Abstract
Muscadinia rotundifolia, the muscadine grape, has been cultivated for centuries in the southeastern United States. M. rotundifolia is resistant to many of the pathogens that detrimentally affect Vitis vinifera, the grape species commonly used for winemaking. For this reason, M. rotundifolia is a valuable genetic resource for breeding. Single-molecule real-time reads were combined with optical maps to reconstruct the two haplotypes of each of the 20 M. rotundifolia cv. Trayshed chromosomes. The completeness and accuracy of the assembly were confirmed using a high-density linkage map. Protein-coding genes were annotated using an integrated and comprehensive approach. This included using full-length cDNA sequencing (Iso-Seq) to improve gene structure and hypothetical spliced variant predictions. Our data strongly support that Muscadinia chromosomes 7 and 20 are fused in Vitis and pinpoint the location of the fusion in Cabernet Sauvignon and PN40024 chromosome 7. Disease-related gene numbers in Trayshed and Cabernet Sauvignon were similar, but their clustering locations were different. A dramatic expansion of the Toll/Interleukin-1 Receptor-like Nucleotide-Binding Site Leucine-Rich Repeat (TIR-NBS-LRR) class was detected on Trayshed chromosome 12 at the Resistance to Uncinula necator 1 (RUN1)/Resistance to Plasmopara viticola 1 (RPV1) locus, which confers strong dominant resistance to powdery and downy mildews. A genome browser, annotation, and Blast tool for Trayshed are available at www.grapegenomics.com.
Keywords: muscadine grapes, vitaceae evolution, optical maps, hybrid genome assembly, full-length cDNA sequencing, disease resistance, powdery mildew
Introduction
Viticulture and winemaking typically use Vitis vinifera fruit. Though V. vinifera wines have desirable organoleptic properties, V. vinifera is extremely sensitive to diverse pathogens. In contrast, Muscadinia is a genus closely related to Vitis (Small 1913; Bouquet 1978; Olmo 1986; Olien 1990) that is resistant to diverse biotic and abiotic stresses. Muscadinia rotundifolia is the foundation of the muscadine grape industry in the USA, where its fruit, juice, and wine are produced (Olien 1990). M. rotundifolia is native to the warm and humid southeastern USA. Its natural habitat includes the states bordering the Gulf of Mexico and extends from northern Arkansas to Delaware (Bouquet 1978; Olmo 1986; Olien 1990; Heinitz et al. 2019).
Vitis and Muscadinia belong to Vitaceae, a family that contains 16 other genera and approximately 950 species (Wen et al. 2018). These genera are estimated to have diverged between 18 and 47 million years ago (Ma) (Wan et al. 2013; Liu et al. 2016; Ma et al. 2018), a process that involved several chromosome rearrangements (Karkamkar et al. 2010; Wen et al. 2018). Vitis has 19 chromosomes (2n = 38) (Olmo 1937), but like other genera in Vitaceae, including Ampelopsis, Parthenocissus, and Ampelocissus, Muscadinia has 20 chromosomes (2n = 40) (Branas 1932; Patil and Patil 1992; Karkamkar et al. 2010; Chu et al. 2018). Though successful crosses yielding fertile hybrids have been obtained, early attempts at producing hybrids of Vitis and Muscadinia often resulted in sterile progeny; this and the graft incompatibility observed between the two genera (Patel and Olmo 1955) are thought to be caused by their difference in chromosome number (Ravaz 1902; Dearing 1917; Patel and Olmo 1955; Olmo 1971; Bouquet 1980; Walker et al. 1991). SSR markers and genetic maps revealed that linkage groups LG7 and LG20 in Muscadinia correspond to the proximal and distal regions of chromosome 7 in Vitis (Blanc et al. 2012; Delame et al. 2019; Lewter et al. 2019). Chromosome 7 in Vitis may be derived from the fusion of its ancestor’s chromosomes 7 and 20. The presence of residual telomeric repeats in chromosome 7 of Vitis (Lewter et al. 2019) could provide additional support for this hypothesis, insight into the evolutionary history of Vitaceae, and understanding of the structure and function of the Muscadinia genome.
M. rotundifolia is a desirable partner with which to hybridize because it is resistant to many stresses, pests, and diseases that adversely affect V. vinifera and cause substantial crop loss (Olmo 1971). M. rotundifolia is resistant to Pierce’s disease (Xylella fastidiosa) (Ruel and Walker 2006), phylloxera (Daktulosphaira vitifolia) (Ravaz 1902; Davidis and Olmo 1964; Firoozabady and Olmo 1982), downy mildew (Plasmopara viticola) (Olmo 1971; Staudt and Kassemeyer 1995), powdery mildew (Erysiphe necator syn. Uncinula necator) (Olmo 1986; Merdinoglu et al. 2018), and other diseases and pests (Ravaz 1902; Olmo 1971; Walker et al. 2014). Several loci associated with resistance to pathogens affecting V. vinifera were identified in M. rotundifolia, including Resistance to Uncinula necator 1 (RUN1) (Pauquet et al. 2001), RUN2 (Riaz et al. 2011), Resistance to Erysiphe Necator 5 (REN5)(Blanc et al. 2012), Resistance to P. viticola 1 (RPV1) (Merdinoglu et al. 2003), and RPV2 (Merdinoglu et al. 2018). However, the lack of a M. rotundifolia reference sequence and gene annotation limits gene discovery and the characterization of resistance loci.
A draft assembly of the M. rotundifolia cv. Trayshed (Trayshed, hereafter) genome using Single-Molecule Real-Time (SMRT) sequencing was instrumental in resolving the genetic basis of sex determination in grapes (Massonnet et al. 2020). Here, we report the phased, chromosome-scale assembly of Trayshed, which was produced improving the previous SMRT assembly with the introduction of the optical maps in a hybrid scaffolding approach and consensus genetic map from multiple wild and cultivated grape species. Full-length cDNA isoforms (Iso-Seq) were also sequenced and used as transcriptional evidence for the annotation of protein-coding genes. This assembly and its annotation were used to identify where M. rotundifolia chromosomes 7 and 20 fused to create V. vinifera chromosome 7 but also to identify genes at the RUN1/RPV1 locus.
Materials and methods
Trayshed chromosome construction
A Saphyr Genome Imaging Instrument was used to generate a Bionano Next-Generation Mapping (NGM) of Trayshed. Ultra-high molecular weight DNA (>500 kbp) was extracted from young leaves by Amplicon Express (Pullman, WA, USA). DNA was then labeled with a DLE-1 non-nicking enzyme (CTTAAG) and stained according to the Bionano Prep™ Direct Label and Stain (DLS) Kit (Bionano Genomics, San Diego, CA, USA) instructions. Labeled DNA was loaded onto the SaphyrChip nanochannel array for imaging on the Saphyr system (Bionano Genomics, San Diego, CA, USA). Imaged molecules longer than 150 kbp were kept. These molecules were then assembled using Bionano Solve v.3.3 (Lam et al. 2012) with the following parameters “haplotype,” “noES,” and “noCut,” significance cutoffs were adopted P < 1e-10 to generate draft consensus contigs, P < 1e-11 for draft consensus contig extension, P < 1e-15 for the final merging of the draft consensus contigs. The complete configuration file used for the assembly is available as Supplemental File S1. This assembly procedure produced a 1.18 Gbp consensus genome map with an N50 of 5.6 Mbp.
PacBio contigs generated with FALCON-Unzip as described in Massonnet et al. (2020) were scaffolded with the genome maps using HybridScaffold v.04122018 (Lam et al. 2012) with options “-B2 -N1” for conflict resolution. The complete parameter configuration is available as Supplemental File S2. The ploidy state of the 2,000 rhAmpseq markers conserved across wild and domesticated grapes (Zou et al. 2020) was evaluated using DISPR (https://github.com/douglasgscofield/dispr) to keep the ones found in a maximum of two copies across different sequences. Hybrid scaffolds were then pre-processed to prevent marker duplication and wrong primary contigs/haplotigs co-placement. Two sets of chromosome-scale pseudomolecules were reconstructed using the pre-processed hybrid scaffolds and HaploSync v.1 tool suite (https://github.com/andreaminio/HaploSync). The hybrid scaffolds were sorted using the curated rhAmpseq markers (Zou et al. 2020). Sequences were treated independently from FALCON Unzip’s classification as primary contigs or haplotigs but, the relationship information between the primary contigs and the corresponding alternate haplotigs was conserved during the process. Then, the two haplotypes of each chromosome were compared to identify and correct assembly errors and fill gaps.
cDNA library preparation and sequencing
Total RNA from Trayshed leaves were isolated using a Cetyltrimethyl Ammonium Bromide (CTAB)-based extraction protocol as described in Blanco-Ulate et al. (2013). RNA purity was evaluated with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Hanover Park, IL, USA), RNA quantity with a Qubit 2.0 Fluorometer and a broad range RNA kit (Life Technologies, Carlsbad, CA, USA), and RNA integrity by electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Total RNA (300 ng, RNA Integrity Number > 8.0) were used for cDNA synthesis and library construction. A cDNA SMRTbell library was prepared. First-strand synthesis and cDNA amplification were accomplished using the NEB Next Single Cell/Low Input cDNA Synthesis & Amplification Module (New England, Ipswich, MA, USA). The cDNAs were subsequently purified with ProNex magnetic beads (Promega, WI, USA) following the instructions in the Iso-Seq Express Template Preparation for Sequel and Sequel II Systems protocol (Pacific Biosciences, Menlo Park, CA, USA). ProNex magnetic beads (86 µL) were used to select amplified cDNA ≥ 2 kbp. At least 80 ng of the size-selected, amplified cDNA were used to prepare the cDNA SMRTbell library. This was followed by DNA damage repair and SMRTbell ligation using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer's protocol. One SMRT cell was sequenced on the PacBio Sequel I platform (DNA Technology Core Facility, University of California, Davis, CA, USA). IsoSeq reads were extracted using IsoSeq v3 protocol with default parameters and low-quality isoform polished with LSC v.2.0 (Au et al. 2012). A cDNA library was also prepared using the Illumina TruSeq RNA sample preparation kit v.2 (Illumina, CA, USA) following Illumina’s low-throughput protocol. This library was evaluated for quantity and quality with the High Sensitivity chip in an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA) and sequenced in 100 bp paired-end reads using an Illumina HiSeq4000 sequencer (DNA Technology Core Facility, University of California, Davis, CA, USA).
Genome annotation
The completeness of the gene space in the hybrid assembly was estimated using BUSCO v.3.0.2 (Waterhouse et al. 2018) with the embryophyta_odb9 conserved gene set and by aligning the PN40024 V1 CDS against the Trayshed assembly using BLAT v.36x2 with default parameters (Kent 2002). PN40024 CDS were filtered before the alignment and included only single-copy genes (i.e., with unique mapping on its genome). The structural annotation of the M. rotundifolia cv. Trayshed genome was performed with a modified version of the pipeline used for the Vitis vinifera cv. Zinfandel genome (Vondras et al. 2019) and fully described here: https://github.com/andreaminio/AnnotationPipeline-EVM_based-DClab. Briefly, high-quality Iso-Seq Trayshed data were used to produce high-quality gene models for training gene predictors in PASA v.2.3.3 (Haas 2003) along with transcript evidence obtained from RNAseq data by performing transcriptome assemblies using Stringtie v.1.3.4d (Pertea et al. 2015) and Trinity v.2.6.5 (Grabherr et al. 2011) and from external databases. Public databases, transcriptome assemblies, and the Iso-Seq data described above were used as transcript and protein evidence. They were mapped on the genome using PASA v.2.3.3 (Haas 2003), MagicBLAST v.1.4.0 (Boratyn et al. 2019), and Exonerate v.2.2.0 (Slater and Birney 2005). Ab initio predictions were generated using BUSCO v.3.0.2 (Waterhouse et al. 2018), Augustus v.3.0.3 (Stanke et al. 2006), GeneMark v.3.47 (Lomsadze 2005), and SNAP v.2006-07-28 (Korf 2004). Repeats were annotated using RepeatMasker v.open-4.0.6 (Smit et al. 2013). Next, EvidenceModeler v.1.1.1 (Haas et al. 2008) used these predictions to generate consensus gene models. The final functional annotation was produced combining blastp v.2.2.28 (Camacho et al. 2009) hits against the Refseq plant protein database (https://ftp.ncbi.nlm.nih.gov/refseq/, retrieved January 17th, 2019) and InterProScan v.5.28-67.0 (Jones et al. 2014) outputs using Blast2GO v.4.1.9 (Gotz et al. 2008). To evaluate hemizygosity, CDS from each haplotype were aligned on the opposite haplotype using GMAP v. 2019-09-12 (Wu and Watanabe 2005) and alignments with at least 80% identity and coverage were considered valid. To prevent over-estimation of the gene content diversity between the two haplotypes due to pseudomolecule reconstruction partiality, unplaced sequences were also searched for copies of the CDS missing in the alternative haplotype.
Genome size quantification by flow cytometry
DNA content was estimated using flow cytometry (n = 3 individual leaves). Lycopersicon esculentum cv. Stupické polní tyčkové rané DNA was selected as the internal reference standard with a genome size of 2 C = 1.96 pg; 1 C = 958 Mbp (Doležel et al. 1992). Nuclei extraction was performed using the Cystain PI absolute P kit (Sysmex America Inc., IL, USA). Approximately 5 mg (0.7 cm2) of young healthy leaves from grapevine and tomato were finely cut with a razor blade in a Petri dish containing 500 µL of extraction buffer. The nuclei suspension was filtered through a 50 µm filter (CellTrics, Sysmex America Inc., IL, USA) and 2 mL of a propidium iodide staining solution was added (Doležel and Bartoš 2005; Bertier et al. 2013). Measurements were acquired using a Becton Dickinson FACScan (Franklin Lakes, New Jersey) equipped with a 488 nm laser. The data were analyzed using FlowJo v.10 (https://www.flowjo.com/solutions/flowjo). DNA content was inferred by linear regression using the tomato DNA reference standard.
Comparison of genome assembly and synteny analysis
The Trayshed and Cabernet Sauvignon (Massonnet et al. 2020) genomes were compared with NUCmer from the MUMmer v.4.0.0beta5 tool suite (Marçais etal. 2018), and the “–mum” parameter set. Descriptive statistics of the alignment were obtained using the MUMmer script dnadiff. For visualization purposes, alignments with at least 90% identity and 7,500 bp long were kept. Blastp v.2.2.28 (Camacho et al. 2009) was used to align the annotated proteins of Trayshed and Cabernet Sauvignon. Alignments with at least 80% reciprocal identity and coverage were retained. Pairwise protein information was associated with genes and processed with McScanX v.11.Nov.2013 (Wang et al. 2012) with default parameters to identify syntenic regions.
Localization of Muscadinia SNP markers
To localize in the Trayshed genomic sequences the corresponding position of each marker identified from Muscadinia populations in Lewter et al 2019, the sequence surrounding each of them was extracted from PN40024 V0 (Lewter et al. 2019) keeping 500 base-pair of overhang upstream and downstream of the SNP position. Sequences were then aligned with BLAT v.36x2 (Kent 2002) over the Trayshed chromosomes to recover the marker position. Alignments with at least 90% identity for both query and reference, 90% of coverage on the query, and a maximum of 50-bp-long gaps were considered as hits. They were then filtered for uniqueness based on the number of bases matching. The previously published consensus map of M. rotundifolia was used to assess the completeness of the chromosome reconstruction (Lewter et al. 2019). Sliding windows (window size = 20 markers, sliding = 10 markers) were designed to move across the uniquely mapped markers on the genome. The percentage of relative marker positions consistent with their genetic position on the high-density genetic map was calculated per window.
Telomeric repeats analysis
Telomeric repeats of “CCCTAA” and its reverse complement were searched along chromosome 7 of Cabernet Sauvignon and PN40024 V2 (Canaguier et al. 2017) using vmatchPattern from the R package Biostrings v.2.56.0 (Pagès et al. 2019). After peaks in the distribution of telomeric repeats were identified in both genomes, genomic regions of Cabernet Sauvignon (chr7: 18,675,000–18,677,000 bp) and PN40024 (chr7: 17,572,000–17,574,000bp) were extracted. Motif enrichment analysis was performed using MEME v.5.1.0 (Bailey and Elkan 1994) and reported as “logo” using the ggseqlogo R package v.0.1 (Wagih 2017).
Identification of NBS-LRR genes
Predicted Trayshed and Cabernet Sauvignon proteins were scanned using hmmsearch from HMMER v.3.3 (http://hmmer.org/) with a significance threshold (sequence E-value) of 0.001 and Hidden Markov Models (HMM) corresponding to the following different Pfam (El-Gebali et al. 2019) domains: NB-ARC (Pfam PF00931), TIR (PF01582), and LRR (PF00560, PF07723, PF07725, and PF12799). The proteome was also scanned with NLR-annotator (Steuernagel et al. 2020). Coiled-coil (CC) domain-containing proteins were identified by COILS (Lupas et al. 1991) during the InterProScan annotation. The identified proteins were then divided into six protein classes according to their domain composition: CC-NBS-LRR, CC-NBS, TIR-NBS-LRR, TIR-NBS, NBS-LRR, and NBS. To capture the largest number of potential NBS-LRR related genes, genes lacking the NB-ARC domain but with “NBS-LRR” functional annotations were also selected and divided into two classes: TIR-X and CC-X. For these eight protein classes, Multiple EM for Motif Elicitation (MEME) analysis was performed with the flags “-mod anr -nmotifs 20” to identify conserved domains for each class. Then, FIMO v.5.1.0 (Find Individual Motif Occurrences) (Grant et al. 2011) was run on each protein class using the corresponding MEME results; proteins with at least five conserved domains were kept. NBS-LRR gene clusters were defined as groups of at least two NBS-LRR genes, each separated by no more than eight non NBS-LRR genes (Richly et al. 2002) in a region spanning a maximum of 200 kbp (Holub 2001).
RUN1/RPV1 locus analysis and NBS-LRR genes phylogeny
Boundaries of the RUN1/RPV1 locus were defined by mapping two SSR markers, VMC4f3.1 and VMC8g9 (Barker et al. 2005), on Trayshed and Cabernet Sauvignon haplotypes using GMAP v. 2019-09-12 (Wu and Watanabe 2005). Trayshed chromosome 12 was split into 50-kbp blocks and queried with the genomic region corresponding to the locus in Cabernet Sauvignon using tblastx (BLAST v.2.2.29) (Camacho et al. 2009). Four-way pairwise comparisons between haplotypes were performed, results were filtered keeping alignments with at least 90% of identity, at least 100 bp long, and located within the boundaries of the resistance locus in the query and the reference. Hits overlapping annotated LTR retrotransposons were discarded. Proteins corresponding to the first alternative spliced variant of each NBS-LRR gene at the RUN1/RPV1 locus were aligned using MUSCLE v.3.8.31 (Edgar 2004) with default parameters and by including GAPDH (VIT_17s0000g10430) ortholog sequences as an outgroup. Distances between the alignments were extracted using the R package seqinr v.4.2.4 (Charif and Lobry 2007). The estimation of the corresponding neighbor-joining tree, its rooting, and bootstrapping (1,000 replicates) were performed using the R package ape v.5.4.1 (Paradis and Schliep 2019). The tree was drawn using ggtree v.2.2.4 (Yu et al. 2017). Species-specific clusters were identified as five or more grouped proteins from the same NBS-LRR class resulting from the phylogenetic analysis with a maximum of one protein coming from the other species in the same tree branch. Coding sequences of the Trayshed-specific protein cluster were aligned per class using MACSE v.2.03 (Ranwez et al. 2018) and trimmed using the option “min_percent_NT_at_ends = 0.80”. Synonymous substitution rates (dS) were obtained using yn00 from PAML v.4.9f (Yang 2007).
Data analysis and visualization
Data were parsed with R v.4.0.3 (R Core Team 2020) in RStudio v.1.4.869 (RStudio Team 2020) using tidyverse v.1.3.0 (Wickham et al. 2019), GenomicFeatures v.1.40.1 for genomic ranges manipulation (Lawrence et al. 2013), and rtracklayer for GFF files v.1.48.0 (Lawrence et al. 2009).
Results and discussion
Construction of the twenty phased M.rotundifolia cv. Trayshed chromosomes
The haploid genome size of M. rotundifolia cv. Trayshed evaluated by flow cytometry was estimated at 483.4 ± 3.1 Mbp, a size comparable to PN40024 (487 Mbp; Jaillon et al. 2007), but smaller than Cabernet Sauvignon (557.0 ± 2.4 Mbp; Supplementary Figure S1).
Optical maps were generated using the BioNano Genomics technology at 2,057x coverage to further scaffold the Trayshed genome assembly produced previously from Single-Molecule Real-Time (SMRT; Pacific Biosciences) long reads (N50 = 1,479,367 bp; Massonnet et al. 2020). The BioNano consensus genome map was combined with the contigs to produce an 896.0 Mbp hybrid assembly nearly twice the expected haploid genome size of Trayshed determined by flow cytometry. Genome sizes estimated by sequencing are known to be underestimates, while the flow cytometry approach tends to produce overestimates (Elliott and Gregory 2015). In the hybrid assembly, ∼70% of the complete BUSCO genes were duplicated and, 2.16 ± 1.07 copies of each PN40024 gene were present. Altogether, the size of the assembly, the amount of duplicated BUSCO genes (Supplementary Table S1), and the doubled representation of PN20024 genes are strong evidence that the draft assembly represents the diploid genome of Trayshed. Finally, we reconstructed two sets of phased chromosome-scale pseudomolecules (Haplotype 1, Hap1, and Haplotype 2, Hap2; Table 1) using a consensus grape genetic map as a guide (Zou et al. 2020). The chromosome-scale assemblies were 400.4 Mbp for Hap1 and 369.9 Mbp for Hap2. Based on the flow cytometry data, the whole assembly represents 86.2% of the estimated diploid genome size (2x 483.4 Mbp). On average, chromosome size was 89.09 ± 8.87% conserved between the two haplotypes (Supplementary Table S2). It was not possible to recover the positional information for regions lacking genetic markers, which were therefore excluded from the pseudomolecule assembly (Table 1). The unplaced scaffolds represented only 7.5% of the genome assembly, which is much less than in PN40024 V1 (12.4%; Jaillon et al. 2007) and similar to that of the V2 assembly (6%; Canaguier et al. 2017). The unplaced scaffolds were significantly more repetitive than the rest of the genome (52% vs. 42%) and carried about 7% of the total protein-coding genes (3,564 genes). The Trayshed assembly also showed a lower level of gaps overall (∼1 Mbp per haplotype) than PN40024 (∼10 Mbp).
Table 1.
Genome assembly statistics
| Haplotype 1 | Haplotype 2 | Unplaced | |
|---|---|---|---|
| Assembly length (bp) | 400,450,509 | 369,985,741 | 62,961,436 |
| Number of scaffolds | 20 | 20 | 1,690 |
| Average length (bp) | 20,022,525 | 18,499,287 | 37,255 |
| Maximum length (bp) | 34,098,201 | 26,289,366 | 1,487,392 |
| N50 length (bp) | 20,338,664 | 18,578,327 | 46,805 |
| Total gap length (bp) | 645,752 | 1,182,032 | 125,041 |
| Repetitive content (%) | 42.54 | 42.35 | 51.99 |
| Number of protein-coding genes | 25,706 | 23,673 | 3,564 |
| Complete BUSCO copies (%) | 90.6 | 86.2 | 9.6 |
The Trayshed assembly structure was confirmed by comparing positions along each chromosome with their linkage group positions within a high-density M. rotundifolia genetic map (Lewter et al. 2019) (Supplementary Figures S2–S4). More than 80% of markers’ positions were consistent between the genetic and physical maps, with strong overall correlation (R > 0.90) along all chromosomes for both haplotypes.
Analysis of chromosome structure supports a fusion of chromosomes 7 and 20 in Vitis
Each set of Trayshed’s twenty chromosomes was compared with the chromosome-scale Cabernet Sauvignon assembly (Massonnet et al. 2020), which is the most complete and contiguous diploid genome assembly of grapevine currently available. The two genomes were highly syntenic (Figure 1A, Supplementary Figure S5). Based on the four haplotype comparisons, colinear regions were 2,433 ± 28 bp long and shared 92.19 ± 0.37% identity (Supplementary Table S3). On average, short insertions (<50bp) were 40-bp long, long insertions were 1,501-bp long, short deletions were 40-bp, and long deletions were 998-bp. The longest deletion (∼50 kbp) was detected on Cabernet Sauvignon chromosome 8 and the longest insertion (∼79 kbp) was observed on chromosome 2 (Supplementary Table S4).
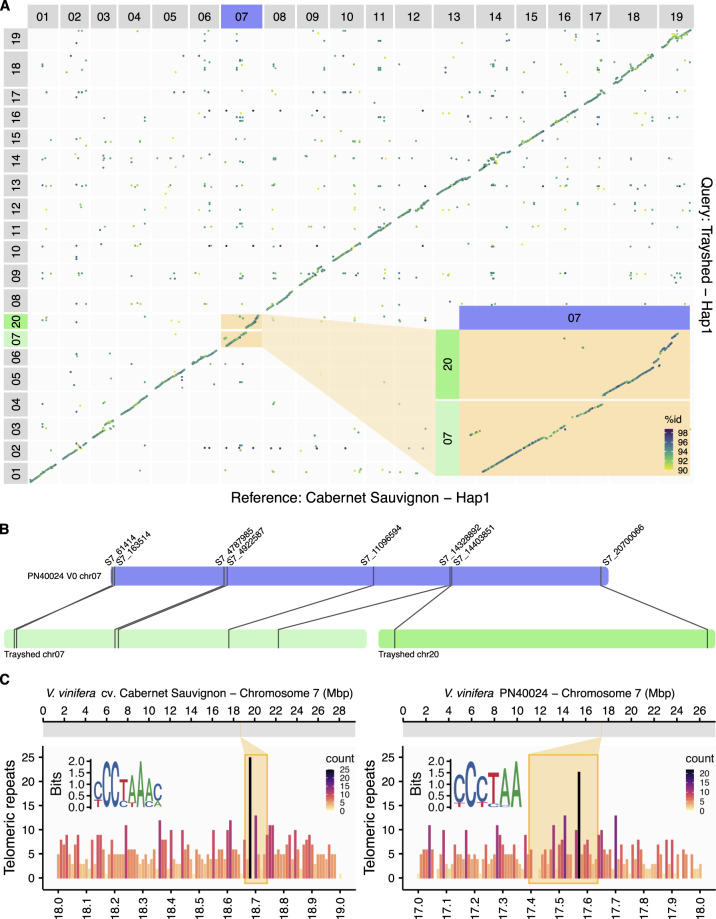
Figure 1.
Chromosome fusion localization in Vitis vinifera. (A) Whole-genome alignment of Trayshed Hap1 (y-axis) on Cabernet Sauvignon Hap1 (x-axis). The putative fusion of Trayshed chromosomes 7 and 20 (green) and Cabernet Sauvignon chromosome 7 (purple) is inset. The percentage identity (% id) between the alignments is displayed as a color gradient. (B) The positions of markers on Trayshed chromosomes 7 and 20 (green) and their corresponding location on PN40024 V0 chromosome 7. (C) The distribution of telomeric repeats in 1 Mbp containing the expected chromosome fusion in Cabernet Sauvignon (left panel) and PN40024 (right panel). The frequency of telomere repeats is represented as a color gradient. The most enriched motif in the 2-kbp region surrounding the peak of telomeric repeats is inset for each genotype.
Trayshed chromosomes 7 and 20 aligned at the beginning and end of Cabernet Sauvignon chromosome 7, respectively. This supports the fusion of these two chromosomes in Vitis reported previously using genetic maps (Blanc et al. 2012; Lewter et al. 2019; Figure 1A). The marker-containing regions used for Trayshed chromosome structure validation also support this hypothesis. Markers on Trayshed LG 7 and 20 were associated with V.vinifera chromosome 7 (Figure 1B). Telomeric repeats can be used as indicators of chromosome number reduction rearrangements (Sousa and Renner 2015). Enrichment of these sequences is expected in a genomic region where a chromosome fusion occurred. To determine the position of chromosome fusion in Cabernet Sauvignon, we searched for telomeric repeats in the region of chromosome 7 that should contain the fusion point based on whole-genome alignments. We examined the genomic region of Cabernet Sauvignon chromosome 7 that spanned Trayshed chromosomes 7 and 20 (chr 7: 18,661,816–18,740,440 bp; Figure 1C, top panel) and found an enrichment of telomeric repeats in this region (Figure 1C, bottom panel). A similar pattern was detected in the hypothetical fusion region of PN40024 (chr 7: 17,392,916–17,636,181 bp; Figure 1C). Together, these data support a chromosome number reduction by fusion in V. vinifera when compared with M. rotundifolia.
Full-length cDNA sequencing and annotation of the protein-coding genes
Together with the public data and RNA-Seq data already available, high-quality full-length cDNA sequences (Iso-Seq; Pacific Biosciences, Supplementary Figure S6) were generated as transcriptional evidence to support the gene model predictions. From leaf tissues, 336,932 raw reads were sequenced, clustered, and error corrected with LSC (Au et al. 2012) to generate 34,558 high-quality isoforms, 260 low-quality isoforms, and 111,672 singletons (Supplementary Figure S6). After obtaining consensus gene models, alternative splice variants were predicted if supported by Iso-Seq data, with 2.87 transcripts per gene on average (Supplementary Figure S6). BUSCO genes (Hap1: 92.3%, Hap2: 85.2%) were well-represented in the diploid Trayshed transcriptome. The structural annotation included 52,943 protein-coding gene loci (Table 1) and 83,873 proteins (including the alternative forms). There were 25,706 and 23,673 genes on Hap1 and Hap2, respectively (Table 1). The repetitive content composed 42.54 and 42.35% of Hap1 and Hap2, respectively, and was higher in the unplaced sequences (51.99%). The coding sequences of each haplotype were aligned on its alternative to evaluate hemizygosity. We found 2,586 and 1,034 hemizygous genes on Hap1 (10.06%) and Hap2 (4.37%), respectively. These results are consistent with V. vinifera cv. Zinfandel (4.56%; Vondras et al. 2019) but lower than the levels found in V. vinifera cv. Chardonnay (14.6%; Zhou et al. 2019).
Based on blastp results (80% identity, 80% coverage), 33,822 (63.88%) Trayshed genes are homologous with 31,471 (54.83%) genes of Cabernet Sauvignon. The 22,755 proteins that did not have a potential ortholog in Cabernet Sauvignon were significantly enriched in functional categories related to cell communication, signal transduction, signaling, regulation of biological and cellular processes, and cellular responses to stimulus and biological regulation (P < 0.01; Fisher’s exact test). On average, colinear blocks of homologous genes were ∼152 genes long (block extension interrupted with more than 25 unmatched genes in a row, Supplementary Table S5) based on the synteny results with Cabernet Sauvignon. The longest block of colinear genes was found on chromosome 14 and contained 613 genes (Supplementary Table S5).
NBS-LRR intracellular receptors encoded in the Trayshed genome
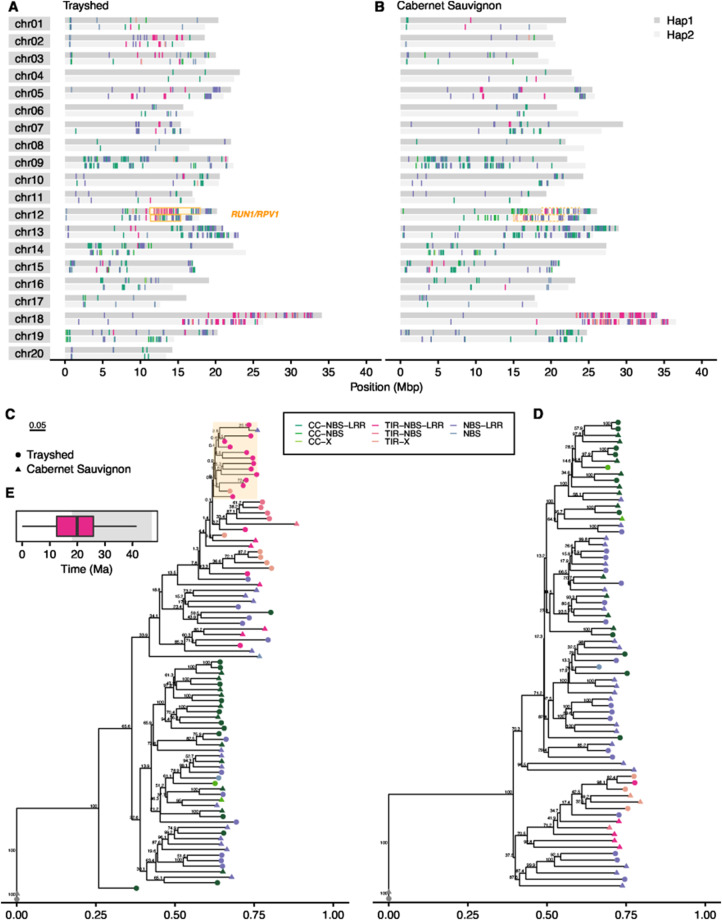
Trayshed is attractive to grape breeding programs because it is resistant to pathogens that affect V.vinifera. In plants, resistance to pathogens is primarily attributed to the activity of resistance genes (R genes). Nucleotide-binding site leucine-rich repeat (NBS-LRR) genes constitute the largest family of R genes. NBS-LRR genes are associated with resistance to powdery and downy mildew pathogens in several species, including Arabidopsis (Warren et al. 1998), wheat (He et al. 2018), barley (Wei et al. 1999; Zhou et al. 2001), pepper (Jo et al. 2017), and grapes (Riaz et al. 2011; Zini et al. 2019). We divided the NBS-LRR genes into eight different classes depending on the domains detected in the proteins: CC-NBS-LRR, CC-NBS, CC-X, TIR-NBS-LRR, TIR-NBS, TIR-X, NBS-LRR, NBS (called hereafter NBS-LRR when described as a whole). Overall, the Trayshed genome shows a slightly higher number of NBS-LRR genes (1,158) than Cabernet Sauvignon (1,013). Both species showed the highest number of NBS-LRR localized on chromosomes 9, 12, 13, and 18 (Figure 2A, Supplementary Table S6). NBS-LRR genes can occur in clusters that could be critical to their function as resistance loci (Hulbert et al. 2001; Meyers et al. 2003). The average number of clusters detected in Trayshed per haplotype (122) was similar to Cabernet Sauvignon (120), but their arrangement in the genome varied substantially (Supplementary Figure S7). Among the haplotypes, in Cabernet Sauvignon, NBS-LRR clusters accumulated the most on chromosome 18 Hap1 while the highest number was observed on chromosome 12 in Trayshed Hap1. These differences could plausibly contribute to disparities in disease resistance.
Figure 2.
NBS-LRR genes in Trayshed and Cabernet Sauvignon RUN1/RPV1 loci. Location of the different classes of NBS-LRR along the different chromosomes of the two haplotypes (Hap1: dark grey, Hap2: light grey) in Trayshed (A) and Cabernet Sauvignon (B). The locations of RUN1/RPV1 are indicated with an orange box. Phylogenetic trees of NBS-LRR proteins at RUN1/RPV1 locus in Trayshed and Cabernet Sauvignon, Hap1 (C) and Hap2 (D). GAPDH was used to root the tree and is colored in grey. (E) The boxplot represents an approximation of the expansion time of the protein cluster highlighted in orange in panel (C). The approximation of the divergence time between Vitis and Muscadinia genera is highlighted in grey. The same color coding is used throughout the figure to distinguish the NBS-LRR classes.
Expansion of NBS-LRR genes at the RUN1/RPV1 locus in Trayshed
The clusters of NBS-LRR genes on chromosome 12 in Trayshed are fewer and scattered in Cabernet Sauvignon (Figure 2, A and B). This chromosome contains a known powdery mildew resistance locus, RUN1/RPV1 (Pauquet et al. 2001), with boundaries defined by the VMC4f3.1 and VMC8g9 SSR markers (Barker et al. 2005). This region was studied in Trayshed (Hap1—chr12: 11,270,914—17,943,644 bp; Hap2—chr12: 11,341,795—15,346,610 bp) and Cabernet Sauvignon (Hap1—chr12: 18,746,148—23,727,933 bp; Hap2—chr12: 15,055,893—21,663,411); it included 321 and 201 annotated genes in Trayshed Hap1 and Hap2, respectively, and 287 and 312 genes in Cabernet Sauvignon haplotypes. Within RUN1/RPV1, 52 and 33 NBS-LRR were identified in Trayshed Hap1 and Hap2, respectively. For Cabernet Sauvignon, 33 and 40 NBS-LRR were identified in Hap1 and Hap2, respectively.
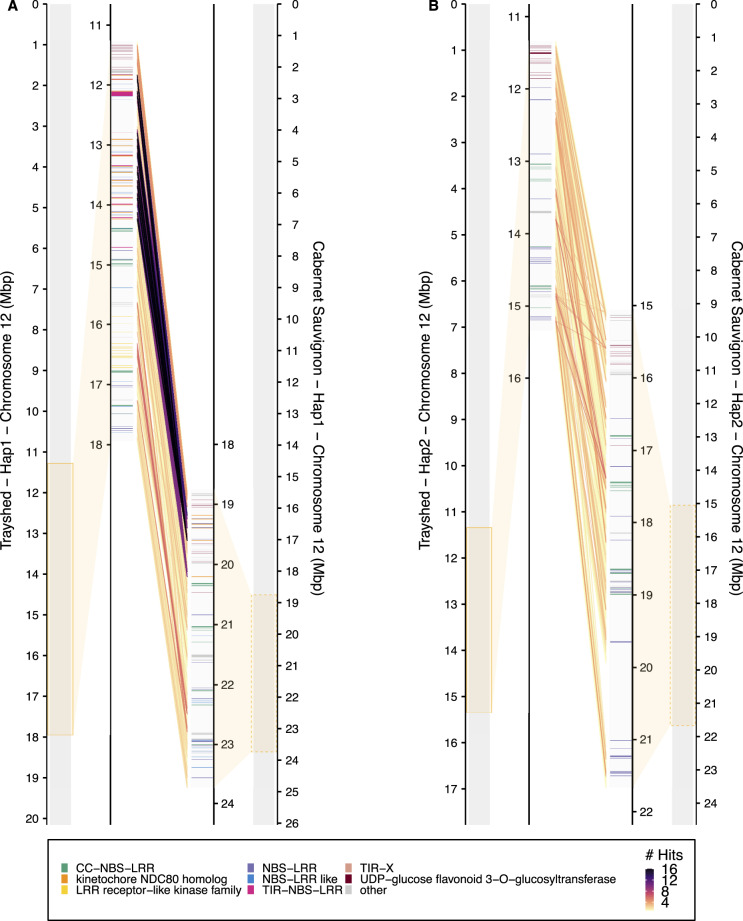
Notably, most of the NBS-LRR classes are present for both species in this region. A phylogenetic analysis of the NBS-LRR proteins at RUN1/RPV1 identified a protein cluster specific to Trayshed Hap1 (Figure 2C, Supplementary Figure S8), absent from the Hap2 (Figure 2D). After aligning the coding sequences of those proteins, the synonymous mutation rate was calculated. We estimated that TIR-NBS-LRR genes diverged at nearly 19 Ma (Figure 2E). This is one of the latest estimates of the divergence between the Muscadinia and Vitis genera found in the literature (∼18–47 Ma)(Wan et al. 2013; Liu et al. 2016; Ma et al. 2018). Thus, a recent expansion of TIR-NBS-LRR genes might have occurred in only Muscadinia and other classes may have expanded prior. To characterize gene duplication in the RUN1/RPV1 alleles, Cabernet Sauvignon haplotypes were aligned on Trayshed haplotypes to evaluate how many homologous regions are detected using tblastx. Within the resistance locus boundaries of both genomes (and excluding LTR retrotransposons), the highest number of alignments was observed when Trayshed Hap1 was used as the target (Figure 3, Supplementary Figure S9). There were 10,270 regions aligned with Cabernet Sauvignon Hap1 (Figure 3A) and 8,372 alignments between Hap2 (Figure 3B). On average, one region in Cabernet Sauvignon Hap1 had 1.75 hits in Trayshed Hap1 and 1.18 hits in Hap2. For the most duplicated region, a single sequence from Cabernet Sauvignon Hap1 matched up to 16 different regions in Trayshed Hap1 (Figure 3A). The increased number of hits per window that was observed in Trayshed Hap1 evidenced the expansion of the locus through an increase of the copy number of homologous regions. The regions in Trayshed Hap1 with the highest number of homologs (with at least 10 hits matching a single Cabernet Sauvignon region) were positioned between ∼11.83 and 14.24 Mbp on chromosome 12 (Figure 3A) and is similar to the genomic region identified by Feechan et al. (2013). It includes UDP-glucosyltransferase-coding genes followed by R gene analogs. In Cabernet Sauvignon, highly duplicated regions were mostly composed of kinetochore NDC80 complex genes; these matched genes with similar or NBS-LRR functions in Trayshed. It is important to note that the assembly of the two RUN1/RPV1 loci of Trayshed was complex. Despite the currently available long-read sequencing technologies, the highly duplicated content at RUN1/RPV1 still presents several technical limitations.
Figure 3.
Synteny between the RUN1/RPV1 alleles in Trayshed and Cabernet Sauvignon. Chromosome 12 of Trayshed (left, target) and Cabernet Sauvignon (right, query) Hap1 (A) or Hap2 (B) are compared with tblastx of 50Kbp blocks. The RUN1/RPV1 resistance locus is highlighted with an orange box. Syntenic gene content within the locus is represented as lines in the center of each panel. Duplications of Cabernet Sauvignon sequences are shown using a color gradient. Highly duplicated sequences in Trayshed are dark purple. The functional annotations depicted include categories represented by at least 5 genes. Categories represented by less than 10 genes and not related to disease resistance genes are included in the category, “other”.
Between Trayshed and Cabernet Sauvignon, functionally annotated disease-related genes were differently clustered along the chromosomes. An expansion of TIR-NBS-LRR was identified, which likely occurred after the divergence of their genera. Similar research could be undertaken to characterize the genes or features at other known resistance loci in M. rotundifolia, like RUN2 and REN5 (Riaz et al. 2011; Blanc et al. 2012). Acquiring this understanding could be expedited with the availability of high-quality reference sequences for resistant selections, like Trayshed, and susceptible cultivars, like Cabernet Sauvignon and others (Massonnet et al. 2020).
Data availability
Sequencing data are accessible at the NCBI repository under the accessions PRJNA635946 and PRJNA593045. Raw optical maps are available at Zenodo under the DOI 10.5281/zenodo.3866087. The supplemental files are available at figshare: https://doi.org/10.25387/g3.13370819. The pipeline for the gene annotation is available at https://github.com/andreaminio/AnnotationPipeline-EVM_based-DClab. Assembly and annotation files are available at www.grapegenomics.com, which also hosts a genome browser and a blast tool for Trayshed.
Funding
This work was funded by the NSF grant #1741627 and partially supported by funds to Dario Cantu from the E. & J. Gallo Winery and the Louis P. Martini Endowment in Viticulture.
Conflict of Interest: The authors declare that they have no competing interests.
Literature cited
- Au KF, Underwood JG, Lee L, Wong WH.. 2012. Improving pacbio long read accuracy by short read alignment. PLoS ONE 7:e46679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C.. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36. [PubMed] [Google Scholar]
- Barker CL, Donald T, Pauquet J, Ratnaparkhe MB, Bouquet A, et al. 2005. Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor Appl Genet. 111:370–377. [DOI] [PubMed] [Google Scholar]
- Bertier L, Leus L, D’hondt L, de Cock AWAM, Höfte M.. 2013. Host adaptation and speciation through hybridization and polyploidy in Phytophthora. PLoS ONE 8:e85385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc S, Wiedemann-Merdinoglu S, Dumas V, Mestre P, Merdinoglu D.. 2012. A reference genetic map of Muscadinia rotundifolia and identification of Ren5, a new major locus for resistance to grapevine powdery mildew. Theor Appl Genet. 125:1663–1675. [DOI] [PubMed] [Google Scholar]
- Blanco-Ulate B, Vincenti E, Powell ALT, Cantu D.. 2013. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front Plant Sci. 4:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, Thierry-Mieg J, Thierry-Mieg D, Busby B, Madden TL.. 2019. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinformatics 20:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquet A. 1978. La muscadine (Vitis rotundifolia michx) et sa culture aux Etats-Unis. OENO One. 12:1. 10.20870/oeno-one.1978.12.1.1413 [DOI] [Google Scholar]
- Bouquet A. 1980. Vitis x Muscadinia hybridization: a new way in grape breeding for disease resistance in France. Proceedings of the 3rd International Symposium on Grape Breeding, Department of Viticulture and Enology, University of California, Davis.p. 42–61.
- Branas MM. 1932. Sur la caryologie des Ampélidées. C R Acad Sci Paris. 194:121–123. [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaguier A, Grimplet J, Gaspero GD, Scalabrin S, Duchene E, et al. 2017. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom Data 14:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charif D, Lobry JR.. 2007.SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. In: U Bastolla, M Porto, HE Roman, M Vendruscolo, editors. Structural Approaches to Sequence Evolution: Molecules, Networks, Populations. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 207–232. [Google Scholar]
- Chu Z-F, Wen J, Yang Y-P, Nie Z-L, Meng Y.. 2018. Genome size variation and evolution in the grape family Vitaceae: Genome size variation in Vitaceae. Jnl of Sytematics Evolution 56:273–282. [Google Scholar]
- Davidis UX, Olmo HP.. 1964. The Vitis vinifera x V. rotundifolia hybrids as phylloxera resistant rootstocks. Vitis 4:129–143. [Google Scholar]
- Dearing C. 1917. Muscadine grape breeding. J Hered. 8:409–424. [Google Scholar]
- Delame M, Prado E, Blanc S, Robert-Siegwald G, Schneider C, et al. 2019. Introgression reshapes recombination distribution in grapevine interspecific hybrids. Theor Appl Genet. 132:1073–1087. [DOI] [PubMed] [Google Scholar]
- Doležel J, Bartoš J.. 2005. Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot. 95:99–110. [10.1093/aob/mci005] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S.. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol Plant 85:625–631. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott TA, Gregory TR.. 2015. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philos Trans R Soc Lond B Biol Sci. 370:20140331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Anderson C, Torregrosa L, Jermakow A, Mestre P, et al. 2013. Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J. 76:661–674. [DOI] [PubMed] [Google Scholar]
- Firoozabady E, Olmo HP.. 1982. Resistance to grape phylloxera in Vitis vinifera x V. rotundifolia grape hybrids. Vitis 21:1–4. [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36:3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Zhu S, Zhao R, Jiang Z, Ji Y, et al. 2018. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol Plant 11:879–882. [DOI] [PubMed] [Google Scholar]
- Heinitz CC, Uretsky J, Dodson Peterson JC, Huerta-Acosta KG, 2019. Crop Wild Relatives of Grape (Vitis vinifera L.) Throughout North America. In: SL Greene, KA Williams, CK Khoury, MB Kantar, MA Walker, LF. Marek, editors. North American Crop Wild Relatives, Volume 2: Important Species. Cham: Springer International Publishing. p. 329–351. [Google Scholar]
- Holub EB. 2001. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet. 2:516–527. [DOI] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q.. 2001. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 39:285–312. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–468. [10.1038/nature06148] [DOI] [PubMed] [Google Scholar]
- Jo J, Venkatesh J, Han K, Lee H-Y, Choi GJ, et al. 2017. Molecular mapping of PMR1, a novel locus conferring resistance to powdery mildew in pepper (Capsicumannuum). Front Plant Sci. 8:2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkamkar SP, Patil SG, Misra SC.. 2010. Cyto–morphological studies and their significance in evolution of family Vitaceae. Nucleus 53:37–43. [Google Scholar]
- Kent WJ. 2002. BLAT : the blast-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, et al. 2012. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 30:771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Gentleman R, Carey V.. 2009. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25:1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, et al. 2013. Software for computing and annotating genomic ranges PLoS Comput Biol. 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewter J, Worthington ML, Clark JR, Varanasi AV, Nelson L, et al. 2019. High-density linkage maps and loci for berry color and flower sex in muscadine grape (Vitis rotundifolia). Theor Appl Genet. 132:1571–1585. [DOI] [PubMed] [Google Scholar]
- Liu X-Q, Ickert-Bond SM, Nie Z-L, Zhou Z, Chen L-Q, et al. 2016. Phylogeny of the Ampelocissus–Vitis clade in Vitaceae supports the New World origin of the grape genus. Mol Phylogenet Evol. 95:217–228. [DOI] [PubMed] [Google Scholar]
- Lomsadze A. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33:6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J.. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- Ma Z-Y, Wen J, Ickert-Bond SM, Nie Z-L, Chen L-Q, et al. 2018. Phylogenomics, biogeography, and adaptive radiation of grapes. Mol Phylogenet Evol. 129:258–267. [DOI] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, et al. 2018. MUMmer4: a fast and versatile genome alignment system PLoS Comput Biol. 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M, Cochetel N, Minio A, Vondras AM, Lin J, et al. 2020. The genetic basis of sex determination in grapes. Nat Commun. 11:2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdinoglu D, Schneider C, Prado E, Wiedemann-Merdinoglu S, Mestre P.. 2018. Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One 52:203–209. [Google Scholar]
- Merdinoglu D, Wiedeman-Merdinoglu S, Coste P, Dumas V, Haetty S, et al. 2003. Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hortic. 603:451–456. [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW.. 2003. Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15:809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olien WC. 1990. The muscadine grape: botany, viticulture, history, and current industry. Am SocHortic Sci. 25:732–739. [Google Scholar]
- Olmo HP. 1937. Chromosome numbers in the European grape (Vitis vinifera). CYTOLOGIA Fujii Jubilae. FujiiJubilaei:606–613. [Google Scholar]
- Olmo HP. 1986. The potential role of (vinifera x rotundifolia) hybrids in grape variety improvement. Experientia 42:921–926. [Google Scholar]
- Olmo HP. 1971. Vinifera rotundifolia hybrids as wine grapes. Am J Enol Vitic. 22:87–91. [Google Scholar]
- Pagès H, Aboyoun P, Gentleman R, DebRoy S.. 2019. Biostrings: Efficient manipulation of biological strings.
- Paradis E, Schliep K.. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R Schwartz. Bioinformatics 35:526–528. [DOI] [PubMed] [Google Scholar]
- Patel GI, Olmo HP.. 1955. Cytogenetics of Vitis: I. The hybrid V. vinifera x V. rotundifolia. Am J Bot. 42:141–159. [Google Scholar]
- Patil SG, Patil VP.. 1992. Karyomorphology of Vitis vinifera, V. rotundifolia and their hybrid. Cytologia 57:91–95. [Google Scholar]
- Pauquet J, Bouquet A, This P, Adam-Blondon A-F.. 2001. Establishment of a local map of AFLP markers around the powdery mildew resistance gene Run1 in grapevine and assessment of their usefulness for marker assisted selection. Theor Appl Genet. 103:1201–1210. [10.1007/s001220100664] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, et al. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing Austria. [Google Scholar]
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F.. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35:2582–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravaz L. 1902. Les Vignes américaines. Porte greffes et producteurs directs, caractères, aptitudes. Publishers: Coulet, Masson. Locations: Montpellier and Paris, France.
- Riaz S, Tenscher AC, Ramming DW, Walker MA.. 2011. Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor Appl Genet. 122:1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Kurth J, Leister D.. 2002. Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol. 19:76–84. [DOI] [PubMed] [Google Scholar]
- RStudio T. 2020. RStudio: Integrated Development Environment for R. Boston, MA: RStudio, Inc. [Google Scholar]
- Ruel JJ, Walker MA.. 2006. Resistance to pierce’s disease in Muscadinia rotundifolia and other native grape species. Am J Enol Vitic. 57:158–165. [Google Scholar]
- Slater G, Birney E.. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small JK. 1913. Flora of the Southeastern United States: being Descriptions of the Seed-Plants, Ferns and Fern-Allies Growing Naturally in North Carolina, South Carolina, Georgia, Florida, Tennessee, Alabama, Mississippi, Arkansas, Louisiana and in Oklahoma and Texas East of the One Hundredth Meridian. New York, NY: J.K.Small. [Google Scholar]
- Smit A, Hubley R, Green P.. 2013. RepeatMasker Open-4.0. http://www.repeatmasker.org 2013-2015.
- Sousa A, Renner SS.. 2015. Interstitial telomere-like repeats in the monocot family Araceae: chromosome evolution in the Araceae. Bot J Linn Soc. 177:15–26. [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt G, Kassemeyer HH.. 1995. Evaluation of downy mildew resistance in various accessions of wild Vitis species. Vitis 34:225–228. [Google Scholar]
- Steuernagel B, Witek K, Krattinger SG, Ramirez-Gonzalez RH, Schoonbeek H, et al. 2020. The NLR-annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. 183:468–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vondras AM, Minio A, Blanco-Ulate B, Figueroa-Balderas R, Penn MA, et al. 2019. The genomic diversification of grapevine clones. BMC Genomics 20:972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O. 2017. ggseqlogo: A “ggplot2” Extension for Drawing Publication-Ready Sequence. Bioinformatics 33:3645–3647. [DOI] [PubMed]
- Walker MA, Lider LA, Goheen AC, Olmo HP.. 1991. VR 039-16 Grape Rootstock. Hortic Sci. 26:1224–1225. [Google Scholar]
- Walker MA, Lund K, Agüero C, Riaz S, Fort K, et al. 2014. Breeding grape rootstocks for resistance to phylloxera and nematodes - it’s not always easy. Acta Hortic. 1045:89–97. [Google Scholar]
- Wan Y, Schwaninger HR, Baldo AM, Labate JA, Zhong G-Y, et al. 2013. A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change. BMC Evol Biol. 13:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, DeBarry JD, Tan X, Li J, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW.. 1998. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10:1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, et al. 1999. The Mla (Powdery Mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of Barley. Genetics 153:1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Lu L-M, Nie Z-L, Liu X-Q, Zhang N, et al. 2018. A new phylogenetic tribal classification of the grape family (Vitaceae): Tribal classification of Vitaceae. Jnl of Sytematics Evolution 56:262–272. [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, et al. 2019. Welcome to the Tidyverse. JOSS 4:1686. [Google Scholar]
- Wu TD, Watanabe CK.. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y.. 2017. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8:28–36. [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Valè G, et al. 2001. Cell-autonomous expression of Barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13:337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Minio A, Massonnet M, Solares E, Lv Y, et al. 2019. The population genetics of structural variants in grapevine domestication. Nat Plants 5:965–979. [DOI] [PubMed] [Google Scholar]
- Zini E, Dolzani C, Stefanini M, Gratl V, Bettinelli P, et al. 2019. R-loci arrangement versus downy and powdery mildew resistance level: a Vitis hybrid survey. Int JMol Sci. 20:3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Karn A, Reisch B, Nguyen A, Sun Y, et al. 2020. Haplotyping the Vitis collinear core genome with rhAmpSeq improves marker transferability in a diverse genus. Nat Commun. 11:413.[10.1038/s41467-019-14280-1] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequencing data are accessible at the NCBI repository under the accessions PRJNA635946 and PRJNA593045. Raw optical maps are available at Zenodo under the DOI 10.5281/zenodo.3866087. The supplemental files are available at figshare: https://doi.org/10.25387/g3.13370819. The pipeline for the gene annotation is available at https://github.com/andreaminio/AnnotationPipeline-EVM_based-DClab. Assembly and annotation files are available at www.grapegenomics.com, which also hosts a genome browser and a blast tool for Trayshed.