Abstract
By the end of year 2019, the new virus SARS-CoV-2 appeared, causing the Coronavirus Disease 2019 (COVID-19), and spread very fast globally. A continuing need for diagnostic tools is a must to contain its spread. Till now, the gold standard method, the reverse transcription polymerase chain reaction (RT-PCR), is the precise procedure to detect the virus. However, SARS-CoV-2 may escape RT-PCR detection for several reasons. The development of well-designed, specific and sensitive serological test like enzyme immunoassay (EIA) is needed. This EIA can stand alone or work side by side with RT-PCR.
In this study, we developed several EIAs including plates that are coated with either specially designed SARS-CoV-2 nucleocapsid or surface recombinant proteins. Each protein type can separately detect anti-SARS-CoV-2 IgM or IgG antibodies. For each EIAs, the cut-off value, specificity and sensitivity were determined utilizing RT-PCR confirmed Covid-19 and pre-pandemic healthy and other viruses-infected sera. Also, the receiver operator characteristic (ROC) analysis was performed to define the specificities and sensitivities of the optimized assay. The in-house EIAs were validated by comparing against commercial EIA kits. All in-house EIAs showed high specificity (98–99%) and sensitivity (97.8–98.9%) for the detection of IgG/IgM against RBD and N proteins of SARS-CoV-2.
From these results, the developed Anti-RBD and anti-N IgG and IgM antibodies EIAs can be used as a specific and sensitive tool to detect SARS-CoV-2 infection, calculate the burden of disease and case fatality rates.
Keywords: SARS-CoV-2, Covid-19, ELISA, Spike protein, Nucleocapsid protein
1. Introduction
The Coronavirus Disease 2019 (COVID-19) is brought about by a recently identified virus during 2019. Subsequently, the factor causing that disease is named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Ahn et al., 2020). The genomic construction of SARS-CoV-2 is now well-known enabling the quick generation of diagnostic tools to detect its spread and isolate infected persons (Mousavizadeh and Ghasemi, 2020). Nowadays, there are several new strains or mutations being reported (Chen et al., 2020, Issa et al., 2020). Viral genome reverse-transcription followed by an amplification utilizing the polymerase chain reaction (RT-PCR), is considered as the gold standard procedure to discover SARS-CoV-2 during the acute phase (Babiker et al., 2020) where there are no antibody markers and over the incubation interval (Lauer et al., 2020). It is very important to identify the prevalence, transmission pattern, and the existing burden of Covid-19 and the resulting case fatality rates (Onder et al., 2020). These can be achieved via RT-PCR and antibody tests. Antibody testing plays a limited role in the incubation period and at the very early onset of the disease (Lauer et al., 2020).However, antibody testing can be used for the detection of current and past infection and as a confirmatory assay for PCR results. Antibody tests (e.g. ELISA and rapid tests), can be used as a primary test before choosing the expensive method (RT-PCR) in low-income and remote areas. Antibody test methods are also helpful for vaccination evaluation and therapeutic antibody development.
Among the α-, β-, γ- and δ-Coronaviruses (CoVs) genera, SARS-CoV-2 is one of the β-CoV genus. The RNA genome of enveloped CoVs is ~ 30 kb (positive-sense and single-stranded) and has about 6 open reading frames (ORFs). SARS-CoV-2 genome has likeness with SARS-CoV (about 79%) and bat SARS-like CoVs (about 95.6%) and has at least 6 ORFs (Zhu et al., 2020). Two polyproteins, found on the first 60% of the genome, are expressed and then turned into almost 16 non-structural type proteins. Four proteins (nucleocapsid (N), membrane (M), spike (S) and envelope (E)) and other accessory proteins are encoded by the rest of the genome (Zhang and Holmes, 2020). Among these four structural proteins, S and N proteins are the highly immunogenic ones (Ahmed et al., 2020).
Here, we developed and validated an indirect serological assay (ELISA) that is capable to determine the immune status of persons. The assay was validated via comparing sera of COVID-19 RT-PCR confirmed patients, normal non-infected healthy controls, and other viruses-infected sera.
2. Material and methods
2.1. Samples
This work was reviewed and approved by The Research Ethics Committee at King Khalid University (HAPO-06-B-001) and agreed on this study. All investigations were performed as per the previously mentioned board rules and guidelines. We used stored coded sera so patient informed consent was not among the requirements.
Coded sera that were diagnosed positive for Covid-19 via RT-PCR (n = 100) were used. In our laboratory, we have stored coded sera obtained from random blood donor volunteers several years (2015) before the onset of the SARS-CoV-2 pandemic (https://journals.ekb.eg/article_13699.html). From these samples (Table 1 ), we randomly chose virus-negative control sera (n = 100, HCV, HBV, HIV, HTLV and CMV-free), virus-infected sera including HBV-positive (surface antigen and PCR confirmed, n = 50) HCV-positive (antibody and PCR confirmed, n = 5), and CMV-positive (antibody confirmed, n = 50).
Table 1.
Type of samples used in the study.
| Sample type | Number |
|---|---|
| SARS-CoV-2-RT-PCR positive | 100 |
| Normal uninfected | 100 |
| HBV-positive (HBsAg+; PCR Confirmed) | 50 |
| HCV-positive (antibody and PCR confirmed | 5 |
| CMV-positive (antibody confirmed) | 50 |
| Total | 305 |
Detection of anti- SARS-CoV-2 IgM&IgG antibodies were done for the above mentioned sera utilizing COVID-19 Human IgG/IgM Assay Kit (Abnova) which either containing microplate coated with the spike protein (Cat: KA5826) or microplate coated with spike RBD + nucleocapsid proteins.
2.2. Cloning of SARS-CoV-2 N and S protein genes
Part of the nucleoprotein (N) of SARS-CoV-2 gene (accession: MW307301, a.a.: 1–286) and Part of the of surface (spike) protein (receptor-binding domain, RBD) genes (accession: MW512912, a.a.: 319–541) were used to design synthetic genes (N and S). Synthetic genes were separately cloned into pET28a expression vector 1.2, propagated in TOP-10 cloning bacteria, expressed in BL21-DE3 expression bacteria and recombinant protein purification and identification were done using the same materials and methods described by Ibrahim et al. (Ibrahim et al., 2018).
2.3. In-house ELISA procedure
For the preparation of in-house ELISA assay plates, we used N recombinant protein prepared above (but not S protein) and commercially available S-RBD proteins (Cusabio Technology LLC., China). The horse-radish (HRP) enzyme-conjugated secondary antibodies (Abs), goat anti-Fc fragment of human IgG and goat F(ab')2 anti-human IgM Abs (Novus Biologicals), were used to detect human anti- SARS-CoV-2 IgM and IgG antibodies respectively. Different recombinant viral proteins were diluted into 4 µg/mL in carbonate/bicarbonate buffer (12.5 mM Sodium bicarbonate, 87.5 mM Sodium carbonate, pH 9.6, Techno Pharmchem) and 50 µL of these preparations were added to separate high binding 96-well ELISA plates (Flat bottom, UltraCruz® ELISA Plate, Santa Cruz Biotechnology). All plates were incubated overnight at 4 °C, washed twice in wash buffer (0.05% (v/v) Tween-20 in phosphate-buffered saline (PBS, 0.01 M Na2HPO4, 0.137 M NaCl, 0.0018 M KH2PO4 and 0.0027 M KCl, pH 7.4 (PBST), all from Sigma-Aldrich) and then blocked with 220 µL blocking buffer (PBST with 2% serum albumin (bovine, BSA), Sigma-Aldrich) for 3 h at ambient temperature. Normal sera (n = 5) and SARS-CoV-2 reactive sera (n = 20) were diluted the same way as indicated in the commercial ELISA kit (1: 200) in blocking buffer and 100 µL of each dilution were loaded into wells of different coated plates. Plates were kept at 37 °C for 50 min, washed thrice with PBST, 100 µL of either goat anti-human IgG Fc fragment HRP-conjugated secondary Abs or goat F(ab')2 anti-human IgM secondary Abs (each 1:20,000 diluted) were added separately to each well and kept at 37 °C for 50 min. Plates were subjected to repeated wash (5 times) utilizing PBST before the addition of 3,3′,5,5′-Tetramethylbenzidine (TMP, 100 µL/well, Sigma-Aldrich) and kept in the dark for 18 min. A 100 µL/well 1 N HCl were added and color intensity of each well was measured at 450 nm via a spectrophotometer (800™ TS, BioTek).
2.4. Checking of the assay specificity
To check the specificity of the in-house prepared ELISA assay procedure of all used recombinant proteins, sera that are confirmed positive to SARS-CoV-2, HCV, HBV, HIV, and CMV viruses as well as normal sera were diluted and assayed as indicated above.
2.5. Checking of the ELISA sensitivity
All serum samples (negative and positive) were serially diluted in blocking buffer (1:2 – 1:2,048) and examined for the existence of human anti- SARS-CoV-2 IgM & IgG Abs following the same way indicated above.
3. Statistical analysis
Cut-off value was calculated according to Wiederschain (Wiederschain, 2009) as the mean OD value at a wavelength of 450 nm of negative controls plus 3 value of their standard deviations (SD).
The sensitivity of all in-house made ELISA was determined according to Ráez-Bravo et al. (Ráez-Bravo et al., 2016) as follows:
The specificity of all in-house made ELISAs was determined as follows:
Using the SPSS V20 software (SPSS Inc., Chicago, Ill., USA), the receiver operating characteristic (ROC) analysis and the area under the curve (AUC) were figured to determine the specificity and sensitivity.
4. Results
4.1. Expression of recombinant N and S proteins
The target part of N gene was successfully expressed in BL21 bacteria and purified using Ni-NTA affinity chromatography column. The purified protein showed high purity. In contrary, target part of S gene, at the time it could be cloned in DH5-α cloning bacteria, it failed to be expressed in BL21 bacteria. Expressed N protein was successfully detected using anti-His antibodies.
4.2. Anti-SARS-CoV-2 IgG/IgM Abs in all sera
Sera that were diagnosed positive for Covid-19 via RT-PCR were checked for anti- SARS-CoV-2 IgG/IgM Abs using a commercial ELISA kit. Results (Table 2 ) show that all the tested samples have IgG, IgM or both IgM and IgG in RT-PCR Covid-19 positive sera with a few negative results.
Table 2.
Assessment of anti- SARS-CoV-2 IgM & IgG Abs in RT-PCR confirmed Covid-19 sera using a commercial ELISA kit.
| Coating SARS-CoV-2 protein | ||||
|---|---|---|---|---|
| S | S + N | |||
| Antibody type | IgG | IgM | IgG | IgM |
| Positive | 95 | 97 | 97 | 98 |
| Negative | 5 | 3 | 3 | 2 |
| Total | 100 | 100 | 100 | 100 |
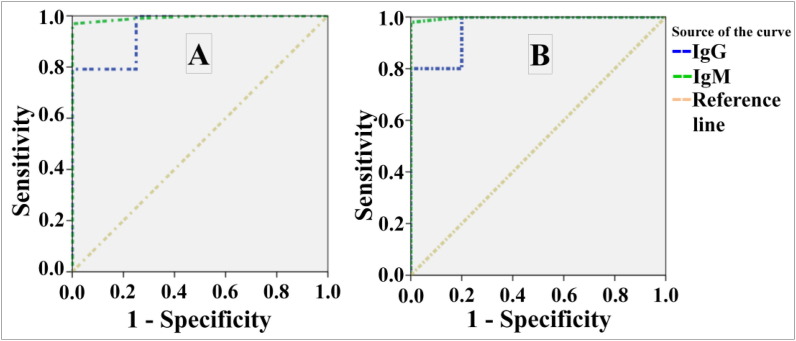
ROC analysis (Fig. 1 ) and AUC (Table 3 &Table 4. ) revealed high specificity/sensitivity of the procedure and its results for detecting anti- SARS-CoV-2 IgM & IgG Abs using S and N proteins.
Fig. 1.
ROC curves of anti- SARS-CoV-2 IgG/IgM Abs utilizing the commercial ELISA. Where A: antibodies against N + S protein; B: antibodies against S protein.
Table 3.
Area under the curve (AUC) after performing ROC analysis for anti-SARAS-CoV-2 spike/Nucleoprotein IgG/IgM Abs utilizing the commercial ELISA kit.
| Test Results Variable(s) | Area | Std. Error | AsymptoticSig. | Asymptotic 95% Confidence Interval | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| IgG | 0.948 | 0.047 | 0.002 | 0.856 | 1.000 |
| IgM | 0.993 | 0.007 | 0.001 | 0.979 | 1.000 |
Table 4.
Area under the curve after performing ROC analysis for anti-SARAS-CoV-2 spike IgG/ IgM Abs utilizing the commercial ELISA kit.
| Test Results Variable(s) | Area | Std. Error | AsymptoticSig. | Asymptotic 95% Confidence Interval | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| IgG | 0.960 | 0.037 | 0.001 | 0.887 | 1.000 |
| IgM | 0.998 | 0.003 | 0.000 | 0.991 | 1.000 |
All control sera (normal uninfected and other viruses-infected) showed no reactivity against S and N antigens using the commercial ELISA kit.
4.3. Cut-off value of in-house ELISA
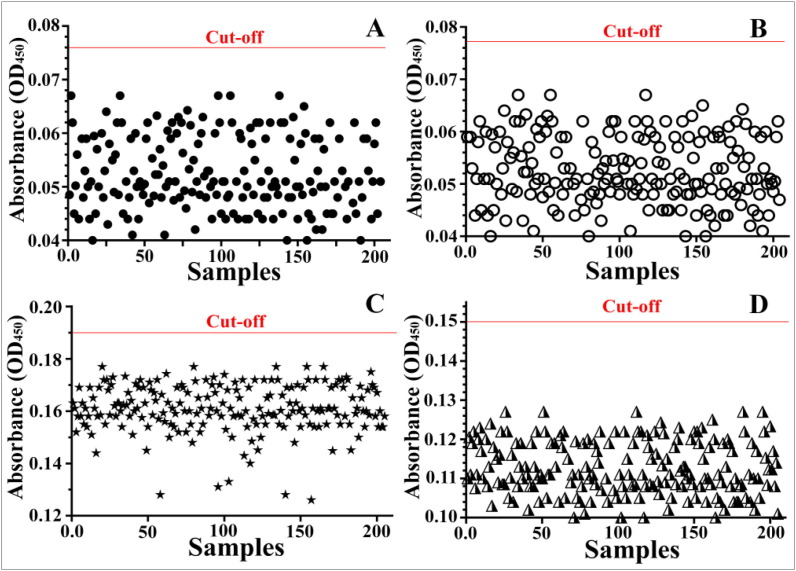
All control serum samples that are confirmed non-reactive to SARS-CoV-2 S and N proteins by the commercial ELISA kit were used to determine the cut-off value of the in-house ELISA (Fig. 3 ). The cut-off value of the IgG against S protein (Fig. 2A) was calculated as mean + 3StDev (0.051 + 3[0.0083] = 0.0759). While the cut-off value of the IgM against S protein (Fig. 2B) was calculated to be 0.077 (0.051 + 3[0.0088] = 0.0774). The cut-off value of the IgG against N protein (Fig. 2C) was calculated to be 0.15 (0.112 + 3[0.0124] = 0.149). The cut-off value of the IgM against N protein (Fig. 2D) was calculated to be 0.199 (0.162 + 3[0.0123] = 0.149).
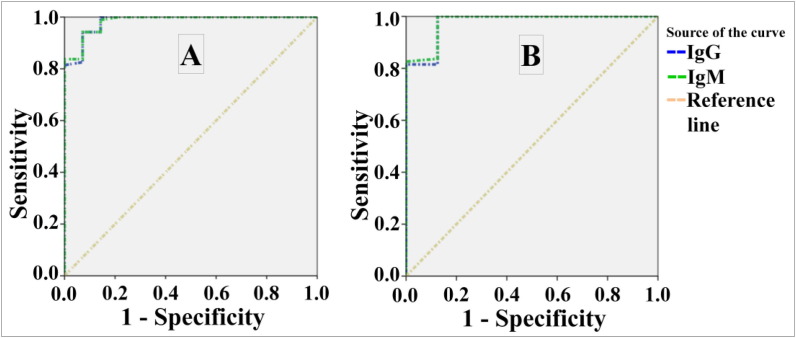
Fig. 3.
ROC curves of anti- SARS-CoV-2 IgM & IgG Abs utilizing the in-house ELISAs. Where A: antibodies against S protein; B: antibodies against N protein.
Fig. 2.
Calculation of in-house cut-off value for the detection of IgG against S protein (A); IgM against S protein (B); IgG against N protein (C) and IgM against M protein. The red lines represent the calculated cut-off value.
4.4. Assessment of anti-SARS-CoV-2 IgM & IgG Abs utilizing in-house ELISA
All sera indicated above were examined for the existence of anti-SARS-CoV-2 IgM & IgG Abs utilizing the in-house ELISA kit. The cut-off for every ELISA type was used to determine the positive/negative samples. Results (Table 5 ) show that all the tested samples have IgG, IgM or both IgG and IgM in Covid-19 sera with a few negative results.
Table 5.
Assessment of anti-SARS-CoV-2 IgM & IgG Abs in RT-PCR confirmed Covid-19 sera.
| Coating SARS-CoV-2 protein | ||||
|---|---|---|---|---|
| S | N | |||
| Antibody type | IgG | IgM | IgG | IgM |
| Positive | 96 | 97 | 97 | 99 |
| Negative | 4 | 3 | 3 | 1 |
| Total | 100 | 100 | 100 | 100 |
ROC analysis (Fig. 3) and AUC (Table 6 &Table 7. ) revealed high specificity/sensitivity of the experiment and its results for detecting anti-SARS-CoV-2 IgM & IgG Abs utilizing S and N proteins.
Table 6.
Area under the curve after performing ROC analysis for anti-SARAS-CoV-2 spike IgM & IgG Abs utilizing the in-house ELISAs.
| Test Results Variable(s) | Area | Std. Error | AsymptoticSig. | Asymptotic 95% Confidence Interval | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| IgG | 0.983 | 0.014 | 0.000 | 0.956 | 1.000 |
| IgM | 0.984 | 0.013 | 0.000 | 0.959 | 1.000 |
Table 7.
Area under the curve after performing ROC analysis for anti-SARAS-CoV-2 Nucleoprotein IgM & IgG Abs utilizing the in-house ELISAs.
| Test Results Variable(s) | Area | Std. Error | AsymptoticSig. | Asymptotic 95% Confidence Interval | |
|---|---|---|---|---|---|
| Lower Bound | Upper Bound | ||||
| IgG | 0.977 | 0.023 | 0.000 | 0.932 | 1.000 |
| IgM | 0.979 | 0.021 | 0.000 | 0.938 | 1.000 |
All control sera (normal uninfected and other viruses-infected) showed no reactivity against S and N antigens using the in-house ELISA.
4.5. The specificity of the in-house ELISAs
The specificity of in-house ELISAs that detect anti-SARSA-Cov-2 S and N IgM and IgG antibodies were tested. All in-house ELISAs reacted only to sera that are confirmed positive to SARS-CoV-2, and did not react to other sera that are confirmed positive for HCV, HBV, HIV, and CMV viruses. Also, the assays did not react with normal sera.
4.6. The sensitivity of the in-house ELISAs
All Covid-19 positive sera showed positive reaction up to the 1: 1024 dilution. The color intensity at 1: 1024 dilution was the least for IgG Abs reacted to both S and N proteins of SARS-CoV-2. At dilution 1:2048, IgG Abs towards S and N proteins turned negative, but IgM Abs towards S and N proteins showed a positive reaction.
5. Discussion
Covid-19 started in 2019 and continues spreading till this moment. The continuous need for diagnostic tool always exists. In the current investigation, we developed and assessed in-house ELISAs for discovering anti-RBD Abs (IgM and IgG) and anti-N Abs (IgM & IgG), and examined their specificity and sensitivity. To evaluate the developed in-house ELISAs, 100 SARSA-Cov-2 PCR-confirmed samples and normal and other viruses infected sera were used.
In the current work, we designed recombinant genes encoding for parts of the N and S proteins. SARS-CoV-2 has several structural proteins (e.g. E, S, M, and N proteins) (Dutta et al., 2020, Walls et al., 2020). Among these four proteins, S and N proteins are most immunogenic ones, so they are widely utilized in building up of immunoassays (Liu et al., 2020). The N protein of SARS-CoV-2 has high likeness with the N protein of MERS-CoV (90%) and alpha and beta coronaviruses (28–49%) (Zhao et al., 2020). The similarity of SARS-CoV-2 N protein with that of the SARS-CoV is the most at the conserved residues at the N-terminal domain of the N protein. It was described that the anti-N protein Abs develops earlier than the spike Abs (Burbelo et al., 2020). We did not use the full length of the S protein and used only the RBD part of it as many studies confirmed its specificity in detecting SARS-CoV-2 infection without cross reactivity with any other corona virus (Chia et al., 2020, Haselmann et al., 2020, Whitman et al., 2020). Also, we used the C-terminus of N protein which shows the lowest similarity with other corona viruses.
In constructing the In-house ELISAs, we used the in-house produced N protein. We used a commercially available RBD as it could not be produced in BL21 bacteria. The product of induced RBD gene seems to be highly toxic the BL21 expression bacteria host. We prepared two types of ELISA plates, one coated with recombinant RBD protein and the other coated with recombinant N protein, both were used to discover IgM and IgG against SARS-CoV-2. Both preparations were examined for their specificity and sensitivity to detect these antibodies. Specificity and sensitivity of the assays were explained as their true negative and true positive rates, respectively. The positive group positivity status was paralleled to confirmed positive sera by RT-PCR. We used the commercial antibody detection kit to first evaluate the possibility of detecting the antibodies in the RT-PCR confirmed sera. ROC analysis revealed high sensitivity and specificity of the kit to detect both anti-S and anti-N Abs. Area under the curve (AUC) after performing ROC analysis for anti-SARAS-CoV-2 spike/Nucleoprotein IgM and IgG Abs utilizing the commercial ELISA kit showed good results (95% and 99% respectively). Also, AUC after performing ROC analysis for anti-SARAS-CoV-2 spike IgM and IgG Abs utilizing the commercial ELISA kit showed good results (96% and 99% respectively). These results indicate that the commercial kit can be used as reference for the in-house ELSAs to compare their outcomes. Using the same analysis criteria (ROC and AUC), in-house ELSAs reflected good results. Detection of IgG and IgM using the RBD and N as target antigens using in-house ELSAs reflected high sensitivity and specificity. AUC after performing ROC analysis for anti-SARAS-CoV-2 spike IgM and IgG Abs utilizing the in-house ELISAs showed good results (98% and 98% respectively) which are comparable with the commercial ELISA kit. In addition, AUC after performing ROC analysis for anti-SARAS-CoV-2 N IgM and IgG Abs utilizing the in-house ELISAs also showed excellent results (97% and 98% respectively) which are comparable with the commercial ELISA kit. These obtained results reflect the excellent suitability and capability of the in-house developed ELISAs to detect IgM and IgG towards both S and N proteins of SARS-CoV-2 with high sensitivity and specificity.
Regarding the specificity of the in-house ELSAs, we used sera infected with different viruses. None of these sera interacted with the coated proteins (S and N) with any degree. These sera showed the same readings of the negative control samples. The negative control samples used here were collected long time before the onset of the SARS-CoV-2 pandemic to avoid the possibility of obtaining the virus by any degree. The commercial ELISA kit recommends a serum dilution to be 1:200. To test the sensitivity of the in-house ELISAs, we diluted the up to 1:1048. The in-house ELISAs could detect the antibodies at high dilution indicting its high sensitivity over the commercial kit.
Here, we designed ELISAs to detect both anti-S and anti-N IgM and IgG Abs. At the time that anti-S antibodies are more specific for SARS-CoV-2 detection (Chia et al., 2020), the detection of anti-N antibodies using ELISA favor the increase of the assay sensitivity if the samples being collected in early disease stage.
In our study, it is noted that nearly all of the SARS-CoV-2 RT-PCT confirmed samples have both IgG and IgM antibody types. These results are in agreement with other investigators (Shu et al., 2020, Xiang et al., 2020).
6. Conclusions
In this investigation we have created, optimized and assessed the performance of in-house ELISAs for the detection of anti-SARS-CoV-2 S and N IgM and IgG Abs and compared these in-house ELISAs to the commercially available ELISA kits. All of the developed in-house ELISAs showed high sensitivity and specificity in their performance and obtained results without any indeterminate results. As these in-house ELISAs showed a high specificity and sensitivity, we recommend using it to detect SARS-CoV-2 infections without the need for RT-PCR confirmation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors extend their appreciation to King Abdulaziz City for Science and Technology (KACST) for funding this work through the Research Project (5-20-01-010-0009).
Footnotes
Peer review under responsibility of King Saud University.
References
- Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12 doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.G., Shin H.J., Kim M.H., Lee S., Kim H.S., Myoung J., Kim B.T., Kim S.J. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) J. Microbiol. Biotechnol. 2020;30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiker A., Myers C., Hill C., Guarner J. SARS-CoV-2 testing: trials and tribulations. Am. J. Clin. Pathol. 2020;153:706–708. doi: 10.1093/ajcp/aqaa052/5813763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Wang M., Wei G.W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W.N., Tan C.W., Foo R., Kang A.E.Z., Peng Y., Sivalingam V., Tiu C., Ong X.M., Zhu F., Young B.E., Chen M.I.C., Tan Y.J., Lye D.C., Anderson D.E., Wang L.F. Serological differentiation between COVID-19 and SARS infections. Emerg. Microbes Infect. 2020;9:1497–1505. doi: 10.1080/22221751.2020.1780951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Gordy J.T. The Nucleocapsid Protein of SARS–CoV-2: a Target for Vaccine Development. J. Virol. 2020;94 doi: 10.1128/jvi.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselmann V., Kittel M., Gerhards C., Thiaucourt M., Eichner R., Costina V., Neumaier M. Comparison of test performance of commercial anti-SARS-CoV-2 immunoassays in serum and plasma samples. Clin. Chim. Acta. 2020;510:73–78. doi: 10.1016/j.cca.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim E.H., Asiri R., Al Syaad K. Genetic fusion of tetanus toxin fragment C (Hc) gene to cholera toxin subunit B (CTB) gene as a preparatory step for double vaccine production. Gene Reports. 2018;10:90–96. doi: 10.1016/j.genrep.2017.11.008. [DOI] [Google Scholar]
- Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral. Pathogenesis. 2020;mSystems 5 doi: 10.1128/msystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (CoVID-19) from publicly reported confirmed cases: Estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-Fatality Rate and Characteristics of Patients Dying in Relation to COVID-19 in Italy. JAMA - J. Am. Med. Assoc. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Ráez-Bravo A., Granados J.E., Serrano E., Dellamaria D., Casais R., Rossi L., Puigdemont A., Cano-Manuel F.J., Fandos P., Pérez J.M., Espinosa J., Soriguer R.C., Citterio C., López-Olvera J.R. Evaluation of three enzyme-linked immunosorbent assays for sarcoptic mange diagnosis and assessment in the Iberian ibex. Capra pyrenaica. Parasites and Vectors. 2016;9 doi: 10.1186/s13071-016-1843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H., Wang S., Ruan S., Wang Y., Zhang J., Yuan Y., Liu H., Wu Y., Li R., Pan S., Ouyang Y., Yuan S., Zhou P., Shang Y. Dynamic Changes of Antibodies to SARS-CoV-2 in COVID-19 Patients at Early Stage of Outbreak. Virol. Sin. 2020;35:744–751. doi: 10.1007/s12250-020-00268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman, J.D., Hiatt, J., Mowery, C.T., Shy, B.R., Yu, R., Yamamoto, T.N., Rathore, U., Goldgof, G.M., Whitty, C., Woo, J.M., Gallman, A.E., Miller, T.E., Levine, A.G., Nguyen, D.N., Bapat, S.P., Balcerek, J., Bylsma, S.A., Lyons, A.M., Li, S., Wong, A.W. yi, Gillis-Buck, E.M., Steinhart, Z.B., Lee, Y., Apathy, R., Lipke, M.J., Smith, J.A., Zheng, T., Boothby, I.C., Isaza, E., Chan, J., Acenas, D.D., Lee, J., Macrae, T.A., Kyaw, T.S., Wu, D., Ng, D.L., Gu, W., York, V.A., Eskandarian, H.A., Callaway, P.C., Warrier, L., Moreno, M.E., Levan, J., Torres, L., Farrington, L.A., Loudermilk, R.P., Koshal, K., Zorn, K.C., Garcia-Beltran, W.F., Yang, D., Astudillo, M.G., Bernstein, B.E., Gelfand, J.A., Ryan, E.T., Charles, R.C., Iafrate, A.J., Lennerz, J.K., Miller, S., Chiu, C.Y., Stramer, S.L., Wilson, M.R., Manglik, A., Ye, C.J., Krogan, N.J., Anderson, M.S., Cyster, J.G., Ernst, J.D., Wu, A.H.B., Lynch, K.L., Bern, C., Hsu, P.D., Marson, A., 2020. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 38, 1174–1183. https://doi.org/10.1038/s41587-020-0659-0 [DOI] [PMC free article] [PubMed]
- Wiederschain, G.Y., 2009. The ELISA guidebook: 2nd Edn., Crowther, J. R. (ed.) in Series Springer Protocols. Methods in Molecular Biology, Vol. 516 (Walker, J., Series ed.) Humana Press, New Jersey, 2009, 566 p., $129, The ELISA guidebook: 2nd Edn., Crowther, J. R. (ed.) in Series Springer Protocols. Methods in Molecular Biology, Vol. 516 (Walker, J., Series ed.) Humana Press, New Jersey, 2009, 566 p., $129. https://doi.org/10.1134/S000629790909017X
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H., Wei X., Cai P., Ma W.L. Antibody Detection and Dynamic Characteristics in Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]