Summary
The mammalian main olfactory pathway detects myriad volatile chemicals using >1,000 odorant receptor (OR) genes, which are organized into two phylogenetically distinct classes (class I and class II). An important question is how these evolutionarily conserved classes contribute to odor perception. Here, we report functional inactivation of a large number of class I ORs in mice via identification and deletion of a local cis-acting enhancer in the class I gene cluster. This manipulation reduced expression of half of the 131 intact class I genes. The resulting class I-depleted mice exhibited a significant reduction in the number of glomeruli responding to carboxylic acids—chemicals associated with microbial action and body odors. These mice also exhibit a change in odor perception marked by a selective loss of behavioral aversion to these compounds. Together, our data demonstrate that class I ORs play a critical role in representing a class of biologically relevant chemosignals.
Keywords: Olfaction, odorant receptor, olfactory epithelium, olfactory bulb, monoallelic expression, odor aversion, carboxylic acids, mouse, enhancer
eTOC Blurb
Cichy et al. show that deleting a cis-acting enhancer in the class I odorant receptor cluster reduces class I receptor gene choice. The resulting class I depleted mice exhibit reduced functional and behavioral responses to carboxylic acids. The results demonstrate a role for class I odorant receptors in detecting an important class of odorants.
Introduction
For many species, the sense of smell is essential for survival. The olfactory system detects and discriminates a broad range of environmental chemicals (odorants) using a large repertoire of olfactory receptors (ORs) that are expressed by olfactory sensory neurons (OSNs) in the main olfactory epithelium. Each OSN expresses a single allele of one OR gene—so-called “singular” expression [1-4]. OSNs expressing the same OR project to defined glomeruli in the olfactory bulb [5]. The OR gene family is the largest in the mouse genome and comprises more than 1,000 genes, divided into two phylogenetic groups, class I and class II ORs [6-9]. Class II ORs are found across vertebrates, and are greatly expanded in amphibians and mammals [8]. Mammalian class I ORs are distinct from, but closely related to, class I OR genes found in fish and may be specialized to detect water-soluble/polar odorants [8, 9]. In mammals, carboxylic acids preferentially activate the dorsomedial olfactory bulb [10-16], a region that corresponds to the projections of class I ORs [17-20]. Class I ORs tend to respond to acids [21-23]. However, there is little information regarding how class I ORs affect the perception of specific chemical classes.
One approach to assess how class I ORs contribute to olfactory perception is via gene deletion. However, deleting >100 intact class I ORs is challenging because the extensive ~3 Mb class I gene cluster is interspersed with other non-olfactory genes, including the β-globin cluster [20, 24, 25]. An alternate approach would be to identify and inactivate enhancer elements that are essential for class I OR gene expression.
OR expression relies on cis-acting enhancers located near and within OR gene clusters [2]. Genetic deletion of several such enhancer elements (named H, P and Lipsi) reduces expression of neighboring class II OR genes [26-30], in some cases ~200 kb away [26]. All three enhancer regions share two features—they are highly conserved across vertebrates and are enriched with a specific homeodomain binding motif that is known to increase the probability of OR gene choice [31-33].
To study class I OR function, we identified and genetically deleted putative enhancer elements within the class I OR gene cluster in mice. Deleting these elements significantly reduced the expression of neighboring class I OR genes, with one of the elements (element A) having the largest impact. Element A is a subregion of a recently identified class I enhancer (the J element) [34] and thus further defines the critical core for this regulatory element. Deleting element A significantly reduced the number of OSNs expressing about half of the class I OR gene family, resulting in a marked reduction in the number of class I glomeruli in the olfactory bulb. Using functional imaging and behavior, we show that the reduction in class I OSNs/glomeruli results in a selective loss of glomerular responses, and behavioral aversion, to carboxylic acids—compounds that are associated with microbial growth and body odors [12, 35, 36]. Together, our results demonstrate that deletion of an OR regulatory element can impact odor perception, and that class I ORs play a critical role in encoding information about carboxylic acids.
Results
Identification of candidate enhancers in the class I OR cluster
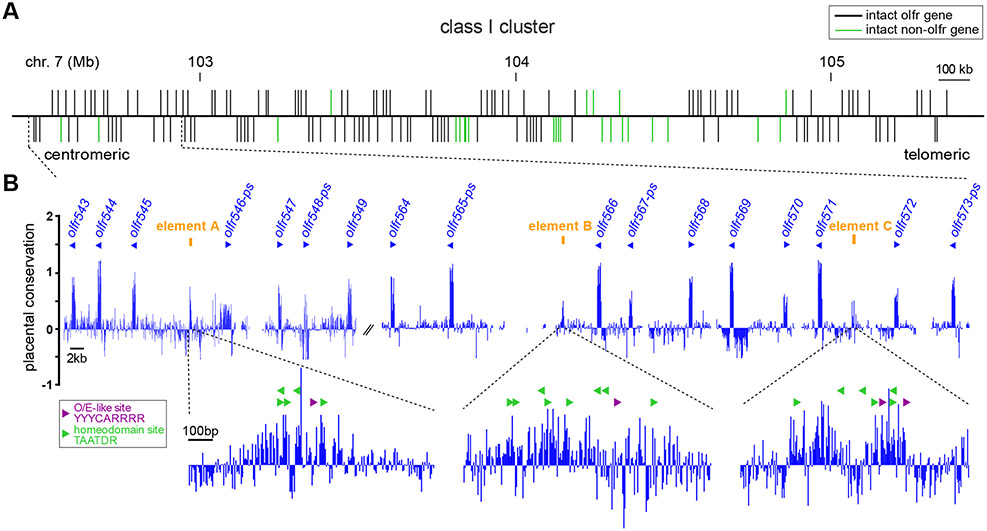
To identify candidate class I OR enhancer elements, we searched the mouse class I OR cluster for intergenic regions that 1) are highly conserved across vertebrates, and 2) contain multiple copies of a homeodomain sequence that is known to promote OR gene expression [31, 33]. To search for conservation, we used a basewise sequence comparison of whole genome alignments for 40 placental mammals (phyloP, UCSC database). We found two highly conserved intergenic regions that contain multiple homeodomain sites (Figure 1). Upon further examination, these regions also contained O/E like sites that are often associated with OR gene promoters [37, 38]. We named these regions “element A” (eA) and “element B” (eB). Both elements are located within the most centromeric 500 kb of the class I cluster, with the closest intact OR genes being olfr545 and olfr566, respectively. In addition, we found a region that is annotated as a possible transcription start site (TSS) of olfr571, but is more conserved than is typically seen for OR promoters. Thus, we also included this region, “element C” (eC) into our analysis.
Figure 1. Identification of class I OR gene regulatory elements by conservation and DNA motif analysis.
A) Organization of the class I OR gene cluster. Intact olfr genes are depicted as black vertical lines. Intact non-olfr genes are shown in green. Genes marked above the horizontal line are transcribed from centromeric to telomeric direction, genes marked below the line are transcribed telomeric to centromeric. B) Evolutionary conservation of the class I OR gene cluster measured by basewise comparison for 40 placental mammals. Positive values indicate conserved DNA regions, negative values indicate regions with acceleration (faster evolution than expected under neutral drift). Transcriptional direction of olfr genes (blue triangles) is indicated above the track. Potential enhancer elements A-C (orange) are defined as highly conserved intergenic regions. Magnified regions show the location of known OR regulatory motifs, O/E-like sites (purple) and homeodomain sites (green), within the potential enhancer elements.
eA is necessary for normal class I OR gene expression
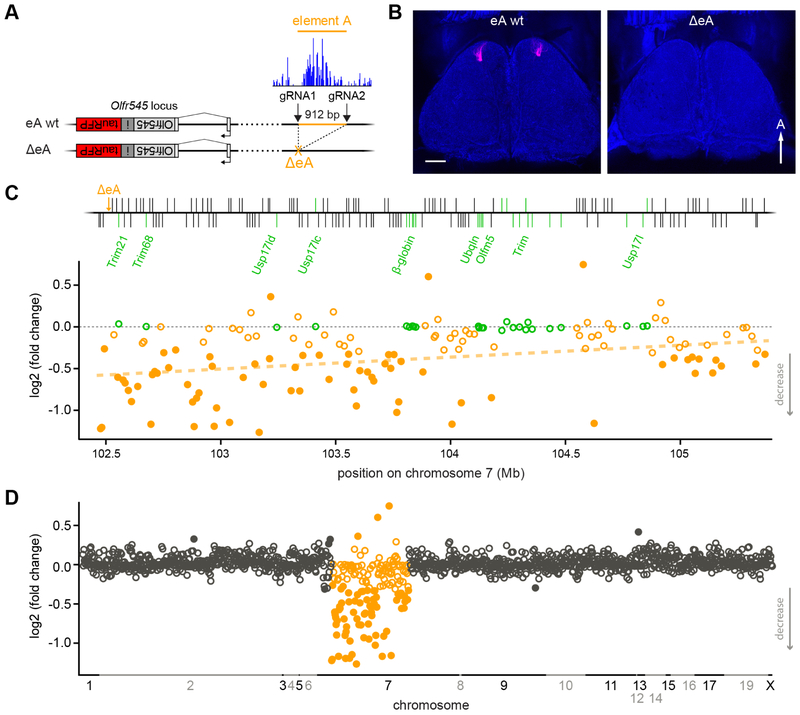
To test if the elements function as enhancers, we deleted each individually using CRISPR/Cas9 in mouse zygotes. We performed gene editing in mice that carry a targeted knock-in of RFP into the olfr545 (S50) OR gene, S50-IRES-tauRFP (Figure 2A, S1A,B), allowing us to visually monitor expression of olfr545, the closest OR gene to eA. In olfr545-rfp mice, RFP-tagged axons converged to glomeruli in the anteromedial portion of the olfactory bulb (Figure 2B, S1A). In olfr545-rfp mice with the deletion of eC (ΔeC) or eB (ΔeB), olfr545 glomeruli were readily visible using confocal fluorescence microscopy (Figure S2). In mice with the deletion of eA (ΔeA), RFP-tagged olfr545 glomeruli were not visible in the olfactory bulb, indicating that deletion of eA had significantly reduced expression of olfr545 (Figure 2B, S1B). In the olfactory epithelium of ΔeA mice, virtually no olfr545-rfp-expressing OSNs were detected (Figure S1): eA wild-type (wt) 298.7 ± 10.7, ΔeA 3 ± 1.9 (mean ± SEM, p = 1 × 10−5, Student’s t-test).
Figure 2: Class I OR expression is reduced in ΔeA mice.
A) Diagram indicating the 912 bp deletion of the conserved eA region (blue) obtained in cis with the olfr545-rfp allele. Location of CRISPR gRNAs is shown. B) Confocal images of the dorsal olfactory bulbs in eA wt (left) and homozygous ΔeA mice (right). Glomeruli (red) formed by olfr545-rfp labeled axons are absent in ΔeA mice. C) Differential expression of genes in the class I cluster in eA wt and ΔeA homozygous mice measured by RNAseq. Top, schematic of the class I cluster. Intact OR genes (black) and non-OR genes (green) are shown. Bottom, log2 fold change in expression for each intact class I OR gene (orange) and non-OR genes (green) vs location on chromosome 7. Filled circles indicate genes with a significant change in expression. n = 6 mice for each genotype. Least-squares fit for OR expression is indicated (orange dashed line). Pearson correlation r = 0.35, p = 3 × 10−5, y = 2×10−7x - 0.5842. D) Differential expression of class I (orange) and class II (black) OR genes in eA wt and homozygous ΔeA mice. The x-axis indicates the location of OR genes on each chromosome. Filled circles depict genes with a significant change in mRNA expression. n = 6 mice for each genotype. Scale bar = 500 μm. A= anterior. See also Figures S1-S4.
Next, we quantified expression of all OR genes using RNA sequencing (RNAseq) from olfactory mucosa of ΔeA, ΔeB and ΔeC homozygous mice and their respective wild-type littermates. For ΔeC, six class I OR gene showed significantly reduced expression (Figure S2F), five of which could be attributed to strain effects (see Methods). Thus, eC may act as an enhancer/promoter for olfr571, which is immediately downstream of eC. In ΔeB mice, we also observed reduced expression of a single class I OR, olfr577 (Figure S2C), which is distant from this element (10 intact genes away). Therefore, eB may represent a distant enhancer for olfr577 (see Discussion).
In contrast to the other elements, deletion of eA had a robust effect on expression of class I OR genes over a large portion of the cluster. Homozygous ΔeA mice exhibited significantly reduced expression of 70 out of 131 intact class I OR genes (Figure 2C; adjusted p <0.05; Wald test). Across the downregulated genes, expression levels were reduced to 30.7 ± 18.1 % (mean ± SD) of control levels. Of these, expression was completely abolished (normalized reads of < 10) for 18 class I OR genes. The effect was indistinguishable for genes transcribed from the positive or negative strand: positive strand = 31 downregulated with expression reduced to 27.7 ± 17.5 %; negative strand = 39 downregulated with expression reduced to 33.2 ± 18.2 % (mean ± SD) of control mRNA levels (p = 0.72, Mann-Whitney U test). Notably, mRNA expression of olfr545 (the closest OR) were reduced to 3 ± 2 % (mean ± SEM) of expression levels in control mice (Figure S1E; adjusted p < 0.001; Wald test). Importantly, this matched the reduction in the number of olfr545-rfp expressing OSNs in the epithelium of ΔeA mice (Figure S1D, 1.0 ± 0.5 %, mean ± SEM, p = 1 × 10−5, Student’s t-test), indicating reduced mRNA levels reflect loss of OSNs (see below).
Two ventrally-expressed class I OR genes [20], olfr556 and olfr686, were not significantly changed. We note that three genes (olfr653, olfr65, olfr594) showed increased expression (p < 0.05, Wald test) with a mean increase of 182.3 ± 26.8 % (mean ± SD) of control mRNA levels (Figure 2C).
Interestingly, the overall effect on expression correlated with genomic distance from ΔeA (log2-fold change vs. genomic distance; Pearson correlation coefficient r = 0.35, p = 3 × 10−5, y = 2E−7x - 0.5842), with genes in proximity to eA being more affected. Overall, the most prominent effect was on the same side of the globin cluster, which divides the class I cluster in half [24]. Moreover, expression of non-OR genes in the class I cluster was unaffected (Figure 2C).
Deletion of eA had no discernable effect on OR genes outside of the class I cluster, including TAAR genes. Two class I genes that are located outside of the class I cluster, olfr503 and olfr504, were not expressed, even in wild-type mice. Two class II ORs showed reduced expression, while four showed increased expression (Figure 2D). However, based on SNP analysis, it is likely that these are attributable to strain differences between B6 and 129-derived alleles (from the olfr545-rfp strain) at the loci in question [39]. Taken together, the RNAseq data show that deleting eA selectively decreases the expression of a large fraction of class I ORs.
In addition to single element deletions, we also obtained a ~408 kb large deletion (ΔL) that extends from eA to eC (removing all three elements) on the background of the olfr545-rfp allele (Figure S3A). The deleted interval includes 22 intact class I ORs from olfr547 to olfr571 and 2 non-receptor genes (trim21 and trim68). In homozygous ΔL mice, RFP-tagged, olfr545 glomeruli were not visible in the olfactory bulb (Figure S3B). RNAseq analysis revealed that expression of 77 intact class I ORs was significantly reduced in ΔL mice (Figure S3 C-E; adjusted p < 0.05; Wald test), 12 of which were unchanged in ΔeA mice (Figure S3F). This suggests that removal of all three elements (and perhaps other unidentified sequences) might lead to combinatorial effects that affect additional class I OR genes.
eA regulates the probability of class I OR gene choice
The decrease in class I OR expression observed in ΔeA mice could be caused by a reduction in OR transcripts per OSNs, as well as a reduction in the number of OSNs that express a given OR. To examine effects on OR gene choice probability, we performed in situ hybridizations (ISH) for six different ORs and counted the number of labeled cells in ΔeA mice and control littermates (Figure S4 A, B). For all tested genes, the proportional change in the number of cells detected by ISH closely matches the proportional change in mRNA expression levels detected by RNAseq (Figure S4 C, % change mRNA expression versus % change in cell number; Pearson correlation coefficient r = 0.99, p = 3 × 10−4). In addition, all class I expressing OSNs were found in the dorsal epithelium, demonstrating no shift in zonal expression in ΔeA mice (not shown). Thus, the change in OR expression observed by RNAseq can be attributed primarily to changes in the number of OSNs that choose to express a given OR.
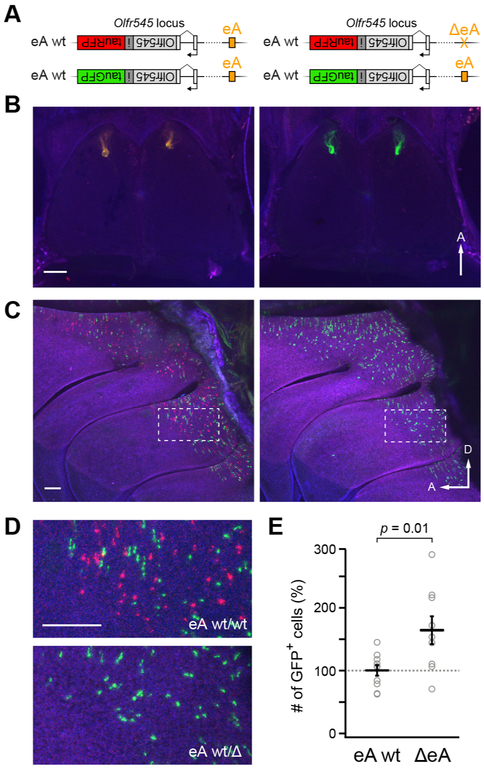
eA acts in cis
Class II OR enhancers act in cis [26-29], but have been hypothesized to interact across chromosomes in trans [30, 40]. Given that expression of the olfr545-rfp allele was abolished in heterozygous ΔeA mice (Figure S1), eA appears to act in cis. To examine whether eA also has an effect in trans, we generated mice with an olfr545-gfp (S50-IRES-tauGFP) allele [18] on one chromosome, and an olfr545-rfp allele, with or without eA, on the other (Figure 3A). We then compared the number of GFP expressing OSNs in mice that have eA in trans of the GFP allele (olfr545-gfp;eA/olfr545-rfp;eA) and those that lack eA in trans (olfr545-gfp;eA/olfr545-rfp;ΔeA). If ΔeA reduces expression of class I genes in trans, we would expect to see a decrease in the number of GFP-expressing OSNs.
Figure 3: eA acts on olfr545 in cis.
A) Genotype of compound heterozygous olfr545-gfp / olfr545-rfp mice in which the RFP allele is in cis with wild-type eA (left) or ΔeA (right). B) Confocal images of the dorsal olfactory bulbs of eA wt mice (left) showing RFP- and GFP-labeled axons converging to anteromedial glomeruli. In ΔeA mice (right), RFP-labeled axons are not seen. GFP-labeled axons converge to anteromedial glomeruli. C) Wholemounts of the olfactory epithelium in eA wt mice (left) showing a punctuate pattern of intermingled GFP- and RFP-expressing OSNs. In ΔeA mice (right), only GFP-expressing OSNs are seen, and their number is higher than in eA wt mice. D) Higher magnification view of epithelium from the region indicated in panel D (dotted lines). E) Number of olfr545-gfp-expressing OSNs is higher when eA is deleted in trans. Counts of GFP-positive neurons were normalized to a control gene olfr160 (M72), and expressed as a percentage of eA wt mice. Plot shows mean ± SEM. Student’s t-test. n = 5 mice for each genotype. Scale bars = 500 μm. A=anterior, D=dorsal.
Contrary to this prediction, the number of GFP-expressing OSNs was actually higher in mice lacking eA in trans of the GFP-tagged allele (Figure 3 B-D). To quantify this difference, we counted the number of GFP-positive olfr545-expressing OSNs relative to a control gene olfr160 (M72)—a dorsally-expressed class II OR gene that is not affected by the eA deletion (see Methods). We observed that the proportion of olfr545-gfp-expressing OSNs was significantly increased in mice lacking eA in trans : eA wt = 100 % ± 8 %; ΔeA = 164 % ± 22 % (mean ± SEM; n = 5, p = 0.01, Student’s t-test) (Figure 3E). This increase in expression of the trans allele is unlikely a direct effect of eA, but rather a consequence of class-specific gene choice restriction, as has been previously reported [18, 41](see Discussion). As expected, expression of olfr545-rfp was abolished by the deletion of eA (Figure 3B-D; Figure S1), supporting the idea that eA acts exclusively in cis.
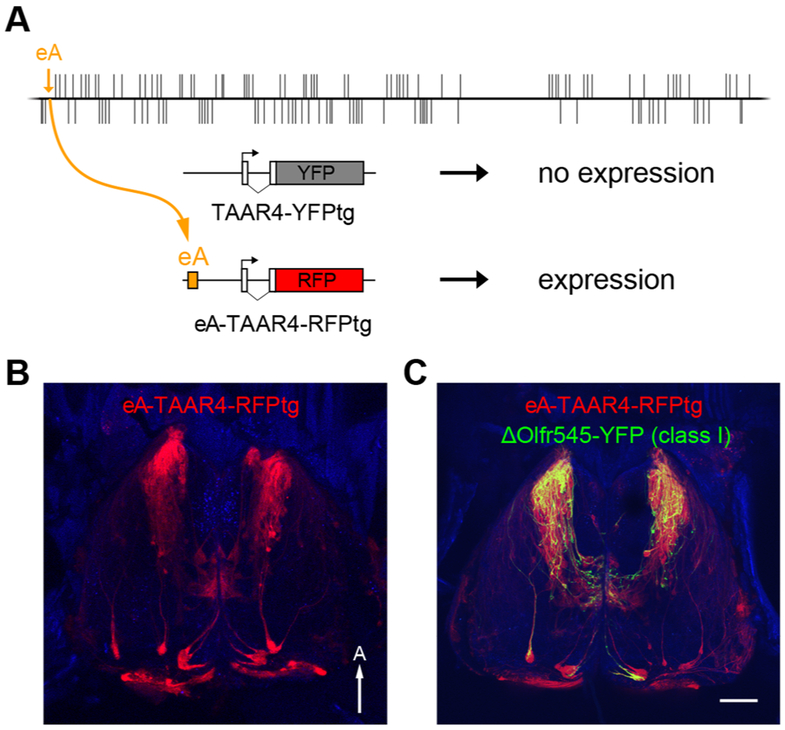
eA is sufficient to drive OR gene choice
Class II olfactory enhancers permit or increase OR gene choice [18, 26, 29]. To examine whether eA is sufficient to drive choice, we asked whether this element could rescue expression of an enhancer-less olfactory transgene. We generated a transgene (TAAR4-YFPtg) consisting of a fragment of the taar4 locus, including 2 kb upstream of the transcription start site, in which the coding sequence is replaced with YFP (Figure 4 A). This base transgene was chosen because it lacks critical enhancers that are required for expression (unpublished data).
Figure 4: eA is sufficient to drive OR gene choice.
A) Adding eA to an enhancer-less transgene drives expression. Diagram of a taar4 transgene backbone in which the receptor coding sequence is replaced with YFP. No expression was observed (0/5 founder lines). The eA sequence was placed on the 5’ end of the transgene, which includes a red fluorescent protein (mRuby3). All founder lines (9/9) exhibited punctate expression in the epithelium. B) Confocal image of the olfactory bulb showing RFP-labeled projections to the anteromedial portion of the dorsal bulb. C) Similar view in mice that harbor the eA-transgene as well as the Δolfr545-yfp allele that labels class I OR expressing OSNs. RFP-tagged axons co-localize with YFP-labeled projections to the class I domain. Scale bar is 500 μm. A= anterior.
For the base transgene, none of the transgenic lines (0 of 5) exhibited reporter gene expression. We therefore generated a transgene (eA-TAAR4-RFPtg) with eA placed on the 5’ end, and including an RFP reporter (Figure 4A). This transgene showed robust reporter gene expression in all transgenic lines (9 of 9). Interestingly, the majority of labeled axons in the dorsal bulb project to an anteromedial domain, resembling the projections of class I ORs (Figure 4B) [17, 18]. To verify that these projections indeed correspond to the class I domain, we crossed the eA-TAAR4tg with an allele encoding a YFP insertion into the S50 locus (Δolfr545-yfp), which labels projections to the class I domain [18]. YFP-labeled axons co-localized with the transgene projections in class I domain (Figure 4C). Taken together, these data demonstrate that eA exhibits choice promoting activity and is capable of supporting gene choice in class I expressing OSNs.
Deletion of eA reduces the number of class I glomeruli
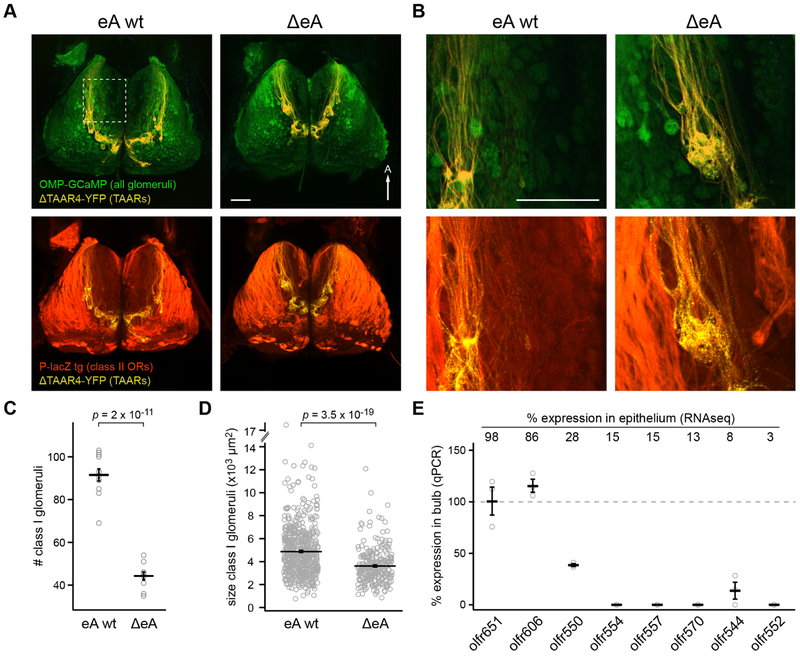
The eA deletion reduces expression of 70 intact class I OR genes. Because glomerular formation requires a minimum number of converging axons [42], even modest decreases in OR expression might result in glomerular loss. To quantify this, we counted class I glomeruli in ΔeA and control littermate mice, restricting our counts to lateral glomeruli (based on axon trajectories) to simply the analysis. To identify class I glomeruli, we used mice in which all glomeruli are labeled with a green fluorescent reporter (GCaMP3), and the TAAR and class II domains are highlighted with YFP and β-galactosidase respectively. This was done by crossing three alleles: omp-gcamp3, Δtaar4-yfp, and P-LacZ [18, 41, 43] on a homozygous ΔeA or wild-type eA background (Figure 5A,B). GCaMP+, YFP−, LacZ− class I glomeruli were visualized via confocal microscopy. ΔeA mice have ~47 fewer lateral class I glomeruli per bulb compared to eA wt littermates: eA wt = 91.5 ± 2.8 glomeruli (n = 5 bulbs); ΔeA = 44.3 ± 1.9 glomeruli (n = 6 bulbs); mean ± SEM; p = 2 × 10−11, Student’s t-test) (Figure 5C). In addition, the remaining glomeruli were significantly smaller in ΔeA mice: mean glomerular area = 4,891 ± 92 μm2 for eA wt and 3,619 ± 95 μm2 for ΔeA (mean ± SEM; n = 3 bulbs; p = 3.5 × 10−19 Mann-Whitney U test (Figure 5D). Overall, there was a corresponding reduction in the size of the class I domain in ΔeA mice: mean area = 1.15 ± 0.3 mm2 for eA wt, and 0.55 ± 0.04 mm2 for ΔeA (mean ± SEM; n = 3; p = 6.35 × 10−7, Student’s t-test). This was also evident by a pronounced anteromedial shift of the adjacent TAAR domain (Figure 5A). No differences were observed in the numbers of TAAR or class II glomeruli (Figure S5).
Figure 5: ΔeA mice have significantly fewer class I glomeruli.
A) Confocal images of the dorsal olfactory bulb of eA wt (left) and ΔeA (right) mice. Top, All glomeruli are labeled green (OMP-GCaMP3) and TAAR glomeruli are yellow (ΔTAAR4-YFP). Bottom, the same bulbs with class II glomeruli labeled in red (P-LacZ transgene). In these mice, class I glomeruli show up as green, but not labeled with YFP or LacZ. Scale bar is 500 μm. B) Higher magnification images of region indicated in panel A (dotted box). C) Number of class I glomeruli in eA wt (91.5 ± 2.8 glomeruli) and ΔeA (44.3 ± 1.9 glomeruli) mice. Student’s t-test. n = 10-12 bulbs for each genotype. D) Size of class I glomeruli expressed as mean glomerular area = 4,891 ± 92 μm2 for eA wt and 3,619 ± 95 μm2 for ΔeA. Mann-Whitney U test. n = 560 glomeruli for eA wt mice, and 263 glomeruli for ΔeA mice (6 bulbs for each genotype). E) OR expression measured by qPCR from olfactory bulbs of ΔeA mice (% of wild-type) for 8 class I ORs that are differentially affected in ΔeA mice. Percent expression of ORs in ΔeA is shown above graph. Plots show mean ± SEM. A= anterior. See also Figure S5.
To further connect reduced class I gene expression and loss of identified class I glomeruli, we used reverse transcription qPCR to assess presence or absence of genetically-defined glomeruli. Because OR mRNA is found in OSN presynaptic terminals, glomerular loss should result in OR transcript loss in the bulb. We measured bulbar expression of 8 representative class I ORs that were either unaffected, or reduced to varying degrees, in ΔeA mice (Figure 5E). All 6 of the affected ORs showed decreased bulb expression in deletion mice (n=3 bulbs per receptor per genotype; p < 0.05; Generalized Linear Model), with 4 of these ORs being undetectable. In contrast, expression of two ORs that were relatively unaffected in ΔeA mice persisted in ΔeA bulbs. These data further link the loss of class I gene expression to the loss of class I glomeruli. Taken together, these findings indicate that ΔeA creates a substantial deficit in class I input to the olfactory bulb.
Deletion of eA reduces the number of acid-responsive glomeruli
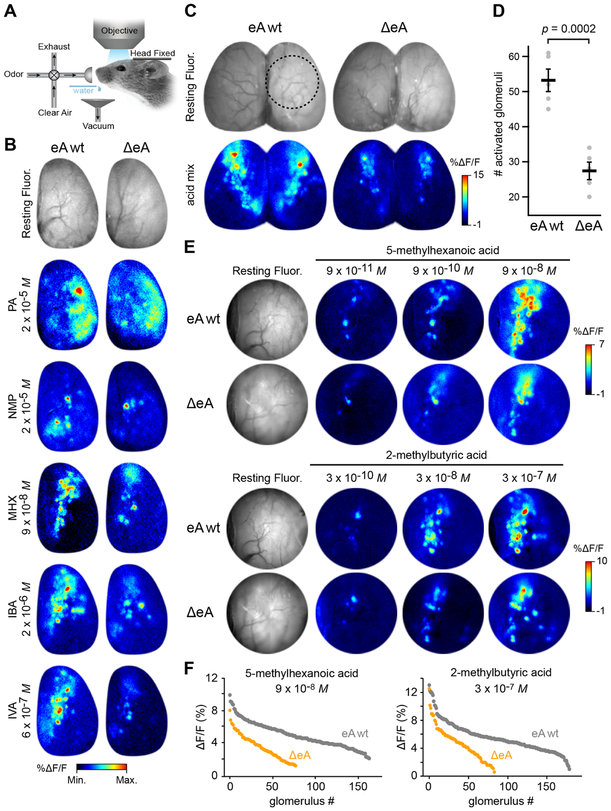
To examine functional deficits caused by ΔeA, we measured odor-evoked glomerular responses using calcium imaging in awake animals that express the genetically-encoded calcium sensor GCaMP3 in all mature OSNs, omp-gcamp3wt/+ (Figure 6A and S6). Consistent with previous findings [18, 19], we found that short chain carboxylic acids evoke responses preferentially in the anteromedial bulb (Figure 6 B,C). Moreover, we observed dramatically reduced numbers of activated glomeruli in ΔeA vs eA wt bulbs for 18 different individual carboxylic acids (Figure 6B). No difference was seen for odorants that predominantly activate class II glomeruli. Consistent with our anatomical findings, response patterns evoked by TAAR ligands (amines) were shifted anteriorly (Figure 6B). To estimate how many glomeruli were functionally missing, we used a mix of 10 carboxylic acids that activates a large number of class I glomeruli (Figure 6C). The number of responsive glomeruli in ΔeA mice was reduced by ~50% compared to eA wt mice (Figure 6D): eA wt = 53.2 ± 3.2 glomeruli vs ΔeA = 27.4 ± 2.5 glomeruli (mean ± SEM; n = 5 bulbs; p = 0.0002, Student’s t-test).
Figure 6: ΔeA mice have significantly fewer carboxylic acid-responsive glomeruli.
A) Experimental setup for in vivo calcium imaging in awake mice. B) Top, resting GCaMP fluorescence in olfactory bulb of eA wt (left) and ΔeA (right) mice. Bottom, odor-evoked GCaMP responses in eA wt (left) and ΔeA (right) mice to single odorants. Propyl acetate (PA) preferentially activates the lateral class II domain. N-methylpiperidine (NMP) selectively activates TAAR glomeruli. Note that NMP responses are shifted anteriorly in ΔeA mice. 5-methylhexanoic acid (MHX), isobutyric acid (IBA) and isovaleric acid (IVA) activate the anteromedial portion of the dorsal bulb corresponding to the class I domain. All acids activate fewer glomeruli in ΔeA mice. Pseudocolor lookup table maps %ΔF/F (bottom scale): PA 0 to 6; NMP −1 to 12; MHX −1 to 5; IBA −1 to 10; IVA −1 to 9. C) Top, resting GCaMP fluorescence in eA wt (left) and ΔeA (right) bulbs. Circle indicates region imaged at higher magnification in panel E. Bottom, Odor-evoked GCaMP responses in eA wt (left) and ΔeA (right) mice to a mix of 10 carboxylic acids. D) Number of glomeruli activated by the acids mix in eA wt (53.2 ± 3.2 glomeruli) and ΔeA (27.4 ± 2.5 glomeruli) mice. Student’s t-test. n = 5 mice for each genotype. Plot shows mean ± SEM. E) Higher magnification imaging of odor-evoked activity to increasing concentrations of MHX and 2-methylbutyric acid (MBA) in eA wt and ΔeA mice. Left, Resting GCaMP fluorescence. Right, Odor evoked responses. Midline is left. F) Distribution of response amplitudes across all responsive glomeruli in all mice tested (n=4 for each genotype) using the highest concentration of MHX (left) and MBA (right) in eA wt and ΔeA mice. See also Figure S6.
To further quantify changes in glomerular sensitivity across concentration, we compared glomerular responses in ΔeA and control mice using 14 different carboxylic acids in 10-fold concentration steps. As with the acids mix, the functional deficit became most evident at higher concentrations (Figure 6E, S6A). Both the overall number and the amplitude of glomerular responses were lower in ΔeA mice (Figure 6F).
Changes in behavioral sensitivity have been linked to loss of the highest affinity glomeruli for a particular odor [44]. Interestingly, we could not resolve differences in overall glomerular sensitivity between genotypes despite having removed ~50% of responsive glomeruli. At the lowest concentrations, fewer glomeruli responded in ΔeA vs. control mice, but the thresholds of the most sensitive glomeruli were the same (Figure 6E, S6 E,F). One possible explanation is that the highest affinity responses are mediated by one or a few broadly-tuned and sensitive ORs (glomeruli) that are spared by the deletion. This does not appear to be the case, since we observed 33 unique glomeruli responding at threshold concentrations of 8 acids in control mice, and 18 in ΔeA mice (Figure S6 C,D). Taken together, these data demonstrate that ΔeA mice have a significant and selective functional deficit in responding to carboxylic acids, but that at least some of the highest-sensitivity glomeruli persist (see Discussion).
Deletion eA mice fail to avoid carboxylic acids
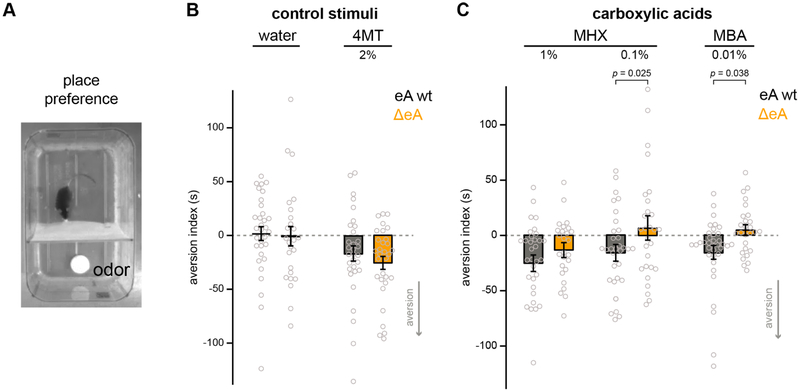
Some carboxylic acids have been shown to elicit aversion in mice [17]. To determine whether class I OR depleted mice have a deficit in behavioral responses to carboxylic acids, we tested ΔeA and eA wt mice in a two-chamber place preference assay to measure valence responses to individual odorants. In this assay, mice choose to occupy an odorized or a non-odorized compartment (Figure 7A), occupancy is quantified via video tracking, and a valence index is calculated based on time spent in each chamber with negative values indicating aversion [44].
Figure 7: ΔeA abolishes behavioral aversion to low concentrations of carboxylic acids.
A) Video image of the place-preference setup. Mice move freely between two compartments divided by a plastic curtain, one containing an odor source (white dish). B) Aversion index reflects the difference in time spent in the odorized chamber in the presence of water (pre-trial) vs odor. Negative values indicate aversion. Control odors are water and the aversive odor 4-methylthiazole (4MT). C) Aversion index values for eA wt and ΔeA mice in response to 0.01% 5-methylhexanoic acid (MHX) and 0.01% 2-methylbutyric acid (MBA), both of which elicit significantly higher aversion in wt than in ΔeA. 1% MHX elicits aversion in both wt and ΔeA. Plots show mean ± SEM.
Mice of both genotypes exhibited aversion to the previously characterized aversive odorant 4-methylthiazole (4MT) [45, 46]: Aversion Index = −16.2 ± 7.3 for eA wt; −24.4 ± 5.8 for ΔeA (mean ± SEM; n = 27-31 mice; p =0.43, generalized linear mixed model; Figure 7B). Moreover, both strains showed no response to a neutral stimulus, water: eA wt = 1.8 ± 6.5; ΔeA =−0.3 ± 8.7 (mean ± SEM; n = 26-32 mice; p = 0.95, generalized linear mixed model; Figure 7B). In contrast, low concentrations of the class I OR ligands 5-methylhexanoic acid (0.1 %, MHX) and 2-methylbutyric acid (0.01%, MBA) elicited strong aversion in eA wt mice, but not in ΔeA mice: MHX; eA wt = −16.0 ± 7.5 vs ΔeA= 6.3 ± 9.9 (n = 26-27 mice, p = 0.025) MBA; eA wt = −16 ± 6.5 vs ΔeA = 4.7 ± 4.6 (n = 25-28 mice, mean ± SEM; p = 0.038, generalized linear mixed model; Figure 7C). At higher concentrations (1%), MHX elicited aversion in both genotypes with no significant difference between strains: eA wt = −25.3 ± 7.4 vs ΔeA = −13.3 ± 5.7 (mean ± SEM; n = 24-26 mice, p =0.35, generalized linear mixed model; Figure 7C). Other parameters (distance traveled, mean speed, number of chamber crossings, and latency to cross) were examined, but were not different between wild-type and deletion mice. Together, these results show that aversion to low concentrations of specific carboxylic acids depends on class I ORs.
Discussion
Our data demonstrate that class I OR genes contribute significantly to the perception of a specific class of odorants, volatile carboxylic acids. Deleting a class I-specific enhancer element, eA, reduced expression of about half of the class I OR repertoire. We show that this manipulation markedly reduced functional class I inputs to the olfactory bulb and behavioral responses to carboxylic acids. Together, our data indicate that class I ORs may serve as primary receptors for this class of compounds.
Function of class I regulatory elements
The deletion of eA reduces expression of around half of the intact class I OR genes, having a pronounced effect on ORs in close proximity, but also effecting expression of the most telomeric class I gene, olfr692, which is located ~3 Mb downstream. Thus, eA acts over a significant fraction of the cluster. This is in contrast to previously described class II enhancer elements which act locally within ~200 kb [26-29].
In addition to the dependence on proximity, there was gene-to-gene variability in the enhancer effect. The reasons for this are not clear. Expression of some class I OR genes may be independent of long-range enhancers. If some class I OR genes are enhancer-independent, there may be sequence motifs in their proximal promoters that support independent expression. Recent work has characterized specific features of class I OR promoters [38]. Alternatively, there may be other enhancers in addition to those we deleted. The recently described J element [34] comprises our eA sequence, and the eA and J deletions affect the same subpopulation of class I ORs. Thus, eA likely defines the essential core of a single enhancer element. In addition, genome-wide searches for OR enhancers based on epigenetic signatures and transcription factor binding identified 63 potential elements [29, 40], two of which (Milos and Pontikonisi) are located within the class I cluster [40]. The functions of Milos and Pontikonisi have not been previously tested. Our ~400 kb deletion (ΔL) removed all three of our identified enhancers and both Milos and Pontikonisi. Compared with ΔeA alone, only a handful of additional class I genes was down regulated in the larger deletion. This indicates that Milos and Pontikonisi likely have a modest effect on local OR gene expression.
The elements eA, eB and eC were all chosen to contain at least one copy of a homeodomain sequence (TAATGA) that is found in identified class II enhancers (e.g. H element), and has been shown to enhance OR gene choice [31, 33]. Interestingly, the known functional enhancers eA/J, H and P all contain an extended homeodomain sequence, YTTTTAATGA [33, 34] which is known to affect OR expression [32, 33]. This sequence is absent in eB, eC and Pontikonisi, and Milos lacks the TAATGA homeodomain sequence altogether. Thus, the strong influence of eA/J on class I OR expression, as compared with the other local elements, may in part be attributable to the presence of this extended homeodomain site.
ΔeA and a class I cell type
In heterozygous ΔeA mice, expression of the S50 allele on the intact chromosome was significantly higher than in eA wt mice. This is most likely the result of a restriction in class I OR gene expression [18] rather than a direct effect of the enhancer in trans. Restricted choice has been best illustrated for the TAARs. Deletion of the TAAR cluster on one allele reduces the number of TAAR alleles by half, which doubles the number of OSNs that express any one of the remaining TAARs [41]. This is expected if a fixed subset of OSNs is restricted to choose from the TAAR cluster. This effect was not observed in heterozygous H and P deletions—expression of enhancer-dependent class II genes was the expected 50% of wild-type [27, 32]. In our model, the effect is negligible for class II elements like H and P because only a small fraction of the >900 class II OR genes is affected by each enhancer deletion. The magnitude of the effect was somewhere in between for class I ORs. In heterozygous ΔeA mice, expression of ~50% of class I ORs on one cluster (1/4 of all class I alleles) was reduced. We observed that the probability of expression of olfr545 from the intact allele increased by ~50% (rather than doubling). Thus, our data are consistent with biases in gene choice among OSNs choosing class I ORs, class II ORs, and TAARs [18, 41].
Transgene expression directed from eA labels axonal projections to the class I domain, despite the transgene backbone (transcriptional unit) being from a TAAR locus. A similar pattern of expression was imparted by transgenes driven by the full J element [34]. In contrast, class II enhancers direct expression in OSNs that preferentially project to the ventral bulb and the dorsal class II domain [18, 31, 32]. Together, these results indicate that long-range enhancers such as eA/J play a role in specifying expression in cell-type restricted populations of OSNs.
OR expression and glomerular number
The number of glomeruli lost (in the lateral projection) of ΔeA mice (~47) was considerably more than the number of genes (18) with abolished expression. Coalescence of olfactory axons into glomeruli is thought to be cooperative [42, 47]. Therefore, it is likely that glomeruli corresponding to some of the 52 downregulated ORs fail to form due to reduced OSN number. In this view, moderate down-regulation of OR gene expression can lead to complete loss of functional inputs.
The absolute numbers of total (and missing) class I glomeruli might be underestimated. Glomeruli were visualized via confocal microscopy in dorsal olfactory bulb whole-mounts, and some glomeruli were difficult to resolve through the dense axonal trajectories overlying a portion of the class I domain. We counted ~92 lateral class I glomeruli, fewer than one might predict from the total of annotated intact class I genes (~130). However, it is unknown how many of the presumed intact class I ORs encode functional receptors or are expressed at sufficient levels to form glomeruli. Our RNAseq results show that ~20 intact class I genes are expressed at low levels, indicating that the expected number of class I glomeruli may be closer to 110. In any case, our findings emphasize that functional consequences of reduced OR expression may be more severe than one might predict based solely on OR gene expression analysis.
Role of class I ORs in olfaction
We show a functional link between reduced class I OR gene expression, loss of responsive glomeruli, and behavioral deficits, when using volatile carboxylic acids as stimuli. Our data are consistent with previous findings showing that ablation of all dorsal glomeruli reduces behavioral aversion to several chemicals including acids [17]. Short chain carboxylic acids are produced by microbial carbohydrate metabolism and are components of spoiled food and body odors [12, 35, 36]. Our data indicate that class I ORs (and the class I domain) may be in part specialized to process information about carboxylic acids and odors associated with microbial action [48].
While there was a significant reduction in the number of acid-responsive glomeruli in class I depleted mice, we were unable to observe complete loss of the most sensitive glomeruli for specific acids—the number of responsive glomeruli was reduced, but response threshold of the remaining glomeruli was unchanged. Given that half of the class I glomeruli were removed, the persistence of highest affinity responses for all tested odors would seem improbable. Our imaging data rule out the possibility that there are relatively few high-sensitivity acid-responsive glomeruli. We cannot rule out the possibility that the most sensitive acid-ORs are not class I. However, it should be noted that our experiment was designed to detect at least a 10-fold difference in glomerular sensitivity, a concentration difference that is readily detectable using behavioral thresholding. Consequently, another explanation is that we simply did not have the resolution to reveal a smaller shift in glomerular sensitivity between strains.
We were able to observe a loss of behavioral aversion to carboxylic acids. It is possible that a subpopulation of class I ORs that elicits aversion to carboxylic acids via specialized downstream circuits is functionally lost in ΔeA mice [17, 46, 49, 50]. Alternatively, the decreased number of responsive glomeruli in ΔeA mice might alter valence by reducing perceived intensity of the carboxylic acids. Increasing odor concentration correlates with the number of activated glomeruli [51-53] and perceived intensity [54], while perceived intensity correlates with negative valence for some odors [55]. Overall, our data argue that class I ORs play a critical role in the representation of carboxylic acid, and suggest that the class I domain in the olfactory bulb is specialized to process information from a chemically distinct subset of odorants.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Thomas Bozza (bozza@northwestern.edu). Mouse lines generated in this study are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All mouse strains were maintained on a 14h light, 10h dark cycle and were provided ad libitum access to food and water. Animals used for awake imaging were housed in a 12:12 light/dark cycle and water restricted. All procedures involving animals were approved by the Northwestern University Institutional Animal Care and Use Committee. Experiments were performed on both male and female mice, except RNAseq data which were collected from males only. Mice were assigned randomly to experimental groups within genotypes.
METHOD DETAILS
Generation of mouse strains
olfr545-rfp (S50-IRES-tauRFP):
An Asc I cassette containing auto-excising Cre/neomycin sequences, followed by an internal ribosome entry site and the coding sequence for a bovine tau-mCherry fusion, was inserted just downstream of the OR coding sequence in a targeting vector for the olfr545 locus [18]. The targeting vector was electroporated into 129S6 ES cells, and homologous recombinant clones were identified by long-range PCR, and used to generate chimeras by aggregation with C57BL/6J morulae.
Germline animals were bred
ΔeA, ΔeB, ΔeC mice:
Two guide RNAs were designed to target a PAM sequence 5’ and 3’ of each putative enhancer element. The recognition sequence was cloned into pX458 (Addgene #48138) and the gRNA was transcribed in vitro using T7 RNA polymerase. The gRNAs and wild-type Cas9 RNA were injected into homozygous olfr545-rfp zygotes. Founders were screened by PCR and sequencing. The mutant alleles were isolated with the following deletions: 912 bp (chr7:102,509,701-102,510,612) for eA; 1,155 bp (chr7: 102,847,692-102,848,846) for eB; 1,005 bp (chr7:102,916,999-102,918,004) for eC. In addition to the single element deletions, we also isolated an allele with a large deletion (ΔL) that extends from eA to eC (408,217 bp, chr7: 102,509,797-102,918,014). All of the alleles were backcrossed for three generations on a C57BL/6J background. Homozygous deletion mice appear healthy and show normal fertility. To fix the genetic background of the class I cluster (129) for RNA sequencing and behavioral experiments, ΔeA, ΔeB and ΔL mice were crossed to olfr545-rfp mice and then intercrossed. ΔeC mice were only backcrossed on a C57BL/6J background. For in vivo imaging, we isolated an allele by random recombination in which the linked ΔeA and OMP-GCaMP3 mutations were obtained in cis on the same chromosome.
Taar4-YFP transgene:
A transgene consisting of a 7.5 kb fragment of the mouse taar4 locus, including 2.3 kb upstream from the transcription start site, was modified by replacing the coding sequence with Venus YFP.
eA-Taar4-YFP transgene:
The transgene was modified by (1) insertion of eA (chr7:102,509,833-102,510,379) placed ~2 kb upstream of the transcription start site as above, (2) replacing Venus-YFP [56] with mRuby3 [57].
The transgenes were column purified and injected into C57BL/6J zygotes to obtain transgenic founders. Experimental animals were maintained on a C57BL/6J background.
Nucleotide sequence conservation and motif analysis
The nucleotide sequence of the mouse class I OR cluster (chr7: 102,476,773-105,369,317, mm10) was compared with the class I OR cluster sequence in other species using the UCSC genome browser (https://genome.ucsc.edu). Evolutionary conservation for 40 placental mammals was measured by phyloP from the PHAST package which uses a basewise sequence comparison of whole genome alignments. Sequences were visually inspected for homeodomain sites (TAATDR) and O/E-like sites (YYYCARRRR) [31-33].
Quantitative PCR
Quantitative RT-qPCR was performed using 20-22 week old mice of both sexes. RNA was extracted from olfactory mucosa or bulb samples using RNeasy RNA isolation kit (Qiagen). Samples were treated with DNAse I to eliminate genomic DNA contamination. RNA quality was determined using the 2100 Bioanalyzer (Agilent Technologies). All samples had an RNA Integrity Number (RIN) >9. Reverse transcription was performed using Superscript III Reverse Transcriptase (Invitrogen) with 300 ng of total RNA for mucosa samples, and 2 μg of total RNA for olfactory bulb.
Samples were amplified using a Bio-Rad CFX384 Touch™ Real-Time PCR Detection System using iQ™ SYBR® Green Supermix (Bio-Rad). Cycling conditions were: 95°C for 3 minutes, followed by 40 cycles of 95°C for 15s and 59°C for 45s. Reactions were 10 μL, containing 5 μL of SYBR Green Supermix, 200-400 nM of each primer, and cDNA equivalent to 2 ng of total RNA for mucosa samples and 30 ng of total RNA for olfactory bulb samples. All reactions were run in triplicate. Melt curve analysis was performed at the end of each qPCR run to verify amplicon specificity. No-template controls were run for every primer set on every plate. Genomic DNA was not detected in no-reverse-transcription controls. Expression of OR genes was measured against gusb as a reference gene. Primer efficiencies were measured by constructing standard curves across template concentrations. All primers had measured efficiencies of 96-103%.
RNA sequencing
RNA sequencing was performed using P30 male mice that were homozygous for the ΔeA, ΔeB, ΔeC or ΔL alleles, and corresponding wild-type littermates. Mice were euthanized and the olfactory epithelium was immediately collected for RNA extraction using the RNeasy Kit (Qiagen). Isolation was performed according to the manufacturer’s protocol. RNA was treated with Qiagen’s on-column RNase-Free DNase Set to eliminate genomic DNA contamination. The quality and concentration of the RNA was determined (2100 Bioanalyzer; Agilent Technologies) prior to cDNA synthesis. Libraries were prepared with the TruSEQ RNA Library Prep Kit (Illumina), multiplexed and 100 bp PE sequencing was performed on the Illumina HiSeq 4000. RNAseq reads were aligned to the GENCODE M18 genome using the STAR aligner [58]. Differential expression was quantified using DESeq2 [59]. We note that two class I OR genes (olfr587 and olfr588) that are annotated as pseudogenes in B6 have intact open reading frames and are robustly expressed in 129 (the source of our class I cluster allele).
In Situ Hybridization
In situ hybridizations of the olfactory epithelium were performed as described previously [60], with minor modifications. The probes for class I ORs were as previously described [18, 41]. DIG-labeled probes were synthesized directly from restriction digested PCR products containing the coding sequence of the ORs. All probes were hybridized at 60°C and washed at 65°C. Fluorescence labeled OSNs were counted on representative sections from eA wt, and homozygous ΔeA mice using confocal microscopy.
Histological preparations
Olfactory epithelium sections were prepared from P30 homozygous ΔeA mice and eA wt littermates. Mice were anesthetized and transcardially perfused with 4% paraformaldehyde. The nose was dissected and postfixed at 4 °C overnight followed by 0.5 M EDTA decalcification and 30% sucrose cryoprotection (both overnight at 4 °C). OCT-embedded noses were cryosectioned at 12 μm.
For the whole-mount analysis of olfactory bulbs and epithelia, genetically-encoded fluorescent markers were visualized without fixation using a Leica SP8 or a Zeiss LSM 880 confocal microscope. Wholemount images were collected as z-stacks and displayed as flattened projections.
For class I glomerular counts, P16-21 mice that were heterozygous for OMP-GCaMP3 (to label all glomeruli), ΔTAAR4-YFP (to label the TAAR glomeruli), and hemizygous for the P-LacZ transgene (to label the dorsal class II glomeruli) were used to identify class I glomeruli. β-galactosidase expression was visualized using Fast Red Violet (FRV)/Xgal [47]. Images of GCaMP and YFP fluorescence were taken before and after staining with FRV and before and after images were overlaid using Fiji software [61]. CCaMP3+, YFP−, FRV− glomeruli were classified as class I. To avoid double-counting isofunctional medial and lateral class I glomeruli corresponding to the same OR, medial glomeruli were excluded based on orientation of axonal trajectories. For each animal, glomeruli were counted from the left and right bulbs and averaged.
For the analysis of olfr545-expressing OSN number, GFP+ (olfr545) and RFP+ (olfr160) OSNs were counted in mice that have (or do not have) eA in trans with the olfr545-gfp allele (olfr545-gfp;eA/olfr545;eA vs. olfr545-gfp;eA/olfr545-rfp;ΔeA), with both genotypes being heterozygous for olfr160-rfp mutation. Cell counts were taken from the same area of the medial turbinates from confocal images, and the ratio GFP+/RFP+ was calculated and averaged across animals.
Awake in vivo imaging
Imaging was performed in head-fixed, wheel-running mice as described previously [44], with the following modifications. Animals were 2-5 month old (male and female) homozygous ΔeA or wild-type, both heterozygous for the OMP-GCaMP3 allele. An optical window and headbar were implanted over the olfactory bulbs. After recovery, fluorescence signals were recorded using a wide-field fluorescence microscope. Response maps were obtained by subtracting a temporal average preceding the stimulus from a temporal average at the maximum response. For individual odorants, response maps from 3-4 trials with the same odor concentration were averaged to improve signal-to-noise ratio. Individual glomerular responses were measured from manual regions of interest (ROIs). Response amplitude was measured as the mean of a 2s window following odor onset, subtracted from the mean of a 6s window prior to stimulus onset (Figure S6). Traces were background subtracted from a non-responsive ROI. Responses were called when the mean signal exceeded 3x SD from baseline. All responses were visually validated in movie format to rule out baseline shifts (e.g. from movement, intrinsic signal / blood vessel artifacts etc.) that may be present in awake imaging.
Images were analyzed and processed using either Neuroplex or Turbo-SM (RedShirtImaging) and ImageJ software. All odorants were purchased from Millipore-Sigma and dilutions were made in distilled water. The odor concentration in the vapor phase was calculated using the vapor pressure for the respective odorant:
Where C is the concentration in ppm, Pvap is the vapor pressure of the odor and Patm is the atmospheric pressure in mmHg at 25°C. The concentration in ppm was then converted to molarity through division by molar volume.
The mix of 10 carboxylic acids contained isobutyric acid, pyruvic acid, isovaleric acid, butyric acid, 5-methylhexanoic acid, 2-methylpentanoic acid, propionic acid, valeric acid, hexanoic acid and acetic acid in equal dilutions. Dose response functions for 5-methylhexanoic acid and 2-methylbutyric acid were fitted to a Hill function:
where R is the response amplitude, C is odor concentration, EC50 is the concentration at half-maximal response, and n is the Hill coefficient.
Behavioral analysis
The place preference assay was performed as described previously [50] with minor modifications. Mice were 10-16 week old males and females that were either homozygous for the ΔeA allele or corresponding wild-type littermates. Briefly, each mouse was first habituated to the experimenter for two days, followed by a two-day habituation period to the experimental set up (pre-trial). During pre-trials mice underwent the same experimental procedure, but no odorants were used. On experimental days, mice were placed into a cage that was divided by a curtain into a larger non-odorized (2/3) and a smaller odorized (1/3) compartment. After a 3 min re-habituation period, mice were exposed to the water stimulus for 3 min, followed by a 3 min odorant exposure. For stimulus presentation, 20 μl of either water or diluted odorant was pipetted on a filter paper that was placed into a plastic dish. The dish was perforated to allow odorants to escape, but to avoid direct contact. Control odorants were water and the aversive odorants 4MT (2% v/v; 6 × 10−6 M saturated vapor) [45, 46]. Class I-activating odorants were 2-methylbutyric acid (0.01 % v/v; 3 × 10−9 M saturated vapor) and 5-methylhexanoic acid (1 % and 0.1% v/v; 9×10−8 M and 9×10−9 M saturated vapor). Saturated vapor estimates give an upper limit for stimulus concentrations, which are effectively lower given diffusion in the open cage. All odorants were diluted in water. Mice were video tracked using Limelight 3.0 software (Actimetrics). The aversion index was calculated as the time difference that mice spent in the odorized compartment with water as a stimulus vs. with odorant as a stimulus. Data were analyzed using a generalized linear mixed model by SPSS 25 (IBM).
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes were determined based on the number of animals required to obtain statistically significance in previous experiments. Data collection and analysis were performed blind to the conditions of the experiment. Means were compared via t-test or Mann-Whitney U test based on normality of the data (Shapiro-Wilk test). Differences in behavioral data (e.g. aversion index) were analyzed using a Generalized Linear Mixed Model (with contrasts) using genotype, odor and genotype * odor as fixed effects, trial as a random effect, and an identity link function. Differential expression in RNAseq data was estimated using the Wald test correcting for multiple comparisons using the method of Benjamini and Hochberg. Statistical tests were performed using SPSS 25 (IBM) or R (www.r-project.org). All statistical tests were two-tailed.
DATA AND CODE AVAILABILITY
RNAseq data are available in the Geo database (Geo Accession numbers pending). Other data are available from the Lead Contact upon request.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2-methylbutyric acid | Sigma #193070 | CAS 116-53-0 |
| 5-methylhexanoic acid | Bedoukian #474 | CAS 628-46-6 |
| 4-methylthiazole | Sigma #W371602 | CAS 693-95-8 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| olfr545-gfp, S50-IRES-tauGFP (Olfr545tm1Mom/MomJ) | [18] | Jax: 006712 |
| Δolfr545-venus, ΔS50-YFP (Olfr545tm4Mom/MomJ) | [18] | Jax: 006716 |
| Δtaar4-venus, ΔT4-YFP (Taar4tm3.1Tboz) | [41] | MGI:5517471 |
| olfr160-rfp, M72-IRES-tauRFP (Olfr160tm1.1Tboz/J) | [43] | Jax: 029637 |
| omp-gcamp3 (Omptm1.1Tboz/J) | [43] | Jax: 029581 |
| P-LacZ transgene | [18] | |
| ΔeA | this paper | |
| ΔeB | this paper | |
| ΔeC | this paper | |
| Taar4-YFP transgene | this paper | |
| eA-Taar4-YFP transgene | this paper | |
| olfr545-rfp (S50-IRES-tauRFP) | this paper | |
| Oligonucleotides | ||
| Olfr651 fwd, TGTGAACCAAGAGAGTCCAG | this paper | |
| Olfr651 rev, CTGGGTTGTAACAGCTTAGG | this paper | |
| Olfr606 fwd, CTGCTAACAGGAGACAAAGAG | this paper | |
| Olfr606 rev, GAGTCCACTGTTACTCTCTAG | this paper | |
| Olfr550 fwd, TTACGAACACAGAAACAGATGAG | this paper | |
| Olfr550 rev, CCAGGAGTGCTGGTATTGC | this paper | |
| Olfr554 fwd, CTTCTAGCTGATGAGGATGAG | this paper | |
| Olfr554 rev, AGAGCTGGGTACTGAACATAC | this paper | |
| Olfr557 fwd, GAGAAGTCTGTCATCAAGCAG | this paper | |
| Olfr557 rev, CCAAACTTCCATTTGGGGTG | this paper | |
| Olfr570 fwd, TTGTGTTGAAAGCCAGGTCC | this paper | |
| Olfr570 rev, CTCGGAGGTATTGAGAATCATC | this paper | |
| Olfr544 fwd, ACTTGACCAGTCTTTACTTCC | this paper | |
| Olfr544 rev, TGGGTGTAGACATTAACTACAG | this paper | |
| Olfr552 fwd, CAGCCTTCCCAGTGACATTG | this paper | |
| Olfr552 rev, CTATCAGCAGAGTCAGTGTTC | this paper | |
| Gusb fwd, GCTGATCACCCACACCAAAG | this paper | |
| Gusb rev, CACAGATAACATCCACGTACG | this paper | |
| Recombinant DNA | ||
| Software and Algorithms | ||
| STAR Aligner | [58] | NA |
| DEseq2 | [59] | NA |
| Other | ||
Highlights.
Deletion of an enhancer reduces expression of class I odorant receptors in mice
Class I receptor depletion selectively reduces acid responses in the olfactory bulb
Loss of class I receptors and glomeruli reduces behavioral aversion to acids
Class I odorant receptors are critical for the perception of carboxylic acids
Acknowledgements
This work was supported by grants from NIH/NIDCD, R01DC013576 (TB), R03DC014565 (AD) and the Deutsche Forschungsgemeinschaft CI 222/1-1 (AC). We thank Bill Kath for help with bioinformatics, Amanda Menzie and Maddie Ratkowski for technical support, Rodrigo Pacifico for generation of early transgenic constructs, Lynn Doglio and the Transgenic and Targeted Mutagenesis Facility at Northwestern University for transgenic mouse production, Pieter Faber and The Genomics Facility at University of Chicago for high-throughput sequencing, and Paul Feinstein for input and comments on the manuscript.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chess A, Simon I, Cedar H, and Axel R (1994). Allelic inactivation regulates olfactory receptor gene expression. Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- 2.Serizawa S, Miyamichi K, and Sakano H (2004). One neuron-one receptor rule in the mouse olfactory system. Trends Genet 20, 648–653. [DOI] [PubMed] [Google Scholar]
- 3.Mombaerts P (2004). Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol 14, 31–36. [DOI] [PubMed] [Google Scholar]
- 4.Nagai MH, Armelin-Correa LM, and Malnic B (2016). Monogenic and Monoallelic Expression of Odorant Receptors. Mol Pharmacol 90, 633–639. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P (2006). Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol 22, 713–737. [DOI] [PubMed] [Google Scholar]
- 6.Glusman G, Yanai I, Rubin I, and Lancet D (2001). The complete human olfactory subgenome. Genome Res 11, 685–702. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, and Firestein S (2002). The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5, 124–133. [DOI] [PubMed] [Google Scholar]
- 8.Niimura Y, and Nei M (2005). Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A 102, 6039–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freitag J, Ludwig G, Andreini I, Rossler P, and Breer H (1998). Olfactory receptors in aquatic and terrestrial vertebrates. J Comp Physiol A 183, 635–650. [DOI] [PubMed] [Google Scholar]
- 10.Imamura K, Mataga N, and Mori K (1992). Coding of odor molecules by mitral/tufted cells in rabbit olfactory bulb. I. Aliphatic compounds. J Neurophysiol 68, 1986–2002. [DOI] [PubMed] [Google Scholar]
- 11.Uchida N, Takahashi YK, Tanifuji M, and Mori K (2000). Odor maps in the mammalian olfactory bulb: domain organization and odorant structural features. Nat Neurosci 3, 1035–1043. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi YK, Nagayama S, and Mori K (2004). Detection and masking of spoiled food smells by odor maps in the olfactory bulb. J Neurosci 24, 8690–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson BA, Woo CC, Hingco EE, Pham KL, and Leon M (1999). Multidimensional chemotopic responses to n-aliphatic acid odorants in the rat olfactory bulb. J Comp Neurol 409, 529–548. [PubMed] [Google Scholar]
- 14.Slotnick BM, Panhuber H, Bell GA, and Laing DG (1989). Odor-induced metabolic activity in the olfactory bulb of rats trained to detect propionic acid vapor. Brain Res 500, 161–168. [DOI] [PubMed] [Google Scholar]
- 15.Bozza TC, and Kauer JS (1998). Odorant response properties of convergent olfactory receptor neurons. J Neurosci 18, 4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guthrie KM, and Gall C (2003). Anatomic mapping of neuronal odor responses in the developing rat olfactory bulb. J Comp Neurol 455, 56–71. [DOI] [PubMed] [Google Scholar]
- 17.Kobayakawa K, Kobayakawa R, Matsumoto H, Oka Y, Imai T, Ikawa M, Okabe M, Ikeda T, Itohara S, Kikusui T, et al. (2007). Innate versus learned odour processing in the mouse olfactory bulb. Nature 450, 503–508. [DOI] [PubMed] [Google Scholar]
- 18.Bozza T, Vassalli A, Fuss S, Zhang JJ, Weiland B, Pacifico R, Feinstein P, and Mombaerts P (2009). Mapping of class I and class II odorant receptors to glomerular domains by two distinct types of olfactory sensory neurons in the mouse. Neuron 61, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto H, Kobayakawa K, Kobayakawa R, Tashiro T, Mori K, Sakano H, and Mori K (2010). Spatial arrangement of glomerular molecular-feature clusters in the odorant-receptor class domains of the mouse olfactory bulb. J Neurophysiol 103, 3490–3500. [DOI] [PubMed] [Google Scholar]
- 20.Tsuboi A, Miyazaki T, Imai T, and Sakano H (2006). Olfactory sensory neurons expressing class I odorant receptors converge their axons on an antero-dorsal domain of the olfactory bulb in the mouse. Eur J Neurosci 23, 1436–1444. [DOI] [PubMed] [Google Scholar]
- 21.Malnic B, Hirono J, Sato T, and Buck LB (1999). Combinatorial receptor codes for odors. Cell 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 22.Saito H, Chi Q, Zhuang H, Matsunami H, and Mainland JD (2009). Odor coding by a Mammalian receptor repertoire. Sci Signal 2, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, and Touhara K (2006). Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron 52, 857–869. [DOI] [PubMed] [Google Scholar]
- 24.Bulger M, van Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, Felsenfeld G, Axel R, and Groudine M (1999). Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci U S A 96, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciavatta DJ, Ryan TM, Farmer SC, and Townes TM (1995). Mouse model of human beta zero thalassemia: targeted deletion of the mouse beta maj- and beta min-globin genes in embryonic stem cells. 92, 9259–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, and Sakano H (2003). Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302, 2088–2094. [DOI] [PubMed] [Google Scholar]
- 27.Khan M, Vaes E, and Mombaerts P (2011). Regulation of the probability of mouse odorant receptor gene choice. Cell 147, 907–921. [DOI] [PubMed] [Google Scholar]
- 28.Fuss SH, Omura M, and Mombaerts P (2007). Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell 130, 373–384. [DOI] [PubMed] [Google Scholar]
- 29.Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, Mosley CP, Ahituv N, and Lomvardas S (2014). Enhancer interaction networks as a means for singular olfactory receptor expression. Cell 159, 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monahan K, Horta A, and Lomvardas S (2019). LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature 565, 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vassalli A, Feinstein P, and Mombaerts P (2011). Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci 46, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishizumi H, Kumasaka K, Inoue N, Nakashima A, and Sakano H (2007). Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc Natl Acad Sci U S A 104, 20067–20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Hulst C, Mina RB, Gershon Z, Jamet S, Cerullo A, Tomoiaga D, Bai L, Belluscio L, Rogers ME, Sirotin Y, et al. (2016). MouSensor: A Versatile Genetic Platform to Create Super Sniffer Mice for Studying Human Odor Coding. Cell Rep 16, 1115–1125. [DOI] [PubMed] [Google Scholar]
- 34.Iwata T, Niimura Y, Kobayashi C, Shirakawa D, Suzuki H, Enomoto T, Touhara K, Yoshihara Y, and Hirota J (2017). A long-range cis-regulatory element for class I odorant receptor genes. Nat Commun 8, 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jha SK, and Hayashi K (2015). A quick responding quartz crystal microbalance sensor array based on molecular imprinted polyacrylic acids coating for selective identification of aldehydes in body odor. Talanta 134, 105–119. [DOI] [PubMed] [Google Scholar]
- 36.Natsch A, Derrer S, Flachsmann F, and Schmid J (2006). A broad diversity of volatile carboxylic acids, released by a bacterial aminoacylase from axilla secretions, as candidate molecules for the determination of human-body odor type. Chem Biodivers 3, 1–20. [DOI] [PubMed] [Google Scholar]
- 37.Vassalli A, Rothman A, Feinstein P, Zapotocky M, and Mombaerts P (2002). Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 35, 681–696. [DOI] [PubMed] [Google Scholar]
- 38.Hoppe R, Weimer M, Beck A, Breer H, and Strotmann J (2000). Sequence analyses of the olfactory receptor gene cluster mOR37 on mouse chromosome 4. Genomics 66, 284–295. [DOI] [PubMed] [Google Scholar]
- 39.Ibarra-Soria X, Nakahara TS, Lilue J, Jiang Y, Trimmer C, Souza MA, Netto PH, Ikegami K, Murphy NR, Kusma M, et al. (2017). Variation in olfactory neuron repertoires is genetically controlled and environmentally modulated. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monahan K, Schieren I, Cheung J, Mumbey-Wafula A, Monuki ES, and Lomvardas S (2017). Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pacifico R, Dewan A, Cawley D, Guo C, and Bozza T (2012). An olfactory subsystem that mediates high-sensitivity detection of volatile amines. Cell Rep 2, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ebrahimi FA, and Chess A (2000). Olfactory neurons are interdependent in maintaining axonal projections. Curr Biol 10, 219–222. [DOI] [PubMed] [Google Scholar]
- 43.Arneodo EM, Penikis KB, Rabinowitz N, Licata A, Cichy A, Zhang J, Bozza T, and Rinberg D (2018). Stimulus dependent diversity and stereotypy in the output of an olfactory functional unit. Nat Commun 9, 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dewan A, Cichy A, Zhang J, Miguel K, Feinstein P, Rinberg D, and Bozza T (2018). Single olfactory receptors set odor detection thresholds. Nat Commun 9, 2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Root CM, Denny CA, Hen R, and Axel R (2014). The participation of cortical amygdala in innate, odour-driven behaviour. Nature 515, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito H, Nishizumi H, Suzuki S, Matsumoto H, Ieki N, Abe T, Kiyonari H, Morita M, Yokota H, Hirayama N, et al. (2017). Immobility responses are induced by photoactivation of single glomerular species responsive to fox odour TMT. Nature Communications 8, 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feinstein P, and Mombaerts P (2004). A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell 117, 817–831. [DOI] [PubMed] [Google Scholar]
- 48.Mori K, and Sakano H (2011). How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci 34, 467–499. [DOI] [PubMed] [Google Scholar]
- 49.Horio N, Murata K, Yoshikawa K, Yoshihara Y, and Touhara K (2019). Contribution of individual olfactory receptors to odor-induced attractive or aversive behavior in mice. Nat Commun 10, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dewan A, Pacifico R, Zhan R, Rinberg D, and Bozza T (2013). Non-redundant coding of aversive odours in the main olfactory pathway. Nature 497, 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wachowiak M, and Cohen LB (2001). Representation of odorants by receptor neuron input to the mouse olfactory bulb. Neuron 32, 723–735. [DOI] [PubMed] [Google Scholar]
- 52.Rubin BD, and Katz LC (1999). Optical imaging of odorant representations in the mammalian olfactory bulb. Neuron 23, 499–511. [DOI] [PubMed] [Google Scholar]
- 53.Bozza T, McGann JP, Mombaerts P, and Wachowiak M (2004). In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron 42, 9–21. [DOI] [PubMed] [Google Scholar]
- 54.Sirotin YB, Shusterman R, and Rinberg D (2015). Neural Coding of Perceived Odor Intensity. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rouby C, Pouliot S, and Bensafi M (2009). Odor hedonics and their modulators. Food Quality and Preference 20, 545–549. [Google Scholar]
- 56.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, and Miyawaki A (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20, 87–90. [DOI] [PubMed] [Google Scholar]
- 57.Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, and Chu J (2016). Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep 6, 20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishii T, Omura M, and Mombaerts P (2004). Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocytol 33, 657–669. [DOI] [PubMed] [Google Scholar]
- 61.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data are available in the Geo database (Geo Accession numbers pending). Other data are available from the Lead Contact upon request.