Abstract
Puromycin-sensitive aminopeptidases are found across phyla and are known to regulate the cell-cycle and play a protective role in neurodegenerative disease. PAM-1 is a puromycin-sensitive aminopeptidase important for meiotic exit and polarity establishment in the one-cell Caenorhabditis elegans embryo. Despite conservation of this aminopeptidase, little is known about its targets during development. In order to identify novel interactors, we conducted a suppressor screen and isolated four suppressing mutations in three genes that partially rescued the maternal-effect lethality of pam-1 mutants. Suppressed strains show improved embryonic viability and polarization of the anterior–posterior axis. We identified a missense mutation in wee-1.3 in one of these suppressed strains. WEE-1.3 is an inhibitory kinase that regulates maturation promoting factor. Although the missense mutation suppressed polarity phenotypes in pam-1, it does so without restoring centrosome–cortical contact or altering the cortical actomyosin cytoskeleton. To see if PAM-1 and WEE-1.3 interact in other processes, we examined oocyte maturation. Although depletion of wee-1.3 causes sterility due to precocious oocyte maturation, this effect was lessened in pam-1 worms, suggesting that PAM-1 and WEE-1.3 interact in this process. Levels of WEE-1.3 were comparable between wild-type and pam-1 strains, suggesting that WEE-1.3 is not a direct target of the aminopeptidase. Thus, we have established an interaction between PAM-1 and WEE-1.3 in multiple developmental processes and have identified suppressors that are likely to further our understanding of the role of puromycin-sensitive aminopeptidases during development.
Keywords: polarity, oocyte maturation, puromycin-sensitive aminopeptidase, suppressors, Myt1 kinase, C. elegans
Introduction
The M1 family of zinc metalloproteases is a highly conserved aminopeptidase group that regulates peptide processing, protein degradation, and the cell cycle. Members are characterized by a HEXXH[18X]E, Zinc coordination site and the GXMXN catalytic domain. Family members can be cytoplasmic or membrane associated and have been shown to cleave either a single or series of N-terminal amino acids, often from small peptides (reviewed in Peer 2011).
Within this family are the puromycin-sensitive aminopeptidases (PSAs) that are involved in cell-cycle regulation in many species. PAM-1 is a highly conserved PSA in Caenorhabditis elegans with homologs regulating fertility and meiosis in numerous organisms (Osada et al. 2001; Brooks et al. 2003; Sánchez-Morán et al. 2004). In C. elegans, PAM-1 and other metalloproteases in the family interact in the gonad to ensure fertility and fecundity (Althoff et al. 2014). Both mammalian cells and Dictylostelium require functional PSA for proper progression through the cell cycle (Constam et al. 1995; Poloz et al. 2012). In Dictylostelium, PSA is known to associate with Cdk5, and loss of Cdk5 shows similar phenotypes to inhibition of PSA (Sharma et al. 2002; Huber and O’Day 2011; Huber et al. 2013). Despite this interaction, the mechanism by which these aminopeptidases control fertility and cell-cycle progression is unknown. In addition to cell-cycle control, recent work with a human homolog, NPEPPS, suggest that the protein may play a neuroprotective role by degrading Tau and SOD1 (Yanagi et al. 2009; Kudo et al. 2011; Ren et al. 2011). Due to the many phenotypes associated with loss of these aminopeptidases, it is likely there are unexplored targets.
Caenorhabditis elegans provides a unique system for studying this aminopeptidase family and identifying interacting proteins (Brooks et al. 2003; Lyczak et al. 2006). The cytoplasmic aminopeptidase, PAM-1 is highly expressed in the early embryo and regulates timely exit from meiosis (Lyczak et al. 2006; Fortin et al. 2010). In the absence of PAM-1, defects in chromosome segregation are observed, and the timing of meiotic exit is significantly extended. In addition, PAM-1 regulates centrosome positioning at the posterior cortex of the embryo to ensure establishment of the anterior–posterior axis (Fortin et al. 2010; Saturno et al. 2017).
In wild-type, the centrosome cues polarization of the one-cell embryo, resulting in cortical flows, pseudocleavage, localization of PAR proteins to anterior and posterior domains, and an asymmetric cell division (reviewed in Rose and Gönczy 2014). In pam-1 mutants, reduced centrosome–cortical contact prevents polarity establishment resulting in reduced cortical flows and pseudocleavage. This leads to mislocalization of the cortical PAR proteins and a symmetric first cleavage and embryonic lethality (Lyczak et al. 2006; Fortin et al. 2010; Saturno et al. 2017). When centrosome contact is maintained with the posterior cortex, these polarity defects are rescued, leading to the model that PAM-1 regulates polarity through the centrosome (Fortin et al. 2010). However, nothing is known about the targets and protein interactors PAM-1 may work with to regulate these developmental processes.
To learn more about the targets and protein interactors of PAM-1, we implemented a suppressor screen as an unbiased approach to identify genetic interactors (reviewed in Hodgkin 2005). Here, we describe the isolation and initial characterization of suppressors of pam-1 (spam mutants), that increase the embryonic viability and partially rescue polarity defects in the early embryo. We have identified a mutation in the gene wee-1.3 in one of these suppressors. In order to understand how the cell-cycle regulator WEE-1.3 and the aminopeptidase PAM-1 interact, we looked at suppression phenotypes and for genetic interactions in polarity establishment and oocyte maturation. Here we report evidence that PAM-1 and WEE-1.3 work together in both of these processes.
Materials and methods
Strains and maintenance
Strains were propagated on NGM plates at 15°C as described (Brenner 1974). The N2 Bristol strain was used as wild-type and the CB4856 strain was used for mapping. The alleles used in this study were: pam-1(or347), pam-1(or403), spam-2/wee-1.3(lz5), wee-1.3(syb1738), wee-1.3(q89eb60), wee-1.3 (ana8 [wee-1.3::gfp]) (Fernando et al. 2021), unc-24(e138), spam-1(lz3), spam-1(lz4); spam-3(lz6), nmy-2(cp13[nmy-2::gfp + LoxP]). The following transgenes were used in this study: ddls6[tbg-1::GFP + unc119 (+)], axls1327 [par-1::GFP].
Creation of CRISPR strain
To recreate the mutation identified in wee-1.3(lz5), SunyBiotech Corporation was contracted to produce a CRISPR mutation that would result in the identical amino acid change in WEE-1.3. The strain constructed by SunyBiotech was designed to have the lz5 missense as well as three silent mutations necessary to perform the CRISPR edit and prevent recutting (Supplementary Figure S1). The following guide RNAs were used: Sg1: CCACCAAACGCGCAACGCCGTTT and Sg2: CCGGAGAGTCCGCCGAGAATGAA. The edits were made with a donor plasmid containing the missense mutation as well as changes to prevent recutting of the sequence were included (Supplementary Figure S1). The sequence of the new strain (PHX1738: wee-1.3(syb1738)) was confirmed by SunyBiotech Corporation as well as independently by Sanger sequencing once the strain was received using primers RL01-seq-s: TGCTTGACTCTGATCCGAGG and RL01-seq-a: TCTTCTACGTGGCGATTCCG or WEE-1F: TCTGATCCGAGGATTCGTCC and WEE-1R: GGCATTCTCGGTAGATCACG.
RNAi
Worms were fed RNA interference (RNAi) bacteria as described (Kamath and Ahringer 2003). Worms were placed at 25°C for 20 hours prior to imaging or for other times indicated in time-course experiments. Phenotypes of wee-1.3(RNAi) worms were compared with those treated with an empty RNAi vector (L4440).
For time-course experiments, worms were placed on RNAi bacteria for 24 hours at 25°C. Worms were then transferred to individual RNAi plates every 2 hours and the number of embryos laid was recorded at each transfer.
Suppressor screen
We conducted a suppressor screen to identify proteins that may interact with PAM-1 in the early embryo. Worms with a missense mutation in pam-1(or347), were mutagenized with EMS as previously described (Encalada et al. 2000). Mutagenized F2 worms were grown at a nonpermissive temperature of 25°C where nearly all embryos laid fail to hatch. Worms surviving on plates after 2 weeks were single picked and tested for embryonic viability. Worms with progeny that showed a consistently elevated viability were outcrossed for further analysis.
Embryonic viability
L4 worms were single picked onto plates at 20°C overnight. The next day the adult was removed and the embryos laid were counted. The following day, hatching progeny were counted and embryonic viability calculated (number of hatched progeny/total embryos laid).
Brood analysis
L4 worms were single plated to 20°C and transferred to fresh plates every 24 hours for 72 hours. After worms were removed from each plate, the number of embryos laid was counted and the total from 72 hours of laying were totaled. Data from 20 worms were averaged and standard deviation determined.
SNP mapping
unc-24(e138), pam-1(or347); spam worms were mated to the polymorphic Hawaiian strain CB4856. F2 worms were screened by testing for the presence of pam-1 by screening for N2 SNPs flanking the locus as well as the presence of the closely linked unc-24 marker. These worms were then tested for embryonic viability at levels comparable to the suppressed strain. Each rehomozygozed strain became a mapping line for SNP mapping. SNP mapping of lines was performed as described (Davis et al. 2005).
Sequencing
Whole-Genome Sequencing
Six mapping lines for the spam allele lz5 were created using the above methods. DNA was isolated from these lines for whole-genome sequencing (WGS) as described (O’Rourke et al. 2011; Lowry et al. 2015). Data analysis was performed using a local Galaxy installation running the CloudMap pipeline (modified from Minevich et al. 2012). Mapping regions were identified by the absence of CB4856 SNPs as determined by plotting allele frequency versus chromosomal position. Thirteen variants (10 that changed coding regions) were identified in the area. Each was tested individually for suppressing ability. Based on the combination of our SNP mapping data and WGS data, the location of the lz5 mutation was determined to be located in the gene wee-1.3. Sanger sequencing was then performed on the wee-1.3(lz5) allele (forward primer: TCTGATCCGAGGATTCGTCC; reverse primer: GGCATTCTCGGTAGATCACG) to further confirm the mutation.
Microscopy
For gonad imaging, worms were anesthetized in 0.1% tricaine/tetramisole as described (McCarter et al. 1999) and imaged on a 3% agarose pad or were immobilized on a 6% agarose pad with 2 µl of a 50% solution of 0.1 µm diameter polystyrene microspheres (Kim et al. 2013). For embryo imaging, embryos were released on a coverslip and imaged on a 3% agarose pad.
For DIC and time-lapse imaging, a Nikon DS-Fi3 microscope camera using NIS Elements imaging software version 4.60 was used and acquisition was performed as described (Lyczak et al. 2006). For confocal and time-lapse imaging, a EZ-C3 Nikon Confocal microscope and NIS Elements software was used. PAR-1, centrosome, and NMY-2 fluorescence imaging was performed as previously described (Saturno et al. 2017).
Scoring and analysis
Gonads examined with DIC microscopy were scored for the presence of nucleoli, checking through multiple focal planes. The oocytes were numbered using distance from the spermatheca, with the closest oocyte at the -1 position and the furthest oocyte counted being the -5 position.
For scoring early embryo polarity, DIC and confocal analysis were used. DIC polarity landmarks, pseudocleavage, and asymmetric cell division were scored for each time-lapse. Centrosome distance from the cortex and PAR-1 domains were measured as described (Saturno et al. 2017). Size and number of NMY-2 foci were determined using NIS-Elements EZ-C2 confocal software. Maximum projections were created from a Z-stack of five images (0.5 µm apart) of the cortex on one side of the embryo (closest to the coverslip). Foci >0.5µm2 were counted at -700 and -500 seconds prior to nuclear envelope breakdown (NEBD) and their area was determined with ROI statistics.
For WEE-1.3 levels, confocal images were taken of wee-1.3 (ana8 [wee-1.3::gfp]), and pam-1(or403); wee-1.3 (ana8 [wee-1.3::gfp]) with identical settings. A Z-stack of 5 images 0.5 µm apart were taken for analysis. To calculate WEE-1.3 levels, Z-stacks were turned into maximum intensity projections. The cytoplasm of each oocyte was traced and ROI statistics in the confocal software was used to determine the mean pixel intensity of GFP in each oocyte.
Statistical analysis
Chi Square analysis was used to test for significant differences in embryomnic viability, oocyte maturation, and polarity landmarks. In order to determine differences in WEE-1.3 GFP intensity, a weighted one-way ANOVA was performed. For NMY-2 foci size and sterility, a two-tailed, homoscedastic Student’s T Test was used. P-values were considered significant at the 0.05 alpha level.
Results
Suppressors of pam-1 improve the embryonic viability and rescue polarity phenotypes
In a screen for suppression of the maternal-effect embryonic lethality of pam-1, we identified four suppressing alleles. Each suppressor was confirmed to act recessively and improved the embryonic viability of a missense allele of pam-1(or347) to varying extents (Table 1). pam-1(or347) has a missense allele in the active site required for aminopeptidase activity. Phenotypically it is identical to our null alleles of pam-1 (Lyczak et al. 2006). Each suppressor was mapped to a broad chromosomal region using SNP mapping (Davis et al. 2005) away from pam-1’s position on LG IV. lz3, lz4, and lz6 all mapped to LG I, whereas lz5 mapped to LG II. Complementation testing was performed on suppressors that mapped to LG I. Trans-heterozygotes of lz6 with lz3 or lz4 produced less suppression than for either suppressor mutation alone, indicating that they are unique suppressors. However, the lz3 and lz4 alleles were determined to be mutations in the same gene, as evidenced by a high rescue of embryonic viability in the F1 generation (Table 2). Thus, we named the suppressors, spam-1(lz3), spam-1(lz4), spam-2(lz5), and spam-3(lz6).
Table 1.
Suppressors act recessively to rescue the embryonic lethality of pam-1 worms
| Genotype | Embryonic viability |
|---|---|
| pam-1(or347) | 7.1% |
| pam-1(or347); lz3 | 51.42% |
| pam-1(or347); lz3/+ | 0.57% |
| pam-1(or347); lz4 | 72.06% |
| pam-1(or347); lz4/+ | 5.52% |
| pam-1(or347); lz5 | 20.09% |
| pam-1(or347); lz5/+ | 7.96% |
| pam-1(or347); lz6 | 78.53% |
| pam-1(or347); lz6/+ | 8.5% |
Embryonic viability was determined at 20°C.
Table 2.
Complementation tests of suppressors mapping to LG I: three alleles of two suppressor loci
| Genotype | Embryonic viability |
|---|---|
| pam-1(or347); lz3/lz4 | 58.59% |
| pam-1(or347); lz3/+ lz6/+ | 20.38% |
| pam-1(or347); lz4/+ lz6/+ | 37.30% |
Embryonic viability of F1 progeny at 20°C. Although all alleles showed an interaction, only lz3/lz4 fully failed to complement.
Since pam-1 embryos lack early signs of polarization, we used DIC microscopy to test if these polarity defects were rescued by each suppressor. Each suppressed strain was imaged during the first cell division and signs of polarization were scored. In wild-type, pseudocleavage prior to the first asymmetric cell division is a sign of cortical polarization in the embryo. In pam-1(or347) mutants, fewer embryos undergo pseudocleavage and divide asymmetrically due to defects in polarity establishment (Lyczak et al. 2006). However, each suppressor mutation increased the prevalence of both pseudocleavage and an asymmetric division, suggesting a partial rescue of polarity establishment in suppressed embryos (Figure 1). For instance, pam-1(or347); spam-2(lz5) embryos increased the presence of pseudocleavage from 28% to 61% and the asymmetric cell division from 55% to 79% as compared with pam-1(or347) embryos alone. Similar rescue was observed for other suppressor mutations as well (Figure 1).
Figure 1.
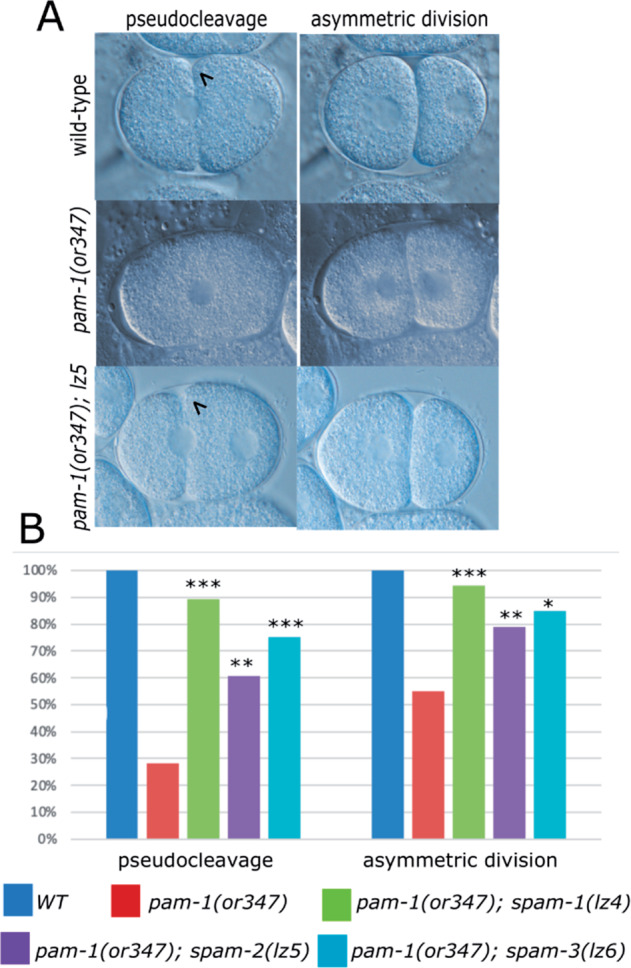
spam mutations restore polarity to pam-1 embryos (A) DIC images show that wild-type embryos exhibit pseudocleavage (arrowhead; n = 19) during axis polarization and divide asymmetrically (n = 44). Many pam-1(or347) embryos do not exhibit pseudocleavage (n = 39) or an asymmetric division (n = 79), but the presence of a suppressor mutation improves both pseudocleavage (arrowhead) (lz4 n = 28; lz5 n = 46; lz6 n = 20) and the asymmetric division (lz4 n = 54; lz5 n = 85; lz6 n = 20) in many embryos. Anterior to the left. (B) Quantification of DIC time-lapse data. Chi square analysis was used to compare pam-1 embryos to each suppressed strain. *P < 0.05; **P < 0.01; ***P < 0.001
wee-1.3(lz5) is a suppressor of pam-1
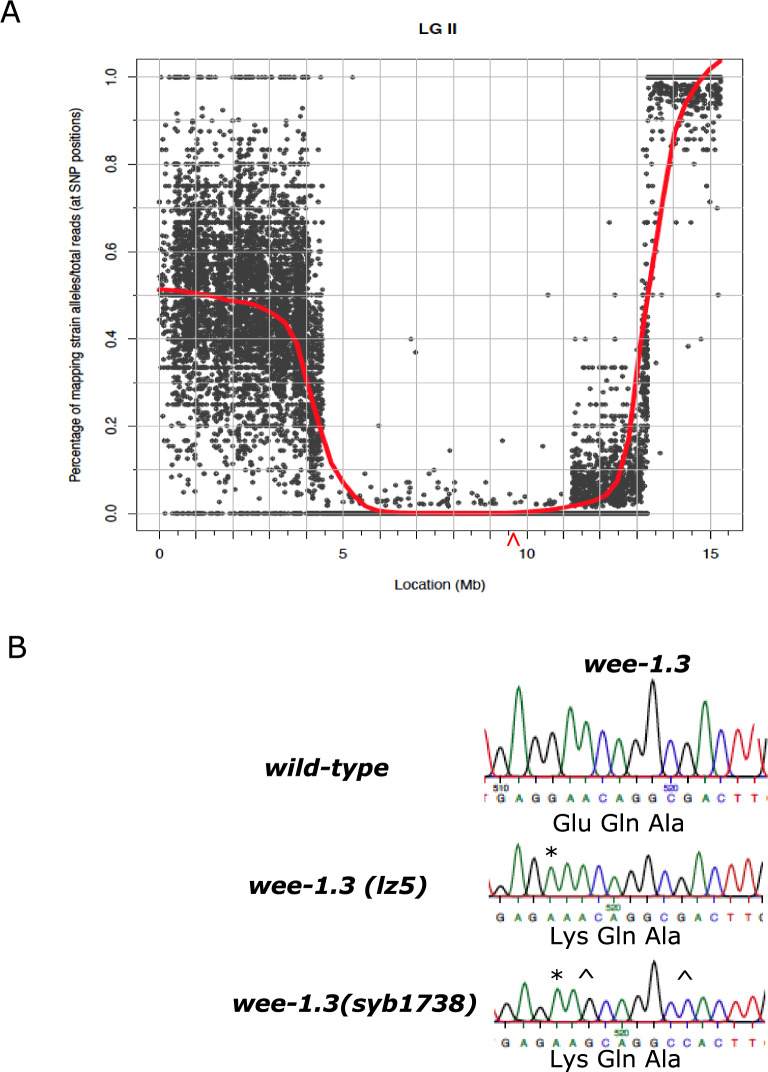
We conducted further work on one of our suppressors, spam-2(lz5). We crossed spam-2(lz5) into a nonsense and missense allele of pam-1 to determine if the suppression of pam-1 was allele specific. spam-2 improved embryonic viability of both pam-1 alleles, but to different extents (Tables 1 and 3). Suppression of the nonsense allele (or403) was also observed, increasing viability from 12% to 44%. In comparison, the missense allele (or347) showed an increased viability from 7% to 20% (Tables 1 and 3). The difference in suppression between the two pam-1 alleles was significant (P < 0.001). Using SNP mapping, we localized spam-2(lz5) to LG II. This was subsequently confirmed by WGS (Figure 2A). In this region, a G to A point mutation at position 1459 in wee-1.3 was discovered, which resulted in a E487K missense mutation (Figure 2B and Figure S1). This region of the protein is not highly conserved among different species (Lamitina and L’Hernault 2002) and has no domains known to be important for protein function. The amino acid change is more C terminal to the ATP binding, kinase, and transmembrane domains. Despite this, a link between a cell-cycle regulator and PAM-1 could provide important insights into its function. We refer to spam-2(lz5) as wee-1.3(lz5) below.
Table 3.
Mutations in wee-1.3 suppress pam-1 lethality but have no effect on their own
| Genotype | Embryonic viability at 20°C |
|---|---|
| +/+ | 94.94% |
| pam-1(or403) | 12.4% |
| pam-1(or403); wee-1.3(lz5) | 43.9%*** |
| pam-1(or403); wee-1.3(syb1738) | 43.0%*** |
| wee-1.3(lz5) | 93.13% |
| wee-1.3(q89eb60) | 5.33% |
| wee-1.3(q89eb60)/wee-1.3(lz5) | 67.49% |
| wee-1.3(syb1738) | 95.0% |
Chi square analysis was used to compare the embryonic viability of pam-1(or403) with the wee-1.3 mutations.
P < 0.001.
Figure 2.
spam-1(lz5) maps to LG II and has a missense mutation in wee-1.3. (A) Plotting the allele frequency of known Hawaiian SNPs against chromosomal position reveals an ∼6.5 Mb region on LG II that contains spam-2(lz5). Allele frequencies were obtained by WGS of mapping lines generated by crossing pam-1(or347); spam-2(lz5) worms to Hawaiian males. A missense variant was detected in the gene wee-1.3. Red line = LOESS regression trendline. The approximate position of wee-1.3 is marked by a red arrowhead. (B) Sanger sequencing was used to confirm the presence of the wee-1.3 missense mutation in our strains. The wee-1.3(lz5) allele has a G1459A(E487K) mutation (*). The CRISPR recreated wee-1.3(syb1738) strain had the identical change (*), as well as some silent mutations( ^) that were introduced to prevent recutting by Cas9; A1461G and G1467C are shown here. All sequences are from strains in the pam-1(or403) background.
To determine if the mutation in wee-1.3 affected its function, we crossed the wee-1.3(lz5) mutation away from pam-1 to see if the worms had any phenotypes associated with wee-1.3 loss or gain-of-function. Loss of WEE-1.3 function is known to affect oocyte maturation and embryonic survival, whereas gain-of-function mutations affect spermatogenesis and male fertility (Lamitina and L’Hernault 2002; Burrows et al. 2006). Embryos produced by wee-1.3(lz5) worms had viability similar to wild-type and oocyte maturation appeared unaffected (Table 3 and see below). In addition, the brood size of wee-1.3(lz5) worms (287 ± 52) was comparable to wild-type (281 ± 62). We then created a compound heterozygote with wee-1.3(lz5) and wee-1.3(q89eb60), an allele known to be recessive maternal-effect lethal (Lamitina and L’Hernault 2002). This strain showed an intermediate level of lethality, but worms heterozygous for wee-1.3(q89eb60) show no lethality on their own. This suggests that wee-1.3(lz5) may partially disrupt its function (Table 3).
In order to confirm that the change in wee-1.3 was in fact the suppressing mutation, we created a CRISPR strain with an identical missense mutation G1459A(E487K), wee-1.3(syb1738) (Figure 2B and Figure S1). Similar to wee-1.3(lz5), wee-1.3(syb1738) exhibited no differences from wild-type in embryonic viability, oocyte maturation, or brood size (298 ± 62) (Table 3 and see below). In addition, males were fertile and used for numerous matings. When crossed into pam-1(or403), wee-1.3(syb1738) suppressed the embryonic lethality of pam-1 (Table 3), thus confirming wee-1.3(lz5) as the suppressing mutation.
wee-1.3(lz5) suppresses the polarity defects of pam-1
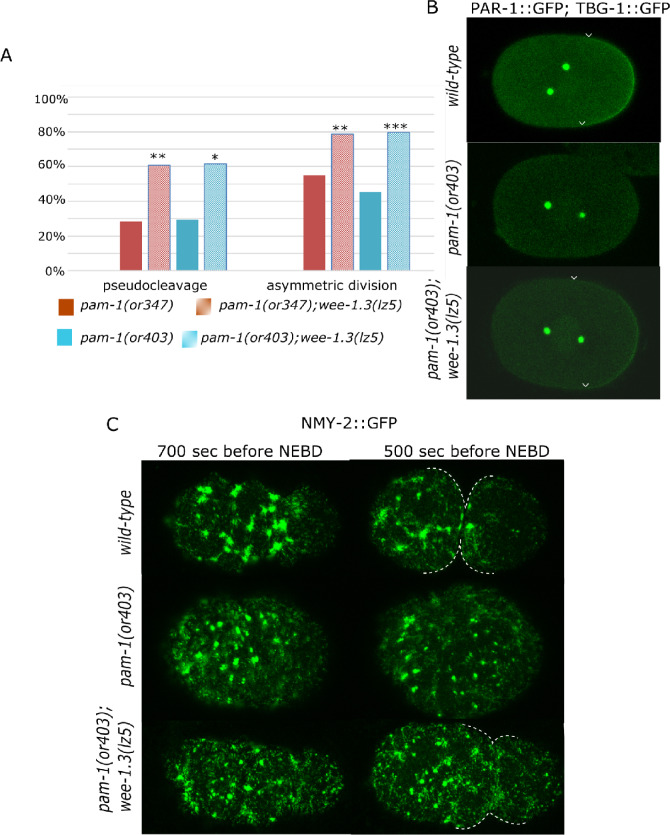
Since wee-1.3(lz5) rescued viability of embryos laid by pam-1 worms, we decided to look in more detail at polarity establishment in the suppressed strains. Similar to what we observed for suppression of pam-1(or347), polarity was rescued in many pam-1(or403); wee-1.3(lz5) mutants. Although only 45.6% of pam-1(or403) embryos divided asymmetrically, 79.6% of pam-1(or403); wee-1.3(lz5) did (Figure 3A). In wild-type, the PAR proteins localize to anterior or posterior cortical domains during polarization (reviewed in Nance and Zallen 2011). To look at this, we then examined the localization and size of the posterior PAR-1 domain. Previously, we found that a little over half of pam-1 embryos had posterior localization of PAR-1 and that this domain was on average smaller than wild-type (31% as compared with 42% embryo length; Saturno et al. 2017). In the presence of the wee-1.3(lz5) suppressor, we found that 100% of the embryos now showed posterior PAR-1 localization and that PAR-1 was restored to a wild-type domain size of 43% embryo length (Figure 3B and Table 4). Thus, the increase in embryonic viability was accompanied by a restoration of polarity establishment in many embryos.
Figure 3.
Polarity is improved in pam-1; wee-1.3(lz5) embryos. (A) wee-1.3(lz5) suppresses polarity defects of two pam-1 alleles. The presence of pseudocleavage in pam-1(or347) (n = 39) and pam-1(or403) (n = 27) is increased in pam-1(or347); wee-1.3(lz5) (n = 46) and pam-1(or403); wee-1.3(lz5) (n = 34). Similarly, while many pam-1(or347) (n = 78) and pam-1(or403) (n = 46) embryos divide symmetrically, the presence of an asymmetric first cleaveage is improved in pam-1(or347); wee-1.3(lz5) (n = 85) and pam-1(or403); wee-1.3(lz5) (n = 54) embryos. (B) PAR-1 and gamma-tubulin GFP confocal images. PAR-1 localizes to the posterior in wild-type during polarization. Arrowheads mark the edges of the PAR-1 domain. Although many pam-1(or403) embryos lack posterior PAR-1 localization (Saturno et al. 2017), 100% of pam-1(or403); wee-1.3(lz5) embryos have posterior localization of PAR-1 in a domain size similar to wild-type (n = 46). C) NMY-2::GFP images show clearing from the posterior in wild-type, 700 seconds prior to NEBD and full pseudocleavage 200 seconds later (n = 18). Both pam-1(or403) (n = 18) and pam-1(or403); wee-1.3(lz5) (n = 13) embryos often lack posterior clearing. Shown here is reduced clearing in both strains, but pseudocleavage is restored in the presence of the suppressor. Pseudocleavage is marked by dotted lines. Anterior to the left in all panels. *P < 0.05; **P < 0.01; ***P < 0.001
Table 4.
Size of PAR-1 domains
| Genotype | Size (% embryo length) of posterior PAR-1 domain |
|---|---|
| +/+ | 42.03 ± 0.89 (n = 32) |
| pam-1(or403) | 30.56 ± 1.44 (n = 46) *** |
| pam-1(or403); wee-1.3(lz5) | 42.72 ± 1.13 (n = 47) |
PAR-1 domain size was measured for all embryos with posterior localization. Size and standard error is reported. There was a significant difference between wild-type and pam-1(or403) domain size, but not wild-type and pam-1(or403); we e-1.3(lz5) domain sizes.
P < 0.001
wee-1.3(lz5) suppression of pam-1 polarity defects is not mediated by changes to the centrosome or actomyosin network
Our previous work demonstrated that the centrosome is mislocalized in pam-1 embryos and spends less time in close proximity to the posterior cortex, resulting in poor polarity establishment (Saturno et al. 2017). Thus, we measured centrosome–cortex contact in pam-1(or403); wee-1.3(lz5) worms to see if polarity rescue occurred by extending the time of this contact. In pam-1(or403) embryos, the centrosome fails to touch the posterior in 14% of embryos (Saturno et al. 2017). In pam-1(or403); wee-1.3(lz5) embryos, 27% did not have centrosome–cortical contact (n = 26). Of the embryos that exhibited contact, contact time was comparable to pam-1 alone, being 2.02 minutes in pam-1; wee-1.3(lz5) embryos and 2.14 minutes in pam-1 embryos (Saturno et al. 2017). Both suppressed and non-suppressed strains showed shorter centrosome contact times than seen in wild-type (3.25 minutes; Saturno et al. 2017). Thus, the rescue of polarity in pam-1; wee-1.3(lz5) embryos is not a result of sustained centrosome contact with the cortex.
Next, we looked at the cortical network of nonmuscle myosin, NMY-2, that mediates cortical polarization in response to the centrosomal cue. During polarization, NMY-2 foci in the cortex are lost from the posterior and flow toward the anterior (Munro et al. 2004). In pam-1 mutants, NMY-2 puncta often fail to clear from the posterior (Saturno et al. 2017; Figure 3C). In addition, the NMY-2 network is less robust in pam-1 mutant embryos. Previously, we reported defects in that NMY-2 foci appeared more sparse and larger in pam-1 embryos (Saturno et al. 2017). To quantify this, we compared images from wild-type and pam-1 mutant embryos at two time points relative to pronuclear envelope breakdown (NEBD). In wild-type embryos at -700 seconds prior to NEBD, initial clearing of NMY-2 foci at the posterior is evident. Shortly afterward, at -500 seconds before NEBD, full posterior clearing and a pseudocleavage furrow are observed. In contrast, in pam-1 mutants, many embryos do not show these polarity landmarks. Although wild-type embryos show posterior clearing 100% of the time, we observed partial clearing in only 61% of pam-1 embryos (n = 18). We measured the number and size of the NMY-2 foci at these two time points. Although the number of foci was not significantly different, we did observe that pam-1 embryos had smaller foci at both time points as compared with wild-type (Figure 3C and Table 5). These problems with the actomyosin cytoskeleton are likely to contribute to the polarity problems in pam-1 mutants (Saturno et al. 2017).
Table 5.
Number and size of NMY-2 foci
| Genotype | Number of NMY-2 foci |
Size of NMY-2 foci (µm2) |
||
|---|---|---|---|---|
| -700 seconds | -500 seconds | -700 seconds | -500 seconds | |
| +/+ | 39.33 ± 5.49 | 29.17 ± 5.89 | 3.62 ± 0.22* | 2.93 ± 0.43* |
| pam-1(or403) | 29.90 ± 2.57 | 21.30 ± 5.52 | 2.61 ± 0.30 | 1.62 ± 0.21 |
| pam-1(or403); wee-1.3(lz5) | 29.75 ± 1.76 | 27.62 ± 4.47 | 2.69 ± 0.48 | 2.23 ± 0.53 |
Data gathered from confocal images at 700 and 500 seconds prior to pro-NEBD (‘see Materials and methods’). Averages and standard error shown for wild-type (n = 8), pam-1 (n = 10), and pam-1; wee-1.3 (n = 8). T-Tests to compare each strain to pam-1 shows no difference in the number of foci, but a difference in area between wild-type and pam-1.
P < 0.05
To determine if wee-1.3(lz5) improves the actomyosin network of pam-1 embryos, we compared the network at comparable times. pam-1(or403) embryos with and without wee-1.3(lz5) had comparable NMY-2 networks and puncta sizes. Both the number of foci and the sizes of the foci were similar in these strains (Table 5). In addition, 38.5% of embryos failed to clear NMY-2 from the posterior, a number similar to pam-1(or403). Thus, the rescue of polarity landmarks by wee-1.3(lz5) was not due to an improvement of the actomyosin cytoskeleton dynamics.
pam-1 and wee-1.3 interact during oocyte maturation
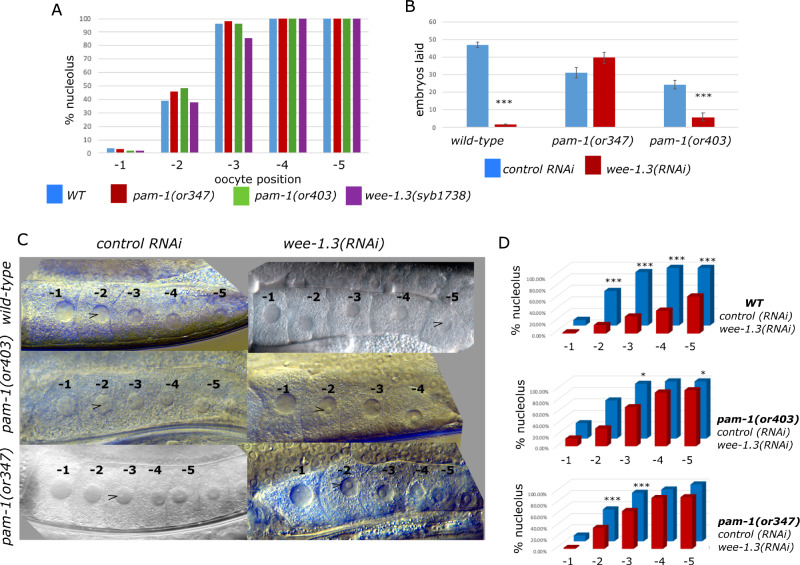
WEE-1.3 plays a well-established role in oocyte maturation (Burrows et al. 2006). When wee-1.3 is inactivated, oocytes precociously mature. Due to the interactions of pam-1 and wee-1.3 in the early embryo, we asked if they also interact during oocyte maturation. First, we compared the maturation state of oocytes in wild-type and pam-1 strains. To do this, we used DIC microscopy to score the presence of the nucleolus in the oocyte nuclei, a sign of immature oocytes. The C. elegans gonad is arranged with a row of maturing oocytes at the promimal end. The most proximal oocyte, closest to the spermatheca is numbered -1 with additional oocytes numbered as -2, -3 as they are arranged distally (McCarter et al. 1999). In wild-type gonads, the -1 oocyte is the most mature and usually does not have a nucleolus, whereas the -2 oocyte has a nucleolus about 40% of the time (Figure 4). A previous study suggested that pam-1 mutants are slower to mature, with more oocytes showing a nucleolus than wild-type (Althoff et al. 2014). However, when we compared wild-type with pam-1(or403) or pam-1(or347), we detected no significant difference in the maturation status of the oocytes (Figure 4A), suggesting that pam-1 on its own does not affect this maturation marker.
Figure 4.
pam-1 and wee-1.3 interact during oocyte maturation. Oocytes are arranged and numbered starting from the most proximal and mature (-1) oocyte near the spermatheca. (A) Oocyte maturation is similar in pam-1 and wee-1.3(syb1738) strains in comparison to wild-type as scored by the presence of the nucleolus. At least 50 gonads were scored for each strain. (B) Although wild-type worms go sterile after treatment with wee-1.3(RNAi), this was reduced in pam-1 strains as seen by continued production of embryos. Data from 20 worms per treatment. (C, D) When wee-1.3 is inactivated by RNAi, precocious oocyte maturation is observed in wild-type worms, but not in pam-1 worms. At least 40 gonads were scored for each treatment. (C) DIC images show one gonad arm starting with the -1 oocyte on the left. Arrowhead points to the first appearance of a nucleolus in the oocytes. (D) Quantification of nucleolus presence in the first five oocytes in RNAi of wee-1.3 in compared with empty vector, L4440. Chi square analysis was performed for each strain between L4440 and wee-1.3(RNAi) *P < 0.05; ***P < 0.001
Next, we compared the maturation state of oocytes in worms with the suppressing missense mutation wee-1.3(syb1738) and observed no difference in the presence of the nucleolus (Figure 4A). The maturation state of wee-1.3(syb1738) oocytes did not differ significantly from wild-type, in contrast to previous findings that wee-1.3(RNAi) oocytes precociously mature (Burrows et al. 2006); therefore, we hypothesize that the missense mutation in wee-1.3 is not a complete loss-of-function mutation.
Similar to past work, when we inactivated wee-1.3 by RNAi in wild-type worms, we saw a dramatic oocyte maturation effect (Burrows et al. 2006). First, we observed sterility (Figure 4B). We compared the number of embryos laid on control and wee-1.3(RNAi) plates between 24 and 28 hours and saw that wild-type worms treated with wee-1.3(RNAi) laid significantly fewer embryos. Wild-type worms on wee-1.3(RNAi) plates laid only 3.8% of the embryos laid on control RNAi plates (Figure 4B). When looking at the oocytes, we saw a great reduction in the number of oocytes that had a nucleolus (Figure 4, C and D). In control RNAi treatment, the nucleolus is normally present in oocytes -2 through -5; however, when wee-1.3 was inactivated, there were significantly fewer nucleoli present (Figure 4, C and D).
To see if wee-1.3 and pam-1 interact in oocyte maturation, we performed the same inactivation studies in two pam-1 backgrounds. When we inactivated wee-1.3 in pam-1 worms, we saw a reduction in the number of sterile worms as well as a much less pronounced oocyte maturation defect (Figure 4). After being placed on wee-1.3(RNAi) plates, pam-1(or403) worms laid 25% of the embryos laid on control RNAi plates. An even stronger interaction was observed for pam-1(or347) worms, which laid a comparable number of embryos on control and wee-1.3(RNAi) plates (Figure 4B). Additionally, the oocyte maturation defect normally induced by wee-1.3(RNAi) was not observed to the same degree in pam-1 strains. When we compared the presence of the nucleolus between pam-1; wee-1.3(RNAi) treated worms with pam-1 control RNAi worms, we observed no differences in oocyte maturation levels in some of the oocytes (Figure 4, C and D). For instance, in pam-1(or347), there was no difference in the presence of nucleoli in the -4 and -5 oocytes. In pam-1(or403) worms, there was no difference observed in the -2 and -4 oocytes on wee-1.3(RNAi) as compared with control RNAi. Thus, the precocious oocyte maturation phenotype in wee-1.3(RNAi) worms was lessened by mutation of pam-1. These data suggest that WEE-1.3 and PAM-1 interact during oocyte maturation. The presence of pam-1 mutations partially protects the worms from sterility and defects in oocyte maturation.
WEE-1.3 localization and levels are unchanged in pam-1 oocytes
As PAM-1 and WEE-1.3 interact during development, we wanted to test if PAM-1 regulates WEE-1.3 protein levels. To test this, we quantified WEE-1.3 fluorescence intensity in developing oocytes using a wee-1.3::gfp strain. This CRISPR tagged locus added GFP to the C terminus of the endogenous wee-1.3 locus (Fernando et al. 2021). Gonads from pam-1 and wild-type worms were compared for WEE-1.3 levels in developing oocytes. The mean intensity of cytoplasmic WEE-1.3::GFP was comparable in wild-type and pam-1 worms (Figure 5). We also did not note any differences in the localization of WEE-1.3 between the strains. These data suggest that PAM-1 does not regulate WEE-1.3 levels in oocytes.
Figure 5.
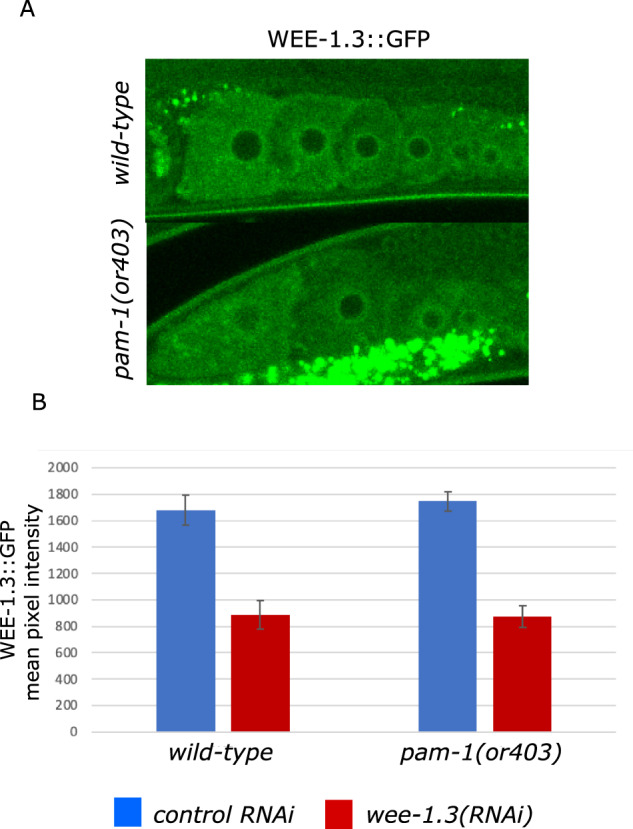
WEE-1.3 levels are comparable in wild-type and pam-1 oocytes. Confocal images of WEE-1.3::GFP show no difference in the localization of WEE-1.3 in wild-type (n = 19) and pam-1 mutant (n = 16) oocytes. The bright dots in the lower panel are autofluorescence of gut granules. (B) When the intensity of WEE-1.3 was compared in each oocyte, (-1 oocyte shown here) no difference was determined between strains; however, RNAi of wee-1.3 (WT n=13 and pam-1 n = 9) significantly reduced the levels. ANOVA P < 0.001.
To see if wee-1.3(RNAi) affected WEE-1.3 levels differently between WT and pam-1 mutants, we compared WEE-1.3::GFP levels after RNAi treatment. Both pam-1(or403) and wild-type strains showed reduced and comparable WEE-1.3::GFP levels after treatment (Figure 5B). Thus, the difference in phenotype between the strains was not a result of differences in WEE-1.3 protein levels in treated strains.
Discussion
Here, we describe the identification of three novel suppressors of pam-1, which partially rescue the maternal-effect embryonic lethality and polarity defects of pam-1 mutants. In one of these suppressors, we identified a missense mutation in wee-1.3 and we showed that pam-1 and wee-1.3 interact in multiple developmental contexts.
Isolation and initial characterization of suppressors
Our genetic approach to identifying factors that may interact with pam-1 yielded four alleles of three suppressor loci. Each suppressor mutation partially rescues the maternal-effect lethality associated with pam-1 mutants and acts in a recessive manner. Although recessive on their own, worms heterozygous for two suppressor mutations do have some improvement in the number of embryos that hatch (Table 2). Although they do not raise hatch rates to levels found for homozygous suppression, some combinations, for instance pam-1(or347); lz4/+ lz6/+, suppress at levels comparable to other suppressors, like pam-1(or347); wee-1.3(lz5). This suggests there may be nonallelic noncomplementation between the suppressors. Thus, it is possible that these suppressors act together in a pathway to suppress pam-1 phenotypes.
Each of our new suppressors works to partially rescue the polarity defects associated with pam-1 mutants (Figure 1). Significantly more embryos underwent pseudocleavage, a sign of cortical polarization (Hird and White 1993) and divided asymmetrically when the suppressor was present. Although we currently do not know the mechanism for this suppression, future work to identify the causative mutations will provide new insights into PAM-1’s function and its role in polarization, and is likely to reveal targets of the aminopeptidase and/or factors that regulate the cytoskeleton, centrosome positioning, and cell-cyle regulation.
Identification of wee-1.3 as a suppressor of pam-1
We identified a missense mutation in wee-1.3 an inhibitory kinase, homologous to Myt1, involved in cell-cycle regulation through inhibition of the maturation promoting factor (MPF). The MPF is made up of CDK-1 and cyclin B which drives entry into M phase of the cell cycle. Phosphorylation of CDK-1 by WEE-1.3 inhibits the complex, whereas the CDC-25 phosphatase removes the inhibitory phosphorylation to reverse the process. Activation of CDK-1 creates a feedback loop by activating CDC-25 and inhibiting WEE-1.3 to ensure quick activation of the complex (reviewed in van den Heuvel 2005). These proteins work together to ensure transient activation of MPF at the G2/M cell-cycle transition. Loss of function of wee-1.3 in C. elegans results in precocious oocyte maturation, whereas gain-of-function mutations cause male sterility due to defects in spermatogenesis (Lamitina and L’Hernault 2002; Burrows et al. 2006). Since our missense mutations in wee-1.3 did not cause any of the previously observed phenotypes on their own, we do not think the mutations in wee-1.3 cause a complete loss of function but are instead specific to suppression of pam-1. The amino acid change is in a region with no known functional domains, so we do not yet understand how it affects WEE-1.3 activity. Based on the reduced viability of embryos produced by wee-1.3(q89eb60)/wee-1.3(lz5) worms, it could be that these wee-1.3 alleles are hypomorphic.
wee-1.3(lz5) partially rescues polarity of pam-1 embryos without affecting the centrosome or the cytoskeleton
The polarization problems in pam-1 were previously tied to shortened centrosome–cortical contact times and defects in the actomyosin cytoskeleton (Saturno et al. 2017). Although we previously reported the NMY-2 foci appear larger in pam-1 mutants, we had not measured the foci or time-matched the embryos. Here we showed that pam-1 embryos have smaller NMY-2 foci when compared with time-matched controls, providing further evidence that PAM-1 regulates the actomyosin cytoskeleton. From phenotypic analysis, it appears that the suppressor does not act at the source of the problem. Suppressed embryos still exhibited very short centrosome–cortex contact and the cortical cytoskeleton looked similar in suppressed and non-suppressed strains. Despite this, pseudocleavage, posterior PAR-1 localization, and the asymmetric first division were largely rescued. Thus, wee-1.3(lz5) acts not to correct the problem of centrosome contact or cytoskeletal organization, but to alleviate the polarity defect by increasing the number of embryos properly localizing the PAR proteins. Whether this interaction is related to WEE-1.3’s role in cell-cycle regulation or a novel function will require further study.
WEE-1.3 and PAM-1 interact in oocyte maturation
WEE-1.3 acts in oocyte maturation in C. elegans. Oocytes are lined up from the -1 position just adjacent to the spermatheca with this oocyte being the most mature. Mature oocytes like the -1 to -2 oocyte have condensed chromosomes and lack a nucleolus. In comparison, immature oocytes instead have a visible nucleolus (Hendzel et al. 1997; McCarter et al. 1999; Hsu et al. 2000). WEE-1.3 acts in immature oocytes to keep MPF inactive and oocytes immature (Burrows et al. 2006). When wee-1.3 is inactivated by RNAi, oocytes precociously mature, whereas inactivation of cdk-1 causes oocyte maturation mature. In the double inactivation, the cdk-1 phenotype is seen, clearly showing that WEE-1.3 acts to inhibit CDK-1 in this system, similar to their interaction during mitosis (Burrows et al. 2006). When we inactivated wee-1.3 via RNAi, we observed these same phenotypes; however, they were not present in wee-1.3(syb1738) or wee-1.3(lz5) worms (Figure 4). This suggests the missense allele does not significantly compromise WEE-1.3 function during oocyte maturation.
Previous work on PAM-1 in the C. elegans gonad suggests that it is one of a few peptidases in this family that regulates reproductive success (Althoff et al. 2014). Although this prior study documented a subtle delay in oocyte maturation in pam-1 worms (Althoff et al. 2014), we did not see the same. There was no significant difference in the presence of the nucleolus in the oocytes of pam-1 as compared with wild-type gonads (Figure 4). This difference between our data and the prior study may be due to differences they report in oocyte maturation in wild-type (Althoff et al. 2014), which varies from our work and other studies (Burrows et al. 2006). When wee-1.3 was inactivated by RNAi, we saw a strong difference in oocyte maturation between wild-type and pam-1 strains. This effect was clearly seen by the prevention of sterility in pam-1 worms in which wee-1.3 is inactivated. The oocytes in pam-1 treated with wee-1.3(RNAi) did not exhibit the same precocious oocyte maturation seen when wee-1.3 was inactivated without the pam-1 mutation. Thus, mutations in pam-1 partially protect worms from precocious oocyte maturation, suggesting a role for the aminopeptidase in the process.
Allele specificity
wee-1.3(lz5) is able to suppress both a missense and nonsense allele of pam-1. Although embryonic viability improved more in the presence of the nonsense allele, pam-1(or403), the extent of polarity rescue was comparable with either pam-1 allele. Our previous work showed that in our nonsense allele, no detectable PAM-1 protein was observed after antibody staining (Fortin et al. 2010). Thus, for polarity rescue, it is unlikely that wee-1.3(lz5) works to restore PAM-1 function but may instead work downstream or bypass the requirement for functional PAM-1. Future work with our additional suppressors will reveal if this is a common trend or if some are allele specific suppressors.
Although rescue of embryonic viability was higher for the suppressed nonsense allele, when looking at interactions in the gonad, we saw something different. The nonsense allele of pam-1 was not as protective as the missense allele when wee-1.3 was inactivated by RNAi. Although both pam-1 strains were less sensitive to precocious oocyte maturation, only the missense allele fully protected against the sterility associated with loss of wee-1.3. Thus, the interactions of wee-1.3 and pam-1 alleles vary in different contexts and may indicate differences in how WEE-1.3 and PAM-1 interact in these different developmental processes. Perhaps partial PAM-1 function is needed in the gonad for the interaction with wee-1.3. Similarly, it could be that the pam-1(or347) and pam-1(or403) alleles differentially regulate WEE-1.3 levels or activity to account for these differences. As we learn more about the interaction, this may become more clear.
Mechanism of action
One important function for aminopeptidases is the degradation of short peptides during the final stage of proteolysis. During certain dynamic cellular processes like cell division or gamete maturation, proteins must be removed from the cell. These proteins are tagged with ubiquitin before being degraded by the proteasome, resulting in short peptide sequences (Glotzer et al. 1991). In order to conserve resources, cells repurpose these peptides by breaking them down into individual amino acids. Protein degradation is also vital for the first cellular division. For example, in order to transition from oocyte meiosis to embryonic mitosis, the microtubule severing complex MEI-1 must be degraded (Lu and Mains 2007). Given the importance of protein degradation in these processes and significance of aminopeptidases in the degradation pathway, it is possible that PAM-1 plays several roles in the transition from oocyte to embryo.
How then do wee-1.3 and pam-1 interact? One possibility for the mechanism of action is that WEE-1.3 may be a direct target of PAM-1. If this were true, we should observe higher levels of WEE-1.3 in pam-1 mutants. However, both the levels and localization of WEE-1.3 in the gonad were unchanged in pam-1 mutants and RNAi reduced WEE-1.3 levels comparably in both control and pam-1 mutant strains (Figure 5). However, WEE-1.3 may be processed by PAM-1 without being totally degraded. Another possibility that we can test in the future is a change in activity. WEE-1.3 may be overactive in pam-1 mutants, and perhaps the point mutation in the protein may alleviate the effect. wee-1.3(lz5) may also reduce the activity of the inhibitory kinase, allowing activation of another protein that bypasses the requirement for PAM-1. Our finding that pam-1 worms are less sensitive to reduction of wee-1.3 function via RNAi, and the possible hypomorphic nature of wee-1.3(lz5) are both consistent with these possibilities.
Another possibility is that PAM-1 regulates another component of the cell-cycle machinery. PAM-1 may positively regulate CDC-25 or MPF components. Work in Dictylostelium has shown that a PSA can associate with Cdk5 and regulate its localization and activity (Huber and O’Day 2011; Huber et al. 2013). This work, coupled with the role of PSAs in regulating mitosis and meiosis suggest that this interaction with WEE-1.3 and components of the MPF may be relevant for more than just C. elegans (Constam et al. 1995; Osada et al. 2001; Lyczak et al. 2006). A further look into the role of PAM-1 in the cell cycle may provide insights.
Still another possibility is that PAM-1 directly or indirectly affects OMA-1/-2, protein implicated in the oocyte to embryo transition. Levels of OMA-1 and the closely related OMA-2 are high in maturing oocytes but the proteins are degraded during mitosis (Detwiler et al. 2001; Shimada et al. 2002). This degradation requires the dual-specificity kinase, MBK-2, as well as CDK-1, and cyclin B3 (Shirayama et al. 2006). OMA-1 regulation spans the oocyte to embryo transition, including polarization of proteins in the one-cell embryo. Likewise, interactions we have seen between PAM-1 and WEE-1.3 work together during oocyte maturation and polarization. Future work will focus on this and other potential interactions to determine the mechanism of PAM-1 and WEE-1.3 interaction. As both PAM-1 and WEE-1.3 are conserved, it is likely that interactions we uncover may be present more broadly. By moving forward to identify the mutations in our additional suppressors we may uncover new proteins involved in cell polarity and cell-cycle regulation.
Data availability
All strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at figshare: https://doi.org/10.25387/g3.14114156.
Acknowledgments
Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Strain WDC08 was kindly provided by Anna Allen, Howard University prior to publication. The following undergraduates worked on aspects of this project over the years: Robin Alsher, Angela Hong, Ethan Kabel, Zachary Klock, Jessica Meeker, Thuy Nguyen, Margaret Williams, and Elizabeth Wolosin.
Funding
This work was funded by NSF: IOS-0918950 to R.L., NIH: GM110614 to R.L., NIH: GM049869 and GM131749 to B.B. and J.L.
Conflicts of interest: None declared.
Literature cited
- Althoff MJ, Flick K, Trzepacz C.. 2014. Collaboration within the M1 aminopeptidase family promotes reproductive success in Caenorhabditis elegans. Dev Genes Evol. 224:137–146. doi:10.1007/s00427-014-0470-3 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DR, Hooper NM, Isaac RE.. 2003. The Caenorhabditis elegans orthologue of mammalian puromycin-sensitive aminopeptidase has roles in embryogenesis and reproduction. J. Biol. Chem. 278:4279–42801. doi:10.1074/jbc.M306216200 [DOI] [PubMed] [Google Scholar]
- Burrows AE, Sceurman BK, Kosinski ME, Richie CT, Sadler PL, et al. 2006. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development. 133:697–709. doi:10.1242/dev.02241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Tobler AR, Rensing-Ehl A, Kemler I, Hersh LB, et al. 1995. Puromycin-sensitive aminopeptidase: sequence analysis, expression, and functional characterization. J Biol Chem. 270:26931–26939. doi:10.1074/jbc.270.45.26931 [DOI] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, et al. 2005. Rapid single nucleotide polymophism mapping in C. elegans. BMC Genomics. 118.6: doi:10.1186/1471-2164-6-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler MR, Reuben M, Li X, Rogers E, Lin R.. 2001. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev Cell. 1:187–199. doi:10.1016/S1534-5807(01)00026-0 [DOI] [PubMed] [Google Scholar]
- Encalada SE, Martin PR, Phillips JB, Lyczak R, Hamill DR, et al. 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev Biol. 228:225–238. doi:10.1006/dbio.2000.9965 [DOI] [PubMed] [Google Scholar]
- Fernando LM, Kyrionna G, Boateng R, Allen AK, 2021. Comparison of N- and C-terminally endogenously GFP-tagged WEE- 1. 3 strains in C. elegans. microPublication Biol. doi: 10.17912/micropub.biology.000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin SM, Marshall SL, Jaeger EC, Greene PE, Brady LK, et al. 2010. The PAM-1 aminopeptidase regulates centrosome positioning to ensure anterior-posterior axis specification in one-cell C. elegans embryos. Dev Biol. 344:992–1000. doi:10.1016/j.ydbio.2010.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW.. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138. doi:10.1038/349132a0 [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, et al. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360. doi:10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Hird SN, White JG.. 1993. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol. 121:1343–1355. doi:10.1083/jcb.121.6.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. 2005. Genetic suppression. WormBook. 1-13. doi: 10.1895/wormbook.1.59.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J-Y, Sun Z-W, Li X, Reuben M, Tatchell K, et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes logical changes on the DNA template in an energy. Cell. 102:279–291. [DOI] [PubMed] [Google Scholar]
- Huber RJ, O’Day DH.. 2011. Nucleocytoplasmic transfer of cyclin dependent kinase 5 and its binding to puromycin-sensitive aminopeptidase in Dictyostelium discoideum. Histochem Cell Biol. 136:177–189. doi:10.1007/s00418-011-0839-6 [DOI] [PubMed] [Google Scholar]
- Huber RJ, Catalano A, O'Day DH.. 2013. Cyclin-dependent kinase 5 is a calmodulin-binding protein that associates with puromycin-sensitive aminopeptidase in the nucleus of Dictyostelium. Biochim Biophys Acta. 1833:11–20. doi:10.1016/j.bbamcr.2012.10.005 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. doi:10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kim E, Sun L, Gabel CV, Fang-Yen C.. 2013. Long-Term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One. 8: e53419.doi:10.1371/journal.pone.0053419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo LC, Parfenova L, Ren G, Vi N, Hui M, et al. 2011. Puromycin-sensitive aminopeptidase (PSA/NPEPPS) impedes development of neuropathology in HPSA/TAUP301L double-transgenic mice. Hum Mol Genet. 20:1820–1833. doi:10.1093/hmg/ddr065 [DOI] [PubMed] [Google Scholar]
- Lamitina ST, L’Hernault SW.. 2002. Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development. 129:5009–5018. [DOI] [PubMed] [Google Scholar]
- Lowry J, Yochem J, Chuang C-H, Sugioka K, Connolly AA, et al. 2015. High-throughput cloning of temperature-sensitive Caenorhabditis elegans mutants with adult syncytial germline membrane architecture defects. G3 Genes. G3 (Bethesda). 5:2241–2255. doi:10.1534/g3.115.021451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Mains PE.. 2007. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev Biol. 302:438–447. doi:10.1016/j.ydbio.2006.09.053 [DOI] [PubMed] [Google Scholar]
- Lyczak R, Zweier L, Group T, Murrow MA, Snyder C, et al. 2006. The puromycin-sensitive aminopeptidase PAM-1 is required for meiotic exit and anteroposterior polarity in the one-cell Caenorhabditis elegans embryo. Development. 133:4281–4292. doi:10.1242/dev.02615 [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T.. 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 205:111–128. doi:10.1006/dbio.1998.9109 [DOI] [PubMed] [Google Scholar]
- Minevich G, Park DS, Blankenberg D, Poole RJ, Hobert O.. 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics. 192:1249–1269. doi:10.1534/genetics.112.144204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR.. 2004. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 7:413–424. doi:10.1016/j.devcel.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Nance J, Zallen JA.. 2011. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 138: 799–809. doi:10.1242/dev.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, et al. 2011. A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS One. e16644.6: doi:10.1371/journal.pone.0016644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada T, Watanabe G, Kondo S, Toyoda M, Sakaki Y, et al. 2001. Male reproductive defects caused by puromycin-sensitive aminopeptidase deficiency in mice. Mol Endocrinol. 15:960–971. doi:10.1210/mend.15.6.0643 [DOI] [PubMed] [Google Scholar]
- Peer WA. 2011. The role of multifunctional M1 metallopeptidases in cell cycle progression. Ann Bot. 107:1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poloz Y, Catalano A, O'Day DH.. 2012. Bestatin inhibits cell growth, cell division, and spore cell differentiation in Dictyostelium discoideum. Eukaryot Cell. 11:545–557. doi:10.1128/EC.05311-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Ma Z, Hui M, Kudo LC, Hui K-S, et al. 2011. Cu, Zn-superoxide dismutase 1 (SOD1) is a novel target of Puromycin-sensitive aminopeptidase (PSA/NPEPPS): PSA/NPEPPS is a possible modifier of amyotrophic lateral sclerosis. Mol Neurodegener. 6: 29.doi:10.1186/1750-1326-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose L, Gönczy P.. 2014. Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryos. WormBook. 1–43. [DOI] [PubMed] [Google Scholar]
- Sánchez-Morán E, Jones GH, Franklin FCH, Santos JL.. 2004. A puromycin-sensitive aminopeptidase is essential for meiosis in Arabidopsis thaliana. Plant Cell. 16:2895–2909. doi:10.1105/tpc.104.024992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saturno DM, Castanzo DT, Williams M, Parikh DA, Jaeger EC, et al. 2017. Sustained centrosome-cortical contact ensures robust polarization of the one-cell C. elegans embryo. Dev Biol. 422:135–145. doi:10.1016/j.ydbio.2016.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Brock DA, Ammann RR, DeShazo T, Khosla M, et al. 2002. The Cdk5 homologue, Crp, regulates endocytosis and secretion in Dictyostelium and is necessary for optimum growth and differentiation. Dev Biol. 247:1–10. doi:10.1006/dbio.2002.0684 [DOI] [PubMed] [Google Scholar]
- Shimada M, Kawahara H, Doi H.. 2002. Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells. 7:933–947. doi:10.1046/j.1365-2443.2002.00570.x [DOI] [PubMed] [Google Scholar]
- Shirayama M, Soto MC, Ishidate T, Kim S, Nakamura K, et al. 2006. The conserved kinases CDK-1, GSK-3, KIN-19, and MBK-2 promote OMA-1 destruction to regulate the oocyte-to-embryo transition in C. elegans. Curr Biol. 16:47–55. doi:10.1016/j.cub.2005.11.070 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S. 2005. Cell-Cycle Regulation. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi K, Tanaka T, Kato K, Sadik G, Morihara T, et al. 2009. Involvement of puromycin-sensitive aminopeptidase in proteolysis of tau protein in cultured cells, and attenuated proteolysis of frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) mutant tau. Psychogeriatrics. 9:157–166. doi:10.1111/j.1479-8301.2010.00307.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at figshare: https://doi.org/10.25387/g3.14114156.