). Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Jung, J. , Kim, S.‐K. , Jung, S.‐H. , Jeong, M.‐J. & Ryu, C.‐M. (2020) Sound vibration‐triggered epigenetic modulation induces plant root immunity against Ralstonia solanacearum . Frontiers in Microbiology, 11, 1978. doi: 19 10.3389/fmicb.2020.01978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanoli, K. , Kokalas, V. , Koveos, D. , Junker, R. , & Farré‐Armengol, G. (2020). Bacteria affect plant–mite interactions via altered scent emissions. Journal of Chemical Ecology, 46, 782–792. [DOI] [PubMed] [Google Scholar]
- Karban, R. , Shiojiri, K. , Ishizaki, S. , Wetzel, W. C. , & Evans, R. Y. (2013). Kin recognition affects plant communication and defence. Proceedings of the Royal Society B: Biological Sciences, 280, 20123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban, R. , Wetzel, W. C. , Shiojiri, K. , Ishizaki, S. , Ramirez, S. R. , & Blande, J. D. (2014). Deciphering the language of plant communication: Volatile chemotypes of sagebrush. The New Phytologist, 204, 380–385. [DOI] [PubMed] [Google Scholar]
- Kessler, A. , & Heil, M. (2011). The multiple faces of indirect defences and their agents of natural selection. Functional Ecology, 25, 348–357. [Google Scholar]
- Khashi u Rahman, M. , Zhou, X. , & Wu, F. (2019). The role of root exudates, CMNs, and VOCs in plant–plant interaction. Journal of Plant Interactions, 14, 630–636. [Google Scholar]
- Klimm, F. S. , Weinhold, A. , & Volf, M. (2020). Volatile production differs between oak leaves infested by leaf‐miner Phyllonorycter harrisella (Lepidoptera: Gracillariidae) and galler Neuroterus quercusbaccarum (Hymenoptera: Cynipidae). European Journal of Entomology, 117, 101–109. [Google Scholar]
- Kollasch, A. M. , Abdul‐Kaf, A. , Body, M. J. A. , Pinto, C. F. , Appel, H. M. , & Cocroft, R. B. (2020). Leaf vibrations produced by chewing provide a consistent acoustic target for plant recognition of herbivores. Oecologia, 194, 1–13. [DOI] [PubMed] [Google Scholar]
- Kong, C. H. , Xu, X. H. , Zhang, M. , & Zhang, S. Z. (2010). Allelochemical tricin in rice hull and its aurone isomer against rice seedling rot disease. Pest Management Science, 66, 1018–1024. [DOI] [PubMed] [Google Scholar]
- Kong, C. H. , Zhang, S. Z. , Li, Y. H. , Xia, Z. C. , Yang, X. F. , Meiners, S. J. , & Wang, P. (2018). Plant neighbor detection and allelochemical response are driven by root‐secreted signaling chemicals. Nature Communications, 9, 3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, H. G. , Song, G. C. , Sim, H. J. , & Ryu, C.‐M. (2020). Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME Journal. 10.1038/s41396-020-00759-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenblum, E. , Dong, Y. , Szymanski, J. , Panda, S. , Jozwiak, A. , Massalha, H. , … Aharoni, A. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root‐to‐root signaling. Proceedings of the National Academy of Sciences, 117, 3874–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroes, A. , Weldegergis, B. T. , Cappai, F. , Dicke, M. , & van Loon, J. J. (2017). Terpenoid biosynthesis in Arabidopsis attacked by caterpillars and aphids: Effects of aphid density on the attraction of a caterpillar parasitoid. Oecologia, 185, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudjordjie, E. N. , Sapkota, R. , Steffensen, S. K. , Fomsgaard, I. S. , & Nicolaisen, M. (2019). Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome, 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, Y. S. , Ryu, C. M. , Lee, S. , Park, H. B. , Han, K. S. , Lee, J. H. , … Bae, D. W. (2010). Proteome analysis of Arabidopsis seedlings exposed to bacterial volatiles. Planta, 232, 1355–1370. [DOI] [PubMed] [Google Scholar]
- Lazazzara, V. , Bueschl, C. , Parich, A. , Pertot, I. , Schuhmacher, R. , & Perazzolli, M. (2018). Downy mildew symptoms on grapevines can be reduced by volatile organic compounds of resistant genotypes. Scientific Reports, 8, 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik, J. , Tibboel, M. , Dicke, M. , & van Loon, J. J. (2017). Inoculation of susceptible and resistant potato plants with the late blight pathogen Phytophthora infestans: Effects on an aphid and its parasitoid. Entomologia Experimentalis et Applicata, 163, 305–314. [Google Scholar]
- Lee, B. , Lee, S. , & Ryu, C. M. (2012). Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non‐pathogenic bacteria in pepper. Annals of Botany, 110, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. M. , Kim, S. K. , Lee, N. , Ahn, C. Y. , & Ryu, C. M. (2020). d‐Lactic acid secreted by Chlorella fusca primes pattern‐triggered immunity against Pseudomonas syringae in Arabidopsis. The Plant Journal, 102, 761–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner, M. , Kaiser, R. , Rasmussen, M. O. , Driguez, H. , Boland, W. , & Mithöfer, A. (2008). Microbial oligosaccharides differentially induce volatiles and signalling components in Medicago truncatula . Phytochemistry, 69, 2029–2040. [DOI] [PubMed] [Google Scholar]
- Li, L.‐L. , Zhao, H.‐H. , & Kong, C.‐H. (2020). (−)‐Loliolide, the most ubiquitous lactone, is involved in barnyardgrass‐induced rice allelopathy. Journal of Experimental Botany, 71, 1540–1550. [DOI] [PubMed] [Google Scholar]
- Li, S. , Zhang, J. , Liu, H. , Liu, N. , Shen, G. , Zhuang, H. , & Wu, J. (2020). Dodder‐transmitted mobile signals prime host plants for enhanced salt tolerance. Journal of Experimental Botany, 71, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Hussain, M. , Avery, P. B. , Qasim, M. , Fang, D. , & Wang, L. (2016). Volatiles from plants induced by multiple aphid attacks promote conidial performance of Lecanicillium lecanii . PLoS One, 11, e0151844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Gresa, M. P. , Paya, C. , Ozaez, M. , Rodrigo, I. , Conejero, V. , Klee, H. , … Lison, P. (2018). A new role for green leaf volatile esters in tomato stomatal defense against Pseudomonas syringe pv. tomato. Frontiers in Plant Science, 9, 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Raez, J. A. , Shirasu, K. , & Foo, E. (2017). Strigolactones in plant interactions with beneficial and detrimental organisms: The Yin and Yang. Trends in Plant Science, 22, 527–537. [DOI] [PubMed] [Google Scholar]
- Maggini, V. , Bandeira Reidel, R. V. , De Leo, M. , Mengoni, A. , Rosaria, G. E. , Miceli, E. , … Pistelli, L. (2020). Volatile profile of Echinacea purpurea plants after in vitro endophyte infection. Natural Product Research, 34, 2232–2237. [DOI] [PubMed] [Google Scholar]
- Mannaa, M. , Han, G. , Jeon, H. W. , Kim, J. , Kim, N. , Park, A. R. , … Seo, Y. S. (2020). Influence of resistance‐inducing chemical elicitors against pine wilt disease on the rhizosphere microbiome. Microorganisms, 8(6), 884. 10.3390/microorganisms8060884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel, J. W. , Alford, A. R. , & Dickens, J. (2007). Evaluation of a novel host plant volatile‐based attracticide for management of Colorado potato beetle, Leptinotarsa decemlineata (Say). Crop Protection, 26, 822–827. [Google Scholar]
- Mirabella, R. , Rauwerda, H. , Allmann, S. , Scala, A. , Spyropoulou, E. A. , Vries, M. , … Schuurink, R. C. (2015). WRKY40 and WRKY6 act downstream of the green leaf volatile E‐2‐hexenal in Arabidopsis. The Plant Journal, 83, 1082–1096. [DOI] [PubMed] [Google Scholar]
- Moisan, K. , Lucas‐Barbosa, D. , Villela, A. , Greenberg, L. O. , Cordovez, V. , Raaijmakers, J. M. , & Dicke, M. (2020). No evidence of modulation of indirect plant resistance of Brassica rapa plants by volatiles from soil‐borne fungi. Ecological Entomology, 45, 1200–1211. [Google Scholar]
- Moreira, X. , Nell, C. S. , Katsanis, A. , Rasmann, S. , & Mooney, K. A. (2018). Herbivore specificity and the chemical basis of plant–plant communication in Baccharis salicifolia (Asteraceae). New Phytologist, 220, 703–713. [DOI] [PubMed] [Google Scholar]
- Munoz‐Parra, E. , Pelagio‐Flores, R. , Raya‐Gonzalez, J. , Salmeron‐Barrera, G. , Ruiz‐Herrera, L. F. , Valencia‐Cantero, E. , & Lopez‐Bucio, J. (2017). Plant–plant interactions influence developmental phase transitions, grain productivity and root system architecture in Arabidopsis via auxin and PFT1/MED25 signalling. Plant, Cell & Environment, 40, 1887–1899. [DOI] [PubMed] [Google Scholar]
- Neal, A. , & Ton, J. (2013). Systemic defense priming by Pseudomonas putida KT2440 in maize depends on benzoxazinoid exudation from the roots. Plant Signaling & Behavior, 8, e22655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerva, L. , Varese, G. C. , Falk, B. W. , & Turina, M. (2017). Mycoviruses of an endophytic fungus can replicate in plant cells: Evolutionary implications. Scientific Reports, 7, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic, V. , Dahlin, I. , Vucetic, A. , Petrovic‐Obradovic, O. , Glinwood, R. , & Webster, B. (2013). Volatile exchange between undamaged plants – A new mechanism affecting insect orientation in intercropping. PLoS One, 8, e69431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okutani, F. , Hamamoto, S. , Aoki, Y. , Nakayasu, M. , Nihei, N. , Nishimura, T. , … Sugiyama, A. (2020). Rhizosphere modeling reveals spatiotemporal distribution of daidzein shaping soybean rhizosphere bacterial community. Plant, Cell & Environment, 43, 1036–1046. [DOI] [PubMed] [Google Scholar]
- Oliveira, M. D. M. , Varanda, C. M. R. , & Félix, M. R. F. (2016). Induced resistance during the interaction pathogen × plant and the use of resistance inducers. Phytochemistry Letters, 15, 152–158. [Google Scholar]
- Orlovskis, Z. , & Reymond, P. (2020). Pieris brassicae eggs trigger inter‐plant systemic acquired resistance against a foliar pathogen in Arabidopsis . New Phytologist, 228, 1652–1661. [DOI] [PubMed] [Google Scholar]
- Paika, S. M. , Jin, E. S. , Sim, S. J. , & Jeon, N. L. (2018). Vibration‐induced stress priming during seed culture increases microalgal biomass in high shear field‐cultivation. Bioresource Technology, 254, 340–346. [DOI] [PubMed] [Google Scholar]
- Paschold, A. , Halitschke, R. , & Baldwin, I. T. (2006). Using ‘mute’ plants to translate volatile signals. Plant Journal, 45, 275–291. [DOI] [PubMed] [Google Scholar]
- Peñaflor, M. F. G. , & Bento, J. M. S. (2019). Red‐rot infection in sugarcane attenuates the attractiveness of sugarcane borer‐induced plant volatiles to parasitoid. Arthropod‐Plant Interactions, 13, 117–125. [Google Scholar]
- Piesik, D. , Lemnczyk, G. , Skoczek, A. , Lamparski, R. , Bocianowski, J. , Kotwica, K. , & Delaney, K. J. (2011). Fusarium infection in maize: Volatile induction of infected and neighboring uninfected plants has the potential to attract a pest cereal leaf beetle, Oulema melanopus . Journal of Plant Physiology, 168, 1534–1542. [DOI] [PubMed] [Google Scholar]
- Piesik, D. , Pańka, D. , Jeske, M. , Wenda‐Piesik, A. , Delaney, K. J. , & Weaver, D. K. (2013). Volatile induction of infected and neighbouring uninfected plants potentially influence attraction/repellence of a cereal herbivore. Journal of Applied Entomology, 137, 296–309. [Google Scholar]
- Pontin, M. , Bottini, R. , Burba, J. L. , & Piccoli, P. (2015). Allium sativum produces terpenes with fungistatic properties in response to infection with Sclerotium cepivorum . Phytochemistry, 115, 152–160. [DOI] [PubMed] [Google Scholar]
- Poveda, J. , Hermosa, R. , Monte, E. , & Nicolás, C. (2019). Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non‐host Brassicaceae roots and increases plant productivity. Scientific Reports, 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qawasmeh, A. , Raman, A. , & Wheatley, W. (2015). Volatiles in perennial ryegrass infected with strains of endophytic fungus: Impact on African black beetle host selection. Journal of Applied Entomology, 139, 94–104. [Google Scholar]
- Qian, Y. , Li, D. , Han, L. , & Sun, Z. (2010). Inter‐ramet photosynthate translocation in buffalograss under differential water deficit stress. Journal of the American Society for Horticultural Science, 135, 310–316. [Google Scholar]
- Quintana‐Rodriguez, E. , Morales‐Vargas, A. T. , Molina‐Torres, J. , Ádame‐Alvarez, R. M. , Acosta‐Gallegos, J. A. , Heil, M. , & Flynn, D. (2015). Plant volatiles cause direct, induced and associational resistance in common bean to the fungal pathogen Colletotrichum lindemuthianum . Journal of Ecology, 103, 250–260. [Google Scholar]
- Rasmann, S. , Kollner, T. G. , Degenhardt, J. , Hiltpold, I. , Toepfer, S. , Kuhlmann, U. , … Turlings, T. C. J. (2005). Recruitment of entomopathogenic nematodes by insect‐damaged maize roots. Nature, 434, 732–737. [DOI] [PubMed] [Google Scholar]
- Ren, L. , Su, S. , Yang, X. , Xu, Y. , Huang, Q. , & Shen, Q. (2008). Intercropping with aerobic rice suppressed Fusarium wilt in watermelon. Soil Biology and Biochemistry, 40, 834–844. [Google Scholar]
- Riedlmeier, M. , Ghirardo, A. , Wenig, M. , Knappe, C. , Koch, K. , Georgii, E. , … Vlot, A. C. (2017). Monoterpenes support systemic acquired resistance within and between plants. Plant Cell, 29, 1440–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas‐San Vicente, M. , & Plasencia, J. (2011). Salicylic acid beyond defence: Its role in plant growth and development. Journal of Experimental Botany, 62, 3321–3338. [DOI] [PubMed] [Google Scholar]
- Rodriguez, P. A. , & Bos, J. I. (2013). Toward understanding the role of aphid effectors in plant infestation. Molecular Plant–Microbe Interactions, 26, 25–30. [DOI] [PubMed] [Google Scholar]
- Roiloa, S. R. , Antelo, B. , & Retuerto, R. (2014). Physiological integration modifies delta15N in the clonal plant Fragaria vesca, suggesting preferential transport of nitrogen to water‐stressed offspring. Annals of Botany, 114, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck, M. J. (2019). Evolutionary and ecological links between plant and fungal viruses. New Phytologist, 221, 86–92. [DOI] [PubMed] [Google Scholar]
- Rudrappa, T. , Czymmek, K. J. , Pare, P. W. , & Bais, H. P. (2008). Root‐secreted malic acid recruits beneficial soil bacteria. Plant Physiology, 148, 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova, D. , Rack‐Wetzlinger, U. , Cernava, T. , Schaefer, A. , Schmuck, M. , & Berg, G. (2017). Aerial warfare: A volatile dialogue between the plant pathogen Verticillium longisporum and its antagonist Paenibacillus polymyxa . Frontiers in Plant Science, 8, 1294. 10.3389/fpls.2017.01294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semchenko, M. , John, E. A. , & Hutchings, M. J. (2007). Effects of physical connection and genetic identity of neighbouring ramets on root‐placement patterns in two clonal species. The New Phytologist, 176, 644–654. [DOI] [PubMed] [Google Scholar]
- Sharifi, R. , Ahmadzadeh, M. , Sharifi‐Tehrani, A. , & Talebi‐Jahromi, K. (2010). Pyoverdine production in Pseudomonas fluorescens UTPF5 and its association with suppression of common bean damping off caused by Rhizoctonia solani (Kühn). Journal of Plant Protection Research, 50, 72–78. [Google Scholar]
- Sharifi, R. , Lee, S. M. , & Ryu, C. M. (2018). Microbe‐induced plant volatiles. New Phytologist, 220, 684–691. [DOI] [PubMed] [Google Scholar]
- Sharifi, R. , & Ryu, C. M. (2017). Chatting with a tiny belowground member of the holobiome: Communication between plants and growth‐promoting rhizobacteria. Advances in Botanical Research, 82, 135–160. [Google Scholar]
- Sharifi, R. , & Ryu, C.‐M. (2018a). Biogenic volatile compounds for plant disease diagnosis and health improvement. The Plant Pathology Journal, 34, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi, R. , & Ryu, C.‐M. (2018b). Revisiting bacterial volatile‐mediated plant growth promotion: Lessons from the past and objectives for the future. Annals of Botany, 122, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi, R. , & Ryu, C.‐M. (2018c). Sniffing bacterial volatile compounds for healthier plants. Current Opinion in Plant Biology, 44, 88–97. [DOI] [PubMed] [Google Scholar]
- Sharifi, R. , & Ryu, C.‐M. (2020). Formulation and agricultural application of bacterial volatile compounds. In Ryu C.‐M. (Ed.), Bacterial volatile compounds as mediators of airborne interactions. Singapore: Springer. [Google Scholar]
- Shulaev, V. , Silverman, P. , & Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature, 385, 718–721. [Google Scholar]
- Simmi, F. Z. , Dallagnol, L. J. , Ferreira, A. S. , Pereira, D. R. , & Souza, G. M. (2020). Electrome alterations in a plant‐pathogen system: Toward early diagnosis. Bioelectrochemistry, 133, 107493. [DOI] [PubMed] [Google Scholar]
- Sobhy, I. S. , Bruce, T. J. , & Turlings, T. C. (2018). Priming of cowpea volatile emissions with defense inducers enhances the plant's attractiveness to parasitoids when attacked by caterpillars. Pest Management Science, 74, 966–977. [DOI] [PubMed] [Google Scholar]
- Song, G. C. , Choi, H. K. , & Ryu, C.‐M. (2015). Gaseous 3‐pentanol primes plant immunity against a bacterial speck pathogen, Pseudomonas syringae pv. tomato via salicylic acid and jasmonic acid‐dependent signaling pathways in Arabidopsis . Frontiers in Plant Science, 6(821). 10.3389/fpls.2015.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, G. C. , Sim, H. J. , Kim, S. G. , & Ryu, C. M. (2016). Root‐mediated signal transmission of systemic acquired resistance against above‐ground and below‐ground pathogens. Annals of Botany, 118(4), 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. Y. , Ye, M. , Li, C. , He, X. , Zhu‐Salzman, K. , Wang, R. L. , … Zeng, R. S. (2014). Hijacking common mycorrhizal networks for herbivore‐induced defence signal transfer between tomato plants. Scientific Reports, 4, 3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. Y. , Zeng, R. S. , Xu, J. F. , Li, J. , Shen, X. , & Yihdego, W. G. (2010). Interplant communication of tomato plants through underground common mycorrhizal networks. PLoS One, 5, e13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis, I. A. , Proietti, S. , Hickman, R. , Van Verk, M. C. , Zamioudis, C. , & Pieterse, C. M. (2018). Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. The Plant Journal, 93, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, J. , & Stencel, A. (2020). A part‐dependent account of biological individuality: Why holobionts are individuals and ecosystems simultaneously. Biological Reviews of the Cambridge Philosophical Society, 95, 1308–1324. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K. , Matsui, K. , Iijima, Y. , Akakabe, Y. , Muramoto, S. , Ozawa, R. , … Akitake, S. (2014). Intake and transformation to a glycoside of (Z)‐3‐hexenol from infested neighbors reveals a mode of plant odor reception and defense. Proceedings of the National Academy of Sciences, 111, 7144–7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, C. , Lakshmanan, V. , & Bais, H. P. (2017). Interplant aboveground signaling prompts upregulation of auxin promoter and malate transporter as part of defensive response in the neighboring plants. Frontiers in Plant Science, 8, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, H. , Ishiga, Y. , Watanabe, S. , Konishi, T. , Egusa, M. , Akiyoshi, N. , … Sakamoto, A. (2016). Allantoin, a stress‐related purine metabolite, can activate jasmonate signaling in a MYC2‐regulated and abscisic acid‐dependent manner. Journal of Experimental Botany, 67, 2519–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigahira, H. , & Yamawo, A. (2019). Competitive responses based on kin‐discrimination underlie variations in leaf functional traits in Japanese beech (Fagus crenata) seedlings. Evolutionary Ecology, 33, 521–531. [Google Scholar]
- Tedersoo, L. , Bahram, M. , & Zobel, M. (2020). How mycorrhizal associations drive plant population and community biology. Science, 367, eaba1223. [DOI] [PubMed] [Google Scholar]
- Toome, M. , Randjärv, P. , Copolovici, L. , Niinemets, U. , Heinsoo, K. , & Luik, A. (2010). Leaf rust induced volatile organic compounds signalling in willow during the infection. Planta, 232, 235–243. [DOI] [PubMed] [Google Scholar]
- Tu, S. , Yang, R. , Xu, X. , Chen, J. , Luo, Q. , Zhu, Z. , … Yan, X. (2017). Flg22‐triggered oxylipin production in Pyropia haitanensis . Phycological Research, 65, 86–93. [Google Scholar]
- Tungadi, T. , Groen, S. C. , Murphy, A. M. , Pate, A. E. , Iqbal, J. , Bruce, T. J. , … Carr, J. P. (2017). Cucumber mosaic virus and its 2b protein alter emission of host volatile organic compounds but not aphid vector settling in tobacco. Virology Journal, 14, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahabi, K. , Reichelt, M. , Scholz, S. S. , Furch, A. C. U. , Matsuo, M. , Johnson, J. M. , … Oelmuller, R. (2018). Alternaria brassicae induces systemic jasmonate responses in arabidopsis which travel to neighboring plants via a Piriformsopora indica hyphal network and activate abscisic acid responses. Frontiers in Plant Science, 9, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam, N. M. , & Bouwmeester, H. J. (2016). Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends in Plant Science, 21, 256–265. [DOI] [PubMed] [Google Scholar]
- van Doan, C. , Züst, T. , Maurer, C. , Zhang, X. , Machado, R. A. R. , Mateo, P. , … Robert, C. A. M. (2020). Volatile‐mediated defence regulation occurs in maize leaves but not in maize root. Plant, Cell & Environment. 10.1111/pce.13919. [DOI] [PubMed] [Google Scholar]
- van Doorn, M. M. , Merl‐Pham, J. , Ghirardo, A. , Fink, S. , Polle, A. , Schnitzler, J. P. , & Rosenkranz, M. (2020). Root isoprene formation alters lateral root development. Plant & Cell Environment, 43(9), 2207–2223. [DOI] [PubMed] [Google Scholar]
- Van West, P. V. , Morris, B. , Reid, B. , Appiah, A. A. , Osborne, M. , Campbell, T. , … Gow, N. A. R. (2002). Oomycete plant pathogens use electric fields to target roots. Molecular Plant–Microbe Interactions, 15, 790–798. [DOI] [PubMed] [Google Scholar]
- Vannier, N. , Bittebiere, A.‐K. , Mony, C. , & Vandenkoornhuyse, P. (2020). Root endophytic fungi impact host plant biomass and respond to plant composition at varying spatio‐temporal scales. Fungal Ecology, 44, 100907. [Google Scholar]
- Vannier, N. , Mony, C. , Bittebiere, A.‐K. , Theis, K. R. , Rosenberg, E. , & Vandenkoornhuyse, P. (2019). Clonal plants as meta‐holobionts. mSystems, 4, e00213–e00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veen, C. , Fry, E. , ten Hooven, F. , Kardol, P. , Morriën, E. , & De Long, J. R. (2019). The role of plant litter in driving plant–soil feedbacks. Frontiers in Environmental Science, 7, 168. [Google Scholar]
- Vucetic, A. , Dahlin, I. , Petrovic‐Obradovic, O. , Glinwood, R. , Webster, B. , & Ninkovic, V. (2014). Volatile interaction between undamaged plants affects tritrophic interactions through changed plant volatile emission. Plant Signaling & Behavior, 9, e29517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. H. , Wu, L. , Wang, Z. , Alabady, M. S. , Parson, D. , Molumo, Z. , & Fankhauser, S. C. (2020). Characterizing changes in soil microbiome abundance and diversity due to different cover crop techniques. PLoS One, 15, e0232453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Liu, J. , Zhan, Y. , & Liu, Y. (2019). A new slow‐release formulation of methyl salicylate optimizes the alternative control of Sitobion avenae (Fabricius) (Hemiptera: Aphididae) in wheat fields. Pest Management Science, 75, 676–682. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Li, Y. , Li, T. , Zhao, T. , & Liao, Y. (2020). Conservation tillage decreases selection pressure on community assembly in the rhizosphere of arbuscular mycorrhizal fungi. Science of the Total Environment, 710, 136326. [DOI] [PubMed] [Google Scholar]
- Waters, M. T. , Gutjahr, C. , Bennett, T. , & Nelson, D. C. (2017). Strigolactone signaling and evolution. Annual Review of Plant Biology, 68, 291–322. [DOI] [PubMed] [Google Scholar]
- Wenig, M. , Ghirardo, A. , Sales, J. H. , Pabst, E. S. , Breitenbach, H. H. , Antritter, F. , … Vlot, A. C. (2019). Systemic acquired resistance networks amplify airborne defense cues. Nature Communications, 10, 3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D. , Qi, T. , Li, W. X. , Tian, H. , Gao, H. , Wang, J. , … Xie, D. (2017). Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Research, 27, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Desurmont, G. , Degen, T. , Zhou, G. , Laplanche, D. , Henryk, L. , & Turlings, T. C. (2017). Combined use of herbivore‐induced plant volatiles and sex pheromones for mate location in braconid parasitoids. Plant, Cell & Environment, 40(3), 330–339. [DOI] [PubMed] [Google Scholar]
- Yang, J. W. , Yi, H.‐S. , Kim, H. , Lee, B. , Lee, S. , Ghim, S.‐Y. , & Ryu, C.‐M. (2011). Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below‐ground microflora. Journal of Ecology, 99, 46–56. [Google Scholar]
- Yang, M. , Zhang, Y. , Qi, L. , Mei, X. , Liao, J. , Ding, X. , … Zhu, S. (2014). Plant–plant–microbe mechanisms involved in soil‐borne disease suppression on a maize and pepper intercropping system. PLoS One, 9, e115052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.‐F. , Li, L.‐L. , Xu, Y. , & Kong, C.‐H. (2018). Kin recognition in rice (Oryza sativa) lines. New Phytologist, 220, 567–578. [DOI] [PubMed] [Google Scholar]
- Ye, M. , Glauser, G. , Lou, Y. , Erb, M. , & Hu, L. (2019). Molecular dissection of early defense signaling underlying volatile‐mediated defense regulation and herbivore resistance in rice. The Plant Cell, 31, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H. S. , Heil, M. , Adame‐Alvarez, R. M. , Ballhorn, D. J. , & Ryu, C. M. (2009). Airborne induction and priming of plant defenses against a bacterial pathogen. Plant Physiology, 151, 2152–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebelo, S. A. , Matsui, K. , Ozawa, R. , & Maffei, M. E. (2012). Plasma membrane potential depolarization and cytosolic calcium flux are early events involved in tomato (Solanum lycopersicon) plant‐to‐plant communication. Plant Science, 196, 93–100. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Kim, M. S. , Krishnamachari, V. , Payton, P. , Sun, Y. , Grimson, M. , … Pare, P. W. (2007). Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis . Planta, 226, 839–851. [DOI] [PubMed] [Google Scholar]
- Zhang, P. J. , Broekgaarden, C. , Zheng, S. J. , Snoeren, T. A. , van Loon, J. J. , Gols, R. , & Dicke, M. (2013). Jasmonate and ethylene signaling mediate whitefly‐induced interference with indirect plant defense in Arabidopsis thaliana . New Phytologist, 197, 1291–1299. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. C. , Zou, Y. N. , Liu, L. P. , & Wu, Q. S. (2019). Common mycorrhizal networks activate salicylic acid defense responses of trifoliate orange (Poncirus trifoliata). Journal of Integrative Plant Biology, 61, 1099–1111. [DOI] [PubMed] [Google Scholar]
- Zhao, M. , Cheng, J. , Guo, B. , Duan, J. , & Che, C.‐T. (2018). Momilactone and related diterpenoids as potential agricultural chemicals. Journal of Agricultural and Food Chemistry, 66, 7859–7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Ma, Z. , Lu, X. , Zhu, L. , & Yan, C. (2020). Pseudomonas fluorescens MZ05 enhances resistance against Setosphaeria turcica by mediating benzoxazinoid metabolism in the maize inbred line Anke35. Agriculture, 10, 32. [Google Scholar]
- Zhou, Y. , Cen, H. , Tian, D. , Wang, C. , & Zhang, Y. (2019). A tomato and tall fescue intercropping system controls tomato stem rot. Journal of Plant Interactions, 14, 637–647. [Google Scholar]
- Zhuang, H. , Li, J. , Song, J. , Hettenhausen, C. , Schuman, M. C. , Sun, G. , … Wu, J. (2018). Aphid (Myzus persicae) feeding on the parasitic plant dodder (Cuscuta australis) activates defense responses in both the parasite and soybean host. New Phytologist, 218, 1586–1596. [DOI] [PubMed] [Google Scholar]


