Abstract
At present, the global pandemic as it relates to novel coronavirus pneumonia is still a very difficult situation. Due to the recent outbreak of novel coronavirus pneumonia, novel chest X-ray (CXR) images that can be used for deep learning analysis are very rare. To solve this problem, we propose a deep learning framework that integrates a convolutional neural network and a capsule network. DenseCapsNet, a new deep learning framework, is formed by the fusion of a dense convolutional network (DenseNet) and the capsule neural network (CapsNet), leveraging their respective advantages and reducing the dependence of convolutional neural networks on a large amount of data. Using 750 CXR images of lungs of healthy patients as well as those of patients with other pneumonia and novel coronavirus pneumonia, the method can obtain an accuracy of 90.7% and an F1 score of 90.9%, and the sensitivity for detecting COVID-19 can reach 96%. These results show that the deep fusion neural network DenseCapsNet has good performance in novel coronavirus pneumonia CXR radiography detection.
Keywords: COVID-19, Chest X-ray, Classification, Deep learning, Capsule neural network
1. Introduction
In December 2019, researchers discovered a new type of febrile respiratory disease caused by severe acute respiratory syndrome coronavirus type 2 (novel coronavirus), which has spread rapidly worldwide. Novel coronavirus pneumonia was officially named coronavirus disease 2019 (novel coronavirus pneumonia) by the World Health Organization (WHO) on February 11, 2020 [1]. By the end of August 2020, the number of novel coronavirus pneumonia cases had exceeded 25 million and the death toll had exceeded 800,000 [2]; these outcomes seriously hindered global economic development and cultural exchanges and posed a serious threat to human health.
However, reverse transcription-polymerase chain reaction (RT-PCR) can identify patients before symptom onset and identify infected patients at an early stage, which is the “gold standard” of the novel coronavirus test [[3], [4], [5]]. However, high-performance detection equipment or platforms are needed, and high-sensitivity RT-PCR instruments are expensive. In addition, nucleic acid detection takes a long time. Considering the transportation of samples and the backlog of samples, the results can usually be reported within 24 h at the earliest.
Compared with nucleic acid detection, radiological examination (computed tomography (CT) or X-ray) also has some visual markers, such as frosted glass opacity and bilateral radiation abnormalities [6,7], which can be used to distinguish novel coronavirus pneumonia infection from non-COVID-19 infection. Because of its high sensitivity, CT is widely used in the auxiliary diagnosis of novel coronavirus pneumonia [5,8]. However, the high infection rate of novel coronavirus has led to its rapid spread worldwide. Low-cost rapid detection technology is crucial to screening novel coronavirus pneumonia, especially in underdeveloped areas. Although chest X-ray is helpful for the early detection of novel coronavirus pneumonia, there are many kinds of viral pneumonia with certain similarities, which makes it difficult for inexperienced radiologists to accurately distinguish other viral pneumonias from novel coronavirus pneumonias. It has been reported that the sensitivity of X-ray examination of novel coronavirus pneumonia is only 69% [9]. If the performance of X-ray detection of novel coronavirus pneumonia can be significantly improved, it will be of great significance.
With the successful application of artificial intelligence technology in the medical field, an excellent method is to use medical image processing and deep learning technology to assist in the detection and diagnosis of medical images [10]. Deep learning technology can reveal many inconspicuous image features in medical images. Convolution neural networks (CNNs) have been widely used in research fields for depth feature extraction. Of course, the deep learning technology of novel coronavirus pneumonia detection has also been actively explored [[11], [12], [13], [14], [15], [16]]. Linda Wang et al. [11] proposed a deep convolution neural network COVID-Net that can be used for COVID-19 detection and achieved an overall accuracy of 93.3% in the dataset COVIDx proposed by the author. COVID-19 detection sensitivity reached 91%.
At present, the CNN, as an excellent feature extractor, is the method used by most researchers who use deep learning technology to detect COVID-19. However, CNNs have some defects, such as being unable to capture the spatial relationships of features and being unable to recognize pictures with rotations or other transformations. In 2017, Geoffrey E. Hinton et al. [17] introduced the capsule network in detail. The capsules in the capsule network are composed of a group of related neurons, which represent various attributes of a specific entity, such as posture, texture features, and tone. Capsule networks can fully capture image features, postures and spatial relationships by means of neuron “packaging”, so capsules can detect certain types of patterns and reduce the dependence of the network on large datasets. The capsule network has been proven to be a good substitute for CNNs. This is truly an exciting achievement. Parnian Afshar et al. [18] aimed to use a capsule network to detect COVID-19 and proposed COVID-CAPS, and they achieved excellent results, with an overall accuracy of 95.7% and sensitivity of 90%. The results proved that capsule network detection of COVID-19 was feasible.
Using deep learning technology to analyze medical images tests the quality and quantity of datasets, and it is difficult to obtain a large number of medical images with good quality when a new epidemic suddenly occurs. In this work, we focus on solving the problem of training excellent detection models on small COVID-19 datasets. This method takes the capsule network as the main body and DenseNet [19] as the feature extractor and proposes a deep learning framework DenseCapsNet for COVID-19 detection. We indicate through experiments that DenseCapsNet can effectively train an excellent COVID-19 detection model using such small datasets. Compared with the COVID-19 detection model using the same data, our proposed network has better overall accuracy and sensitivity and can detect COVID-19 cases more accurately. In addition, this paper also gives the chest X-ray (CXR) image preprocessing operation to solve the problem of data heterogeneity between public datasets and the potential feasibility analysis of the framework for patient shunt and medical diagnosis. The main contributions of this paper are as follows:
-
1)
Based on the capsule neural network (CapsNet) and the dense convolutional network (DenseNet), a deep learning framework, DenseCapsNet, which can realize end-to-end classification of COVID-19 automatic diagnosis, is proposed.
-
2)
DenseCapsNet achieves state of the art performance by increasing the sensitivity of COVID-19 detection from 69% to 96% using X-rays.
-
3)
A set of reasonable preprocessing process is proposed to alleviate the problem of image heterogeneity among different datasets.
-
4)
Analysis of the potential value of the deep learning framework in clinical diagnosis based on DenseCapsNet is presented.
2. Material and methods
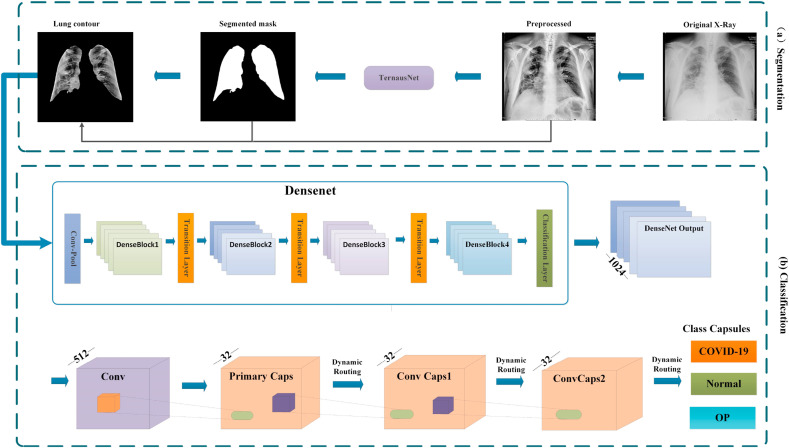
The overall structure of the framework is shown in Fig. 1 . First, to solve the problem of image heterogeneity between different dataset resources, CXR images are preprocessed to realize data normalization. Then, the preprocessed data are fed into the segmentation network to extract the lung region of the CXR image. Finally, the DenseNet part of the classification network extracts rich features from the lung region and transfers the features to the capsule network to infer the disease type of the CXR image. Hereinafter, the network framework will be described in detail.
Fig. 1.
The overall architecture of the proposed neural network. The (a) part is a segmentation network, and the (b) part is DenseCapsNet for feature extraction and classification.
2.1. Data preprocessing methods
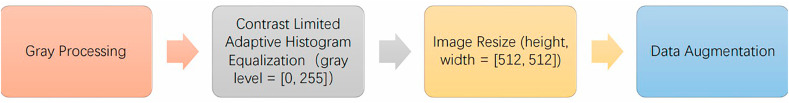
Using different scanning equipment and scanning methods will inevitably lead to certain heterogeneity in data resolution, image size, image definition and other issues. To reduce the impact of data heterogeneity on network performance, we propose some preprocessing steps for CXR, the details of which are shown in Fig. 2 . First, grayscale processing is performed on the original CXR image and then the local contrast of the CXR image is improved by using contrast-limited adaptive histogram equalization (CLAHE) [20] to obtain more image details.
Fig. 2.
CXR image preprocessing steps.
After the image enhancement processing of the CXR image is completed, the data augmentation operation will be carried out. Data augmentation methods should be as random as possible to generate more meaningful training data. In this study, random rotation, vertical flip and random cutting data augmentation are used to train the network to alleviate the overfitting problem. We also normalize the data to speed up the model fitting process and finally improve the generalization ability of the model.
| (1) |
In the formula, x is the pixel intensity value in each channel of the three red-blue-green (RGB) channels of the CXR image, mean is the average pixel intensity value of each channel, and std is the standard deviation of the pixel intensity of each channel.
2.2. Segmentation network
In this work, we need to extract the lung contour of CXR images with a segmented network. The proposed method ultimately selects TernausNet [21] with state-of-the-art (SOTA) performance as the semantic segmentation part. TernausNet uses the pretrained VGG11 as the encoding part and removes the full connection layer, the last max pooling and softmax layer of VGG11 [22]. In the decoding part, upsampling and convolution operations are carried out on the feature map extracted from the encoding, and the high-resolution features of the encoding part are combined through jump connections. The specific structure is shown in Fig. 3 .
Fig. 3.
Semantic segmentation network display.
When training TernausNet, we use three loss functions for back propagation. In Ref. [23], BCEWithLogitsLoss, SoftDiceLoss and InvSoftDiceLoss loss functions are used to calculate the loss value and sum it to obtain the total loss value. The segmentation effect obtained by back propagation using the total loss value is optimal.
2.3. Classification network
1) Using the convolutional neural network for feature extraction: DenseNet can fully flow information, reuse features, strengthen feature propagation and reduce the number of parameters through its densely connected features. These characteristics enable DenseNet to achieve better results than ResNet [24] under the same parameter quantity in less time. We use DenseNet121 after ImageNet [25] pretraining to prevent the effect of random model initialization on the model fitting ability and to ensure that the model can achieve better results on a smaller dataset to the greatest extent possible. DenseNet121 is the simplest type of dense convolution network and has the smallest number of parameters. The network structure is shown in Fig. 4 . We reduced the chest X-ray data to a unified size, enhanced it with data and then converted it into tensors. After normalization, we input it into the DenseNet121 network. The network structure is shown in Fig. 4. We reduced the chest X-ray data to a unified size, enhanced it with data and then converted it into tensors. After normalization, we input it into the DenseNet121 network architecture. Using DenseNet121's excellent feature extraction and feature reuse capabilities, we retained all the kinds of detailed features of the data as much as possible and finally output 1024 features.
Fig. 4.
DenseNet structure diagram. The dense block and transition layer are the main structures of DenseNet. The dense block can make the network structure better for feature transfer and feature reuse. The transition layer can reduce the size of the feature graph and promote feature transfer between adjacent dense blocks. Finally, the information transmission of the network will be more intensive.
2) Capsule Network: The architecture of the capsule network is relatively clear and mainly consists of a series of capsule layers [17]. The capsule layer of the capsule network is composed of multiple capsules, and each capsule is composed of a group of neurons. The primary capsule layer is preceded by a convolutional layer composed of 512 convolutional kernels with the size of 1*1, which filters the output features of DenseNet to 512. After the reshaping operation, a primary capsule layer is formed. Dynamic routing is adopted between the primary capsule layer and the convolution capsule layer to route the output vector to all possible parent nodes.
3) Loss Function: For the loss function of the capsule network, we use the spread loss function, which can reduce the sensitivity of training to the model initialization and superparameters, and is the target class activation. If the distance between the activation error classes and is less than the margin (m), the loss function will penalize the model according to.
The loss function is defined as follows:
| (2) |
The initial value of the margin is set to 0.2. During the training process, the value of the margin increases by 0.1–0.9 per iteration, which can prevent early capsule death.
4) DenseCapsNet Network Structure: The structure of DenseCapsNet that we propose is shown in Fig. 1. As a shallow network, CapsNet cannot extract deep features as does a deep neural network. Above, we briefly introduced DenseNet121 and explained the leading position of DenseNet121 in a CNN. DenseNet121 as feature extractor is obviously a good choice and can compensate for the deficiency of CapsNet to some extent. DenseCapsNet is mainly composed of DenseNet121 and CapsNet so that it fully leverages the advantages of CNNs and capsule networks. First, we retain the feature extraction part of the CNN DenseNet121 (excluding the linear layer) to extract the features from COVID-19 chest X-ray data. We input novel coronavirus pneumonia 3D X-ray data and adjust the data to a unified size. DenseNet 121 outputs 1024 feature maps and transmits them to the first convolution layer of the capsule network, which filters the above features to 512 and reshapes them into vectors to be transmitted to the main capsule layer. Finally, the capsule layer outputs the instantiated parameters containing normal and COVID-19 images, and the vector lengths represent the probability of the occurrence of each category.
3. Experimental results
3.1. Dataset description
The datasets used in this study are all public datasets; these datasets have been studied many times by other COVID-19 researchers, indicating the research value of these data. Statistics of CXR image data used to segment network training are shown in Table 1 , and statistics of the dataset used to classify network training are shown in Table 2 .
Table 1.
Segmentation dataset.
| Dataset | Normal | Tuberculosis | Total |
|---|---|---|---|
| NLMMC [26] | 80 | 58 | 138 |
Table 2.
Classification dataset.
1) Segmentation Network Dataset: To train the segmentation network, we used the Montgomery County Chest X-ray Database [26] collected by the U.S. National Library of Medicine. The NLM-Montgomery County Chest X-ray database (NLMMC) contains 138 CXR images with lung contour masks, including 80 normal cases and 58 tuberculosis cases. All images are stored in PNG format with sizes of 4892*4020 or 4020*4892. The NLMMC database was randomly divided into training and testing at an 8:2 ratio.
2) Classification Dataset: In this study, three COVID-19 datasets were collected, with a total of 9432 CXR images. Drawing on the ideas of COVID-Net, we divided the classified dataset into three categories: normal, other pneumonia (except COVID-19), and COVID-19. Among them, normal CXR images were mainly derived from Refs. [27,29], for a total of 2917 images. The three datasets above contributed to the CXR images of other patients with pneumonia and COVID-19, totaling 5734 and 781 images, respectively. All images are stored in PNG format, and the image sizes are mainly distributed between 2563*1148 and 723*711. Some of the images had lesion locations marked by doctors, so we deleted this part of the images to avoid the disclosure of the information marked to affect the model training.
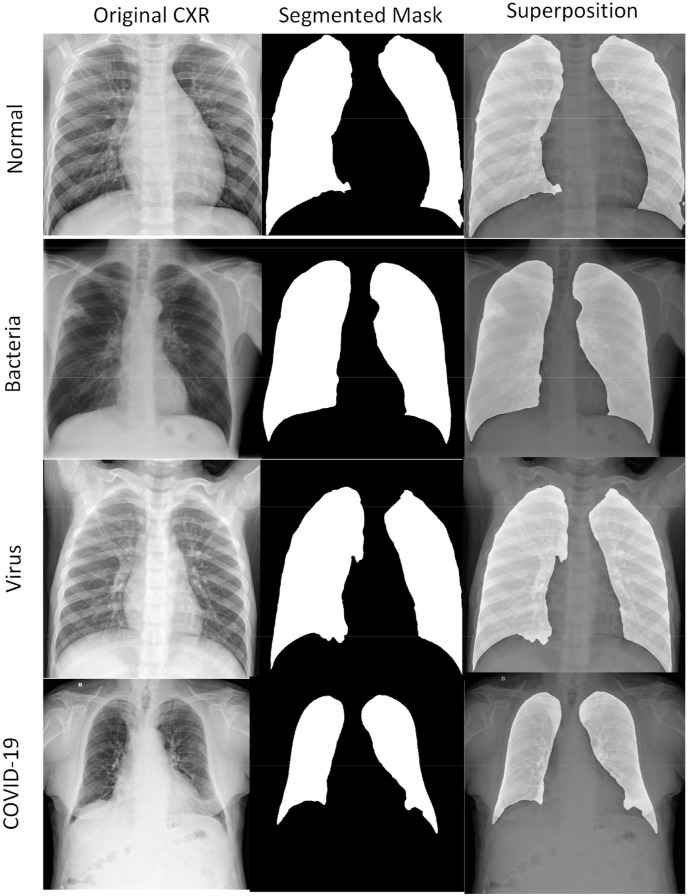
We collected the data uniformly and asked the radiologist to select four representative CXR images (Fig. 5 ) and made a detailed description of different case characteristics. In Fig. 5(a), the two lungs had clear veins and no parenchymal lesion was seen in the lungs. The hilum of the two lungs was not large, and nodules and bump shadows were not seen. There was no abnormality in the size or shape of the heart shadow. The two diaphragms were smooth, and the two costal diaphragms had sharp angles. To summarize, it should be diagnosed as normal. In Fig. 5(b), the two lung textures thickened, and the right lower lung had a small piece of flocculent shadow, uneven density, and fuzzy edges. There was consolidation of the lower lobe of left lung. In summary, a diagnosis of bacterial pneumonia should be made. In Fig. 5(c), lung texture increased and blurred. A focal patchy image was apparent in the lung field. There were diffuse high-density shadows in both lungs. In summary, patients with viral pneumonia should be diagnosed. In Fig. 5(d), the frosted glass-like structure is visible at the margin of the pulmonary vessels, with unclear boundaries. The lesions are asymmetrically patchy, and there is diffuse air turbidity. In summary, a COVID-19 patient should be diagnosed.
Fig. 5.
(a), (b), (c), (d) are CXR images of normal, bacterial, viral pneumonia, and COVID-19, respectively.
After giving the lesion characteristics of typical patients in the public dataset, radiologists also use a computer to randomly select 200 chest X-ray images from a dataset for diagnosis. Radiology experts believe that the labels provided by the public dataset are accurate and that the patient characteristics of various pneumonias in the dataset are representative, further confirming the research value of the dataset.
To test the performance of the neural network framework we proposed, a computer was used to randomly select 750 CXR images from the above dataset, with 250 for normal, 250 for other pneumonia and 250 for novel coronavirus pneumonia. Then, 750 CXR images were randomly divided into training, validation and testing sets. See Table 3 for details of the dataset division.
Table 3.
Detailed information of data split.
| Training | Validation | Test | |
|---|---|---|---|
| Normal | 175 | 25 | 50 |
| Other pneumonia | 175 | 25 | 50 |
| COVID-19 | 175 | 25 | 50 |
3.2. Model Parameter Setting and evaluation method
1) Model Parameter Setting: In the experiment, for the semantic segmentation part, the number of epochs was set to 75, the learning rate was set to 1e-4, and the Adam optimizer was used. For the classification network, the number of epochs is set as 30, the learning rate is 1e-4, the optimizer uses ADAM [30], and the number of dynamic routing iterations of the capsule network is 3. All network frameworks are implemented in PyTorch [31] using two NVIDIA TITAN Xp GPUs.
2) Segmentation Performance Metrics: The Jaccard coefficient and Dice coefficient were used to evaluate the segmentation performance of the segmentation network. All the above evaluation metrics can measure the similarity between the finite sets. If there are two sets A and B, the Jaccard coefficient and Dice coefficient can be used to measure the similarity, which can be defined as:
| (3) |
| (4) |
3) Classification Performance Metrics: In the classification part, according to the confusion matrix, the values of true (TP), true negative (TN), false negative (FN) and false positive (FP) can be obtained. By calculating the above four values, accuracy, precision, recall, specificity and F1 score can be obtained. This study uses these five metrics to evaluate the classification performance of the model. These five measures are used by most researchers on COVID-19 [16,32,33] and are authoritative. The above evaluation metrics are calculated as follows:
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
3.3. Data preprocessing performance
In this study, the pretreatment method mentioned in this paper was first used to preprocess the classified dataset to achieve the effects of CXR image histogram equalization and contrast enhancement. The specific implementation process is implemented using Python and OpenCV. First, grayscale processing is performed on the original three-channel CXR image to obtain a single-channel image. Next, CLAHE is used to improve the image contrast (specific parameter settings: clipLimit = 3.0, tileGridSize =(8,8)) and adjust the image size uniformly to 512*512. Experimental results are shown in Fig. 6 . After pretreatment, CXR image details were more prominent, the lung contour was clearer, and the histogram was more balanced.
Fig. 6.
Image preprocessing effect. The first row is the original CXR and the corresponding histogram; the second row is the image enhanced CXR and the corresponding histogram.
3.4. Semantic segmentation performance
In the experiment, we selected three semantic segmentation networks with excellent performance, namely, GCN [34], SegNet [35] and TernausNet. The NLMMC dataset was randomly divided into training and validation sets at an 8:2 ratio. All the above three semantic segmentation networks were trained and tested with the segmented NLMMC dataset, and the Jaccard and Dice coefficients were used to evaluate the ability of the network model to extract lung contours. The final results are shown in Table 4 . Obviously, TernausNet has achieved the best performance, and the Jaccard coefficient is even improved by nearly 0.1 compared with the second place SegNet, which may be because the encoder part is VGG11: the network depth is relatively shallow, and the pretrained network has a stronger generalization ability and is more suitable for segmentation training tasks with small datasets. Fig. 7 shows the lung contour extracted from the NLMMC data of TernausNet. Due to the lack of a hand-prepared mask for the classified dataset, there is no way to use the classified data to train the segmentation network. Fortunately, the TernausNet segmentation model trained with the NLMMC dataset also achieved good results in extracting the lung contour from the classified data, and the results are shown in Fig. 8 .
Table 4.
Segmentation network performance comparison.
| Methods | Jaccard | Dice |
|---|---|---|
| GCN | 0.826 ± 0.022 | 0.905 ± 0.023 |
| SegNet | 0.843 ± 0.033 | 0.912 ± 0.019 |
| TernausNet | 0.942 ± 0.026 | 0.969 ± 0.021 |
Fig. 7.
Semantic segmentation network TernausNet extracted lung contour results from the NLMMC dataset.
Fig. 8.
The lung contour results were extracted from the classified dataset by TernausNet. Superposition represents the result of overlaying the segmented mask onto the original CXR.
3.5. Classification network performance
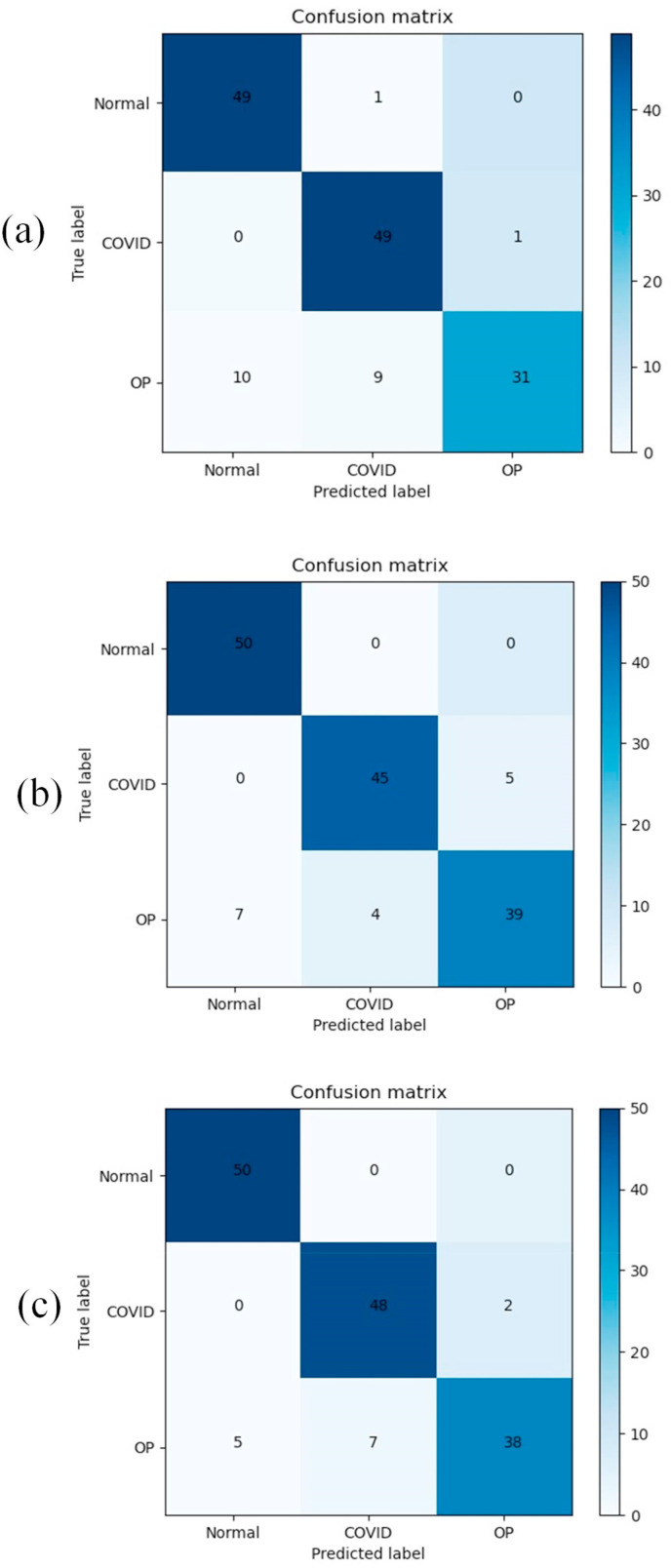
The classification performance of DenseCapsNet, a deep learning framework proposed in this paper, is shown in Table 5 . All experiments were repeated five times and averaged. When training ResNet and DenseNet, we used ImageNet's pretraining parameters to initialize the network parameters and then trained the network with the COVID-19 dataset without freezing any layers. The confusion matrix for ResNet50, DenseNet121, and DenseCapsNet is shown in Fig. 9 . Before DenseCapsNet is formally proposed, the feature extraction part is compared first. In this work, ResNet50 and DenseNet121 were selected as candidate feature extraction parts. It can be clearly found from Table 5 that DenseNet is superior to ResNet50 in all metrics, and the recall and accuracy are even improved by 3.3%. According to the performance, DenseNet121 was selected as the feature extraction part and fed into CapsNet to form the proposed DenseCapsNet deep learning framework. It can also be seen from Table 5 that the detection performance of the fused network is superior to the detection of COVID-19 using CNN alone in all metrics.
Table 5.
Classification network performance comparison.
| Methods | Accuracy | Precision | Recall | F1 score | Specificity |
|---|---|---|---|---|---|
| ResNet50 | 86 ± 1.3 | 87.7 ± 1.3 | 86 ± 1.3 | 86.8 ± 1.3 | 93 ± 0.7 |
| DenseNet121 | 89.3 ± 1.4 | 89.4 ± 1.6 | 89.3 ± 1.4 | 89.3 ± 1.4 | 94.7 ± 0.6 |
| DenseCapsNet | 90.7 ± 2.2 | 91.1 ± 2.2 | 90.7 ± 2.2 | 90.9 ± 2.2 | 95.3 ± 1.1 |
Fig. 9.
(a), (b) and (c) are confusion matrices of ResNet50, DenseNet121 and DenseCapsNet, respectively, where OP is an abbreviation for other pneumonia.
4. Discussion
4.1. The influence of preprocessing on the performance of segmented networks
At present, the global novel coronavirus pneumonia situation is still grim, which makes it difficult to obtain carefully processed CXR image data. The data used in this experimental study come from many different novel coronavirus pneumonia datasets, and the data heterogeneity between different databases is obvious. Novel coronavirus pneumonia datasets lack available manual masks, so we must choose other open CXR datasets with masks to train the segmentation network. To enhance the generalization ability of the segmentation model as much as possible, some necessary preprocessing operations are needed for the public dataset. In this experiment, TernausNet is trained with the NLMMC dataset, and then the lung contours of the original image and preprocessed image of the CXR image of the classified dataset are extracted. The results are shown in Fig. 10 . After the original CXR image is preprocessed, the lung contour is clearer and the contrast is more obvious, which can significantly improve the segmentation capability of the segmentation network on the classified dataset. This is mainly due to the CLAHE operation, which covers the gray distribution range of the original image and histogram in the range [0, 255] (Fig. 6). The gray distribution is more uniform, and the image details are more obvious. Histogram equalization can indeed alleviate the heterogeneity of the image.
Fig. 10.
Influence of preprocessing operation on segmentation performance.
4.2. Classification performance analysis of DenseCapsNet
As seen from Table 5, DenseNet121 is superior to ResNet50 in all five metrics of COVID-19 CXR detected by CNN alone. This is because the trainable parameters of DenseNet121 are smaller than those of ResNet50. When training the model on a small dataset, small parameters have certain advantages, and too large parameters may lead to overfitting and other problems. On the network structure, the dense block structure can realize feature transfer within the module, while the transition layer can realize the feature transfer between different dense blocks. The powerful feature reuse makes DenseNet121 have fewer parameters and more efficient calculations and can also retain some low-level features in the 1024 features finally output. In the confusion matrix of Fig. 9 (a) (b), ResNet50 is better than DenseNet121 in detecting COVID-19. However, ResNet50 is indeed easier to diagnose other pneumonia as normal or COVID-19 in other pneumonia detection, which will still increase the workload of radiologists. In the global pandemic situation of the COVID-19 epidemic, the increase in workload will undoubtedly lead to the failure of COVID-19 patients to receive timely diagnosis and treatment.
The proposed framework integrates DenseNet and CapsNet and uses classified datasets for training and testing. In Table 5, DenseCapsNet's five metrics are all superior to DenseNet121, with an average increase of 1.34%. Through the sensitivity analysis in Table 6 , the sensitivity of DenseCapsNet to detect COVID-19 reached 96% and was significantly better than DenseNet121. The sensitivity of radiologists to diagnose COVID-19 by CXR images is 69%, and even gold standard RT-PCR can only reach 91% sensitivity [36].
Table 6.
Comparison of sensitivity between DenseNet and DenseCapsNet.
| Methods | Sensitivity |
||
|---|---|---|---|
| Normal | Other Pneumonia | COVID-19 | |
| DenseNet | 100 | 78 | 90 |
| DenseCapsNet | 100 | 76 | 96 |
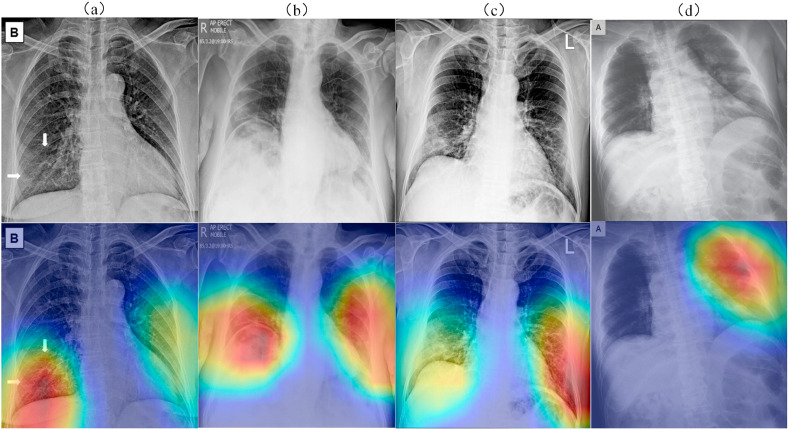
Fig. 11 shows the result of using the grad-CAM [37] algorithm to locate the COVID-19 lesion area, and the red area is the suspicious COVID-19 lesion area located on the network. First, we used the trained model to locate the suspected lesion area in the segmented chest X-ray. Next, we superimposed the resulting heat map onto the original chest X-ray image to fit the radiologist's reading habits. Radiologists gave professional explanations for the CXR image shown in Fig. 11: In image a, the texture of both lungs was thickened, and a few pieces of flocculation shadows were seen in the lower lobe of both lungs, with uneven density and blurred edges, among which the exudation shadow was the most obvious in the lower lobe of the right lung. In image b, the lung in the lower lobe of the right lung is consolidated, with scattered ground glass nodules of both lungs, and multiple plaques and ground glass shadows in the outer lung and subpleural lung. In image c, the shadow exudates from the lower lobe of the right lung, with a larger heart. In image d, there are ground glass nodules on the upper lobe of the left lung with thickened and blurred local lines. The remaining lung has clear texture and no parenchymal lesion was observed. The hilum of both lungs was not large, and the shape of the heart shadow was not abnormal. The a, b and d CXR images with heat maps are basically consistent with the focus areas diagnosed by radiologists. Unfortunately, the heat map of c did not accurately locate the lesion area. In the discussion and analysis with radiology experts, it was found that there were artifacts on both sides of the pleural area of the image, which led to misjudgment of the network.
Fig. 11.
These are four X-ray images of the chest of COVID-19 patients. The first row is the original CXR, and the second row is the suspicious lesion area located by the network.
4.3. Feasibility analysis of DenseCapsNet
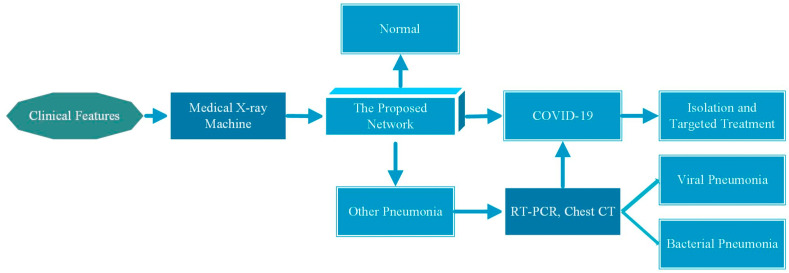
In the context of the global COVID-19 outbreak, many countries are facing a shortage of medical resources to diagnose, isolate and treat COVID-19 patients in a timely manner. Chest CT is more sensitive for detecting and diagnosing COVID-19 than CXR imaging, but CT equipment is expensive and difficult to move, and cross-infection may occur. Therefore, the use of an inexpensive, portable medical X-ray machine to obtain chest X-rays of patients with clinical characteristics such as cold and fever may be more suitable for the current global epidemic situation. Increasing the sensitivity of CXR to COVID-19 diagnosis is valuable and can alleviate some problems, such as shortages of medical resources. In Fig. 12 , we present a flowchart of patient diagnosis with potential application value according to the proposed deep learning framework. The subject still uses the proposed deep learning framework to detect CXR images. According to the confusion matrix in Fig. 9 (c), it can be seen that the proposed deep learning framework may misdiagnose COVID-19 as other pneumonia. After consulting radiologists, we learned that the focal features of CXR images of COVID-19 patients were similar to those of other viral pneumonia lesions. This makes it possible for the network to misdiagnose COVID-19 patients with other viral pneumonias. Fortunately, the framework does not affect the diagnosis of CXR in COVID-19 patients as normal, so in the patient diagnosis flow chart, we recommend that patients diagnosed with other pneumonia be further confirmed through RT-PCR to ensure that COVID-19 infected patients are not missed and treatment is not delayed. Patients diagnosed with COVID-19 should be isolated immediately and targeted for treatment.
Fig. 12.
Flow chart of suspicious patient diagnosis using the proposed framework.
4.4. Comparison with other SOTA methods
DenseCapsNet is a neural network architecture designed based on a capsule network for detecting COVID-19 chest X-rays. We compare a neural network architecture COVID-CAPS, which has excellent performance in diagnosing COVID-19 and is based on a capsule network. To make a fair comparison between DenseCapsNet and COVID-CAPS, we did not use operations such as preprocessing and extracting lung contours and used the same dataset as COVID-CAPS. The comparison results are shown in Table 7 . Without using a large amount of other lung X-ray data for pretraining, our method can be superior to COVID-CAPS after pretraining in accuracy. Compared with COVID-CAPS, DenseCapsNet significantly improves the sensitivity of detecting COVID-19 and avoids missing COVID-19 patients as much as possible, which will greatly reduce the workload of medical workers in screening COVID-19 patients and improve the work efficiency. However, although our proposed framework has significantly improved the detection performance, DenseCapsNet (8 M) is higher than COVID-CAPS (0.29 M) in the number of network trainable parameters. This is mainly because the feature extraction part of DenseCapsNet is much more complicated than that of COVID-CAPS (only four convolution layers). In the future, we will also consider the problem of identifying a lightweight model, reduce trainable parameters and improve model training efficiency.
Table 7.
Comparison of densecapsnet with Covid-caps.
| Methods | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| COVID-CAPS without pretraining | 95.7 | 90 | 95.8 |
| Pretrained COVID-CAPS | 98.3 | 80 | 98.6 |
| Proposed | 98.5 | 97 | 100 |
At the same time, we also compared some classical methods based on the convolutional neural network to detect COVID-19, and the comparison results are shown in Table 8 . Compared with the classic convolutional neural network, DenseCapsNet can achieve better performance. Because Heidari et al.‘s work used 11 times the amount of CXR data as ours, their work outperformed ours in terms of accuracy. However, our method can help radiologists pinpoint the location of the suspected lesion, and we also provide a feasible solution for the proposed method to assist diagnosis (Fig. 12.), which will significantly relieve the pressure on doctors to respond to the epidemic.
Table 8.
Comparison of DenseCapsNet with classical CNN methods.
| Methods | Number of cases | Accuracy |
|---|---|---|
| GoogLeNet Loey et al. [38] | 69 COVID-19 + 79 Normal + 79 BP + 79 VP | 80.56 |
| Ozturk et al. [39] | 125 COVID-19 + 500 Pneumonia+ 500 No finding | 87.02 |
| Mahmud et al. [40] | 305 COVID-19 + 305 VP+ 305 BP | 89.6 |
| Hemdan et al. [14] | 25 COVID-19 (+) 25 Normal | 90.0 |
| Apostolopoulos et al. [41] | 224 COVID-19 + 700 Pneumonia +504 Normal | 93.5 |
| Heidari et al. [42] | 415 COVID-19 + 5179 CAP + 2880 Normal | 94.0 |
| Proposed | 250 COVID-19 + 250 OP + 250 Normal | 90.7 |
| 250 COVID-19 + 250 Non COVID-19 | 98.5 |
Note: BP, OP and VP stand for bacterial pneumonia, other pneumonia and viral pneumonia respectively.
5. Conclusion
To improve the sensitivity of CXR images in diagnosing COVID-19, this paper proposes a deep learning framework for COVID-19 detection based on DenseNet121 and CapsNet. The framework uses DenseNet121 to extract features and CapsNet to package features into capsules to reduce the dependence of neural networks on data volume. Only 250 normal chest radiographs, 250 other pneumonia chest radiographs and 250 COVID-19 chest radiographs were used to achieve remarkable diagnostic results, with accuracy and sensitivity reaching 90.7% and 96%, respectively. We have proven that in the task of using small datasets to detect COVID-19, the performance of the fusion of CNN and CapsNet is better than that of using CNN alone. We also propose the CXR image preprocessing operation, which can alleviate the problem of image heterogeneity between different datasets, and prove that the preprocessing operation can improve the ability of the segmentation network to extract lung contours across datasets. In the context of the novel coronavirus pneumonia pandemic, the in-depth learning framework with high sensitivity for diagnosing COVID-19 may be used for large-scale screening of suspicious COVID-19 patients, so this study also provides a feasibility analysis of large-scale screening and diagnosis of COVID-19 using the proposed neural network framework. We sincerely hope that the epidemic will pass as soon as possible and that people will be able to return to their normal lives.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors would like to thank health care workers worldwide and those in other industries who have contributed to COVID-19 relief efforts since the COVID-19 outbreak, as well as contributors to the public chest X-ray dataset. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. We sincerely wish the people of all countries an early victory over COVID-19.
Footnotes
Hao Quan (First author), Xiaosong Xu, Tingting Zheng, Zhi Li, Mingfang Zhao, Xiaoyu Cui (Member IEEE).
References
- 1.Timeline of WHO’s response to COVID-19 World health organization web site. https://www.who.int/news-room/detail/29-06-2020-covidtimeline [Online]. Available:
- 2.WHO Coronavirus Disease (COVID-19) Dashboard World health organization web site. https://covid19.who.int/ [Online]. Available:
- 3.Wang Wenling, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA The Journal of the American Medical Association. 2020;323:18. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM et al., “Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR,” 2020, Euro Surveill.. 25(3):2000045. [DOI] [PMC free article] [PubMed]
- 5.Ai T., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296 doi: 10.1148/radiol.2020200642. Art. no. 200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiology Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H., Han X., Jiang N., Cao Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20:4. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong Hyf, et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020:201160. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenspan Hayit, et al. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imag. 2016;35(5):1153–1159. [Google Scholar]
- 11.Wang Linda, et al. 2020. COVID-net: a Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-Ray Images.https://arxiv.org/abs/2003.09871 arXiv:2003.09871. [Online]. Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (COVID-19) Using X-Ray Images and Deep Convolutional Neural Networks.https://arxiv.org/abs/2003.10849 arXiv:2003.10849. [Online]. Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Din Hemdan E., Shouman M.A., Esmail Karar M. 2020. COVIDX-Net: A Framework of Deep Learning Classifiers to Diagnose COVID-19 in X-Ray Images.https://arxiv.org/abs/2003.11055 arXiv:2003.11055. [Online]. Available: [Google Scholar]
- 15.Apostolopoulos I.D., Aznaouridis S., Tzani M. 2020. Extracting Possibly Representative COVID-19 Biomarkers from X-Ray Images with Deep Learning Approach and Image Data Related to Pulmonary Diseases.https://arxiv.org/abs/2004.00338v2 arXiv:2004.00338. [Online]. Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farooq M., Hafeez A., Covid-ResNet “. 2020. A Deep Learning Framework for Screening of COVID19 from Radiographs.https://arxiv.org/abs/2003.14395 arXiv:2003.14395. [Online]. Available: [Google Scholar]
- 17.Sabour S, Frosst N, Hinton GE., “Dynamic routing between capsules,” In Advances in Neural Information Processing Systems. pages 3856–3866.
- 18.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A., Covid-Caps “. 2020. A Capsule Network-Based Framework for Identification of COVID-19 Cases from X-Ray Images.http://arxiv.org/abs/2004.02696 arXiv:2004.02696. [Online]. Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017. Densely Connected Convolutional Networks; pp. 2261–2269. [Google Scholar]
- 20.Zuiderveld K. Contrast limited adaptive histogram equalization. Graphics Gems. 1994:474–485. [Google Scholar]
- 21.Iglovikov Vladimir, Shvets A. 2018. TernausNet: U-Net with VGG11 Encoder Pre-trained on ImageNet for Image Segmentation.https://arxiv.org/abs/1801.05746 arXiv:1801.05746. [Online]. Available: [Google Scholar]
- 22.Simonyan Karen, Zisserman A. Very Deep Convolutional Networks for Large-scale Image Recognition. 2014. https://arxiv.org/abs/1409.1556v1 arXiv.org.
- 23.MEDAL-IITB Lung-segmentation. https://github.com/MEDAL-IITB/Lung-Segmentation/tree/master/VGG_UNet [Online]. Available:
- 24.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016:770–778. [Google Scholar]
- 25.Krizhevsky Alex, Sutskever I., Hinton G. NIPS Curran Associates Inc.; 2012. ImageNet Classification with Deep Convolutional Neural Networks. [Google Scholar]
- 26.Jaeger S., Candemir S., Antani S., Wáng Y.-X.J., Lu P.-X., Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imag. Med. Surg. 2014;4(6):475. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Praveen Corona hack: chest X-ray-dataset. https://www.kaggle.com/praveengovi/coronahack-chest-xraydataset [Online]. Available:
- 28.Paul Cohen J., Morrison P., Dao L. 2020. COVID-19 Image Data Collection.http://arxiv.org/abs/2003.11597 arXiv:2003.11597. [Online]. Available: [Google Scholar]
- 29.Rahman Tawsifur, Chowdhury Dr Muhammad, Khandakar Amith. COVID-19 radiography database. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database/data [Online]. Available.
- 30.Kingma Diederik, Ba J. Adam: A Method for Stochastic Optimization. 2014. https://arxiv.org/abs/1412.6980v9 arXiv.org.
- 31.Paszke A., et al. Pytorch: an imperative style, high-performance deep learning library. in Proc. Advances Neural Inf. Process. Syst. 2019:8026–8037. [Google Scholar]
- 32.Chowdhury M.E.H., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 33.Minaee S., Kafieh R., Sonka M., Yazdani S., Soufi G.J. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng Chao, et al. 2017 IEEE Conference on Computer Vision and Pattern Recognition. CVPR) IEEE; 2017. Large kernel matters — improve semantic segmentation by global convolutional network. [Google Scholar]
- 35.Badrinarayanan Vijay, Kendall A., Cipolla R. IEEE Transactions on Pattern Analysis & Machine Intelligence; 2017. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. 1-1. [DOI] [PubMed] [Google Scholar]
- 36.Wong H.Y.F., et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. Mar. 2020 doi: 10.1148/radiol.2020201160. Art. no. 201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D., “Grad-Cam Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. Feb. 2020;128(2):336–359. [Google Scholar]
- 38.Loey M., Smarandache F., M Khalifa N.E. Within the lack of chest covid-19 x-ray dataset: a novel detection model based on gan and deep transfer learning. Symmetry. 2020;12:651. [Google Scholar]
- 39.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heidari M., Mirniaharikandehei S., Khuzani A.Z., Danala G., Qiu Y., Zheng B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inf. 2020;144:104284. doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]