Abstract
Prostate cancer (PCa) is the second leading cause of cancer-related death among men in the United States. Androgen receptor (AR) signaling is the dominant oncogenic pathway in PCa and the main strategy of PCa treatment is to control the AR activity. A large number of patients acquire resistance to Androgen deprivation therapy (ADT) due to AR aberrant activation, resulting in castration-resistant prostate cancer (CRPC). Understanding the molecular mechanisms underlying AR signaling in the PCa is critical to identify new therapeutic targets for PCa patients. The recent advances in high-throughput RNA sequencing (RNA-seq) techniques identified an increasing number of non-coding RNAs (ncRNAs) that play critical roles through various mechanisms in different diseases. Some ncRNAs have shown great potentials as biomarkers and therapeutic targets. Many ncRNAs have been investigated to regulate PCa through direct association with AR. In this review, we aim to comprehensively summarize recent findings of the functional roles and molecular mechanisms of AR-related ncRNAs as AR regulators or targets in the progression of PCa.
Keywords: prostate cancer, androgen receptor, non-coding RNA, microRNA, lncRNA, circRNA
Introduction
Prostate cancer (PCa) is the most frequently diagnosed cancer and the second-highest cause of cancer death among men in the United States, with an estimated 248,530 new cases and 34,130 deaths expected in 2021 in the United States (Siegel et al., 2021). The growth and survival of PCa are mainly dependent on the sex steroid hormone, androgens (Folkerd and Dowsett, 2010). The androgen receptor (AR) is a ligand-activated transcription factor that is vital for both normal prostate development and tumorigenesis. Upon binding by androgen in the cytoplasm, AR dimerizes and translocates to the nucleus, stimulating target gene transcription through association with androgen response elements (AREs) within promoter and enhancer sequences. AR and its downstream signal cascades are critical for the initiation and progression of both localized and advanced metastatic PCa (Scher and Sawyers, 2005). Advances in screening and therapeutic strategies promoted successful treatment of PCa by surgery and/or radiation. The testing of prostate-specific antigen (PSA), a prototypic AR target, has been used for years as a diagnostic biomarker for the disease (Lilja et al., 2008). Androgen deprivation therapy (ADT) by AR antagonists and chemical castration is the standard treatment for patients with biochemical recurrence after primary therapy or with locally advanced or metastatic disease (Feldman and Feldman, 2001). Patients with metastasis-free PCa have a 100% 5-year survival rate (Brawley, 2012). Unfortunately, the majority of primary cancers will eventually acquire ADT resistance and progress to castration-resistant prostate cancer (CRPC) (Kirby et al., 2011). Patients with metastatic PCa have a low 5-year survival rate (Kirby et al., 2011; Brawley, 2012). Generally, CRPC is caused by AR aberrant activation with enhanced AR expression, hypersensitivity to androgens (Waltering et al., 2009), intra-tumoral steroidogenesis (Locke et al., 2008), and abnormal AR splicing variant expression (Sun et al., 2010). Recent studies revealed that the frequency of AR-negative neuroendocrine prostate cancer (NEPC) and AR-Null and Neuroendocrine-Null Prostate Cancer (Double-Negative PCa, DNPC) is elevated due to the application of potent AR antagonists such as enzalutamide (ENZ) and abiraterone (Aparicio et al., 2011; Hu et al., 2015; Bluemn et al., 2017; Labrecque et al., 2019). Therefore, there is an urgent need to elucidate the molecular mechanisms contributing to the development of AR-dependent CRPC as well as advanced NEPC and DNPC for developing alternative therapeutic options for advanced PCa.
Next-generation RNA sequencing (RNA-seq) advances have become overly convenient to measure gene expression levels and explore new transcriptional units across the transcriptome. 2% of the human genome encodes approximately 20,000 protein-coding genes (Lander et al., 2001; Venter et al., 2001; Alexander et al., 2010; Consortium, 2012) and up to 80% of the human genome encodes a large number of non-coding RNAs (ncRNA) (Consortium, 2012; Djebali et al., 2012). Ribosomal RNA (rRNA) (about 80% of the total RNA weight) and transfer RNA (tRNA) (about 15% of the total RNA weight) are two of the most abundant ncRNA types in cells (Palazzo and Lee, 2015). The other ncRNAs are categorized as short ncRNAs (sncRNAs) and long ncRNAs (lncRNAs) based on whether their size is longer than 200 bases (Cech and Steitz, 2014). SncRNAs include microRNA (miRNA) (Lagos-Quintana et al., 2001), small interfering RNA (siRNA) (Hamilton and Baulcombe, 1999), piwi-interacting RNA (piRNA) (Siomi et al., 2011), small nucleolar RNA (snoRNA) (Kiss, 2001; Matera et al., 2007), small nuclear RNA (snRNA) (Matera et al., 2007; Guiro and Murphy, 2017), and tRNA-derived fragments (tRF) (Schimmel, 2018). Unlike linear lncRNAs, circular RNAs (circRNAs) are single-stranded circularized ncRNAs commonly generated from the precursor mRNA (pre-mRNA) back-splicing process (Memczak et al., 2013; Salzman, 2016). Functionally, rRNA, tRNA, snoRNA, and snRNA are housekeeping ncRNAs, while miRNA, siRNA, piRNA, tRF, lncRNA, and circRNA are regulatory transcripts.
Over recent decades, ncRNAs have been emerged as critical regulators instead of junk RNAs in different disease processes, including cancer (Du et al., 2013; Hayes et al., 2014; Vo et al., 2019). Notably, many ncRNAs (miRNA, lncRNA, and circRNA) are aberrantly expressed with significant contribution to PCa initiation and/or progression (Bonci et al., 2008; Prensner et al., 2011; Pickl et al., 2014; Fredsoe et al., 2018; Hua et al., 2018; Chen S. et al., 2019). MiR-101 negatively regulates EZH2 expression by binding to EZH2 3′ untranslated region (UTR), and has a strong negative correlation with PCa progression from benign to localized disease to metastasis (Varambally et al., 2008). LncRNA SChLAP1 is critical for PCa cell invasiveness and metastasis through antagonizing the genome-wide localization and regulatory functions of the SWI/SNF chromatin-modifying complex (Prensner et al., 2013). CircRNA 0005276 (circ0005276), a circular RNA stem from XIAP, is highly expressed in PCa tissues with advanced tumor stage and metastasis. Circ0005276 interacts with FUS to regulate the transcription of XIAP in PCa, thus promoting the tumorigenesis and development of PCa (Feng et al., 2019). Aside from miRNAs, lncRNAs and circRNAs are relatively new players in the ncRNA field and are less well understood. ncRNAs are gaining widespread attention for their abundance in number, expression specificity, functional roles in diseases, and potential clinical applications. Given the critical role of AR in PCa initiation and progression, we will focus on the impact of miRNA, lncRNA, and circRNA, on AR’s function in PCa in this review.
ncRNAs as AR Regulators
AR Is Repressed by miRNAs
MicroRNAs are a family of small untranslated RNAs with ∼21–25 nucleotides in size that control gene expression by mediating target mRNA degradation (He and Hannon, 2004; Bartel, 2009), and repressing (Pillai et al., 2005; Petersen et al., 2006; Mathonnet et al., 2007) or promoting (Vasudevan et al., 2007; Truesdell et al., 2012) target mRNA translation. These regulations often occur through the association of miRNAs with 3′ UTRs of transcripts (Bartel, 2009; Krol et al., 2010), and some miRNAs may also target 5′ UTR and coding regions of transcripts. Many tools have been developed to predict miRNA targets (Chen L. et al., 2019). AR mRNA is comprised of a 1.1 kb 5′ UTR, a 2.7 kb open reading frame (ORF), and an exceptionally long 3′ UTR with a length of approximately 6.8 kb (Ostling et al., 2011; Ebron and Shukla, 2016). Thus AR mRNA is the most miRNA targeted transcript in PCa cells (Hamilton et al., 2016; Table 1).
TABLE 1.
MiRNAs targeting AR mRNA.
| MiRNAs (References) | Expression in PCa | Target region of AR mRNA | Cancer types | Function in cancer cells |
| miR-124 (Shi et al., 2013, 2015; Xiong et al., 2017) | ↓ | 3′UTR | PCa, bladder cancer | Inhibits cell growth and increases apoptosis |
| miR-135b (Ostling et al., 2011; Aakula et al., 2015; Bao S. X. et al., 2020) | ↓ | 3′UTR | PCa, hepatocellular carcinoma (HCC), breast cancer | Inhibits cell growth |
| miR-181c-5p (Wu et al., 2019) | – | 3′UTR | PCa | Increases ENZ sensitivity of PCa cells and represses cell invasion |
| miR-185 (Ostling et al., 2011; Qu et al., 2013; Liu C. et al., 2015; Kalinina et al., 2020) | ↓ | 3′UTR | PCa, breast cancer | Suppresses cell growth, migration, invasion, and tumorigenicity |
| miR-193a-3p (Kumar et al., 2016; Liu et al., 2017) | ↓ | Coding region | PCa | Suppresses cell growth |
| miR-197-3p (Fletcher et al., 2019; Huang et al., 2020) | ↓ | 3′UTR | PCa | Inhibits cell growth and colony formation |
| miR-205 (Boll et al., 2013; Hagman et al., 2013; Coarfa et al., 2016; Kalinina et al., 2020) | ↓ | 3′UTR | PCa, breast cancer | Suppresses cell growth, migration, and invasion |
| miR-297 (Ostling et al., 2011; Fang et al., 2016) | ↓ | 3′UTR | PCa | Suppresses cell growth |
| miR-299-3p (Ostling et al., 2011; Ganapathy et al., 2020) | ↓ | 3′UTR | PCa | Suppresses cell growth, migration, induces cell cycle arrest, and apoptosis |
| miR-30b-3p (Kao et al., 2014; Kumar et al., 2016) | ↓ | 3′UTR | PCa | Suppresses EMT phenotypes and inhibits cell migration and invasion |
| miR-31 (Lin et al., 2013; Coarfa et al., 2016; Wang et al., 2016) | ↓ | Coding region | PCa | Suppresses cell growth, migration and invasiveness |
| miR-320a (Okato et al., 2016; Sato et al., 2016; Lieb et al., 2018) | ↓ | 3′UTR | PCa | Suppresses cell growth, migration and invasiveness |
| miR-320b (Hsieh et al., 2013; Lieb et al., 2018; Dai et al., 2019) | ↓ | 3′UTR | PCa | Suppresses cell growth |
| miR-346 (Fletcher et al., 2019; Lin et al., 2020) | ↑ | 3′UTR | PCa | Promotes cell growth and invasion |
| miR-34a (Liu et al., 2011; Ostling et al., 2011; Kashat et al., 2012; Leite et al., 2015) | ↓ | 3′UTR | PCa | Inhibits prostate cancer stem cell regeneration and metastasis |
| miR-34c (Hagman et al., 2010; Ostling et al., 2011; Walter et al., 2013; Fang et al., 2016) | ↓ | 3′UTR | PCa | Suppresses cell growth |
| miR-361-3p (Fletcher et al., 2019; Liu B. et al., 2020) | ↓ | 3′UTR | PCa | Suppresses cell growth and increases ENZ sensitivity of PCa cells |
| miR-371-3p (Ostling et al., 2011; Leite et al., 2015; Kumar et al., 2016) | ↓ | Coding region, 3′UTR | PCa | Suppresses cell growth |
| miR-421 (Ostling et al., 2011; Meng et al., 2016) | ↓ | 3′UTR | PCa | Suppresses cell growth, induces cell cycle arrest, reduces glycolysis, and inhibits migration |
| miR-449a (Ostling et al., 2011; Zheng et al., 2015; Chen W. et al., 2017; Guo et al., 2018) | ↓ | 3′UTR | PCa, bladder cancer | Suppresses cell growth, invasion, and angiogenesis |
| miR-449b (Ostling et al., 2011; Mortensen et al., 2014) | ↑ | 3′UTR | PCa | Suppresses cell growth |
| miR-488∗ (Sikand et al., 2011; Ebron and Shukla, 2016) | - | 3′UTR | PCa | Suppresses cell growth, increases apoptosis |
| miR-541-3p (Kumar et al., 2016; He et al., 2021) | ↓ | 3′UTR | PCa | Suppresses cell growth and enhances the radiosensitivity of PCa cells |
| miR-634 (Ostling et al., 2011; Kumar et al., 2016) | – | 3′UTR | PCa | Suppresses cell growth |
| miR-646 (Kumar et al., 2016) | – | Coding region 3′UTR | PCa | Suppresses cell growth |
| miR-654-5p (Ostling et al., 2011; Kumar et al., 2016) | – | 3′UTR | PCa | Suppresses cell growth |
| miR-92a-2-5p (Liu G. et al., 2020) | – | 3′UTR | HCC | Increases liver cancer cell invasion |
| miR-9-5p (Ostling et al., 2011; Kumar et al., 2016; Moazzeni et al., 2017; Bandini et al., 2020) | – | 3′UTR | PCa, breast cancer | Suppresses cell growth |
Several groups have systemically explored AR modulatory miRNAs through different strategies (Ostling et al., 2011; Hamilton et al., 2016; Kumar et al., 2016; Fletcher et al., 2019). Ostling et al. (2011) and Kumar et al. (2016) used pre-miR libraries to perform gain-of-function screening for AR-modulatory miRs in PCa cells. Östling et al. identified and validated 13 miRNAs that interact with the AR 3′ UTR region and could significantly reduce AR 3′ UTR activity: miR-135b, miR-185, miR-297, miR-299-3p, miR-34a, miR-34c, miR-371-3p, miR-421, miR-449a, miR-449b, miR-634, miR-654-5p, and miR-9. Among these miRNAs, miR-185 and miR-34a were consistently reported by other groups to show the regulatory function on PCa aggressiveness through directly targeting AR (Kashat et al., 2012; Qu et al., 2013). Similarly, Kumar et al. (2016) identified 15 miRNAs (miR-101-3p, miR-138-5p, miR-149-3p, miR-30b-3p, miR-30c-5p, miR-30d-5p, miR-411-3p, miR-425-5p, miR-488-5p, miR-541-3p, miR-635, miR-646, miR-650, miR-654, and miR-9-5p) that significantly suppressed AR 3′ UTR reporter activity, especially miR-9-5p, miR-30b-3p and miR-541-3p, highlighting the critical role of miR-30 family members in inhibiting AR activity through 3′ UTR association. The study also revealed that 3 miRNAs, miR-371-3p, miR-193-3p, and miR-646 could suppress AR transcriptional activity through binding sites within the coding region of AR mRNA (Kumar et al., 2016). The role of miR-488 in inhibiting AR expression in PCa cells was also confirmed by Sikand et al. (2011). Hamilton et al. (2016) applied photoactivatable ribonucleoside-enhanced cross-linking immunoprecipitation of the Argonaute protein and sequencing (AGO-CLIP-Seq) to broadly explore interactions between miRNAs and miRNA target sites in a panel of PCa cells (Hamilton et al., 2016). Among 22 PCa driver genes, AR 3′ UTR has the most abundant miRNA target sites (71 unique miRNA families at 147 seed sites), including the miR-135, miR-185, miR-34, miR-421, and miR-9 families reported by Ostling et al. (2011).
Recently, using a library of LNA-modified antisense inhibitors against 983 human miRNAs, Fletcher et al. (2019) systematically identified microRNAs modulating AR activity in PCa cells. The application of the miR inhibitors limits off-target or non-specific effects by avoiding targeting endogenous miRNA processing or effector complexes. 78 miRNA inhibitors were found to significantly modulate AR reporter activity, including inhibitors of miR-135b, miR-421, miR-449a, miR-634, and miR-654-5p, which is consistent with Ostling et al.’s (2011) report. Interestingly, inhibition of miR-346, miR-361-3p, and miR-197 significantly reduced AR activity in a dose-dependent manner. Upregulation of AR 3′ UTR activity by miR-346, miR-361-3p, and miR-197 was also confirmed by AR 3′ UTR reporter assay combined with miRNA mimics. Mimics of miR-346, miR-361-3p, and miR-197 also prevented Actinomycin D-induced loss of AR transcript. Previous studies demonstrated that miR-346 binds to 3′ UTR of AGO2 and hTERT. Upon miR-346 binding to the 3′ UTR of AGO2 and hTERT, the middle sequence motif (CCGCAU) of miR-346 recruits G-rich RNA sequence binding factor 1 (GRSF1) to form a “bulge loop,” thus facilitating the recruitment of AGO2 mRNA and hTERT mRNA to ribosomes to promote translation (Guo et al., 2015; Song et al., 2015). MiR-346, miR-361-3p, and miR-197 may enhance AR mRNA stability and promote AR expression through a similar mechanism. In Fletcher et al.’s (2019) study, miR-197 inhibitor increased caspase activity and suppressed cell growth, indicating miR-197 may act as a tumor promoter. However, miR-197’s expression was lower in prostate tumor tissues than in normal tissues (Fletcher et al., 2019), which is consistent with the miR-197 inhibitor’s role in promoting PCa cell growth reported by Huang et al. (2020). Besides, the miR-361-3p inhibitor also suppressed PCa cell growth (Fletcher et al., 2019), while another report showed that miR-361-3p enhanced ENZ sensitivity of PCa cells and inhibited PCa cell growth by suppressing AR splice variant 7 (AR-V7) expression (Liu B. et al., 2020). Thus, the function of miR-197 and miR-361-3p in PCa cell growth from different studies are conflicted. Some miRNAs may be able to either activate or repress mRNA translation, and whether they act as translation activators or repressors highly depends on cell cycle and RNA binding factors (Vasudevan et al., 2007; Truesdell et al., 2012). Further studies are necessary to disentangle these contradictory results.
Many miRNAs targeting AR mRNA might be not reflected in the result from the above systematic analysis (Table 1). MiR-205 is mainly expressed in prostate basal epithelial cells (Zhang et al., 2010; Gandellini et al., 2012), and PCa patients with low expression of miR-205 have poor survival (Hagman et al., 2013). MiR-205 can suppress AR expression by binding to AR 3′ UTR, and also interfere with MAPK and IL-6 signaling pathways in PCa cells (Boll et al., 2013; Hagman et al., 2013), while the promoter region of the miR-205 gene contains ARE and the expression of miR-205 is increased after AR activation by R1881 treatment (Hagman et al., 2013). MiR-212 is downregulated in prostate tumor cells, and it suppresses the transcription of AR and AR-V7 through direct targeting hnRNPH1, which may regulate AR mRNA transcription or splicing. Interestingly, hnRNPH1 can also interact with AR protein and modulate AR binding to target genes (Yang et al., 2016). MiR-31 is regulated by promoter hypermethylation in both triple-negative breast cancer (Augoff et al., 2012) and PCa (Lin et al., 2013), and its expression is negatively correlated with the aggressiveness of the PCa. MiR-31 directly inhibits AR expression through binding to the coding region of AR mRNA. Subsequently, genes related to cell cycle regulation are also repressed by miR-31 as its direct targets. Interestingly, miR-31 can be suppressed by AR as a transcriptional target, forming a negative regulation loop between miR-31 and AR (Lin et al., 2013). MiR-124 was downregulated in PCa (Shi et al., 2013, 2015), breast cancer (Feng et al., 2016), and bladder cancer (Xiong et al., 2017). Intravenous delivery of miR-124 in combination with ENZ sufficiently inhibited prostate tumor growth and increased cell apoptosis (Shi et al., 2015). Mechanistically, miR-124 directly represses AR along with EZH2 and SRC through binding to the 3′ UTR regions of these mRNAs (Shi et al., 2015). HDAC inhibitor, OBP-801, induced miR-320a mediated suppression of AR expression through binding to the 3′UTR of AR (Sato et al., 2016). Both miR-181c-5p and miR-361-3p could regulate the expression of AR-V7 but not wild-type AR in PCa cells via binding to the specific target sequence in the AR-V7 3′UTR (Wu et al., 2019; Liu B. et al., 2020). Besides, exosome transportation of miR-92a-2-5p from macrophages to liver cancer cells could suppress AR expression by directly targeting AR 3′UTR and enhance the invasion capacity of liver cancer cells (Liu G. et al., 2020).
AR Is a Target of lncRNAs
The size of lncRNAs is normally longer than 200 bases, and thus they can fold into complex structures to carry out various functions through interaction with protein, chromatin, and RNA (Schmitt and Chang, 2016; Goodall and Wickramasinghe, 2020). LncRNAs have been reported to regulate gene transcription by recruiting transcription regulators or direct interaction with chromatin, affect protein and mRNA stability through direct binding, and act as sponges for miRNAs (Goodall and Wickramasinghe, 2020). With these diverse regulatory mechanisms, lncRNAs regulate numerous signal pathways and play critical roles in different cellular processes and disease progression. Therefore, lncRNAs have great potentials as biomarkers or therapeutic targets. Currently, many lncRNAs are characterized to participate in PCa progression through direct association with AR protein, DNA, or mRNA (Table 2).
TABLE 2.
LncRNAs regulating AR.
| LncRNAs (References) | Expression in PCa | Molecular mechanisms | Cancer types | Function in cancer cells |
| ARLNC1 (Zhang et al., 2018) | ↑ | Stabilizes the AR transcript via RNA-RNA interaction | PCa | Promotes cell growth |
| GAS5 (Kino et al., 2010; Pickard et al., 2013; Hudson et al., 2014; Luo et al., 2017) | ↓ | Interacts with AR protein and suppresses its transcriptional targets | PCa, breast cancer | Promotes cell apoptosis and decreases viability |
| HOTAIR (Zhang et al., 2015) | ↑ | interacts with AR protein and protects it from degradation | PCa | Promotes cell growth and invasion |
| HOXA11-AS-203 (Schmidt et al., 2020) | – | Interacts with AR protein | Melanoma | - |
| HOXC-AS1 (Takayama et al., 2020) | ↑ | Interacts with U2AF2 and promotes AR mRNA splicing | PCa | Promotes cell growth |
| LBCS (Gu et al., 2019) | ↓ | Interacts with hnRNPK and AR mRNA to suppress AR translation | PCa | Suppresses cell growth |
| MALAT1 (Dai et al., 2019) | ↑ | Sponges miR-320b and activates AR signaling | PCa | Promotes cell growth, metastasis and invasion |
| PCAT1 (Guo et al., 2016) | ↑ | Interacts with AR protein and regulates its chromosome binding | PCa | Promotes cell growth |
| PCGEM1 (Yang et al., 2013; Hung et al., 2014; Prensner et al., 2014c; Zhang et al., 2016) | ↑ | Interacts with AR protein and enhances its transactivation | PCa | Promotes cell growth |
| PlncRNA-1 (Cui et al., 2013; Fang et al., 2016) | ↑ | Sponges miR-34c and miR-297 and protects AR mRNA | PCa | Suppresses apoptosis, promotes cell growth and migration |
| PRKAG2-AS1 (Takayama et al., 2020) | ↑ | Interacts with U2AF2 and promotes AR mRNA splicing | PCa | Promotes cell growth |
| PRNCR1 (Yang et al., 2013; Prensner et al., 2014c) | ↑ | Interacts with AR protein and enhances its transactivation | PCa | Promotes cell growth |
| SARCC (Zhai et al., 2016, 2017) | – | Interacts with AR protein and destabilizes it | RCC | Suppresses cell growth |
| SLNCR1 (Schmidt et al., 2016, 2020) | – | Interacts with AR protein and regulates its chromosome binding | Melanoma | Promotes melanoma invasion |
| SOCS2-AS1 (Misawa et al., 2016) | ↑ | Interacts with AR protein and regulates its cofactor recruitment | PCa | Promotes cell growth, migration, and suppresses apoptosis |
| SRA (Lanz et al., 1999) | – | Interacts with AR protein and enhances its transactivation | Breast cancer | – |
| YY1BM (Wu et al., 2020) | – | Blocks the interaction between YY1 and AR protein | Esophageal squamous cell carcinoma | Suppresses cell growth |
LncRNA HOTAIR was first studied in breast cancer and identified its functionality to reprogram chromatin state by affecting the chromatin occupancy of Polycomb repressive complex 2 (PRC2) and altering histone H3K27 methylation (Wu et al., 2015). Zhang et al. (2015) showed that HOTAIR is upregulated in advanced PCa and could reduce AR degradation through directly binding to AR protein which blocked the interaction between AR and MDM2. Interestingly, miR-34a was reported to suppress HOTAIR expression through direct binding (Chiyomaru et al., 2013). Coincidentally, AR is a target of miR-34a (Ostling et al., 2011), suggesting HOTAIR may also modulate AR mRNA expression by acting as a sponge of miR-34a. LncRNA PCAT1 is a prostate-specific regulator correlated with PCa progression, which was first showed to activate AKT and NF-κB signaling in CRPC through reconfiguring FKPB51-IKKα-PHLPP complex after a direct interaction with FKBP51 (Shang et al., 2019), and suppress BRCA2 expression (Prensner et al., 2014b) and regulate MYC stabilization at the post-transcriptional level (Prensner et al., 2014a). Furthermore, PCAT1 was reported to interact with AR and LSD1 (Guo et al., 2016). This interaction alters the genomic occupancy of the AR-LSD1 complex, which mainly regulates the transcription of AR target genes through interaction with chromatin (Metzger et al., 2005; Guo et al., 2016). AR has also been investigated as an oncogene in human renal cell carcinoma (RCC) (He et al., 2014; Huang Q. et al., 2017), which is consistent with the incidence that RCC is more frequently diagnosed in men than women (Siegel et al., 2021). LncRNA SARCC was reported to suppress RCC through binding and destabilizing AR protein and thus concealing AR’s downstream transcriptional targets (Zhai et al., 2016, 2017). PCGEM1 is a well-known prostate tissue-specific lncRNA associated with high-risk PCa patients (Srikantan et al., 2000; Petrovics et al., 2004; Xue et al., 2013). However, the exact mechanisms of how PCGEM1 is associated with PCa are conflicting. Yang et al. (2013) reported that PCGEM1, together with PRNCR1 could bind to AR protein and increase its activity through forming an AR-bound enhancer-promoter loop, and both contribute to castration resistance in PCa. However, Prensner et al. (2014c) failed to verify the binding of PCGEM1 and PRNCR1 to AR, and they also suggested that neither gene is a component of AR signaling. Then Hung et al. (2014) showed that PCGEM1 regulates PCa metabolism partially through AR activation, but mainly through promoting chromatin recruitment of c-MYC and activating c-MYC signaling via physical interaction between PCGEM1 and c-MYC. Interestingly, Zhang et al. (2016) reported that PCGEM1 could pull down heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and splicing factor U2AF65. The PCGEM1-hnRNP A1 interaction could suppress hnRNP A1 interaction with AR pre-mRNA, while PCGEM1-U2AF65 interaction could promote U2AF65 interaction with AR pre-mRNA, indicating that PCGEM1 may participate in the AR signaling by regulation of AR mRNA splicing (Zhang et al., 2016). The relationship between PCGEM1 and AR, and the clinical significance of PCGEM1 in PCa need further studies.
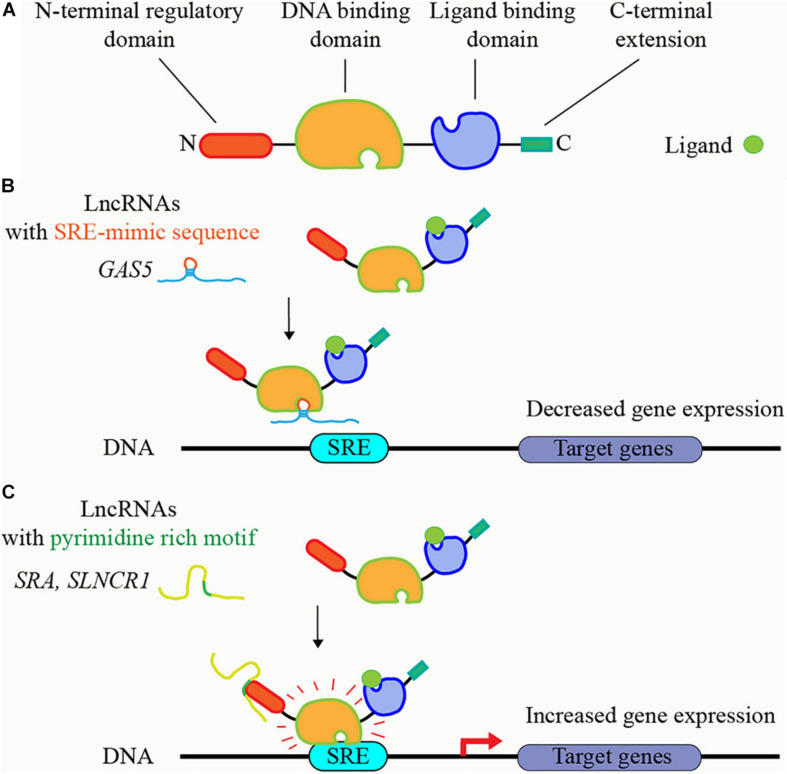
Growth arrest-specific 5 (GAS5) is a lncRNA firstly identified in growth-arrested mammalian cells (Schneider et al., 1988), and overexpression of GAS5 could induce cell apoptosis and cell cycle arrest in PCa cells (Pickard et al., 2013; Luo et al., 2017; Sun et al., 2017). Intriguingly, GAS5 was shown to interact with some steroid receptors which share similar response sequences, including glucocorticoid receptor (GR), mineralocorticoid receptor (MR), progesterone receptor (PR), and AR, at their DNA binding domains through GAS5 contained hairpin RNA glucocorticoid receptor response element (GRE)-mimic and, thereby, inhibit the association of these receptors with their DNA recognition sequence and thus repress the transcriptional activity of these steroid receptors (Kino et al., 2010). These interactions are in a conserved, sequence-specific manner (Hudson et al., 2014). LncRNA steroid receptor RNA activator (SRA) could selectively enhance the transactivation of steroid receptors, such as PR, GR, estrogen receptor (ER), and AR, through interaction with their N-terminal, regulatory domain (NTD) (Lanz et al., 1999). Schmidt et al. (2016, 2019) reported another lncRNA, steroid receptor RNA activator-like non-coding RNA (SLNCR1), recruits AR to MMP9 and EGR1-bound genomic loci to regulate melanoma invasion and proliferation. Interestingly, several confirmed AR bound lncRNAs, such as HOTAIR, SRA, SLNCR1, and even PCGEM1, all include a conserved region with a similar sequence (SLNCR1609–637) which is required for AR-lncRNA interaction (Yang et al., 2013; Zhang et al., 2015; Schmidt et al., 2016). Further investigation revealed that AR NTD binds with short, pyrimidine-rich RNA containing at least one CYUYUCCWS motif, and lncRNA HOXA11-AS-203 which contains such motif was validated to bind with AR NTD (Schmidt et al., 2020). These studies strongly suggested that some lncRNAs containing specific sequences may bind to AR protein and other steroid receptors at the DNA binding domain to compete with the target response elements and suppress their transcriptional activity, or at the N-terminal regulatory domain to modulate their transactivation.
A specific type of lncRNAs called enhancer RNAs (eRNAs) are derived from super-enhancers and have been proven to control mRNA transcription through facilitating enhancer-promoter interaction (Kim et al., 2010; Wang et al., 2011). KLK3 eRNA (KLK3e) is an eRNA produced from the upstream enhancer regions of Kallikrein-related peptidase 3 (KLK3) (Hsieh et al., 2014), a well-known AR regulated gene encoding the protein product PSA. KLK3e’s expression is induced by AR, and KLK3e could scaffold the AR-associated protein complex, the KLK3 enhancer, and the KLK2/3 promoter, resulting in enhanced transcriptional activation of nearby KLK3 and long-distance KLK2. daSilva et al. (2018) further identified numerous lncRNAs bound by AR (ARA-lncRNAs), and many of them are also transcriptionally regulated by AR. Further analysis revealed that protein-coding genes adjacent to these ARA-lncRNAs had a significantly greater androgen-induced change in expression than protein-coding genes neighboring lncRNAs not associated with AR, and suppressing the expression of ARA-lncRNA attenuates androgen-induced expression change of protein-coding genes adjacent to the ARA-lncRNA. These ARA-lncRNAs’ transcription start sites (TSSs) are enriched with epigenetic signatures of active enhancers, highlighting hundreds of AR-bound lncRNAs act as cis-regulatory RNA enhancers to control the androgen regulatory program of PCa cells (daSilva et al., 2018). The exact regulation mechanisms and functions of AR-associated eRNAs in PCa cells are still waiting to be fully discovered.
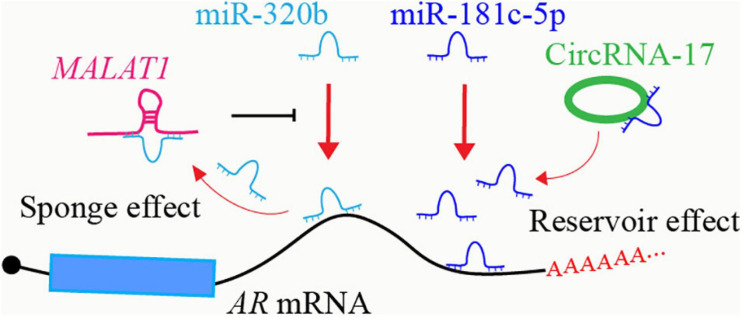
Additionally, several lncRNAs can affect AR signaling through RNA-RNA interaction. LncRNA ARLNC1 was reported to directly bind to AR mRNA 3′ UTR, stabilize AR mRNA, and increase the cytoplasmic fraction of AR mRNA, thus regulating PCa cell growth and apoptosis (Zhang et al., 2018). LncRNA LBCS was also shown to interact with AR mRNA and hnRNPK, forming a complex and suppressing AR translation efficiency (Gu et al., 2019). LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), one of the most studied lncRNAs, was previously reported to bind to EZH2 and enhance EZH2-mediated repression of Polycomb-dependent target genes (Hirata et al., 2015; Wang et al., 2015). MALAT1 could also function by activating AR signaling through sponging miR-320b which targets AR 3′ UTR (Sato et al., 2016; Dai et al., 2019).
Apart from PCGEM1, several AR-regulated lncRNAs (CRPC-lncs) which are highly expressed in CRPC tissues, were also reported to participate in AR mRNA splicing through association with splicing factors (Takayama et al., 2020). Among them, FAM83H-AS1, PRKAG2-AS1, HOXC-AS1, ELFN1-AS1, and ERVK3-1 could interact with U2AF2, which is a component of the U2 complex in spliceosome and regulates AR mRNA splicing (Liu et al., 2014; Takayama, 2019; Takayama et al., 2020). Silencing these lncRNAs reduced the nuclear enrichment of U2AF2 and suppressed the association of U2AF2 with AR pre-mRNA, resulting in decreased AR expression and inhibited PCa cell growth (Takayama et al., 2020).
A few lncRNAs have been reported to encode short peptides which may play roles as proteins (Anderson et al., 2015; Huang J. Z. et al., 2017). Recently, Wu et al. reported that a Y-linked lncRNA, LINC00278, could encode a Yin Yang 1 (YY1)-binding micropeptide, YY1BM. YY1BM could suppress the interaction between YY1 and AR and thus downregulate eEF2K expression and induce apoptosis in human esophageal squamous cell carcinoma (Wu et al., 2020).
The Regulation of AR by circRNAs
CircRNAs have been recognized as regulatory RNAs (Memczak et al., 2013; Guarnerio et al., 2016), and exhibit critical roles through mechanisms like lncRNAs. CircRNA could inhibit miRNA target degradation as sponges (Hansen et al., 2013; Zheng et al., 2016), bind to proteins, RNAs, and DNAs to affect gene transcription (Li et al., 2015; Yang et al., 2017), RNA splicing (Conn et al., 2017), and translation (Li et al., 2020), and serve as protein scaffold containing different binding sites (Du et al., 2017). CircRNA also has its specific regulatory mechanism distinct from lncRNA. CircRNAs’ unique circularization structure lacking open ribonucleotide end may resist the RNA cleavage by miRNA recruited exonuclease, thus stabilizing miRNAs after binding (Piwecka et al., 2017; Chen S. et al., 2019). CircRNA profiling through ribosomal-depleted RNA sequencing has identified many circRNAs that are differentially expressed between normal and cancerous prostate tissues (Zheng et al., 2016; Chen S. et al., 2019). Among them, circRNA-17 is lower expressed in higher grade PCa tissues, and suppressing circRNA-17 could increase the expression of AR-V7, and enhance the resistance to anti-AR therapy. Further investigation revealed that circRNA-17 could bind and stabilize miR-181c-5p, which targets the 3′UTR of AR-V7 (Wu et al., 2019). Since circRNA is a relatively new research field of ncRNAs, more investigations about circRNAs are needed to explore and elucidate their exact roles in tumorigenesis.
ncRNAs as AR Targets
As a critical hormonal transcription factor, AR can exhibit its function through direct binding to ARE located at enhancers and promoters of its targets. Genomic occupation of AR and profiles of androgen-responsive genes have been defined through Chromatin Immunoprecipitation (ChIP)-on-chip, ChIP-seq assays (Jin et al., 2013). AR directly targeted miRNAs (Takayama et al., 2011; Pasqualini et al., 2015) and lncRNAs (Zhang et al., 2018; Takayama et al., 2020) in PCa cells have also been systematically identified by combined analysis of androgen dysregulated miRNA and lncRNA expression data from microarray or RNA sequencing with AR genome-wide binding information (Table 3).
TABLE 3.
AR regulated ncRNAs.
| ncRNAs (References) | Regulation by AR | Molecular mechanisms | Cancer types | Function in cancer cells |
| miRNAs | Targeted genes | |||
| Let-7d (Ramberg et al., 2011) | ↑ | PBX3 | PCa | – |
| miR-1 (Liu Y. N. et al., 2015; Chen W. Y. et al., 2017) | ↑ | ZBTB46, SRC | PCa | Inhibits metastasis |
| miR-101 and miR-26a (Cao et al., 2010) | ↑ | EZH2 | PCa | Inhibits proliferation and invasiveness |
| miR-125b (Rajabi et al., 2011) | ↑ | MUC1, BAK1 | PCa | Promotes cell growth |
| miR-135a (Kroiss et al., 2015) | ↑ | ROCK1, ROCK2 | PCa | Inhibits invasiveness |
| miR-141 (Waltering et al., 2011) | ↑ | - | PCa | Promotes cell growth |
| miR-148a (Fujita et al., 2010; Murata et al., 2010) | ↑ | CAND1 (Murata et al., 2010), MSK1 (Fujita et al., 2010) | PCa | Promotes LNCaP cell growth (Murata et al., 2010), Suppresses PC3 cell growth, and invasiveness (Fujita et al., 2010) |
| miR-185-5p (Huang Q. et al., 2017; Zhu et al., 2019) | ↑ | CSF-1, VEGFC, HIF2α | RCC | Increases RCC cell metastases to lung and liver, while suppresses the lymph nodes metastases |
| miR-193a-3p (Jia et al., 2017) | ↑ | AJUBA | PCa | Promotes cell migration |
| miR-21 (Ribas et al., 2009; Mishra et al., 2014) | ↑ | TGFBR2 | PCa | Promotes cell growth |
| miR-216a (Chen et al., 2012) | ↑ | TSLC1 | HCC | Promotes cell growth and migration |
| miR-22, miR-29a, and miR-17-92 cluster (Pasqualini et al., 2015) | ↑ | LAMC1, MCL1 | PCa | Deceases cell viability and migration |
| miR-29 (Takayama et al., 2015) | ↑ | TET2 | PCa | Promotes cell growth and migration |
| miR-31 (Lin et al., 2013) | ↓ | AR | PCa | Suppresses cell growth |
| miR-32 and miR-148a (Jalava et al., 2012) | ↑ | BTG2, PIK3IP1 | PCa | Reduces apoptosis or promotes cell growth |
| miR-99a/let7c/125b-2 cluster (Sun et al., 2014) | ↓ | IGF1R | PCa | Suppresses cell growth |
| miR-421 (Ostling et al., 2011; Meng et al., 2016) | ↓ | AR (Ostling et al., 2011), NRAS, PRAME, CUL4B, and PFKFB2 (Meng et al., 2016) | PCa | Suppresses cell growth, induces cell cycle arrest, reduces glycolysis, and inhibits migration |
| lncRNAs | ||||
| ARNILA (Yang et al., 2018) | ↓ | Sponges miR-204 to facilitate Sox4 expression | Breast cancer | Promotes migration, invasion and EMT |
| AR-Associated lincRNAs (daSilva et al., 2018) | ↑ | Scaffolds AR-dependent looping complex | PCa | – |
| CTBP1-AS (Takayama et al., 2013) | ↑ | Represses CTBP1 by recruiting PSF together with histone deacetylases | PCa | Promotes cell growth |
| DANCR (Jia et al., 2016) | ↓ | Represses TIMP2/3 expression by mediating the binding of EZH2 on their promoters | PCa | Promotes cell invasion and metastasis |
| DRAIC (Sakurai et al., 2015) | ↓ | - | PCa | Suppresses cell migration and invasion |
| KLK3e (Hsieh et al., 2014) | ↑ | Scaffolds AR-dependent looping complex | PCa | Promotes cell growth |
| Linc00304 (Zhang et al., 2019) | ↓ | Promotes CCNA1 expression | PCa | Promote cell growth and cell cycle progression |
| Linc00844 (Lingadahalli et al., 2018) | ↑ | Indirectly modulates AR binding to chromatin | PCa | Suppresses cell migration and invasion |
| Linc01503 (He et al., 2020) | ↑ | Recruits SFPQ and activates FOSL1 | Nasopharyngeal carcinoma (NPC) | Promotes cell growth, migration, and invasion |
| LncRNA-p21 (Luo et al., 2019b) | ↓ | Interacts with EZH2 and enhances STAT3 methylation | PCa | Promotes ENZ induced neuroendocrine differentiation (NED) |
| PCAT18 (Crea et al., 2014) | ↑ | – | PCa | Promotes cell growth, migration, and invasion |
| PCAT29 (Malik et al., 2014; Sakurai et al., 2015) | ↓ | – | PCa | Suppresses cell migration and invasion |
| POTEF-AS1 (Misawa et al., 2017) | ↑ | Represses Toll-like receptor signaling | PCa | Promotes cell growth and suppresses apoptosis |
| SOCS2-AS1 (Misawa et al., 2016) | ↑ | Interacts with AR protein and regulates its cofactor recruitment | PCa | Promotes cell growth and suppresses apoptosis |
| TMPO-AS1 (Huang et al., 2018) | ↓ | – | PCa | Promotes cell growth and migration |
| circRNAs | ||||
| circAR3 (Luo et al., 2019a) | ↓ | Encoded by AR gene | PCa | No effect on cell growth and invasion |
| AR-circRNAs (Cao et al., 2019) | ↓ | Encoded by AR gene | PCa | – |
| circHIAT1 (Wang k. et al., 2017) | ↓ | Stabilizes miR-195-5p/29a-3p/29c-3p | ccRCC | Suppresses cell migration and invasion |
| circRNA7 (Bao S. et al., 2020) | ↓ | Sponges miR-7-5p and increases VE-cadherin and Notch4 | HCC | Promotes vasculogenic mimicry formation |
| circZMIZ1 (Jiang et al., 2020) | ↑ | Increases expression of AR and AR-V7 | PCa | Promote cell growth and cell cycle progression |
Takayama et al. (2011) integrated 5′-cap analysis of gene expression (CAGE) and ChIP-on-chip analysis and identified a cluster of androgen-inducible miRNAs in LNCaP cells, including miR-100, miR-125b, miR-21, miR-218-1, miR-218-2, miR-221, miR-222, and let-7c, which are all located adjacent to androgen receptor binding sites (Takayama et al., 2011). Among them, miR-21 has been verified as one of 16 AR-responsive miRNAs (Ribas et al., 2009). Interestingly, miR-21 may indirectly increase AR expression by decreasing PTEN, forming a positive regulation loop between AR and miR-21 (Mishra et al., 2014). MiR-125b was shown to be induced by AR and partially involved in AR’s downregulation of MUC1 by targeting MUC1 3′ UTR (Rajabi et al., 2011). Sun et al. (2014) reported that miR-125b-2, let-7c, and miR-99a are a cluster of miRNAs from the same host gene, and were all repressed following androgen activation in LNcaP cells, which is in contrast to Takayama et al.’s result probably due to different conditions of androgen treatment and tissue culture. Pasqualini et al. (2015) performed AR ChIP-seq and miRNA host gene array analysis after AR stimulation in DUCaP cells, and successfully identified 32 miRNA host genes that were significantly regulated and bound by AR. MiR-22 and miR-29a are significantly increased by AR activation in a time-dependent manner, and both of them are higher expressed in benign prostate tissues when compared to tumor tissues. MiR-125b, miR-22, and miR-29a/b were later examined to mediate AR’s repression of TET2 in PCa cells (Takayama et al., 2015). Besides, AR-induced miR-26a together with miR-101 both target EZH2 at the 3′ UTR region and thus are involved in AR’s regulation of EZH2 (Cao et al., 2010). Murata et al. (2010) identified androgen-responsive miRNAs in LNCaP cells through short RNA sequencing and the expression of miR-148a, miR-141, and miR-200a along with miR-125b, miR-22, and miR-29b were all increased after R1881 treatment in LNCaP cells (Murata et al., 2010). The function of miR-148a is complex in prostate cancer progression. MiR-148a was highly expressed in PCa patients and significantly correlated with biochemical recurrence of PCa independent of PSA values (Al-Qatati et al., 2017), and it was shown to promote LNCaP cell growth through targeting CAND1 3′ UTR (Murata et al., 2010). However, overexpression of miR-148a precursor suppressed androgen-refractory PC3 cell growth (Fujita et al., 2010). Another report also suggested miR-148a exhibited tumor suppressor roles in several common cancers (Lujambio et al., 2008). As more AR-regulated miRNAs in PCa and other types of cancers were reported, it is clear that AR’s function is partially mediated by AR-induced oncogenic miRNAs and -inhibited tumor-suppressive miRNAs (Table 3).
Similarly, many AR-regulated lncRNAs have also been identified in PCa cells (Table 3). Misawa et al. (2016) identified 5 lncRNAs induced by androgen through RNA sequencing. Surprisingly, one of these 5 lncRNAs, SOCS2-AS1, was shown to interact with AR protein and modulate AR activity by regulating cofactor recruitment, leading to a positive regulation loop in PCa cells. Zhang et al. (2018) performed an integrative transcriptomic analysis in PCa tissues combined with AR ChIP-seq, resulting in the identification of AR-regulated clinically relevant lncRNAs and ARLNC1 was identified as one of the AR-regulated lncRNAs that regulates AR mRNA stability (Zhang et al., 2018). Another lncRNA in the list is PRCAT38, which was later proven to share enhancers with TMPRSS2, and both of them are activated by AR/FOXA1 binding (Chen Z. et al., 2019). PCAT29 is a PCa-associated lncRNA suppressed by DHT and knocking down PCAT29 increases PCa cell proliferation and migration (Malik et al., 2014). Takayama et al. (2020) identified AR-regulated lncRNAs which are highly expressed in CRPC tissues. Among the list, PRKAG2-AS1 was suppressed by AR activation, while HOXC-AS1 was induced by AR activation, and both lncRNAs play essential roles in AR mRNA splicing through interaction with AR splicing factor, U2AF2.
Several circRNAs directly regulated by AR have also been reported (Table 3). CircRNA-ZMIZ1 is upregulated in PCa patients’ plasma samples than in corresponding normal samples (Jiang et al., 2020). CircRNA-ZMIZ1 expression is increased by androgen activation, and silencing circRNA-ZMIZ1 induces PCa cell growth inhibition and cell cycle arrest. ChIP-seq and luciferase assay confirmed that AR suppresses the expression of circRNA-HIAT1 in clear cell renal cell carcinoma (ccRCC). CircRNA-HIAT1 serves as a “reservoir” to stabilize miR-195-5p/29a-3p/29c-3p that target CDC42, thus indicating AR promotes ccRCC through regulating circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 axis (Wang k. et al., 2017). Interestingly, AR can be transcribed into several circRNAs due to alternative RNA splicing (Cao et al., 2019; Luo et al., 2019a). The expression of these AR-transcribed circRNAs are positively correlated with linear AR transcripts and can be detected in plasma samples from metastatic castration-resistant PCa (mCRPC) patients and may serve as biomarkers of high-risk primary PCa (Cao et al., 2019; Luo et al., 2019a).
Discussion
Crosstalk Between ncRNAs in AR Regulatory Network
Some lncRNAs and circRNAs share similar RNA sequences as the miRNA targeted mRNA, and then they could act as miRNA sponges to diminish miRNA-induced mRNA degradation (Hansen et al., 2013; Goodall and Wickramasinghe, 2020). For example, lncRNA MALAT1 could decrease miR-320b mediated AR mRNA degradation through competitively binding to miR-320b (Dai et al., 2019; Figure 1). Some circRNA-miRNA interactions may form RNA duplex resistant to RNA cleavage, thus stabilizing miRNAs (Piwecka et al., 2017; Chen S. et al., 2019; Wu et al., 2019). CircRNA-17 suppresses the expression of AR-V7 by binding and stabilizing miR-181c-5p which induces the degradation of AR-V7 through targeting its 3′ UTR region (Wu et al., 2019; Figure 1). Both lncRNAs and circRNAs can also serve as scaffolds to mediate interactions between proteins and RNAs. LncRNAs and circRNAs might also mutually affect the binding with the same targets, which may be due to the RNA sequence similarity between lncRNAs and circRNAs, or RNA structure-induced protein conformational change.
FIGURE 1.
Crosstalk between ncRNAs in AR regulatory network. LncRNA MALAT1 acts as a sponge to inhibit miR-320b-targeted AR mRNA degradation. CircRNA-17 binds to and stabilizes miR-181c-5p, enhancing miR-181c-5p-targeted AR mRNA degradation.
Feedbacks Between AR and ncRNAs
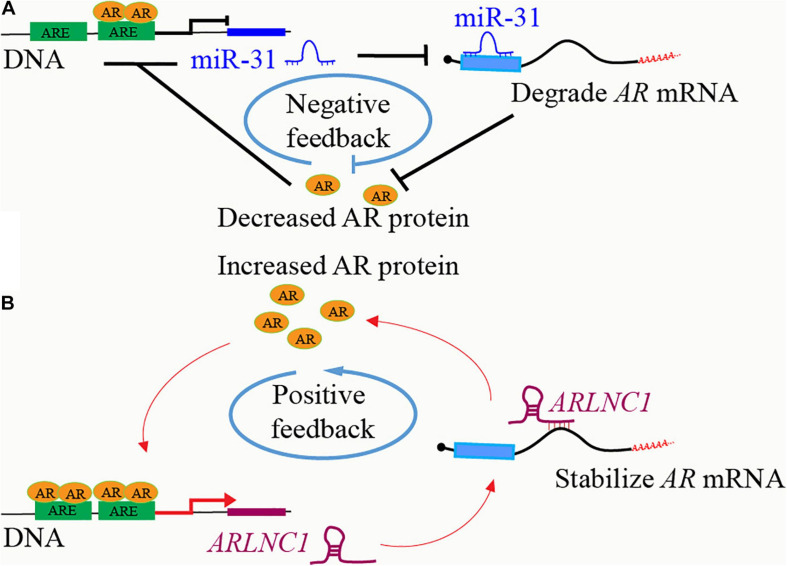
As one of the most important regulators in PCa, the expression of AR is precisely controlled by various factors through different mechanisms in different stages, including feedback regulation loops between AR and ncRNAs. Many ncRNAs regulate AR expression at transcription and post-transcription levels, while they are also regulated by AR. MiR-31 inhibits AR expression by directly targeting the AR mRNA coding region, while miR-31 itself is suppressed as an AR repressive target, thus forming a negative feedback loop to promote PCa (Lin et al., 2013; Figure 2A). AR increases the expression of miR-21, which in turn increase AR expression and activity probably via the down-regulation of PTEN (Ribas et al., 2009; Mishra et al., 2014). Both AR-induced HOXC-AS1 and AR-repressed PRKAG2-AS1 can regulate AR mRNA splicing and promote AR expression (Takayama et al., 2020). The expression of ARLNC1 is increased after AR binding to the ARLNC1 promoter region. ARLNC1 further stabilizes AR mRNA and promotes AR expression through binding to AR mRNA 3′ UTR, thus forming a positive regulation loop in PCa cells (Zhang et al., 2018; Figure 2B).
FIGURE 2.
Feedbacks between AR and ncRNAs. (A) AR binds to the promoter region of lncRNA ARLNC1 and induces its expression. ARLNC1 binds to and stabilizes AR mRNA, which increases AR expression and further activates the transcription of itself, ARLNC1, forming a positive feedback regulation loop. (B) AR binds to the promoter region of miR-31 and suppresses its expression. MiR-31 induces AR mRNA degradation via targeting the coding region of AR mRNA, thus forming a negative feedback regulation loop.
Steroid Receptors and lncRNAs
Several lncRNAs have been shown to regulate steroid receptors, including GR, MR, PR, AR, and other nuclear receptors, through directly binding in a conserved sequence-specific manner (Kino et al., 2010; Hudson et al., 2014; Liu et al., 2016). These lncRNAs bind to these receptors at two protein domains: DNA binding domain, and the N-terminal, regulatory domain (Figure 3A). GAS5 contained a GRE-mimic hairpin RNA sequence and thus can bind to GR DNA domain, block GR’s binding to GRE DNAs, and suppress GR-induced gene transcription (Kino et al., 2010; Hudson et al., 2014; Schmidt et al., 2016; Figure 3B); Figure 3B). On the other hand, several AR-bound lncRNAs share short, pyrimidine-rich RNA motif (CYUYUCCWS) that are required for the interaction with steroid receptors at N-terminal, regulatory domain, such as HOTAIR, SRA, SLNCR1, HOXA11-AS-203, and PCGEM1 (Lanz et al., 1999; Yang et al., 2013; Zhang et al., 2015; Schmidt et al., 2016; Figure 3C); Figure 3C). The discovery of ncRNAs including circRNAs containing steroid receptor responsive element (SRE) mimic RNA sequences, or the pyrimidine-rich RNA motif (CYUYUCCWS) is of great interest to identify new mechanisms in various diseases that ncRNAs regulate steroid receptors through binding to steroid receptors. These regulation mechanisms may also apply to the ncRNAs that bind to other transcription factors.
FIGURE 3.
LncRNAs bind to steroid receptors and regulate their activities. (A) Principal functional domains of steroid receptors. The steroid receptors have N-terminal regulatory domain, central DNA binding domain, ligand-binding domain, and C-terminal extension. (B) LncRNAs containing steroid receptor responsive element (SRE) mimic RNA sequence, such as GAS5, block the binding of steroid receptors to SRE DNA sequence, and suppress steroid receptors’ transcriptional activity. (C) LncRNAs containing short, pyrimidine-rich RNA motif, such as SRA and SLNCR1, bind to steroid receptors at N-terminal, regulatory domain, and increase steroid receptors’ transcriptional activity.
Clinical Implications of ncRNAs in PCa
Increasing research of ncRNAs has greatly revolutionized our understanding of RNA biology. More and more evidence showed that ncRNAs have critical functions in diverse diseases. MiRNA, lncRNA, and circRNA are ubiquitously expressed throughout the body, and they can readily be measured from various human samples, including serum, saliva, and urine (Weber et al., 2010; Iyer et al., 2015; Vo et al., 2019). Many ncRNAs play important roles in PCa and their expressions are correlated with different clinicopathological characteristics of PCa patients. These ncRNAs hold great promises as biomarkers and therapeutic targets in clinical applications. Several single miRNA and panels of miRNAs combinations from plasma or tissue samples of PCa patients have shown more extraordinary diagnostic performance than PSA (Kachakova et al., 2015; Kelly et al., 2015). At the same time, serum miR-210 level is notably correlated with the change in PSA level during treatment among metastatic CRPC patients (Cheng et al., 2013). MRX34, a synthetic miRNA mimic of miR-34a that directly regulates at least 24 known oncogenes including AR, is the first miRNA mimic in clinic application (Bouchie, 2013). In phase 1 clinical trial (NCT01829971), MRX34 was delivered in patients with advanced solid tumors by a liposome technology named Smarticles, which demonstrated exciting proof-of-concept for miRNA-based cancer treatment but unfortunately failed due to serious adverse events (Hong et al., 2020). On the other hand, lncRNA PCA3 is specifically overexpressed in most PCa cancer patients (Bussemakers et al., 1999) and has been approved by the FDA as a PCa diagnostic marker in the urine of PCa patients (de Kok et al., 2002; Deras et al., 2008), but its use for assessing response to ADT in advanced PCa is limited (Martinez-Pineiro et al., 2014). Some other lncRNAs have been identified as biomarkers for metastatic PCa, such as PCAT18 (Crea et al., 2014) and SChLAP1 (Prensner et al., 2014d). Candidate circRNAs were also identified and detected in urine to serve as biomarkers for PCa (Vo et al., 2019).
Antisense oligonucleotides (ASOs) as an RNA-based therapeutic approach can induce gene silencing through RNase H-mediated degradation of target RNAs. It has shown improved target specificity and stability as well as tolerated toxicity after significant advancements in the design, chemical modifications, and delivery (Verma, 2018). Several ASO-based drugs have been approved by the FDA for the treatment of different human diseases (Dhuri et al., 2020). For PCa treatment, ASOs targeting Bcl-2 mRNA (Oblimersen- G3139) (NCT00085228) and Clusterin mRNA (Custirsen- OGX011) (NCT01188187) had been evaluated in PCa human patients in Phase II and III clinical trials, but both failed due to major toxic events or no significant survival improvement (Sternberg et al., 2009; Beer et al., 2017). Additionally, ASOs targeting Hsp27 mRNA (Apatorsen- OGX-427) (NCT01120470) and AR mRNA (ARRx- AZD5312) (NCT03300505) are currently under Phase I clinical trials. In addition to targeting the protein-coding mRNAs, ASO targeting lncRNA MALAT1 dramatically prevented lung cancer metastasis in a pulmonary metastatic mouse model (Gutschner et al., 2013), showing attractive potentials for developing ASO drugs targeting functional ncRNAs to treat PCa. More preclinical investigations for ASO targeting ncRNAs are needed to enable ASO-based prostate cancer treatment in the near future.
LncRNA Studies Through in vivo Mouse Models
Most lncRNAs’ functions and mechanisms were revealed through knocking down strategies from in vitro study until to date, and several lncRNAs are proven to be necessary for life and brain development through the studies in the knockout mouse model (Sauvageau et al., 2013; Nakagawa et al., 2014). On the other hand, accumulating evidence showed that inactivating the same lncRNAs in mouse models resulted in no phenotype, and even opposite effects for some lncRNAs (Bassett et al., 2014; Sun and Ma, 2019). Bassett et al. (2014) summarized the results of in vivo studies of 30 lncRNAs through different inactivation strategies from 17 groups. Among them, lncRNA MALAT1 was inactivated through 3 different strategies: deleting 3kb genomic region covering the 5′ end of MALAT1 and its promoter (Zhang et al., 2012), removing the entire 7kb MALAT1 gene (Eissmann et al., 2012), and premature transcriptional termination by inserting lacZ and polyadenylation sequences downstream of the transcriptional start site of MALAT1 (Nakagawa et al., 2012). All these MALAT1-deficient mice from these 3 studies were viable and fertile without significant changes in mice development and growth, and global gene expression, which argues against the in vitro and xenograft studies that demonstrated MALAT1’s role in promoting cell proliferation and metastases through regulating pre-mRNA splicing (Tripathi et al., 2010), coordinating gene transcription (Yang et al., 2011; Wang et al., 2015), and acting as competitive endogenous RNA (Wang y. et al., 2017; Dai et al., 2019). Recently, Kim et al. (2018) investigated MALAT1’s role in a transgenic mouse model of breast cancer in which the MALAT1 gene was inactivated through premature transcriptional termination by inserting lacZ and polyadenylation sequences as Nakagawa et al. (2012). Targeted inactivation of MALAT1 in this breast cancer mouse model doesn’t affect breast tumor growth, but surprisingly promotes breast cancer lung metastasis (Kim et al., 2018), which is consistent with some other reports that suggest MALAT1 functions as a tumor suppressor (Xu et al., 2015; Cao et al., 2016; Han et al., 2016; Latorre et al., 2016; Kwok et al., 2018). Importantly, the metastatic-promoting effect of MALAT1 insertional inactivation can be reversed by genetical re-expression of MALAT1, and targeted transgenic overexpression of MALAT1 in mice inhibits breast cancer metastasis (Kim et al., 2018), strongly suggesting that the lncRNA MALAT1 suppresses breast cancer metastasis.
There are several possible reasons to explain why some in vivo mouse models failed to validate lncRNAs’ function discovered from in vitro and xenograft studies. First, most lncRNAs were studied in cells through the knocking down methods mediated by short-hairpin RNA (shRNA) or siRNA without appropriate rescue assays. Silencing nuclear lncRNAs requires the nuclear enrichment of AGO2 and RNA interference (RNAi) factors Dicer, TRBP, and TRNC6A/GW182 (Gagnon et al., 2014), while AGO2’s nuclear distribution depends on cell type and tissue context (Sharma et al., 2016), and thus results from knocking down lncRNAs in cells lacking nuclear distribution of AGO2 are questionable. Secondly, several lncRNAs were silenced in cells and mouse through ASO treatment. However, the delivery of ASO to the targeted cells and organs is still a challenge, and it is not clear whether ASO could efficiently degrade the nascent RNAs. Besides, the potential off-target effects of ASO may lead to non-specific results (Deleavey and Damha, 2012). Thirdly, loss of function approaches in cells and mouse models through large size gene deletion or clustered regularly interspaced short palindromic repeats (Crispr)-Cas9 knocking out may also delete the neighboring genes and destroy the regulatory elements for other genes. Some lncRNAs function through cis mechanisms to regulate their neighboring genes. The effect of destructing these regulatory elements located in the lncRNA genome loci and neighboring genes prevails against the effect of lncRNA loss (Yin et al., 2015). LncRNA inactivation induced by transcriptional terminator insertion abrogates lncRNA transcription with minimal disruption of genomic sequences and mice phenotypes induced by insertional inactivation of lncRNAs can be rescued by re-expression of lncRNAs (Bond et al., 2009; Berghoff et al., 2013; Grote et al., 2013). Fourthly, even though lncRNA is inactivated through the same method, different mouse models may display different phenotypes. Insertional inactivation of MALAT1 in MMTV-PyMT mouse which is a transgenic model of metastatic breast cancer, induced significantly increased lung metastasis of breast cancer cells (Kim et al., 2018), while insertional inactivation of MALAT1 in mouse with normal physiological condition showed no apparent phenotype (Nakagawa et al., 2012), suggesting MALAT1 is dispensable for development but plays important roles in suppressing breast cancer metastasis. Taken together, it is critical to choose the proper method to generate lncRNA depleting cells and mouse model based on lncRNA’s cellular distribution, genome localization and its function mechanism, and it is also important to take rescue experiments into consideration when investigating lncRNAs’ function through loss of function methods (Bassett et al., 2014; Kopp and Mendell, 2018).
Conclusion
A growing number of novel discovered ncRNAs and various research has revealed the crucial roles of ncRNAs in different disease processes. Many ncRNAs have been verified to participate in PCa initiation or progression by regulating or mediating AR signaling. These ncRNAs hold great potentials as diagnostic biomarkers and therapeutic targets. Given that ncRNAs consist of the majority of the human transcriptome, a long journey in the understanding of ncRNAs, especially lncRNAs, circRNAs, and other ncRNAs, is yet to be achieved. Further investigations based on high-throughput sequencing technology and integrative bioinformatics analysis will enable the discovering of new functional ncRNAs and their regulation mechanisms, and these works will further promote the development of effective therapeutic strategies for PCa.
Author Contributions
YY and QC developed the concept of the review. YY and KL collected the related publications and drafted this manuscript. QL and QC helped to revise this manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported in part by grants from the United States Department of Defense (W81XWH-17-1-0357, W81XWH-19-1-0563, and W81XWH-20-1-0504 to QC), American Cancer Society (RSG-15-192-01 to QC), NIH/NCI (R01CA208257 and Prostate SPORE P50CA180995 Developmental Research Program to QC), and Northwestern University Polsky Urologic Cancer Institute (to QC).
References
- Aakula A., Leivonen S. K., Hintsanen P., Aittokallio T., Ceder Y., Borresen-Dale A. L., et al. (2015). MicroRNA-135b regulates ERalpha, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol. Oncol. 9 1287–1300. 10.1016/j.molonc.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander R. P., Fang G., Rozowsky J., Snyder M., Gerstein M. B. (2010). Annotating non-coding regions of the genome. Nat. Rev. Genet. 11 559–571. 10.1038/nrg2814 [DOI] [PubMed] [Google Scholar]
- Al-Qatati A., Akrong C., Stevic I., Pantel K., Awe J., Saranchuk J., et al. (2017). Plasma microRNA signature is associated with risk stratification in prostate cancer patients. Int. J. Cancer. 141 1231–1239. 10.1002/ijc.30815 [DOI] [PubMed] [Google Scholar]
- Anderson D. M., Anderson K. M., Chang C. L., Makarewich C. A., Nelson B. R., McAnally J. R., et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160 595–606. 10.1016/j.cell.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio A., Logothetis C. J., Maity S. N. (2011). Understanding the lethal variant of prostate cancer: power of examining extremes. Cancer Discov. 1 466–468. 10.1158/2159-8290.cd-11-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augoff K., McCue B., Plow E. F., Sossey-Alaoui K. (2012). miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 11:5. 10.1186/1476-4598-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini E., Fanini F., Vannini I., Rossi T., Plousiou M., Tumedei M. M., et al. (2020). miR-9-5p as a regulator of the androgen receptor pathway in breast cancer cell lines. Front. Cell. Dev. Biol. 8:579160. 10.3389/fcell.2020.579160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Jin S., Wang C., Tu P., Hu K., Lu J. (2020). Androgen receptor suppresses vasculogenic mimicry in hepatocellular carcinoma via circRNA7/miRNA7-5p/VE-cadherin/Notch4 signalling. J. Cell. Mol. Med. 24 14110–14120. 10.1111/jcmm.16022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S. X., Wang C. H., Jin S., Hu K. W., Lu J. T. (2020). miR-135b-5p suppresses androgen receptor-enhanced hepatocellular carcinoma cell proliferation via regulating the HIF-2alpha/c-Myc/P27 signals in vitro. Oncol. Targets Ther. 13 9991–10000. 10.2147/ott.s268214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Akhtar A., Barlow D. P., Bird A. P., Brockdorff N., Duboule D., et al. (2014). Considerations when investigating lncRNA function in vivo. Elife 3:e03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer T. M., Hotte S. J., Saad F., Alekseev B., Matveev V., Flechon A., et al. (2017). Custirsen (OGX-011) combined with cabazitaxel and prednisone versus cabazitaxel and prednisone alone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel (AFFINITY): a randomised, open-label, international, phase 3 trial. Lancet Oncol. 18 1532–1542. 10.1016/s1470-2045(17)30605-8 [DOI] [PubMed] [Google Scholar]
- Berghoff E. G., Clark M. F., Chen S., Cajigas I., Leib D. E., Kohtz J. D. (2013). Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140 4407–4416. 10.1242/dev.099390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemn E. G., Coleman I. M., Lucas J. M., Coleman R. T., Hernandez-Lopez S., Tharakan R., et al. (2017). Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 32 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll K., Reiche K., Kasack K., Morbt N., Kretzschmar A. K., Tomm J. M., et al. (2013). MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 32 277–285. 10.1038/onc.2012.55 [DOI] [PubMed] [Google Scholar]
- Bonci D., Coppola V., Musumeci M., Addario A., Giuffrida R., Memeo L., et al. (2008). The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 14 1271–1277. 10.1038/nm.1880 [DOI] [PubMed] [Google Scholar]
- Bond A. M., VanGompel M. J. W., Sametsky E. A., Clark M. F., Savage J. C., Disterhoft J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12 1020–1027. 10.1038/nn.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchie A. (2013). First microRNA mimic enters clinic. Nat. Biotechnol. 31:577. 10.1038/nbt0713-577 [DOI] [PubMed] [Google Scholar]
- Brawley O. W. (2012). Trends in prostate cancer in the united states. J. Natl. Cancer Inst. Monogr. 2012 152–156. 10.1093/jncimonographs/lgs035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemakers M. J., van Bokhoven A., Verhaegh G. W., Smit F. P., Karthaus H. F., Schalken J. A., et al. (1999). DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 59 5975–5979. [PubMed] [Google Scholar]
- Cao P., Deng Z., Wan M., Huang W., Cramer S. D., Xu J., et al. (2010). MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol. Cancer. 9:108. 10.1186/1476-4598-9-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Ma T., Ungerleider N., Roberts C., Kobelski M., Jin L., et al. (2019). Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene 38 7060–7072. 10.1038/s41388-019-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Wang Y., Li J., Lv M., Niu H., Tian Y. (2016). Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J. Cancer Res. 6 2561–2574. [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Steitz J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157 77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Chen L., Heikkinen L., Wang C., Yang Y., Sun H., Wong G. (2019). Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 20 1836–1852. 10.1093/bib/bby054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. J., Yeh S. H., Liu W. H., Lin C. C., Huang H. C., Chen C. L., et al. (2012). Androgen pathway stimulates microRNA-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology 56 632–643. 10.1002/hep.25695 [DOI] [PubMed] [Google Scholar]
- Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., et al. (2019). Widespread and functional RNA circularization in localized prostate cancer. Cell 176 831–843. [DOI] [PubMed] [Google Scholar]
- Chen W., Liu Y., Chen H., Ning H., Ding K. (2017). Loss of miR-449a-caused PrLZ overexpression promotes prostate cancer metastasis. Int. J. Oncol. 51 435–444. 10.3892/ijo.2017.4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. Y., Tsai Y. C., Siu M. K., Yeh H. L., Chen C. L., Yin J. J., et al. (2017). Inhibition of the androgen receptor induces a novel tumor promoter, ZBTB46, for prostate cancer metastasis. Oncogene 36 6213–6224. 10.1038/onc.2017.226 [DOI] [PubMed] [Google Scholar]
- Chen Z., Song X., Li Q., Xie L., Guo T., Su T., et al. (2019). Androgen receptor-activated enhancers simultaneously regulate oncogene TMPRSS2 and lncRNA PRCAT38 in prostate cancer. Cells 8:864. 10.3390/cells8080864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. H., Mitchell P. S., Kroh E. M., Dowell A. E., Chery L., Siddiqui J., et al. (2013). Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS One 8:e69239. 10.1371/journal.pone.0069239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T., Yamamura S., Fukuhara S., Yoshino H., Kinoshita T., Majid S., et al. (2013). Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One 8:e70372. 10.1371/journal.pone.0070372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coarfa C., Fiskus W., Eedunuri V. K., Rajapakshe K., Foley C., Chew S. A., et al. (2016). Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 35 2345–2356. 10.1038/onc.2015.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn V. M., Hugouvieux V., Nayak A., Conos S. A., Capovilla G., Cildir G., et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 3:17053. [DOI] [PubMed] [Google Scholar]
- Consortium E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F., Watahiki A., Quagliata L., Xue H., Pikor L., Parolia A., et al. (2014). Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 5 764–774. 10.18632/oncotarget.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Ren S., Lu J., Wang F., Xu W., Sun Y., et al. (2013). The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol. 31 1117–1123. 10.1016/j.urolonc.2011.11.030 [DOI] [PubMed] [Google Scholar]
- Dai X., Liu L., Liang Z., Guo K., Xu S., Wang H. (2019). Silencing of lncRNA MALAT1 inhibits cell cycle progression via androgen receptor signaling in prostate cancer cells. Pathol. Res. Pract. 215 712–721. 10.1016/j.prp.2019.01.011 [DOI] [PubMed] [Google Scholar]
- daSilva L. F., Beckedorff F. C., Ayupe A. C., Amaral M. S., Mesel V., Videira A., et al. (2018). Chromatin landscape distinguishes the genomic loci of hundreds of androgen-receptor-associated lincRNAs from the loci of non-associated lincRNAs. Front. Genet. 9:132. 10.3389/fgene.2018.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok J. B., Verhaegh G. W., Roelofs R. W., Hessels D., Kiemeney L. A., Aalders T. W., et al. (2002). DD3 (PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 62 2695–2698. [PubMed] [Google Scholar]
- Deleavey G. F., Damha M. J. (2012). Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 19 937–954. 10.1016/j.chembiol.2012.07.011 [DOI] [PubMed] [Google Scholar]
- Deras I. L., Aubin S. M., Blase A., Day J. R., Koo S., Partin A. W., et al. (2008). PCA3: a molecular urine assay for predicting prostate biopsy outcome. J. Urol. 179 1587–1592. 10.1016/j.juro.2007.11.038 [DOI] [PubMed] [Google Scholar]
- Dhuri K., Bechtold C., Quijano E., Pham H., Gupta A., Vikram A., et al. (2020). Antisense oligonucleotides: an emerging area in drug discovery and development. J. Clin. Med. 9:2004. 10.3390/jcm9062004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., et al. (2012). Landscape of transcription in human cells. Nature 489 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. W., Fang L., Yang W., Wu N., Awan F. M., Yang Z., et al. (2017). Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell. Death Differ. 24 357–370. 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Fei T., Verhaak R. G., Su Z., Zhang Y., Brown M., et al. (2013). Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20 908–913. 10.1038/nsmb.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebron J. S., Shukla G. C. (2016). Molecular characterization of a novel androgen receptor transgene responsive to MicroRNA mediated post-transcriptional control exerted via 3’-untranslated region. Prostate 76 834–844. 10.1002/pros.23174 [DOI] [PubMed] [Google Scholar]
- Eissmann M., Gutschner T., Hammerle M., Gunther S., Caudron-Herger M., Gross M., et al. (2012). Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. Rna Biol. 9 1076–1087. 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Xu C., Li Y., Cai X., Ren S., Liu H., et al. (2016). A feed-forward regulatory loop between androgen receptor and PlncRNA-1 promotes prostate cancer progression. Cancer Lett. 374 62–74. 10.1016/j.canlet.2016.01.033 [DOI] [PubMed] [Google Scholar]
- Feldman B. J., Feldman D. (2001). The development of androgen-independent prostate cancer. Nat. Rev. Cancer. 1 34–45. [DOI] [PubMed] [Google Scholar]
- Feng T., Shao F., Wu Q., Zhang X., Xu D., Qian K., et al. (2016). miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget 7 16205–16216. 10.18632/oncotarget.7578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yang Y., Zhao X., Fan Y., Zhou L., Rong J., et al. (2019). Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell. Death Dis. 10:792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C. E., Sulpice E., Combe S., Shibakawa A., Leach D. A., Hamilton M. P., et al. (2019). Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene 38 5700–5724. 10.1038/s41388-019-0823-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkerd E. J., Dowsett M. (2010). Influence of sex hormones on cancer progression. J. Clin. Oncol. 28 4038–4044. 10.1200/jco.2009.27.4290 [DOI] [PubMed] [Google Scholar]
- Fredsoe J., Rasmussen A. K. I., Thomsen A. R., Mouritzen P., Hoyer S., Borre M., et al. (2018). Diagnostic and prognostic microRNA biomarkers for prostate cancer in cell-free urine. Eur. Urol. Focus. 4 825–833. 10.1016/j.euf.2017.02.018 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Kojima K., Ohhashi R., Hamada N., Nozawa Y., Kitamoto A., et al. (2010). MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J. Biol. Chem. 285 19076–19084. 10.1074/jbc.m109.079525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon K. T., Li L., Chu Y., Janowski B. A., Corey D. R. (2014). RNAi factors are present and active in human cell nuclei. Cell. Rep. 6 211–221. 10.1016/j.celrep.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy K., Staklinski S., Hasan M. F., Ottman R., Andl T., Berglund A. E., et al. (2020). Multifaceted function of microRNA-299-3p fosters an antitumor environment through modulation of androgen receptor and VEGFA signaling pathways in prostate cancer. Sci. Rep. 10:5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandellini P., Profumo V., Casamichele A., Fenderico N., Borrelli S., Petrovich G., et al. (2012). miR-205 regulates basement membrane deposition in human prostate: implications for cancer development. Cell. Death Differ. 19 1750–1760. 10.1038/cdd.2012.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Wickramasinghe V. O. (2020). RNA in cancer. Nat. Rev. Cancer 21 22–36. [DOI] [PubMed] [Google Scholar]
- Grote P., Wittler L., Hendrix D., Koch F., Wahrisch S., Beisaw A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 24 206–214. 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Chen X., Xie R., Xie W., Huang L., Dong W., et al. (2019). A novel AR translational regulator lncRNA LBCS inhibits castration resistance of prostate cancer. Mol. Cancer. 18:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnerio J., Bezzi M., Jeong J. C., Paffenholz S. V., Berry K., Naldini M. M., et al. (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165 289–302. 10.1016/j.cell.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Guiro J., Murphy S. (2017). Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol. 7:170073. 10.1098/rsob.170073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ahmed M., Zhang F., Yao C. Q., Li S., Liang Y., et al. (2016). Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 48 1142–1150. 10.1038/ng.3637 [DOI] [PubMed] [Google Scholar]
- Guo J., Hu J., Cao R., Chen Q., Li K. (2018). Androgen receptor is inactivated and degraded in bladder cancer cells by phenyl glucosamine via miR-449a restoration. Med. Sci. Monit. 24 2294–2301. 10.12659/msm.906836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Lv J., Liu M., Tang H. (2015). miR-346 Up-regulates argonaute 2 (AGO2) protein expression to augment the activity of other microRNAs (miRNAs) and contributes to cervical cancer cell malignancy. J. Biol. Chem. 290 30342–30350. 10.1074/jbc.m115.691857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Hammerle M., Eissmann M., Hsu J., Kim Y., Hung G., et al. (2013). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73 1180–1189. 10.1158/0008-5472.can-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman Z., Haflidadottir B. S., Ceder J. A., Larne O., Bjartell A., Lilja H., et al. (2013). miR-205 negatively regulates the androgen receptor and is associated with adverse outcome of prostate cancer patients. Br. J. Cancer 108 1668–1676. 10.1038/bjc.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman Z., Larne O., Edsjo A., Bjartell A., Ehrnstrom R. A., Ulmert D., et al. (2010). miR-34c is downregulated in prostate cancer and exerts tumor suppressive functions. Int. J. Cancer 127 2768–2776. 10.1002/ijc.25269 [DOI] [PubMed] [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. 10.1126/science.286.5441.950 [DOI] [PubMed] [Google Scholar]
- Hamilton M. P., Rajapakshe K. I., Bader D. A., Cerne J. Z., Smith E. A., Coarfa C., et al. (2016). The landscape of microRNA targeting in prostate cancer defined by AGO-PAR-CLIP. Neoplasia 18 356–370. 10.1016/j.neo.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wu Z., Wu T., Huang Y., Cheng Z., Li X., et al. (2016). Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell. Death Dis. 7:e2123. 10.1038/cddis.2015.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Hayes J., Peruzzi P. P., Lawler S. (2014). MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 20 460–469. 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- He D., Li L., Zhu G., Liang L., Guan Z., Chang L., et al. (2014). ASC-J9 suppresses renal cell carcinoma progression by targeting an androgen receptor-dependent HIF2alpha/VEGF signaling pathway. Cancer Res. 74 4420–4430. 10.1158/0008-5472.can-13-2681 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5 522–531. 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- He S. W., Xu C., Li Y. Q., Li Y. Q., Zhao Y., Zhang P. P., et al. (2020). AR-induced long non-coding RNA LINC01503 facilitates proliferation and metastasis via the SFPQ-FOSL1 axis in nasopharyngeal carcinoma. Oncogene 39 5616–5632. 10.1038/s41388-020-01388-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Shen F., Qi P., Zhai Z., Wang Z. (2021). miR-541-3p enhances the radiosensitivity of prostate cancer cells by inhibiting HSP27 expression and downregulating beta-catenin. Cell. Death Discov. 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Hinoda Y., Shahryari V., Deng G., Nakajima K., Tabatabai Z. L., et al. (2015). Long Noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 75 1322–1331. 10.1158/0008-5472.can-14-2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D. S., Kang Y. K., Borad M., Sachdev J., Ejadi S., Lim H. Y., et al. (2020). Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 122 1630–1637. 10.1038/s41416-020-0802-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. L., Fei T., Chen Y., Li T., Gao Y., Wang X., et al. (2014). Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. U S A. 111 7319–7324. 10.1073/pnas.1324151111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh I. S., Chang K. C., Tsai Y. T., Ke J. Y., Lu P. J., Lee K. H., et al. (2013). MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis 34 530–538. 10.1093/carcin/bgs371 [DOI] [PubMed] [Google Scholar]
- Hu C. D., Choo R., Huang J. (2015). Neuroendocrine differentiation in prostate cancer: a mechanism of radioresistance and treatment failure. Front. Oncol. 5:90. 10.3389/fonc.2015.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J. T., Ahmed M., Guo H., Zhang Y., Chen S., Soares F., et al. (2018). Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell 174 564–575. 10.1016/j.cell.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Huang J. Z., Chen M., Chen D., Gao X. C., Zhu S., Huang H., et al. (2017). A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell. 68 171–184. 10.1016/j.molcel.2017.09.015 [DOI] [PubMed] [Google Scholar]
- Huang Q., Ma B., Su Y., Chan K., Qu H., Huang J., et al. (2020). miR-197-3p represses the proliferation of prostate cancer by regulating the VDAC1/AKT/beta-catenin signaling axis. Int. J. Biol. Sci. 16 1417–1426. 10.7150/ijbs.42019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Sun Y., Ma X., Gao Y., Li X., Niu Y., et al. (2017). Androgen receptor increases hematogenous metastasis yet decreases lymphatic metastasis of renal cell carcinoma. Nat. Commun. 8:918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Su X., Yan W., Kong Z., Wang D., Huang Y., et al. (2018). Overexpression of AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate 78 1248–1261. 10.1002/pros.23700 [DOI] [PubMed] [Google Scholar]
- Hudson W. H., Pickard M. R., de Vera I. M., Kuiper E. G., Mourtada-Maarabouni M., Conn G. L., et al. (2014). Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat. Commun. 5:5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. L., Wang L. Y., Yu Y. L., Chen H. W., Srivastava S., Petrovics G., et al. (2014). A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. U S A. 111 18697–18702. 10.1073/pnas.1415669112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K., Niknafs Y. S., Malik R., Singhal U., Sahu A., Hosono Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalava S. E., Urbanucci A., Latonen L., Waltering K. K., Sahu B., Janne O. A., et al. (2012). Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 31 4460–4471. 10.1038/onc.2011.624 [DOI] [PubMed] [Google Scholar]
- Jia J., Li F., Tang X. S., Xu S., Gao Y., Shi Q., et al. (2016). Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget 7 37868–37881. 10.18632/oncotarget.9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Gui B., Zheng D., Decker K. F., Tinay I., Tan M., et al. (2017). Androgen receptor-regulated miRNA-193a-3p targets AJUBA to promote prostate cancer cell migration. Prostate 77 1000–1011. 10.1002/pros.23356 [DOI] [PubMed] [Google Scholar]
- Jiang H., Lv D. J., Song X. L., Wang C., Yu Y. Z., Zhao S. C. (2020). Upregulated circZMIZ1 promotes the proliferation of prostate cancer cells and is a valuable marker in plasma. Neoplasma 67 68–77. 10.4149/neo_2019_190213n116 [DOI] [PubMed] [Google Scholar]
- Jin H. J., Kim J., Yu J. (2013). Androgen receptor genomic regulation. Transl. Androl. Urol. 2 157–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachakova D., Mitkova A., Popov E., Popov I., Vlahova A., Dikov T., et al. (2015). Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell. Biol. 34 189–200. 10.1089/dna.2014.2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina T. S., Kononchuk V. V., Yakovleva A. K., Alekseenok E. Y., Sidorov S. V., Gulyaeva L. F. (2020). Association between lymph node status and expression levels of androgen receptor, miR-185, miR-205, and miR-21 in breast cancer subtypes. Int. J. Breast Cancer. 2020:3259393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C. J., Martiniez A., Shi X. B., Yang J., Evans C. P., Dobi A., et al. (2014). miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 33 2495–2503. 10.1038/onc.2013.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashat M., Azzouz L., Sarkar S. H., Kong D., Li Y., Sarkar F. H. (2012). Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am. J. Transl. Res. 4 432–442. [PMC free article] [PubMed] [Google Scholar]
- Kelly B. D., Miller N., Sweeney K. J., Durkan G. C., Rogers E., Walsh K., et al. (2015). A circulating microRNA signature as a biomarker for prostate cancer in a high risk group. J. Clin. Med. 4 1369–1379. 10.3390/jcm4071369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Piao H. L., Kim B. J., Yao F., Han Z., Wang Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50 1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. K., Hemberg M., Gray J. M., Costa A. M., Bear D. M., Wu J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465 182–187. 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3:ra8. 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby M., Hirst C., Crawford E. D. (2011). Characterising the castration-resistant prostate cancer population: a systematic review. Int. J. Clin. Pract. 65 1180–1192. 10.1111/j.1742-1241.2011.02799.x [DOI] [PubMed] [Google Scholar]
- Kiss T. (2001). Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20 3617–3622. 10.1093/emboj/20.14.3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F., Mendell J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroiss A., Vincent S., Decaussin-Petrucci M., Meugnier E., Viallet J., Ruffion A., et al. (2015). Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene 34 2846–2855. 10.1038/onc.2014.222 [DOI] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 597–610. 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- Kumar B., Khaleghzadegan S., Mears B., Hatano K., Kudrolli T. A., Chowdhury W. H., et al. (2016). Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 7 72593–72607. 10.18632/oncotarget.12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok Z. H., Roche V., Chew X. H., Fadieieva A., Tay Y. (2018). A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer 143 668–678. 10.1002/ijc.31386 [DOI] [PubMed] [Google Scholar]
- Labrecque M. P., Coleman I. M., Brown L. G., True L. D., Kollath L., Lakely B., et al. (2019). Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J. Clin. Invest. 129 4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294 853–858. 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., et al. (2001). Initial sequencing and analysis of the human genome. Nature. 409 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lanz R. B., McKenna N. J., Onate S. A., Albrecht U., Wong J., Tsai S. Y., et al. (1999). A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97 17–27. 10.1016/s0092-8674(00)80711-4 [DOI] [PubMed] [Google Scholar]
- Latorre E., Carelli S., Raimondi I., D’Agostino V., Castiglioni I., Zucal C., et al. (2016). The ribonucleic complex HuR-MALAT1 represses CD133 expression and suppresses epithelial-mesenchymal transition in breast cancer. Cancer Res. 76 2626–2636. 10.1158/0008-5472.can-15-2018 [DOI] [PubMed] [Google Scholar]
- Leite K. R., Morais D. R., Florez M. G., Reis S. T., Iscaife A., Viana N., et al. (2015). The role of microRNAs 371 and 34a in androgen receptor control influencing prostate cancer behavior. Urol. Oncol. 33 e15–e22. [DOI] [PubMed] [Google Scholar]
- Li X. X., Xiao L., Chung H. K., Ma X. X., Liu X., Song J. L., et al. (2020). Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol. Cell. Biol. 40 e419–e492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22 256–264. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- Lieb V., Weigelt K., Scheinost L., Fischer K., Greither T., Marcou M., et al. (2018). Serum levels of miR-320 family members are associated with clinical parameters and diagnosis in prostate cancer patients. Oncotarget 9 10402–10416. 10.18632/oncotarget.23781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja H., Ulmert D., Vickers A. J. (2008). Prostate-specific antigen and prostate cancer: prediction, detection and monitoring. Nat. Rev. Cancer. 8 268–278. 10.1038/nrc2351 [DOI] [PubMed] [Google Scholar]
- Lin P. C., Chiu Y. L., Banerjee S., Park K., Mosquera J. M., Giannopoulou E., et al. (2013). Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 73 1232–1244. 10.1158/0008-5472.can-12-2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Cai J., Wang Q. Q. (2020). The significance of circular RNA DDX17 in prostate cancer. Biomed. Res. Int. 2020:1878431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingadahalli S., Jadhao S., Sung Y. Y., Chen M., Hu L., Chen X., et al. (2018). Novel lncRNA LINC00844 regulates prostate cancer cell migration and invasion through AR signaling. Mol. Cancer Res. 16 1865–1878. 10.1158/1541-7786.mcr-18-0087 [DOI] [PubMed] [Google Scholar]
- Liu B., Sun Y., Tang M., Liang C., Huang C. P., Niu Y., et al. (2020). The miR-361-3p increases enzalutamide (Enz) sensitivity via targeting the ARv7 and MKNK2 to better suppress the Enz-resistant prostate cancer. Cell. Death Dis. 11:807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Chen Z., Hu X., Wang L., Li C., Xue J., et al. (2015). MicroRNA-185 downregulates androgen receptor expression in the LNCaP prostate carcinoma cell line. Mol. Med. Rep. 11 4625–4632. 10.3892/mmr.2015.3332 [DOI] [PubMed] [Google Scholar]
- Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu H. T., Zhu N., Shi Y. N., Liu Z., Ao B. X., et al. (2016). Steroid receptor RNA activator: biologic function and role in disease. Clin. Chim. Acta. 459 137–146. 10.1016/j.cca.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Liu G., Ouyang X., Sun Y., Xiao Y., You B., Gao Y., et al. (2020). The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/beta-catenin signaling. Cell. Death Differ. 27 3258–3272. 10.1038/s41418-020-0575-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. L., Xie N., Sun S., Plymate S., Mostaghel E., Dong X. (2014). Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 33 3140–3150. 10.1038/onc.2013.284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu X., Xu X., Li S., Liang Z., Hu Z., et al. (2017). MicroRNA-193a-3p inhibits cell proliferation in prostate cancer by targeting cyclin D1. Oncol. Lett. 14 5121–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. N., Yin J., Barrett B., Sheppard-Tillman H., Li D., Casey O. M., et al. (2015). Loss of androgen-regulated microRNA 1 activates SRC and promotes prostate cancer bone metastasis. Mol. Cell. Biol. 35 1940–1951. 10.1128/mcb.00008-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J. A., Guns E. S., Lubik A. A., Adomat H. H., Hendy S. C., Wood C. A., et al. (2008). Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 68 6407–6415. 10.1158/0008-5472.can-07-5997 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Calin G. A., Villanueva A., Ropero S., Sanchez-Cespedes M., Blanco D., et al. (2008). A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U S A. 105 13556–13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Liu D., Huang C., Wang M., Xiao X., Zeng F., et al. (2017). LncRNA GAS5 Inhibits Cellular Proliferation by Targeting P27 (Kip1). Mol. Cancer Res. 15 789–799. 10.1158/1541-7786.mcr-16-0331 [DOI] [PubMed] [Google Scholar]
- Luo J., Li Y., Zheng W., Xie N., Shi Y., Long Z., et al. (2019a). Characterization of a prostate- and prostate cancer-specific circular RNA encoded by the androgen receptor gene. Mol. Ther. Nucleic Acid. 18 916–926. 10.1016/j.omtn.2019.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Wang K., Yeh S., Sun Y., Liang L., Xiao Y., et al. (2019b). LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat. Commun. 10:2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R., Patel L., Prensner J. R., Shi Y., Iyer M. K., Subramaniyan S., et al. (2014). The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol. Cancer Res. 12 1081–1087. 10.1158/1541-7786.mcr-14-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pineiro L., Schalken J. A., Cabri P., Maisonobe P., de la Taille A. Triptocare Study Group. (2014). Evaluation of urinary prostate cancer antigen-3 (PCA3) and TMPRSS2-ERG score changes when starting androgen-deprivation therapy with triptorelin 6-month formulation in patients with locally advanced and metastatic prostate cancer. BJU Int. 114 608–616. 10.1111/bju.12542 [DOI] [PubMed] [Google Scholar]
- Matera A. G., Terns R. M., Terns M. P. (2007). Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 8 209–220. 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- Mathonnet G., Fabian M. R., Svitkin Y. V., Parsyan A., Huck L., Murata T., et al. (2007). MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317 1764–1767. 10.1126/science.1146067 [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Meng D., Yang S., Wan X., Zhang Y., Huang W., Zhao P., et al. (2016). A transcriptional target of androgen receptor, miR-421 regulates proliferation and metabolism of prostate cancer cells. Int. J. Biochem. Cell. Biol. 73 30–40. 10.1016/j.biocel.2016.01.018 [DOI] [PubMed] [Google Scholar]
- Metzger E., Wissmann M., Yin N., Muller J. M., Schneider R., Peters A. H., et al. (2005). LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437 436–439. 10.1038/nature04020 [DOI] [PubMed] [Google Scholar]
- Misawa A., Takayama K., Urano T., Inoue S. (2016). Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J. Biol. Chem. 291 17861–17880. 10.1074/jbc.m116.718536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa A., Takayama K. I., Fujimura T., Homma Y., Suzuki Y., Inoue S. (2017). Androgen-induced lncRNA POTEF-AS1 regulates apoptosis-related pathway to facilitate cell survival in prostate cancer cells. Cancer Sci. 108 373–379. 10.1111/cas.13151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Deng J. J., Gowda P. S., Rao M. K., Lin C. L., Chen C. L., et al. (2014). Androgen receptor and microRNA-21 axis downregulates transforming growth factor beta receptor II (TGFBR2) expression in prostate cancer. Oncogene 33 4097–4106. 10.1038/onc.2013.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazzeni H., Najafi A., Khani M. (2017). Identification of direct target genes of miR-7, miR-9, miR-96, and miR-182 in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol. Cell. Probes. 34 45–52. 10.1016/j.mcp.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Mortensen M. M., Hoyer S., Orntoft T. F., Sorensen K. D., Dyrskjot L., Borre M. (2014). High miR-449b expression in prostate cancer is associated with biochemical recurrence after radical prostatectomy. BMC Cancer 14:859. 10.1186/1471-2407-14-859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Takayama K., Katayama S., Urano T., Horie-Inoue K., Ikeda K., et al. (2010). miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer P D. 13 356–361. 10.1038/pcan.2010.32 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Ip J. Y., Shioi G., Tripathi V., Zong X., Hirose T., et al. (2012). Malat1 is not an essential component of nuclear speckles in mice. RNA. 18 1487–1499. 10.1261/rna.033217.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., et al. (2014). The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141 4618–4627. 10.1242/dev.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okato A., Goto Y., Kurozumi A., Kato M., Kojima S., Matsushita R., et al. (2016). Direct regulation of LAMP1 by tumor-suppressive microRNA-320a in prostate cancer. Int. J. Oncol. 49 111–122. 10.3892/ijo.2016.3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostling P., Leivonen S. K., Aakula A., Kohonen P., Makela R., Hagman Z., et al. (2011). Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 71 1956–1967. 10.1158/0008-5472.can-10-2421 [DOI] [PubMed] [Google Scholar]
- Palazzo A. F., Lee E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualini L., Bu H., Puhr M., Narisu N., Rainer J., Schlick B., et al. (2015). miR-22 and miR-29a are members of the androgen receptor cistrome modulating LAMC1 and Mcl-1 in prostate cancer. Mol. Endocrinol. 29 1037–1054. 10.1210/me.2014-1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P., Bordeleau M. E., Pelletier J., Sharp P. A. (2006). Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 21 533–542. 10.1016/j.molcel.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Petrovics G., Zhang W., Makarem M., Street J. P., Connelly R., Sun L., et al. (2004). Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene 23 605–611. 10.1038/sj.onc.1207069 [DOI] [PubMed] [Google Scholar]
- Pickard M. R., Mourtada-Maarabouni M., Williams G. T. (2013). Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta. 1832 1613–1623. 10.1016/j.bbadis.2013.05.005 [DOI] [PubMed] [Google Scholar]
- Pickl J. M., Heckmann D., Ratz L., Klauck S. M., Sultmann H. (2014). Novel RNA markers in prostate cancer: functional considerations and clinical translation. Biomed. Res. Int. 2014:765207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Artus C. G., Zoller T., Cougot N., Basyuk E., et al. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309 1573–1576. 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Piwecka M., Glazar P., Hernandez-Miranda L. R., Memczak S., Wolf S. A., Rybak-Wolf A., et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- Prensner J. R., Chen W., Han S., Iyer M. K., Cao Q., Kothari V., et al. (2014a). The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia 16 900–908. 10.1016/j.neo.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Chen W., Iyer M. K., Cao Q., Ma T., Han S., et al. (2014b). PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 74 1651–1660. 10.1158/0008-5472.can-13-3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Iyer M. K., Balbin O. A., Dhanasekaran S. M., Cao Q., Brenner J. C., et al. (2011). Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 29 742–749. 10.1038/nbt.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Iyer M. K., Sahu A., Asangani I. A., Cao Q., Patel L., et al. (2013). The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 45 1392–1398. 10.1038/ng.2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Sahu A., Iyer M. K., Malik R., Chandler B., Asangani I. A., et al. (2014c). The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget 5 1434–1438. 10.18632/oncotarget.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R., Zhao S., Erho N., Schipper M., Iyer M. K., Dhanasekaran S. M., et al. (2014d). RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15 1469–1480. 10.1016/s1470-2045(14)71113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Cui X., Hong Y., Wang J., Li Y., Chen L., et al. (2013). MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Mol. Cell. Biochem. 377 121–130. 10.1007/s11010-013-1576-z [DOI] [PubMed] [Google Scholar]
- Rajabi H., Joshi M. D., Jin C., Ahmad R., Kufe D. (2011). Androgen receptor regulates expression of the MUC1-C oncoprotein in human prostate cancer cells. Prostate 71 1299–1308. 10.1002/pros.21344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramberg H., Alshbib A., Berge V., Svindland A., Tasken K. A. (2011). Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol. Cancer 10:50. 10.1186/1476-4598-10-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas J., Ni X., Haffner M., Wentzel E. A., Salmasi A. H., Chowdhury W. H., et al. (2009). miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 69 7165–7169. 10.1158/0008-5472.can-09-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K., Reon B. J., Anaya J., Dutta A. (2015). The lncRNA DRAIC/PCAT29 locus constitutes a tumor-suppressive nexus. Mol. Cancer Res. 13 828–838. 10.1158/1541-7786.mcr-15-0016-t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J. (2016). Circular RNA expression: its potential regulation and function. Trends Genet. 32 309–316. 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Katsushima K., Shinjo K., Hatanaka A., Ohka F., Suzuki S., et al. (2016). Histone deacetylase inhibition in prostate cancer triggers miR-320-mediated suppression of the androgen receptor. Cancer Res. 76 4192–4204. 10.1158/0008-5472.can-15-3339 [DOI] [PubMed] [Google Scholar]
- Sauvageau M., Goff L. A., Lodato S., Bonev B., Groff A. F., Gerhardinger C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher H. I., Sawyers C. L. (2005). Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J. Clin. Oncol. 23 8253–8261 10.1200/jco.2005.03.4777 [DOI] [PubMed] [Google Scholar]
- Schimmel P. (2018). The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell. Biol. 19 45–58. 10.1038/nrm.2017.77 [DOI] [PubMed] [Google Scholar]
- Schmidt K., Carroll J. S., Yee E., Thomas D. D., Wert-Lamas L., Neier S. C., et al. (2019). The lncRNA SLNCR recruits the androgen receptor to EGR1-bound genes in melanoma and inhibits expression of tumor suppressor p21. Cell. Rep. 27 2493–2507. 10.1016/j.celrep.2019.04.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K., Joyce C. E., Buquicchio F., Brown A., Ritz J., Distel R. J., et al. (2016). The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell. Rep. 15 2025–2037. 10.1016/j.celrep.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K., Weidmann C. A., Hilimire T. A., Yee E., Hatfield B. M., Schneekloth J. S., Jr., et al. (2020). Targeting the oncogenic long non-coding RNA SLNCR1 by blocking its sequence-specific binding to the androgen receptor. Cell. Rep. 30 541–554. 10.1016/j.celrep.2019.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A. M., Chang H. Y. (2016). Long noncoding RNAs in cancer pathways. Cancer Cell. 29 452–463. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., King R. M., Philipson L. (1988). Genes specifically expressed at growth arrest of mammalian cells. Cell 54 787–793. 10.1016/s0092-8674(88)91065-3 [DOI] [PubMed] [Google Scholar]
- Shang Z., Yu J., Sun L., Tian J., Zhu S., Zhang B., et al. (2019). LncRNA PCAT1 activates AKT and NF-kappaB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKalpha complex. Nucleic Acids Res. 47 4211–4225. 10.1093/nar/gkz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N. R., Wang X., Majerciak V., Ajiro M., Kruhlak M., Meyers C., et al. (2016). Cell type- and tissue context-dependent nuclear distribution of human Ago2. J. Biol. Chem. 291 2302–2309. 10.1074/jbc.c115.695049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. B., Ma A. H., Xue L., Li M., Nguyen H. G., Yang J. C., et al. (2015). miR-124 and androgen receptor signaling inhibitors repress prostate cancer growth by downregulating androgen receptor splice variants, EZH2, and Src. Cancer Res. 75 5309–5317. 10.1158/0008-5472.can-14-0795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. B., Xue L., Ma A. H., Tepper C. G., Gandour-Edwards R., Kung H. J., et al. (2013). Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene 32 4130–4138. 10.1038/onc.2012.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2021). Cancer statistics, 2021. CA Cancer J. Clin. 71 7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- Sikand K., Slaibi J. E., Singh R., Slane S. D., Shukla G. C. (2011). miR 488∗ inhibits androgen receptor expression in prostate carcinoma cells. Int. J. Cancer 129 810–819. 10.1002/ijc.25753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M. C., Sato K., Pezic D., Aravin A. A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell. Biol. 12 246–258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Song G., Wang R., Guo J., Liu X., Wang F., Qi Y., et al. (2015). miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Sci. Rep. 5:15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan V., Zou Z., Petrovics G., Xu L., Augustus M., Davis L., et al. (2000). PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc. Natl. Acad. Sci. U S A. 97 12216–12221. 10.1073/pnas.97.22.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg C. N., Dumez H., Van Poppel H., Skoneczna I., Sella A., Daugaard G., et al. (2009). Docetaxel plus oblimersen sodium (Bcl-2 antisense oligonucleotide): an EORTC multicenter, randomized phase II study in patients with castration-resistant prostate cancer. Ann. Oncol. 20 1264–1269. 10.1093/annonc/mdn784 [DOI] [PubMed] [Google Scholar]
- Sun D., Layer R., Mueller A. C., Cichewicz M. A., Negishi M., Paschal B. M., et al. (2014). Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells. Oncogene 33 1448–1457. 10.1038/onc.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Yu Z., Fang X., Liu M., Pu Y., Shao Q., et al. (2017). LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 18 1801–1816. 10.15252/embr.201643668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Sprenger C. C., Vessella R. L., Haugk K., Soriano K., Mostaghel E. A., et al. (2010). Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Invest. 120 2715–2730. 10.1172/jci41824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. T., Ma L. (2019). New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers 11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. (2019). Splicing factors have an essential role in prostate cancer progression and androgen receptor signaling. Biomolecules 9:131. 10.3390/biom9040131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Horie-Inoue K., Katayama S., Suzuki T., Tsutsumi S., Ikeda K., et al. (2013). Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 32 1665–1680. 10.1038/emboj.2013.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Misawa A., Suzuki T., Takagi K., Hayashizaki Y., Fujimura T., et al. (2015). TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat. Commun. 6:8219. [DOI] [PubMed] [Google Scholar]
- Takayama K., Tsutsumi S., Katayama S., Okayama T., Horie-Inoue K., Ikeda K., et al. (2011). Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells. Oncogene 30 619–630. 10.1038/onc.2010.436 [DOI] [PubMed] [Google Scholar]
- Takayama K. I., Fujimura T., Suzuki Y., Inoue S. (2020). Identification of long non-coding RNAs in advanced prostate cancer associated with androgen receptor splicing factors. Commun. Biol. 3:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Ellis J. D., Shen Z., Song D. Y., Pan Q., Watt A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 39 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell S. S., Mortensen R. D., Seo M., Schroeder J. C., Lee J. H., LeTonqueze O., et al. (2012). MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep. 2:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S., Cao Q., Mani R. S., Shankar S., Wang X., Ateeq B., et al. (2008). Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322 1695–1699. 10.1126/science.1165395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Tong Y. C., Steitz J. A. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318 1931–1934. 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., et al. (2001). The sequence of the human genome. Science 291 1304–1351. [DOI] [PubMed] [Google Scholar]
- Verma A. (2018). Recent advances in antisense oligonucleotide therapy in genetic neuromuscular diseases. Ann. Indian Acad. Neurol. 21 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo J. N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176 869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter B. A., Valera V. A., Pinto P. A., Merino M. J. (2013). Comprehensive microRNA profiling of prostate cancer. J. Cancer 4 350–357. 10.7150/jca.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltering K. K., Helenius M. A., Sahu B., Manni V., Linja M. J., Janne O. A., et al. (2009). Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer Res. 69 8141–8149. 10.1158/0008-5472.can-09-0919 [DOI] [PubMed] [Google Scholar]
- Waltering K. K., Porkka K. P., Jalava S. E., Urbanucci A., Kohonen P. J., Latonen L. M., et al. (2011). Androgen regulation of micro-RNAs in prostate cancer. Prostate 71 604–614. 10.1002/pros.21276 [DOI] [PubMed] [Google Scholar]
- Wang D., Ding L., Wang L., Zhao Y., Sun Z., Karnes R. J., et al. (2015). LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget 6 41045–41055. 10.18632/oncotarget.5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., et al. (2011). Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474 390–394. 10.1038/nature10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu W., Black S., Turner O., Daniel J. M., Dean-Colomb W., et al. (2016). Kaiso, a transcriptional repressor, promotes cell migration and invasion of prostate cancer cells through regulation of miR-31 expression. Oncotarget 7 5677–5689. 10.18632/oncotarget.6801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Sun Y., Tao W., Fei X., Chang C. (2017). Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 394 1–12. 10.1016/j.canlet.2016.12.036 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Yang T., Zhao W., Wang N., Li P., et al. (2017). Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget 8 59417–59434. 10.18632/oncotarget.19727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. A., Baxter D. H., Zhang S., Huang D. Y., Huang K. H., Lee M. J., et al. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56 1733–1741. 10.1373/clinchem.2010.147405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Sun Y., Xiang Z., Wang K., Liu B., Xiao G., et al. (2019). Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell. Death Dis. 10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zhang L., Deng J., Guo B., Li F., Wang Y., et al. (2020). A novel micropeptide encoded by Y-linked linc00278 links cigarette smoking and AR signaling in male esophageal squamous cell carcinoma. Cancer Res. 80 2790–2803. 10.1158/0008-5472.can-19-3440 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang L., Zhang L., Wang Y., Li H., Ren X., et al. (2015). Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int. J. Oncol. 46 2586–2594. 10.3892/ijo.2015.2976 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Wang L., Li Y., Chen M., He W., Qi L. (2017). The long non-coding RNA XIST interacted with MiR-124 to modulate bladder cancer growth, invasion and migration by targeting androgen receptor (AR). Cell. Physiol. Biochem. 43 405–418. 10.1159/000480419 [DOI] [PubMed] [Google Scholar]
- Xu S., Sui S., Zhang J., Bai N., Shi Q., Zhang G., et al. (2015). Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 8 4881–4891. [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wang M., Kang M., Wang Q., Wu B., Chu H., et al. (2013). Association between lncrna PCGEM1 polymorphisms and prostate cancer risk. Prostate Cancer Prostatic Dis. 16 139–144. 10.1038/pcan.2013.6 [DOI] [PubMed] [Google Scholar]
- Yang F., Shen Y., Zhang W., Jin J., Huang D., Fang H., et al. (2018). An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell. Death Differ. 25 2209–2220. 10.1038/s41418-018-0123-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Lin C., Jin C., Yang J. C., Tanasa B., Li W., et al. (2013). lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500 598–602. 10.1038/nature12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L. Q., Lin C. R., Liu W., Zhang J., Ohgi K. A., Grinstein J. D., et al. (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147 773–788. 10.1016/j.cell.2011.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Du W. W., Wu N., Yang W., Awan F. M., Fang L., et al. (2017). A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell. Death Differ. 24 1609–1620. 10.1038/cdd.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Jia D., Kim H., Abd Elmageed Z. Y., Datta A., Davis R., et al. (2016). Dysregulation of miR-212 promotes castration resistance through hnRNPH1-mediated regulation of AR and AR-V7: implications for racial disparity of prostate cancer. Clin. Cancer Res. 22 1744–1756. 10.1158/1078-0432.ccr-15-1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., et al. (2015). Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell. Stem. Cell. 16 504–516. 10.1016/j.stem.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Zhai W., Sun Y., Guo C., Hu G., Wang M., Zheng J., et al. (2017). LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor (AR)/miRNA-143-3p signals. Cell. Death Differ. 24 1502–1517. 10.1038/cdd.2017.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W., Sun Y., Jiang M., Wang M., Gasiewicz T. A., Zheng J., et al. (2016). Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2alpha/C-MYC axis under hypoxia. Oncogene 35 4866–4880. 10.1038/onc.2016.19 [DOI] [PubMed] [Google Scholar]
- Zhang A., Zhao J. C., Kim J., Fong K. W., Yang Y. A., Chakravarti D., et al. (2015). LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell. Rep. 13 209–221. 10.1016/j.celrep.2015.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Arun G., Mao Y. S., Lazar Z., Hung G. N., Bhattacharjee G., et al. (2012). The lncRNA Malat1 Is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell. Rep. 2 111–123. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhao W., Valdez J. M., Creighton C. J., Xin L. (2010). Low-density taqman miRNA array reveals miRNAs differentially expressed in prostatic stem cells and luminal cells. Prostate 70 297–304. 10.1002/pros.21064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Lu Y., Kong Z., Zhang Y., Fu F., Su X., et al. (2019). Androgen-responsive lncRNA LINC00304 promotes cell cycle and proliferation via regulating CCNA1. Prostate 79 994–1006. 10.1002/pros.23811 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pitchiaya S., Cieslik M., Niknafs Y. S., Tien J. C., Hosono Y., et al. (2018). Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 50 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhou N., Huang J., Ho T. T., Zhu Z., Qiu Z., et al. (2016). Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget 7 15481–15491. 10.18632/oncotarget.7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Chen J., Ma Z., Liu W., Yang F., Yang Z., et al. (2015). Capsaicin causes inactivation and degradation of the androgen receptor by inducing the restoration of miR-449a in prostate cancer. Oncol. Rep. 34 1027–1034. 10.3892/or.2015.4055 [DOI] [PubMed] [Google Scholar]
- Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Zhao Z., Chou F., Zuo L., Liu T., Yeh S., et al. (2019). Loss of the androgen receptor suppresses intrarenal calcium oxalate crystals deposition via altering macrophage recruitment/M2 polarization with change of the miR-185-5p/CSF-1 signals. Cell. Death Dis. 10:275. [DOI] [PMC free article] [PubMed] [Google Scholar]