Abstract
Agmatine amidinohydrolase, or agmatinase, catalyzes the conversion of agmatine to putrescine and urea. This enzyme is found broadly across kingdoms of life and plays a critical role in polyamine biosynthesis and the regulation of agmatine concentrations. Here we describe the high-resolution X-ray crystal structure of the E. coli agmatinase, SPEB. The data showed a relatively high degree of pseudomerohedral twinning, was ultimately indexed in the P31 space group and led to a final model with eighteen chains, corresponding to three full hexamers in the asymmetric unit. There was a solvent content of 38.5% and refined R/Rfree values of 0.166/0.216. The protein has the conserved fold characteristic of the agmatine ureohydrolase family and displayed a high degree of structural similarity among individual protomers. Two distinct peaks of electron density were observed in the active site of most of the eighteen chains of SPEB. As the activity of this protein is known to be dependent upon manganese and the fold is similar to other dinuclear metallohydrolases, these peaks were modeled as manganese ions. The orientation of the conserved active site residues, in particular those amino acids that participate in binding the metal ions and a pair of acidic residues (D153 and E274 in SPEB) that play a role in catalysis, are similar to other agmatinase and arginase enzymes and is consistent with a hydrolytic mechanism that proceeds via a metal-activated hydroxide ion.
Introduction
Naturally occurring polyamines, linear cationic metabolites with multiple amino groups, are found in all kingdoms of life [1–3] and are essential for critical cellular processes [1–6]. Most organisms from bacteria to humans generate polyamines from ornithine, a product of the urea cycle, through the action of ornithine decarboxylase (ODC). Bacteria can also access polyamines via arginine by multiple routes (Fig 1) involving production of the key polyamine intermediate putrescine. This can occur through decarboxylation of ornithine (ODC), amidohydrolysis of N-carbamoylputrescine (NCP), or a one-step conversion of the metabolite agmatine catalyzed by agmatinase (agmatine ureohydrolase, EC 3.5.3.11). While the biosynthetic route involving agmatine is mainly utilized by bacteria, agmatinase enzymes are also present in plants as well as mammals [2, 7–9].
Fig 1. Biosynthesis of polyamines from agmatine.
Putrescine can be generated from agmatine directly by an agmatinase enzyme (SPEB in E. coli), via two steps by the sequential action of agmatine deiminase and N-carbamoylputrescine amidohydrolase, or from ornithine by ornithine decarboxylase (ODC). Note that, although charges are not shown here for clarity, these metabolites are all expected to carry a positive charge at physiological pH.
Interest in the role agmatine plays in mammals expanded markedly following the discovery that agmatine is an endogenous ligand for imidazoline and adrenergic receptors [10–13]. Subsequent research has revealed a diverse range of cellular functions [14] including a protective role in resisting cellular apoptosis from Ca2+-induced oxidative stress [15], inhibition of nitric oxide synthases [16, 17], modulation of insulin release from pancreatic cells [18], signaling [19, 20] and regulatory roles [1, 2, 21, 22], as well as an intriguing capability to induce antizyme, a negative regulator of cellular polyamine biosynthesis and cellular proliferation [23]. Agmatine is now also considered salient to a number of pathophysiological processes affecting the central nervous system [13, 24]. Cellular homeostasis of agmatine has therefore emerged as an important consideration for several avenues of research. Production of agmatine occurs by decarboxylation of arginine and is catalyzed by the activity of arginine decarboxylase [3, 25, 26]. Agmatine can be consumed by several enzymes, including agmatinase, which catalyzes the one-step conversion to putrescine (Fig 1).
Agmatinase belongs to the family of arginase enzymes [27] and exhibits the characteristic 8-stranded, parallel β-sheet sandwiched between 3 α-helices on either side. As a metallohydrolase, agmatinase is dependent on Mn2+ ions for catalytic activity [28], two of which are bound at the active site in high- and low-affinity sites to serve in activating a nucleophilic water molecule for catalysis [14, 28, 29]. One of the most widely studied bacterial agmatinases is the E. coli enzyme, SPEB, for which kinetic studies have been completed [29–32]. However, while the structure of putative agmatinase enzymes have been reported for several other bacterial species [33–35], no structure of E. coli agmatinase has yet been solved. We report here the crystal structure of agmatinase from E. coli (SPEB) in order to advance our understanding of this important enzyme and the role it plays in regulating agmatine.
Materials and methods
Chemicals and other reagents used
All commercially-sourced reagents were of the highest purity available and were used without further purification. Buffers and salts were sourced from Fisher BioReagents or Fisher Chemical (Fisher Scientific). Molecular biology reagents, including restriction enzymes, T4 ligase, and molecular weight markers were from New England Biolabs. Sparse matrix crystallization screens were from Hampton Research (Crystal Screens, Peg Ion Screens, and SaltRx Screens), Molecular Dimensions (Wizard screens), and Qiagen (JCSG screens). The vector used for expression of SPEB, pTHT, was a gift from Cynthia Kinsland of Cornell University.
Gene synthesis and sub-cloning of SPEB
The E. coli agmatinase amino acid sequence (UniProt P60651) was first codon optimized for expression in an E. coli expression system using the GenSmart codon optimization tool (GenScript) (S1 Fig in S1 File). The gene was synthesized (GenScript) with a 5′ NdeI restriction endonuclease site and a stop codon followed by a XhoI restriction endonuclease site at the 3′ terminus, and delivered in a pUC57 vector (SPEB-pUC57). The gene was subcloned into the pTHT vector, a variant of pET-28 with a tobacco etch virus (TEV) protease site in place of the thrombin site. Briefly, SPEB-pUC57 and pTHT were separately digested with NdeI and XhoI (NEB). The digested gene and plasmid were separated on an agarose gel, gel purified, and then ligated using T4 DNA ligase (NEB). The ligated product was used to transform 5-alpha E. coli cells (NEB) and plated on LB agar supplemented with 50 μg/mL kanamycin. Selected colonies were cultured and the resulting plasmids (SPEB-THT) isolated for sequence verification by Sanger sequencing.
Expression and purification of SPEB
Sequence verified SPEB-THT (S2 Fig in S1 File) was used to transform BL21(DE3) E. coli cells (NEB) and plated on LB agar supplemented with 50 μg/mL kanamycin. An overnight culture was prepared by inoculating 10 mL of LB media, supplemented with 50 μg/mL kanamycin, with a single colony of SPEB-THT in BL21 (DE3). The culture was incubated at 37°C, with vigorous shaking, for 16 hours. A large-scale culture, 1 L of LB media with 50 μg/mL kanamycin, was then inoculated with the 10 mL overnight culture. This culture was grown at 37°C, with vigorous shaking, until the OD600 reached 0.6. The temperature was reduced to 18°C and the culture was allowed to continue shaking for an additional 1 hour. Protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. The induced culture was further incubated, with vigorous shaking, for an additional 16 hours. The cells were then harvested by centrifugation at 6,000g at 4°C for 20 minutes. The pellet was resuspended in 35 mL of lysis buffer (50 mM sodium phosphate, pH 7.4, 300 mM sodium chloride, 10 mM imidazole and 10% glycerol) and lysed by sonication. The lysate was cleared by centrifugation at 25,000 rcf at 4°C for 60 min. The protein was purified from cleared lysate by immobilized metal affinity chromatography using Ni-NTA resin (Qiagen) followed by size exclusion chromatography on an AKTA Pure system (GE). Briefly, the protein was loaded onto a pre-equilibrated 1.0 mL Ni-NTA Superflow cartridge (Qiagen), washed successively with 40 mL of lysis buffer then 20 mL of wash buffer (lysis buffer supplemented with a final concentration of 50 mM imidazole) before being eluted with 15 mL of elution buffer (lysis buffer with a final concentration of 300 mM imidazole). The elution fractions were pooled and further purified by size exclusion chromatography on a Superdex 200 10/300 GL column (GE Life Sciences) running 30 mM Tris, pH 7.0 and 50 mM sodium chloride. Fractions containing pure (>95% by SDS PAGE, S3 Fig in S1 File) protein were pooled, exchanged into buffer containing 10 mM Tris, pH 7.4, and 30 mM NaCl, concentrated to 45 mg/mL, and flash frozen in liquid nitrogen for storage at -80°C.
Crystallization and data collection
Initial crystallization screens were conducted with commercial sparse matrix screen kits Crystal Screen I and II, Peg Ion I and II, and SaltRx I and II (Hampton Research) using hanging drop vapor diffusion. The trials were set up using 1.5 μL of protein (45 mg/mL in 10 mM Tris, pH 7.4, and 30 mM NaCl) and 1.5 μL of well solution and trays were stored at 18°C. After 4–8 days incubation time, crystals appeared in 5 separate conditions. Crystal growth conditions included: 1.0 M ammonium citrate tribasic, and 0.1 M bis-Tris propane, pH 7.0; 0.7 M sodium citrate tribasic dihydrate, and 0.1 M Bis-Tris propane, pH 7.0; 0.7 M sodium citrate tribasic dihydrate, and 0,1 M Tris, pH 8.25; 1.2 M sodium citrate tribasic dehydrate, and 0.1 M Bis-Tris propane, pH 7.0; 3.5 M sodium formate and 0.1 M Tris, pH 8.5. The resulting crystals were rod-shaped in all cases. Crystals from the latter condition were harvested and crushed for seeding using a seed bead (MiTeGen). With microseeding, diffraction-quality crystals grew in overnight in 3.5 M sodium formate and 0.1 M Tris, pH 8.5, using 24 mg/mL of SPEB protein. Crystals were flash frozen in liquid nitrogen without addition of further cryoprotectants, and stored in liquid nitrogen for data collection. Data were collected at 100 K at the North Eastern Collaborative Access team (NE-CAT) beamline, 24-ID-C, at the Advanced Photon Source, using a Pilatus 6M detector. Data collection statistics are provided in Table 1.
Table 1. Data collection and refinement statistics.
| Data Collectiona | |
| PDB ID | 7LBA |
| Beamline | NE-CAT 24-ID-C |
| Resolution range (Å) | 2.20–82.0 |
| Wavelength (Å) | 0.97918 |
| Space Group | P31 |
| Unit Cell Dimensions | |
| a, b, c (Å) | a = b = 139.81, c = 222.99 |
| α, β, γ | α = β = 90°; γ = 120° |
| Measured reflections | 1,457,187 |
| Unique reflections | 247,407 |
| Mean I/σ | 11.2 (3.4) |
| Completeness | 99.9 (100) |
| Redundancy | 5.8 (5.9) |
| Rmerge (%) | 11.3 (55.7) |
| CC1/2 | 0.99 (0.79) |
| Data Refinement | |
| Resolution Range (Å) | 2.20–82.0 (2.20–2.26) |
| Total reflections | 230,595 |
| Test set | 12,411 |
| Rwork | 17.9 |
| Rfree | 21.3 |
| No. of protein atoms | 42,227 |
| No. of metal atoms | 27 |
| No. of water atoms | 1,248 |
| RMSD from ideal | |
| Bonds (Å) | 0.002 |
| Angles (°) | 1.21 |
| Mean B factor (Å2) | 26.4 |
| Ramachandran | |
| Favored (%) | 96.07 |
| Additionally allowed (%) | 3.85 |
| Outliers (%) | 0.07 |
| Clashscoreb | 3.42 (99) |
a Numbers in parentheses correspond to values for the highest resolution shell.
b Value calculated by MolProbity–value in parentheses corresponds to percentile (100% is best) when compared to a representative set of structures of comparable resolution.
Structure solution, refinement and model building
Data were initially indexed, integrated and scaled using the automated processing pipeline RAPD at the North Eastern Collaborative Access Team (NE-CAT) facility. RAPD is a modular package of programs that automates initial data processing using XDS (to integrate and scale the data, and then uses the CCP4 [36] programs Pointless [37] and Scala [37] to analyze the data. After identifying pseudosymmetry and twinning, the data were reprocessed using Mosflm [38], Pointless [37], Aimless/Scala [37] and Ctruncate [39, 40] to index, integrate, and merge in a lower symmetry space group (see Results and Discussion for details). Molecular replacement was conducted using MolRep [41] and completed in two stages. The first stage used the default parameters with the structure of Burkholderia thailandensis hypothetical agmatinase (4DZ4) [34] as the search model and identified a partial solution containing 2 complete SPEB hexamers (12 total SPEB protomers). A round of restrained refinement was carried out using REFMAC5 [42] to generate initial maps. The second stage of molecular replacement, also with MolRep, used the fast rotation function and a phased translation function using structure factors from the masked map of the partial solution. This step enabled the placement of a third SPEB hexamer for a total of 18 protomers. The final model was built through iterative rounds of restrained refinement with intensity-based twin refinement, using REFMAC5, and manual model building using Coot [43]. Non-crystallographic symmetry (NCS) restraints were used during early rounds of refinement and removed for later rounds. Water molecules were added once the refinement converged. Data collection and refinement statistics are provided in Table 1.
Structure analysis
Sequence alignments were completed using the BLAST-P suite [44] or Clustal Omega [45]. The reported sequence identity and conservation were from standard protein BLAST (BLASTP) against the Protein Data Bank proteins database. The protein structure search was completed using the Dali server [46] using the refined structure of SPEB. All known structures with >10% sequence identity, using the PDB25 subset, were reported. To calculate surface areas and free energies for the assembly, the PDBePISA server [47] was used with the refined structure of SPEB. Structural superpositions were generated using the global fit in PyMol (Schrodinger). To place the ligand in the SPEB active site, the structure of the D. radiodurans agmatinase with bound hexane-1,6-diamine (PDB code 1WOG) [33] was first superimposed on the refined SPEB structure. The agmatine ligand (PDB code AG2) was then superimposed on the 1WOG ligand as a starting point. The ligand position was then adjusted so that the two nitrogen atoms of the amidino group of agmatine were placed on the terminal nitrogen atom of the hexane-1,6-diamine ligand and a Mn-bound water atom, respectively, while the terminal amino group of agmatine was positioned over the other terminal amino group of hexane-1,6-diamine. The agmatine ligand was then regularized according to the restraints for the AG2 ligand (agmatine) from the protein data bank (PDB) definition (CIF) file. Figures were generated using PyMol v 2.3.3 (Schrodinger) and ChemDraw Prime v 16.0 (PerkinElmer). The sequence alignment was carried out using Clustal Omega [45] and the alignment figure was generated with ESPript 3 [48].
Results and discussion
Expression, purification, crystallization
The E. coli agmatine ureohydrolase (agmatinase, SPEB) is a 306 residue protein responsible for converting agmatine to putrescine. To define the structural basis for enzyme activity and to complement the existing body of published kinetic and functional characterization of E. coli agmatinase, we set out to solve the X-ray crystal structure of SPEB. The protein was expressed as an N-terminally hexahistidine-tagged construct from a codon-optimized gene. The protein expressed well in LB media, yielding 10–12 mg of purified agmatinase protein per L of media. The protein was purified in two steps using conventional immobilized metal affinity chromatography with Ni-NTA media followed by size exclusion chromatography (SEC) on a Superdex 200 10/300 GL column (GE). The elution profile from SEC suggested that SPEB was a hexamer, as expected (S3 Fig in S1 File). After purification, the protein was estimated to be greater than 90% pure by SDS-PAGE (S4 Fig in S1 File). Prior to initiating crystallization trials, the protein was exchanged into a minimal buffer and concentrated to 45 mg/mL. The initial crystallization trials were conducted by hanging drop vapor diffusion using commercially available sparse matrix screening kits. Within one week, rod-shaped SPEB crystals appeared in several conditions, all containing either sodium formate or sodium citrate. Microseeding at a lower protein concentration was used to generate diffraction-quality crystals of sufficient size for data collection. Crystals for data collection were grown in 3.5 M sodium formate and 0.1 M Tris, pH 8.5, and were used without any additional cryoprotectant added. The data collection statistics are provided in Table 1.
Structure solution
Several complete data sets were collected for SPEB crystals with diffraction visible to 2 Å or better. An inspection of the diffraction patterns revealed a relatively large dimension of the unit cell in one direction and a moderate-high level of mosaicity. The data were initially processed using the RAPD pipeline, which conducts automatic processing with XDS, Pointless and Aimless/Scala. All data sets indexed in a hexagonal space group by pointless, and a number of hexagonal space groups were analyzed. A first round of molecular replacement (MR) was run, using MolRep with the structure of the B. thailandensis hypothetical agmatinase (4DZ4) [34] as a search model, to identify the most probable space group candidate from the Laue class. A likely MR solution was found in the P61 space group (unit cell dimensions a = b = 139.6 Å, c = 222.6 Å, α = β = 90, γ = 120), where one complete hexamer of SPEB was placed. Iterative rounds of refinement and manual model building, using REFMAC5 and Coot, respectively, improved the model, however the Rfree value plateaued at approximately 30%. Analysis of the solvent content yielded a Matthews coefficient of 2.98 and solvent content of 58.8% for a single hexamer (6 chains) of SPEB. The predicted number of chains by analysis of the asymmetric unit (Matthews_Coeff, CCP4) was 8, with an expected solvent content of 45%. Inspection of the maps also revealed a large region of electron density that was unaccounted for, suggestive of the presence of additional chains in the asymmetric unit. Additional rounds of MR were attempted using the SPEB hexamer, protomer and different oligomeric variants. All attempts to place additional chains of SPEB, however, failed.
An inspection of the processed data for obvious pathologies was then conducted. A common warning sign for twinning is when the space group appears to be hexagonal, but is, in fact, a lower space group that is twinned. From twinning tests [39, 49, 50], it was determined that the SPEB dataset showed that the data were likely twinned (pseudomerohedral) with a twin fraction of 0.12–0.13 from the L-test and 0.41 from the H-test. Re-refinement of the P61 data using intensity-based twin refinement improved the R-factors but MR still failed to yield a solution for the remaining density. As the twinning analysis suggested that the Laue symmetry may be too high, the data were reprocessed in several lower space groups using iMosflm [38]. A partial solution, in the P31 space group with two complete hexamers in the asymmetric unit, was identified by MR using the partially refined SPEB structure. In the P31 space group Matthews analysis predicted 12–18 molecules per asymmetric unit with 38–59% solvent content. As with the P61 data set, attempts to fit additional chains using the conventional search approach failed. To identify the location of the additional chains of SPEB, we used an MR protocol in MolRep [41] consisting of using the fast rotation function against modified structure factors followed by a phased translation function. This approach, using the partially refined SPEB hexamer, yielded a clear solution with three complete SPEB hexamers in the asymmetric unit. Refinement of this solution using intensity-based twin refinement yielded high quality maps with clear density for all three SPEB hexamers. The final model, with 18 chains in the asymmetric unit, had a Matthews number of 2.0, 38.5% solvent content, and converged at R-work and R-free values of 17.9 and 21.3%, respectively. The data refinement statistics are given in Table 1. The electron density maps (S5 Fig in S1 File) were of sufficient quality to build 5,479 residues in the 18 chains of the model and to unambiguously place 27 of 36 metal ions and 1,248 water molecules. From prior studies of SPEB enzyme kinetics [14, 28, 29] and the analysis of our structural data, we expect that all of the metal binding sites are fully occupied by metal ions and are most likely to be occupied by manganese. Weaker density in parts of the third hexamer (MNOPQR), however, made placement of some of the metal atoms difficult. As such, we only placed metals in our model for those sites with definitive electron density (S5 Fig in S1 File).
Overall structure of SPEB
The protomer of SPEB comprises an α/β/α sandwich with an eight-stranded parallel β-sheet between two sets of three α-helices (Fig 2A). An additional α-helix runs roughly perpendicular to the plane of the sandwich structure. The strands of the central β-sheet are organized in the order β2- β1- β3- β8- β7- β4- β5- β6. This same fold is conserved within the agmatine ureohydrolase family and, more generally, within the arginase_HDAC superfamily [27]. These proteins are all metal-dependent enzymes that catalyze hydrolysis of an amide bond. A structure similarity search of the PDB25 subset using DALI [51] yielded several members of the arginase superfamily, including agmatinase, arginase and HDAC proteins (Table 2). As anticipated, SPEB showed the highest level of structural similarity to the hypothetical agmatinase from Burkholderia thailandensis (PDB 4DZ4), solved as part of a structural genomics pipeline [34], with an RMSD of 1.5 Å.
Fig 2. Overall structure of SPEB.

The protomer of SPEB (A) shows the conserved α/β/α sandwich that is characteristic of the agmatine ureohydrolase family. The presumed biologically relevant unit, a hexamer with D3 symmetry (B), was observed in the crystal structure and was consistent with SEC data (S3 Fig in S1 File) and the prediction by PDBePISA (47).
Table 2. Dali protein structure comparison results (protein matches to PDB25 with >10% identity).
| PDB ID | RMSD | % ID | Num. res. | Description |
|---|---|---|---|---|
| 4DZ4 | 1.5 | 48 | 317 | Hypothetical agmatinase |
| 1XFK | 2.4 | 21 | 324 | Formimidoylglutamase |
| 4Q3V | 2.5 | 23 | 340 | Arginase |
| 4RHI | 2.3 | 22 | 316 | Arginase |
| 4G3H | 2.8 | 17 | 310 | Arginase (ROCF) |
| 6ODC | 3.3 | 11 | 367 | Histone deacetylase 8 |
| 6HRQ | 3.4 | 10 | 419 | Histone deacetylase |
| 5EDU | 3.6 | 10 | 723 | Histone deac./maltose-binding periplasmic protein |
As expected from SEC (S3 Fig in S1 File), the crystal structure revealed a clear hexameric structure of SPEB (Fig 2B). The hexamer has D3 symmetry, with very little structural variation between protomers. The RMSD for an all-atom superposition of the SPEB protomers onto one another range from 0.162 to 0.181 Å. While eukaryotic proteins of the arginase family are typically trimeric [52], their bacterial counterparts tend to be hexameric [33, 35]. Superposition of SPEB with the hypothetic agmatinase from B. thailandensis (4DZ4), Pseudomonas aeruginosa guanidinopropionase (3NIP), and Deinococcus radiodurans agmatinase (1WOG) show a high degree of conservation in the protomer structure (Fig 3A). However, while there is a relatively conserved hexameric arrangement of the protomers, the overall conservation of structure is much worse in the hexamer with RMSD values ranging from 29.0 to 37.6 Å (Fig 3B; S6 Fig in S1 File). These large RMSD values arise due to slight differences in inter-subunit distances that lead to overall poor overlap of individual atoms when the full hexamers are aligned. Superposition of the structure of SPEB with the eukaryotic agmatinase from Schistosoma mansoni (4Q3V) shows a reasonably well-conserved fold (S7A Fig in S1 File, RMSD of 1.4 Å for the protomer) and a similar 3-fold symmetric arrangement of the chains (S7B Fig in S1 File, RMSD of 4.6 Å for the trimer), however the orientation of the protomers, with respect to one another, is different than that of SPEB.
Fig 3. Comparison of SPEB to other arginase family proteins.
The protomer of SPEB (A, green) superimposes well with the putative agmatinase from B. thailandensis (4DZ4, orange; RMSD 0.555 Å), the guanidinopropionase from P. aeruginosa (3NIP, blue; RMSD 0.793 Å), and the agmatinase from D. radiodurans (1WOG, gray; RMSD 1.13 Å). While the overall orientation of the protomers in the hexamer is similar, the structural similarity is much lower for the hexamers with an RMSD of 37.7 Å between SPEB and 4DZ4 (B, SPEB in yellow, red and blue helices for top 3 chains, bottom 3 chains shown in cyan, green and purple surface representation; 4DZ4 in green helices, see S6 Fig in S1 File for superposition with 3NIP and 1WOG).
For the structure of SPEB, as solved in the P31 space group, there were 3 complete hexamers in the asymmetric unit (S8 Fig in S1 File). As expected, the third hexamer added to the model after accounting for twinning (Chains M, N, O, P, Q and R) had the poorest fit, showed the highest B-factors, and had the largest number of real-space R-value Z-score (RSRZ) outliers. Overall, however, the three hexamers superimposed well, giving RMSD values that ranged from 0.165 to 0.238 Å, with the higher values for hexamer MNOPQR. Analysis of the SPEB assembly by PDBePISA [47] predicted a hexamer, as expected. The calculated surface area of the SPEB hexamer was 47,870 Å2 while the buried surface area was 26,250 Å2. The total calculated solvent free energy gain (ΔGint) upon formation of the hexameric assembly was -244.5 kcal/mol. Each molecule of SPEB makes surface contacts with four other protomers (Fig 2B). The average interface area, per protomer of SPEB, was 1,329.5 Å2, and the predicted average ΔGint was -21.7 kcal/mol.
Metal binding site and active site of SPEB
In the first two hexamers of SPEB that were built into the model (ABCDEF and GHIJKL), a pair of unambiguous spheres of electron density, presumed to be metal ions, were visible in each of the chains (S5 Fig in S1 File). The last hexamer (MNOPQR) also showed likely metal ions at these sites in each protomer, but the density was not as clear. A pair of manganese ions (Mn2+) is a conserved feature of this class of enzyme, and is proposed to activate a water molecule for nucleophilic attack of the substrate [14, 28, 29, 53, 54]. Superposition of SPEB with homologous agmatinase/arginase enzymes showed that the conserved Mn2+ ions in the homologues superimposed well with the observed spheres of electron density in the SPEB structure. Manganese has been shown to be essential for catalytic activity of SPEB [14, 28, 29]. As such, these regions were modeled as a dinuclear Mn2+ site in the structure of SPEB. The metal ions are octahedrally coordinated by a pair of histidine residues (H126 and H151), and four aspartate residues (D149, D153, D230, and D232) (Figs 4 and 5; S5 Fig in S1 File). Two water molecules are also clearly visible occupying the remaining coordination sites (Fig 5; S5 Fig in S1 File). The metal binding residues are a highly conserved feature of this class of enzymes, speaking to the presumed functional importance of these metals (S9 Fig in S1 File) [48].
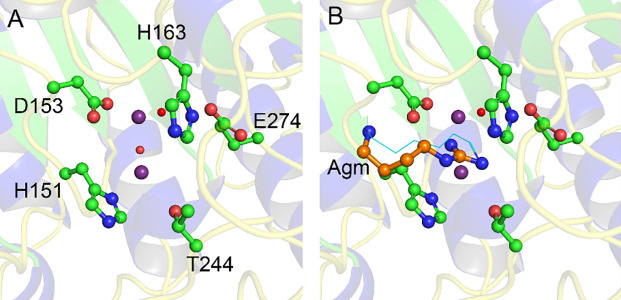
Fig 4. Active site of SPEB.
The conserved active site residues of SPEB (A, chain A) are depicted in ball and stick representation with carbon atoms colored green, oxygen atoms colored red, and nitrogen atoms colored blue. Mn2+ ions are shown as purple spheres, while two active site water molecules are included as red spheres. A putative model of agmatine binding to SPEB (B, depicted with carbon atoms colored orange and nitrogen atoms colored blue) was generated by superposition to the hexane-1,6-diamine ligand in 1WOG (B, cyan line drawing, see structure analysis in Materials and Methods). The position of agmatine illustrates that protein-ligand binding is likely governed predominantly by polar contacts with the guanidinium group. Refer to Fig 5B and 5C for details about hydrogen bonding pattern and distances in the active site.
Fig 5. Metal binding in the SPEB active site.
Two clear peaks of electron density were observed in the structure and were modeled as Mn2+ ions (A, represented as purple spheres). Also shown are well-defined water molecules that may play a role in catalysis (represented as red spheres). A schematic (B) shows the coordination of the metal ions by active site residues, and the corresponding bond distances. The relative positions, and distances, of the water molecules (red spheres) is similarly depicted in a schematic (C) of the same site that was rotated 90°, relative to B, as shown.
Since the arginase_HDAC superfamily catalyzes metal-dependent hydrolysis reactions, the position of the metal(s) in the protein marks the position of the active site. We identified the active site of SPEB using the metal positions in SPEB, and by superposition with the structure of homologous arginases. We then superimposed the ligand, agmatine, onto the hexane-1,6-diamine inhibitor present in the structure of the D. radiodurans agmatinase structure (Fig 4B) [33]. Note that the position of the ligand shown in Fig 4B is not based on experimental results, but is derived solely from superposition with the D. radiodurans structure. From this analysis, it appears that the ligand makes important polar contacts with two of the amino acid side chains that coordinate the metals (H151 and D153) as well as with conserved threonine (T244), glutamate (E274), and histidine (H163) residues (Fig 5; S8 Fig in S1 File). The H163, E274, and T244 sidechains are all proposed to interact with the guanidinium group of the substrate [31, 33].
Metallohydrolases with two metal ions catalyze a diverse range of reactions and make use of several distinct enzyme mechanisms [54]. While there is, as yet, no definitive description of the SPEB mechanism, based on the conserved active site and metal-binding residues, the mechanism of SPEB is expected to be similar to that of the structurally similar-arginase enzyme [28–33, 53, 55, 56]. In this mechanism, an activated water molecule attacks the carbon of the guanidinium group, yielding a tetrahedral intermediate [53, 56]. In the case of SPEB, the substrate would likely interact with D153, T244, and E274 predominantly through polar contacts with the guanidinium group (Fig 4B), and the substrate would not directly interact with the metal ions. The activated water, coordinated between the metal ions (Fig 5C), would attack the carbon of the guanidinium group, forming a tetrahedral intermediate. After a proton transfer to the leaving amino group, the carbonyl of urea forms while the electrons of the carbon-nitrogen bond are pushed to the terminal nitrogen of what becomes the putrescine product. Similar to the proposed arginase mechanism, an aspartate-glutamate pair (D153 and E274 in SPEB, Fig 4) play important roles in properly positioning the substrate, coordinating to the metal ions, and participating in the proton transfer step.
The agmatinase enzymes, which catalyze the hydrolysis of agmatine to putrescine, are key components of the polyamine biosynthetic pathway. These enzymes also appear to play a potential role in regulating neurotransmission in higher organisms by modulating agmatine concentrations. Because of their central importance in these processes, agmatinases and agmatinase-like proteins are emerging drug targets, both in microbes and in humans [5, 13, 24, 57–59]. Structural elucidation of the overall fold and key active site residues, as described here for SPEB from E. coli, provide important insights into substrate binding interactions and enzyme mechanism, both of which provide a solid foundation for future drug development efforts.
Supporting information
(DOCX)
Acknowledgments
Note that this work was carried out as part of a hybrid research/educational structural genomics pipeline hosted by the French lab. Using a crowdsourcing approach, the long-term goal of this program is to engage groups of undergraduate researchers to complete the structural assignment of soluble E. coli proteins. The majority of the experiments and analysis are conducted by teams of undergraduate and high school level researchers under the supervision of experienced crystallographers.
Data Availability
Data are available in the Protein Data Bank under accession number 7LBA. A validation report for this data, generated by the PDB deposition system, has been included as supplementary material with this submission.
Funding Statement
This work was supported by the National Science Foundation through award number MCB-1750637 (JBF) and by a Cottrell Scholar Award to JBF from the Research Corporation for Science Advancement (24413). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grillo MA, Colombatto S (2004) Metabolism and function in animal tissues of agmatine, a biogenic amine formed from arginine. Amino Acids 26: 3–8. 10.1007/s00726-003-0030-z [DOI] [PubMed] [Google Scholar]
- 2.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M (2015) Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J Mol Biol 427: 3389–3406. 10.1016/j.jmb.2015.06.020 [DOI] [PubMed] [Google Scholar]
- 3.Tabor CW, Tabor H (1985) Polyamines in microorganisms. Microbiol Rev 49: 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilad GM, Gilad VH (1996) Brain polyamine stress response: recurrence after repetitive stressor and inhibition by lithium. J Neurochem 67: 1992–1996. 10.1046/j.1471-4159.1996.67051992.x [DOI] [PubMed] [Google Scholar]
- 5.Gilad GM, Salame K, Rabey JM, Gilad VH (1996) Agmatine treatment is neuroprotective in rodent brain injury models. Life Sci 58: PL 41–46. 10.1016/0024-3205(95)02274-0 [DOI] [PubMed] [Google Scholar]
- 6.Tkachenko AG, Akhova AV, Shumkov MS, Nesterova LY (2012) Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res Microbiol 163: 83–91. 10.1016/j.resmic.2011.10.009 [DOI] [PubMed] [Google Scholar]
- 7.Benitez J, Garcia D, Romero N, Gonzalez A, Martinez-Oyanedel J, et al. (2018) Metabolic strategies for the degradation of the neuromodulator agmatine in mammals. Metabolism 81: 35–44. 10.1016/j.metabol.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 8.Iyer RK, Kim HK, Tsoa RW, Grody WW, Cederbaum SD (2002) Cloning and characterization of human agmatinase. Mol Genet Metab 75: 209–218. 10.1006/mgme.2001.3277 [DOI] [PubMed] [Google Scholar]
- 9.Morris SM Jr., (2003) Vertebrate agmatinases: what role do they play in agmatine catabolism? Ann N Y Acad Sci 1009: 30–33. 10.1196/annals.1304.003 [DOI] [PubMed] [Google Scholar]
- 10.Atlas D, Burstein Y (1984) Isolation and partial purification of a clonidine-displacing endogenous brain substance. Eur J Biochem 144: 287–293. 10.1111/j.1432-1033.1984.tb08462.x [DOI] [PubMed] [Google Scholar]
- 11.Atlas D, Burstein Y (1984) Isolation of an endogenous clonidine-displacing substance from rat brain. FEBS Lett 170: 387–390. 10.1016/0014-5793(84)81350-2 [DOI] [PubMed] [Google Scholar]
- 12.Li G, Regunathan S, Barrow CJ, Eshraghi J, Cooper R, et al. (1994) Agmatine: an endogenous clonidine-displacing substance in the brain. Science 263: 966–969. 10.1126/science.7906055 [DOI] [PubMed] [Google Scholar]
- 13.Laube G, Bernstein HG (2017) Agmatine: multifunctional arginine metabolite and magic bullet in clinical neuroscience? Biochem J 474: 2619–2640. 10.1042/BCJ20170007 [DOI] [PubMed] [Google Scholar]
- 14.Uribe E, Reyes MB, Martinez I, Mella K, Salas M, et al. (2020) Functional analysis of the Mn(2+) requirement in the catalysis of ureohydrolases arginase and agmatinase—a historical perspective. J Inorg Biochem 202: 110812. 10.1016/j.jinorgbio.2019.110812 [DOI] [PubMed] [Google Scholar]
- 15.Arndt MA, Battaglia V, Parisi E, Lortie MJ, Isome M, et al. (2009) The arginine metabolite agmatine protects mitochondrial function and confers resistance to cellular apoptosis. Am J Physiol Cell Physiol 296: C1411–1419. 10.1152/ajpcell.00529.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auguet M, Viossat I, Marin JG, Chabrier PE (1995) Selective inhibition of inducible nitric oxide synthase by agmatine. Jpn J Pharmacol 69: 285–287. 10.1254/jjp.69.285 [DOI] [PubMed] [Google Scholar]
- 17.Galea E, Regunathan S, Eliopoulos V, Feinstein DL, Reis DJ (1996) Inhibition of mammalian nitric oxide synthases by agmatine, an endogenous polyamine formed by decarboxylation of arginine. Biochem J 316 (Pt 1): 247–249. 10.1042/bj3160247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satriano J, Cunard R, Peterson OW, Dousa T, Gabbai FB, et al. (2008) Effects on kidney filtration rate by agmatine requires activation of ryanodine channels for nitric oxide generation. Am J Physiol Renal Physiol 294: F795–800. 10.1152/ajprenal.00392.2007 [DOI] [PubMed] [Google Scholar]
- 19.Berkels R, Taubert D, Grundemann D, Schomig E (2004) Agmatine signaling: odds and threads. Cardiovasc Drug Rev 22: 7–16. 10.1111/j.1527-3466.2004.tb00128.x [DOI] [PubMed] [Google Scholar]
- 20.Bernstein HG, Derst C, Stich C, Pruss H, Peters D, et al. (2011) The agmatine-degrading enzyme agmatinase: a key to agmatine signaling in rat and human brain? Amino Acids 40: 453–465. 10.1007/s00726-010-0657-5 [DOI] [PubMed] [Google Scholar]
- 21.Gilad GM, Wollam Y, Iaina A, Rabey JM, Chernihovsky T, et al. (1996) Metabolism of agmatine into urea but not into nitric oxide in rat brain. Neuroreport 7: 1730–1732. 10.1097/00001756-199607290-00007 [DOI] [PubMed] [Google Scholar]
- 22.Mistry SK, Burwell TJ, Chambers RM, Rudolph-Owen L, Spaltmann F, et al. (2002) Cloning of human agmatinase. An alternate path for polyamine synthesis induced in liver by hepatitis B virus. Am J Physiol Gastrointest Liver Physiol 282: G375–381. 10.1152/ajpgi.00386.2001 [DOI] [PubMed] [Google Scholar]
- 23.Satriano J, Kelly CJ, Blantz RC (1999) An emerging role for agmatine. Kidney Int 56: 1252–1253. 10.1046/j.1523-1755.1999.00697.x [DOI] [PubMed] [Google Scholar]
- 24.Barua S, Kim JY, Kim JY, Kim JH, Lee JE (2019) Therapeutic Effect of Agmatine on Neurological Disease: Focus on Ion Channels and Receptors. Neurochem Res 44: 735–750. 10.1007/s11064-018-02712-1 [DOI] [PubMed] [Google Scholar]
- 25.Buch JK, Boyle SM (1985) Biosynthetic arginine decarboxylase in Escherichia coli is synthesized as a precursor and located in the cell envelope. J Bacteriol 163: 522–527. 10.1128/JB.163.2.522-527.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris DR, Pardee AB (1966) Multiple pathways of putrescine biosynthesis in Escherichia coli. J Biol Chem 241: 3129–3135. [PubMed] [Google Scholar]
- 27.Dowling DP, Di Costanzo L, Gennadios HA, Christianson DW (2008) Evolution of the arginase fold and functional diversity. Cell Mol Life Sci 65: 2039–2055. 10.1007/s00018-008-7554-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvajal N, Lopez V, Salas M, Uribe E, Herrera P, et al. (1999) Manganese is essential for catalytic activity of Escherichia coli agmatinase. Biochem Biophys Res Commun 258: 808–811. 10.1006/bbrc.1999.0709 [DOI] [PubMed] [Google Scholar]
- 29.Salas M, Lopez V, Uribe E, Carvajal N (2004) Studies on the interaction of Escherichia coli agmatinase with manganese ions: structural and kinetic studies of the H126N and H151N variants. J Inorg Biochem 98: 1032–1036. 10.1016/j.jinorgbio.2004.02.022 [DOI] [PubMed] [Google Scholar]
- 30.Carvajal N, Olate J, Salas M, Lopez V, Cerpa J, et al. (1999) Evidence that histidine-163 is critical for catalytic activity, but not for substrate binding to Escherichia coli agmatinase. Biochem Biophys Res Commun 264: 196–200. 10.1006/bbrc.1999.1505 [DOI] [PubMed] [Google Scholar]
- 31.Carvajal N, Orellana MS, Salas M, Enriquez P, Alarcon R, et al. (2004) Kinetic studies and site-directed mutagenesis of Escherichia coli agmatinase. A role for Glu274 in binding and correct positioning of the substrate guanidinium group. Arch Biochem Biophys 430: 185–190. 10.1016/j.abb.2004.07.005 [DOI] [PubMed] [Google Scholar]
- 32.Salas M, Rodriguez R, Lopez N, Uribe E, Lopez V, et al. (2002) Insights into the reaction mechanism of Escherichia coli agmatinase by site-directed mutagenesis and molecular modelling. Eur J Biochem 269: 5522–5526. 10.1046/j.1432-1033.2002.03255.x [DOI] [PubMed] [Google Scholar]
- 33.Ahn HJ, Kim KH, Lee J, Ha JY, Lee HH, et al. (2004) Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily. J Biol Chem 279: 50505–50513. 10.1074/jbc.M409246200 [DOI] [PubMed] [Google Scholar]
- 34.Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, et al. (2013) Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS One 8: e53851. 10.1371/journal.pone.0053851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkinson CP, Grody WW, Cederbaum SD (1996) Comparative properties of arginases. Comp Biochem Physiol B Biochem Mol Biol 114: 107–132. 10.1016/0305-0491(95)02138-8 [DOI] [PubMed] [Google Scholar]
- 36.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242. 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82. 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- 38.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67: 271–281. 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans PR (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr 67: 282–292. 10.1107/S090744491003982X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.French S, Wilson K (1978) On the treatment of negative intensity observations. Acta Cryst A 34: 517–525. [Google Scholar]
- 41.Vagin A, Teplyakov A (2010) Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66: 22–25. 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography 60: 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 45.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm L, Sander C (1995) Dali: a network tool for protein structure comparison. Trends Biochem Sci 20: 478–480. 10.1016/s0968-0004(00)89105-7 [DOI] [PubMed] [Google Scholar]
- 47.Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 48.Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Britton D (1972) Estimation of twinning parameter for twins with exactly superimposed reciprocal lattices. Acta Cryst A28: 296–297. [Google Scholar]
- 50.Yeates TO (1988) Simple statistics for intensity data from twinned specimens. Acta Cryst A 44: 142–144. 10.1107/s0108767387009632 [DOI] [PubMed] [Google Scholar]
- 51.Holm L (2020) DALI and the persistence of protein shape. Protein Sci 29: 128–140. 10.1002/pro.3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanyo ZF, Scolnick LR, Ash DE, Christianson DW (1996) Structure of a unique binuclear manganese cluster in arginase. Nature 383: 554–557. 10.1038/383554a0 [DOI] [PubMed] [Google Scholar]
- 53.Christianson DW, Cox JD (1999) Catalysis by metal-activated hydroxide in zinc and manganese metalloenzymes. Annu Rev Biochem 68: 33–57. 10.1146/annurev.biochem.68.1.33 [DOI] [PubMed] [Google Scholar]
- 54.Mitic N, Miraula M, Selleck C, Hadler KS, Uribe E, et al. (2014) Catalytic mechanisms of metallohydrolases containing two metal ions. Adv Protein Chem Struct Biol 97: 49–81. 10.1016/bs.apcsb.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 55.Colleluori DM, Reczkowski RS, Emig FA, Cama E, Cox JD, et al. (2005) Probing the role of the hyper-reactive histidine residue of arginase. Arch Biochem Biophys 444: 15–26. 10.1016/j.abb.2005.09.009 [DOI] [PubMed] [Google Scholar]
- 56.Cox JD, Cama E, Colleluori DM, Pethe S, Boucher JL, et al. (2001) Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase. Biochemistry 40: 2689–2701. 10.1021/bi002318+ [DOI] [PubMed] [Google Scholar]
- 57.Kwon DH, Lu CD (2006) Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob Agents Chemother 50: 1623–1627. 10.1128/AAC.50.5.1623-1627.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uzbay TI (2012) The pharmacological importance of agmatine in the brain. Neurosci Biobehav Rev 36: 502–519. 10.1016/j.neubiorev.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 59.Shek R, Dattmore DA, Stives DP, Jackson AL, Chatfield CH, et al. (2017) Structural and Functional Basis for Targeting Campylobacter jejuni Agmatine Deiminase To Overcome Antibiotic Resistance. Biochemistry 56: 6734–6742. 10.1021/acs.biochem.7b00982 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Data are available in the Protein Data Bank under accession number 7LBA. A validation report for this data, generated by the PDB deposition system, has been included as supplementary material with this submission.