Abstract
The microtubule-associated protein tau has been implicated in the pathogenesis of Alzheimer's disease (AD) and other neurodegenerative disorders. Reducing tau levels ameliorates AD-related synaptic, network, and behavioral abnormalities in human amyloid precursor protein (hAPP) transgenic mice. We used mass spectrometry to characterize the post-translational modification of endogenous tau isolated from wildtype and hAPP mice. We identified seven types of tau modifications at 63 sites in wildtype mice. Wildtype and hAPP mice had similar modifications, supporting the hypothesis that neuronal dysfunction in hAPP mice is enabled by physiological forms of tau. Our findings provide clear evidence for acetylation and ubiquitination of the same lysine residues; some sites were also targeted by lysine methylation. Our findings refute the hypothesis of extensive O-GlcNAc modification of endogenous tau. The complex post-translational modification of physiological tau suggests that tau is regulated by diverse mechanisms.
Abnormal modification and accumulation of the microtubule-associated protein tau is associated with Alzheimer's disease (AD) and other neurodegenerative disorders collectively referred to as tauopathies1. A large body of experimental evidence suggests that cerebral accumulation of amyloid-β (Aβ) peptides contributes causally to AD2 and that tau mediates the adverse effects of Aβ on cognition3–5. However, the Aβ and tau species responsible remain a topic of intense investigation.
Transgenic mice with neuronal overexpression of the human amyloid precursor protein (hAPP) bearing familial AD-linked mutations have pathologically elevated Aβ levels in the brain and develop a range of other AD-like abnormalities2. Although most lines of hAPP mice do not spontaneously develop abnormal modification or accumulation of endogenous mouse tau, genetic ablation of tau prevents or reduces behavioral impairments, synaptic deficits, neural network dysfunction, and related molecular alterations in several lines of hAPP mice without affecting their levels of soluble Aβ, amyloid plaques, or neuritic dystrophy3–7. In addition, tau reduction markedly increases resistance to seizure induction in various models of epilepsy3,5,8–11.
Two principal mechanisms have been proposed by which tau may enable Aβ-induced impairments. According to one hypothesis, physiological forms of tau enable aberrant neural network activity caused by diverse pathogenic triggers3,5. An alternative hypothesis is that Aβ changes the posttranslational modification (PTM) or distribution of tau, causing an adverse gain-of-function that turns tau into an active mediator of Aβ-induced neuronal dysfunction12,13. These possibilities are not mutually exclusive2. Characterizing PTMs of tau may help identify strategies to block the pathological processes it enables or mediates.
Most studies of tau PTMs focused on the phosphorylation of abnormal tau or were done under cell-free conditions or in cultured cell lines. These studies provided important information on abnormal tau. However, much less is known about the type and extent of physiological tau modifications that occur in vivo. Furthermore, many tau modifications have not yet been studied as widely as phosphorylation, including O-GlcNAc modification, ubiquitination, methylation, and acetylation. Methylation of tau on lysine and arginine residues has been described very recently14–16, but the functional effects are mostly unknown. O-GlcNAc modification has been proposed to prevent tau phosphorylation by occupying many serines and threonines17–19, thereby preventing dissociation of tau from microtubules and tau aggregation. However, only one O-GlcNAc site has been found on endogenous tau20, in contrast to 51 phosphorylation sites14,21. Ubiquitination of tau has been identified by mass spectrometry of paired helical filaments isolated from AD brains22,23. Tau acetylation has been studied primarily under cell-free conditions and in cell culture and is increased in AD brains by western blot analysis and immunohistochemistry24–26. Acetylation may oppose ubiquitination and degradation of phosphorylated tau26, but may also serve alternate regulatory functions. Physiological tau probably exists as a complex mixture of differentially modified molecules, but little is known about its baseline ubiquitination or acetylation state.
Further complicating the interpretation of tau modifications, the quality of tandem mass spectrometry (MS/MS) information on which the assignments are based was often suboptimal or unreported21,24,26. Assignment of tau phosphorylation sites is especially difficult because of the clustering of potential phosphorylation sites and the abundance of peptides with multiple phosphoryl moieties21. Recent advances in mass spectrometry have enabled the calculation of confidence scores for modification site assignments, such as the SLIP score or Ascore27,28. However, tau modification sites in online databases do not yet include this information, making it difficult to assess the quality of the information provided29,30.
Here we used mass spectrometry to extensively investigate physiological modifications of endogenous tau in brain tissues from untreated wildtype and hAPP mice. We focused on mouse tau because working with mice made it possible to standardize peri-mortem variables and optimize sample preservation. Moreover, the sequence of mouse tau is similar to human tau (89% identical, 92% similar, blastp vs. human tau 441), and MS allowed reliable quantitative comparison of tau modifications in wildtype and hAPP mice.
RESULTS
Investigation of endogenous tau modifications in wildtype mice
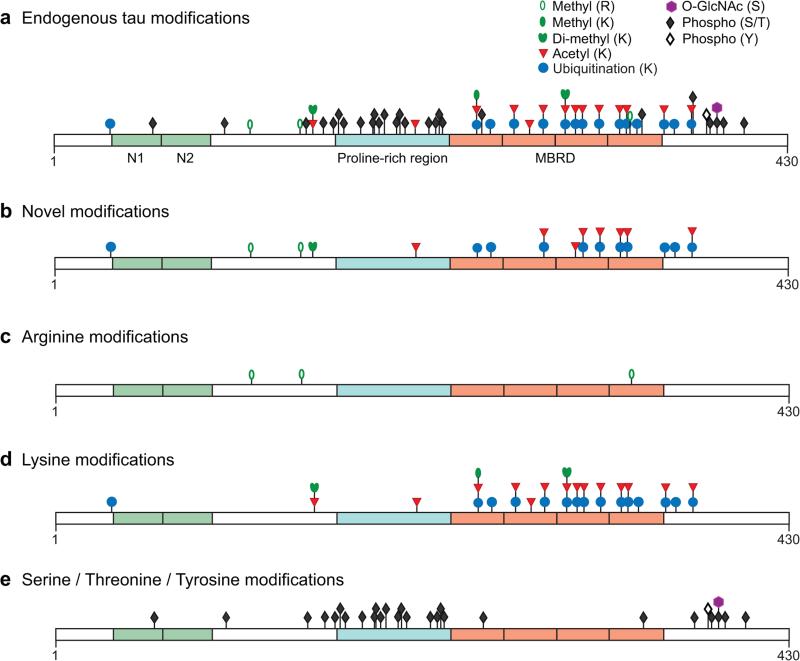
Using endogenous tau samples isolated from wildtype mouse hippocampus and cortex (experiments listed in Supplementary Table 1), we identified and made site assignments for seven types of post-translational modifications: arginine mono-methylation, lysine acetylation, lysine mono-methylation, lysine di-methylation31, lysine ubiquitination, serine O-GlcNAc modification, and serine/threonine/tyrosine phosphorylation (Fig. 1a, Tables 1 and 2, and Supplementary Table 2). Lysine ubiquitination was inferred from the detection of GlyGly-modified lysine, which results from trypsin cleavage of ubiquitin. Our experiments covered 96% of the tau sequence and permitted the assignment of 63 unique post-translational modifications, which are listed in Table 2 by position within mouse tau 430 and the homologous position in human tau 441. As is the convention in tau literature, all amino acid numbering in the text refers to the homologous site in human tau 441. To our knowledge, 22 of the 63 modifications have not been previously reported, and all of these novel modifications occurred on arginine or lysine residues that are conserved in human tau (Table 1, bold and red font in Table 2, and Fig. 1b–d). To search for O-GlcNAcylated tau species, digests of large-scale tau preparations were subjected to lectin weak-affinity chromatography enrichment (Supplementary Table 1) in a strategy optimized to identify O-GlcNAc peptides (Online Methods). O-GlcNAc was identified at a single site (S400) on tau (Fig. 1e), along with 122 O-GlcNAcylated peptide species from co-purifying proteins (Supplementary Table 3). The presence of O-GlcNAc modification at only one site on tau contrasted with the prevalence of other modification types.
Figure 1.
PTMs identified in endogenous mouse tau. Mouse tau was isolated from brains of wildtype mice, and PTMs were assigned by mass spectrometry. Modifications are shown on the longest tau isoform expressed in the mouse central nervous system (430 amino acids). All assigned endogenous tau modifications are shown in the top panel (a). The lower panels show subsets of the same modifications, indicating those that are novel (b) or separating them by the amino acid modified: arginine (c), lysine (d), and serine/threonine/tyrosine (e). The N-terminal exons expressed in mouse tau 430 are shown in green, the proline-rich region in teal, and the four microtubule-binding repeats in orange. Only PTMs with unambiguously assigned sites from wildtype mice (Table 2) are indicated, and all sites are positioned to scale. Note that some amino acid residues can be alternately modified and that these modifications are mutually exclusive (e.g., ubiquitination, mono-/di-methylation, and acetylation). The N-terminal exons N1 and N2 are subject to alternative splicing.
Table 1.
Summary of unambiguously assigned tau modifications in wildtype mice.
| Modification | Sites assigned | Previously reported sitesa | Residue conserved in human tau 441 | Sites in the MBRD | Sites with alternate modifications |
|---|---|---|---|---|---|
| Arginine methylation | 3 | 1 (0.33)b | 3 (1.0) | 1 (0.33) | 0 (0) |
| Lysine acetylation | 14 | 6 (0.42) | 14 (1.0) | 10 (0.71) | 12 (0.86) |
| Lysine Mono-methylation | 1 | 1 (1.0) | 1 (1.0) | 1 (1.0) | 1 (1.0) |
| Lysine Di-methylation | 2 | 1 (0.5) | 2 (1.0) | 1 (0.5) | 2 (1.0) |
| Lysine ubiquitination | 15 | 4 (0.27) | 15 (1.0) | 11 (0.73) | 11 (0.73) |
| Serine O-GlcNAcylation | 1 | 1 (1.0) | 1 (1.0) | 0 (0) | 1 (1.0) |
| Serine/threonine/tyrosine phosphorylation | 27 | 27 (1.0) | 24 (0.89) | 2 (0.07) | 1 (0.04) |
Based on in vitro or in vivo experiments.
Numbers in parentheses indicate the proportion of total for each modification.
Table 2.
Endogenous tau modifications in wildtype and hAPP mice.
| Modification site(s) | PTM | No. of PTMs | Modified residue (mouse 430 isoform) | Homologous residue (human 441 isoform) |
|---|---|---|---|---|
| DHGLKESPP | GlyGly | 1 | 33a | 44 |
| DHGLKAEEA | GlyGly | 1 | 33b | 44 |
| PGSETSDAK | Phospho | 1 | 50|52|53a | 61|63|64 |
| SDAKSTPTAE | Phospho | 1 | 57|58a | 68|69 |
| DAKSTPTAE | Phospho | 1 | 58a | 69 |
| GIGDTPNQE | Phospho | 1 | 100c | 111 |
| VTQARVASK | Methyl | 1 | 115c | 126 |
| IATPRGAAS | Methyl | 1 | 144 | 155 |
| RGAASPAQK | Phospho | 1 | 148 | – d |
| RGAASPAQKGTSNATRIPA | Phospho | 2 | 148,158e | –d,169 |
| SPAQKGTSN | Acetyl | 1 | 152 | 163 |
| SPAQKGTSN | Dimethyl | 1 | 152 | 163 |
| IPAKTTPSPKTP | Phospho | 2 | 164|165, 167 | 175|–,–d |
| IPAKTTPSPKTPPGS | Phospho | 3 | 164, 167, 170 | 175,–d,181 |
| KTTPSPKTP | Phospho | 1 | 167 | – d |
| KTTPSPKTPPGS | Phospho | 2 | 167, 170 | –d,181 |
| PSPKTPPGS | Phospho | 1 | 170 | 181 |
| PSPKTPPGSGEPPKSGERS | Phospho | 2 | 170, 180 | 181, 191 |
| RSGYSSPGSPGTP | Phospho | 2 | 187, 191 | 198, 202 |
| RSGYSSPGSPGTPGSRSRTPS | Phospho | 3 | 187|188|191|194|199 | 198|199|202|205|210 |
| SGYSSPGSP | Phospho | 1 | 188 | 199 |
| SGYSSPGSPGTP | Phospho | 2 | 188, 191 | 199,202 |
| SSPGSPGTP | Phospho | 1 | 191 | 202 |
| SSPGSPGTPGSR | Phospho | 2 | 191, 194 | 202, 205 |
| GSPGTPGSR | Phospho | 1 | 194 | 205 |
| PGSRSRTPSLPTPPTR | Phospho | 2 | 199|201|203|206 | 210|212|214|217 |
| SRSRTPSLP | Phospho | 1 | 201 | 212 |
| SRTPSLPTP | Phospho | 1 | 203 | 214 |
| PSLPTPPTR | Phospho | 1 | 206 | 217 |
| REPKKVAVV | Acetyl | 1 | 214 | 225 |
| AVVRTPPKS | Phospho | 1 | 220 | 231 |
| AWRTPPKSPSASKSRL | Phospho | 2 | 220|224|226|228 | 231|235|237|239 |
| AVVRTPPKSPSAS | Phospho | 2 | 220, 224 | 231, 235 |
| AVVRTPPKSPSASKSRL | Phospho | 2 | 220, 228f | 231, –d |
| PKSPSASKS | Phospho | 1 | 226 | 237 |
| NVRSKIGST | Acetyl | 1 | 248 | 259 |
| NVRSKIGST | GlyGly | 1 | 248 | 259 |
| NVRSKIGST | Methyl | 1 | 248 | 259 |
| SKIGSTENL | Phospho | 1 | 251 | 262 |
| TENLKHQPG | GlyGly | 1 | 256 | 267 |
| IINKKLDLS | Acetyl | 1 | 270 | 281 |
| IINKKLDLS | GlyGly | 1 | 270 | 281 |
| IINKKLDLS | Methyl | 1 | 270g | 281 |
| NVQSKCGSK | Acetyl | 1 | 279 | 290 |
| NVQSKCGSK | GlyGly | 1 | 279g | 290 |
| KDNIKHVPG | Acetyl | 1 | 287 | 298 |
| KDNIKHVPG | GlyGly | 1 | 287 | 298 |
| QIVYKPVDL | Acetyl | 1 | 300 | 311 |
| QIVYKPVDL | Dimethyl | 1 | 300 | 311 |
| QIVYKPVDL | GlyGly | 1 | 300 | 311 |
| VDLSKVTSK | Acetyl | 1 | 306 | 317 |
| VDLSKVTSK | GlyGly | 1 | 306 | 317 |
| KVTSKCGSL | Acetyl | 1 | 310 | 321 |
| KVTSKCGSL | GlyGly | 1 | 310 | 321 |
| SKCGSLGNI | Phospho | 1 | 313g | 324 |
| NIHHKPGGG | Acetyl | 1 | 320 | 331 |
| NIHHKPGGG | GlyGly | 1 | 320 | 331 |
| NIHHKPGGG | Methyl | 1 | 320g | 331 |
| VKSEKLDFK | Acetyl | 1 | 332 | 343 |
| VKSEKLDFK | GlyGly | 1 | 332 | 343 |
| KLDFKDRVQ | Acetyl | 1 | 336 | 347 |
| KLDFKDRVQ | GlyGly | 1 | 336 | 347 |
| DFKDRVQSK | Methyl | 1 | 338 | 349 |
| RVQSKIGSL | GlyGly | 1 | 342 | 353 |
| SKIGSLDNI | Phospho | 1 | 345 | 356 |
| GGGNKKIET | Acetyl | 1 | 358 | 369 |
| GGGNKKIET | GlyGly | 1 | 358 | 369 |
| KKIETHKLTFREN | Phospho | 1 | 362|366 | 373|377 |
| IETHKLTFR | GlyGly | 1 | 364 | 375 |
| NAKAKTDHG | Acetyl | 1 | 374 | 385 |
| NAKAKTDHG | GlyGly | 1 | 374 | 385 |
| AKAKTDHGAEIVYKSPVVS | Phospho | 2 | 375, 385g | 386, 396 |
| AKAKTDHGAEIVYKSPVVSGDTSPRHL | Phospho | 3 | 375, 385, 392|393 | 386, 396, 403|404 |
| AEIVYKSPVVS | Phospho | 2 | 383, 385g | 394, 396 |
| AEIVYKSPVVSGDTSPRHL | Phospho | 3 | 383, 389, 392|393 | 394, 400, 403|404 |
| AEIVYKSPVVSGDTSPRHL | Phospho | 3 | 383, 385, 392|393g | 394, 396, 403|404 |
| IVYKSPVVS | Phospho | 1 | 385 | 396 |
| IVYKSPVVSGDTS | Phospho | 2 | 385, 389 | 396, 400 |
| IVYKSPVV SGDTSPRHL | Phospho | 2 | 385, 392|393 | 396, 403|404 |
| IVYKSPVVSGDTSPRHL | Phospho | 3 | 385, 389, 393 | 396, 400, 404 |
| IVYKSPVVSGDTSPRHL | Phospho | 3 | 385, 389, 392|393 | 396, 400, 403|404 |
| SPVVSGDTS | HexNAc | 1 | 389 | 400 |
| SPVVSGDTS | Phospho | 1 | 389 | 400 |
| SPVVSGDTSPRHL | Phospho | 2 | 389, 392|393 | 400, 403|404 |
| SGDTSPRHL | Phospho | 1 | 393g | 404 |
| PRHLSNVSSTGSIDMV | Phospho | 2 | 398|401|402|403|405 | 409|412|413|414|416 |
| PRHLSNVSSTGSIDMV | Phospho | 3 | 398|401|402|403|405g | 409|412|413|414|416 |
| SSTGSIDMV | Phospho | 1 | 405h | 416 |
Endogenous PTM sites identified in this study are numbered according to the mouse 430 isoform (unless otherwise indicated). Identified sites conserved in mice and humans are also numbered according to the human 441 isoform and are indicated in Fig. 1. Novel modification sites found in wildtype mice are in bold and red font and are highlighted in Fig. 1b. Confirmed modification sites previously identified only by in vitro studies are in bold and blue. Sequences for PTM sites found only in hAPP mice are italicized and gray; these PTMs were excluded from Fig. 1. Site numberings for which unambiguous assignment was not possible are indicated with a vertical bar between alternative assignments. References for previously identified PTMs are provided in Supplementary Table 9.
Mouse tau isoform A peptide. Because this site and modification occurred in alternate isoforms, it was counted only once.
Mouse tau isoform B, C, or D peptide.
Mouse tau isoform A, B, C, or D peptide.
Not conserved in human tau.
Site identified in PSD experiments.
This site was previously reported only in mouse lung.
Modified species was observed only in hAPP samples.
Peptide site was unambiguously assigned with AspN digestion.
The majority of phosphorylation sites were outside the microtubule-binding repeat domain (MBRD) (Fig. 1e). In some instances, multiply phosphorylated peptides did not yield MS/MS spectra of sufficient quality to identify the modified residues unambiguously. To better identify modification sites in these cases, we generated tau samples with higher levels of multiply phosphorylated peptides by subjecting wildtype mice to anesthesia/hypothermia. Results in this model (Supplementary Tables 1 and 2) suggested that endogenous di-phosphorylation in the 210–224 region likely represents phosphorylation of T212 and T217. However, this approach did not resolve doubly and triply phosphorylated sites in the tryptic peptide representing residues 407–438. Because this peptide contained 10 candidate serine/threonine sites for phosphorylation and showed partial methionine oxidation, we could unambiguously assign only one phosphorylation site (S409).
Lysine modifications tended to be enriched in the MBRD, though the enrichment was not statistically significant. Most lysines targeted by acetylation (86%) were also targeted by alternative modifications, usually ubiquitination (Fig. 1d). Conversely, 73% of lysines targeted by ubiquitination were also targeted by acetylation. Acetylation and ubiquitination targeted the same lysine more often than would be predicted by chance (P = 0.00002 by Pearson's chi-squared test), and methylation (both mono- and di-) targeted lysines that were targeted by both acetylation and ubiquitination (P = 0.0005 by likelihood ratio test).
Similar tau modifications in wildtype and hAPP transgenic mice
We next queried our extensive dataset to investigate whether tau modifications are altered by neuronal overexpression of hAPP/Aβ in vivo. We identified in hAPP mice nine tau modification species not found in wildtype mice (italicized in Table 2, see also Supplementary Table 2): five singly modified PTM sites, and four peptides with multiple phosphorylations in residues 386–416. Two of the single modifications, both methylations, are novel. However, their abundance approached the lower limit of detection, so it is uncertain whether they are specific to hAPP mice or also present at undetectable levels in wildtype mice.
Our combined list of tau modifications in wildtype and hAPP mice (Table 2 and Supplementary Table 2) was therefore used to develop a targeted mass spectrometry analysis for a quantitative comparison between hAPP mice and wildtype controls. Because some mass spectra could not be unambiguously assigned in all mice, modified peptides with the same parent mass that could be attributed to modification within a given sequence but without confident site assignment were quantified as a single species (Table 3). Thirty-two different peptide modifications were compared in two independent experiments (Table 3 and Supplementary Tables 4–6), but none differed significantly and consistently between hAPP and wildtype mice (Supplementary Fig. 1a). Because of the potential relevance of OGlcNAc modification of tau to AD17,32–34, the presence of this modification was also assessed by western blot analysis with the site-specific antibody Ab392535. Again, the levels of tau O-GlcNAcylated on S400 did not differ significantly in hAPP and wildtype mice (Supplementary Fig. 2a,b).
Table 3.
Tau modifications quantified in hAPP and wildtype mice.
| Modification site(s) | PTM | No. of modified residues | Modified residue (mouse 430 isoform) | Homologous human residue (441 isoform) | Quantified in whole lysate | Quantified in PSD |
|---|---|---|---|---|---|---|
| DHGLKESPP | GlyGly | 1 | 33a | 44 | + | – |
| DHGLKAEEA | GlyGly | 1 | 33b | 44 | + | – |
| PGSETSDAK | Phospho | 1 | 52a | 63 | + | – |
| SDAKSTPTAE | Phospho | 1 | 57|58a | 68|69 | + | – |
| GIGDTPNQE | Phospho | 1 | 100c | 111 | + | – |
| IATPRGAAS | Methyl | 1 | 144 | 155 | + | – |
| RGAASPAQKGTSNATRIPA | Phospho | 2 | 148, 158d | –e, 169 | – | + |
| IPAKTTPSPKTPPGSGEPPKSGERS | Phospho | 1 | 164|170|174|180 | 175|181|185|191 | + | + |
| IPAKTTPSPKTPPGS | Phospho | 2 | 164|167|170 | 175|–e| 181 | + | – |
| IPAKTTPSPKTPPGS | Phospho | 3 | 164, 167, 170 | 175, –e, 181 | + | – |
| PSPKTPPGSGEPPKSGERS | Phospho | 2 | 170, 180 | 181, 191 | – | + |
| SGERSGYSSPGSPGTPGSR | Phospho | 1 | 184|187|191|194 | 195|198|202|205 | + | + |
| SGERSGYSSPGSPGTPGSR | Phospho | 2 | 184|187|188|191|194 | 195|198|199|202|205 | + | + |
| RSGYSSPGSPGTPGSRSRTPS | Phospho | 3 | 187|188|191|194|199 | 198|199|202|205|210 | + | + |
| PGSRSRTPSLPTPPTR | Phospho | 1 | 199|201|203|206 | 210|212|214|217 | + | + |
| PGSRSRTPSLPTPPTR | Phospho | 2 | 199|201|203|206 | 210|212|214|217 | + | – |
| AVVRTPPKSPSASKS | Phospho | 1 | 220|224|226 | 231|235|237 | + | + |
| AVVRTPPKSPSASKSRL | Phospho | 2 | 220|224|226|228 | 231|235|237|239 | + | – |
| NVRSKIGST | Acetyl | 1 | 248 | 259 | + | – |
| SKIGSTENL | Phospho | 1 | 251 | 262 | + | – |
| IINKKLDLS | Acetyl | 1 | 270 | 281 | + | + |
| IINKKLDLS | GlyGly | 1 | 270 | 281 | + | – |
| VKSEKLDFK | Acetyl | 1 | 332 | 343 | + | – |
| KLDFKDRVQ | Acetyl | 1 | 336 | 347 | + | – |
| KLDFKDRVQ | GlyGly | 1 | 336 | 347 | + | – |
| SKIGSLDNI | Phospho | 1 | 345 | 356 | + | + |
| IETHKLTFR | GlyGly | 1 | 364 | 375 | + | – |
| NAKAKTDHG | Acetyl | 1 | 374 | 385 | + | – |
| AKAKTDHGAEIVYKSPVVSGDTSPRHL | Phospho | 2 | 375|385|389|392|393 | 386|396|400|403|404 | + | + |
| AKAKTDHGAEIVYKSPVVSGDTSPRHL | Phospho | 3 | 375|385|389|392|393 | 386|396|400|403|404 | + | + |
| IVYKSPVVS | Phospho | 1 | 385 | 396 | + | + |
| SPVVSGDTSPRHL | Phospho | 1 | 389|392|393 | 400|403|404 | + | + |
| PRHLSNVSSTGSIDMV | Phospho | 2 | 398|401|402|405 | 409|412|413|416 | + | + |
| SSTGSIDMV | Phospho | 1 | 405f | 416 | + | + |
Sites for which unambiguous assignment was not possible are indicated with a vertical bar between alternative assignments.
Peptide from mouse isoform A.
Peptide from mouse isoforms B, C, or D.
Peptide from mouse isoforms A, B, C, or D.
Identified in PSD experiments.
Not conserved in human tau.
Site unambiguously assigned with AspN digestion.
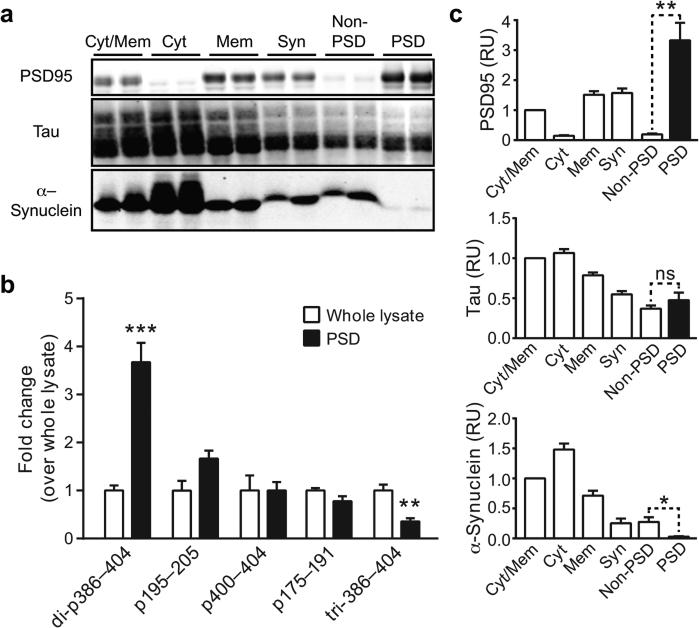
Even small amounts of abnormal tau may be highly pathogenic when associated with the post-synaptic density (PSD)5,12,13,36. Therefore, we also compared tau modifications in PSD fractions isolated from the hippocampus and cortex of hAPP and wildtype mice (Fig. 2a,b). Because of the low amount of tau isolated from PSD fractions, only the most abundant modifications could be compared. Sixteen modified peptides were identified in three independent experiments (Table 3, Supplementary Fig. 1b, and Supplementary Tables 6–8). Acetylated K281 was detected in a single experiment; all other peptide modifications in the PSD were phosphorylations. In a side-by-side comparison of tau isolated from the PSD or from whole hippocampal and cortical lysates, two of the five modified peptides we quantified differentiated PSD tau from whole-lysate tau. In the 386–404 region, di-phosphorylation was higher, and tri-phosphorylation was lower, in PSD tau than in whole-lysate tau (Fig. 2c). None of the tau modifications in PSD fractions were significantly and consistently different in hAPP mice and wildtype controls (Supplementary Fig. 1b). Western blot analysis with PHF1 antibody to identify PSD tau phosphorylated at residues 396 and 404 revealed no significant difference between hAPP and wildtype mice either (Supplementary Fig. 3a,b). Tau phosphorylation at S416 in the PSD was detected at higher frequency in hAPP mice (35%) than in wildtype mice (16%), although the difference was not statistically significant. This site merits further investigation.
Figure 2.
Differential modification of tau in the PSD. (a) Western blot showing PSD95, tau, and α-synuclein levels in two different mice from a replicate cohort at each step of PSD fractionation. Full-length blots are presented in Supplementary Fig. 4. (b) Quantification of western blot signals. n = 8 mice, 5–6 months of age. Values were normalized to the average level of the respective protein in the cytosolic/membrane fraction (arbitrarily defined as 1.0). (c) Quantification of the five most common tau modifications in the PSD fraction relative to average levels in hippocampal and cortical whole lysate (arbitrarily defined as 1.0). Cohort B: n = 10 or 12 mice per group, 7–10 months of age (Supplementary Table 4). Because of site ambiguity, peptide sequences containing modified sites are indicated; di- and tri- denote doubly and triply modified peptides, respectively. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 vs. whole lysate or as indicated by brackets (paired (b) or unpaired (c) t test with Holm correction). Welch's correction was applied in (c) to the analysis of peptides di-p386–404 and tri-p386–404 due to unequal variance. Cyt/Mem, cytosolic- and membrane-containing fraction; Cyt, cytosolic fraction; Mem, membrane fraction; Non-PSD, fraction remaining after PSD extraction; ns, not significant; RU, relative units; Syn, synaptosomal fraction. Quantitative values are means ± SEM. Some error bars in (b) are too small to see.
DISCUSSION
In this study, we identified seven types of physiological PTMs at 63 sites on endogenous mouse tau, one third of which were novel. To our knowledge, this increases to 100 the number of site-assigned PTMs that have been identified on endogenous, wildtype tau from human, mouse, or rat (Supplementary Table 9). Forty-five of the 63 tau PTMs we detected were previously identified in wildtype animals without pathological phenotypes and/or in human brain tissues lacking neurodegeneration. Of the 23 PTMs we did not detect, 15 had been identified only in human brain tissue, and eight were identified in mouse or rat brain by modification-specific enrichment (Supplementary Table 9)14. Of the 16 human PTM sites, six are located in poorly conserved sequences or low-abundance splice variants of mouse tau37,38. The remaining nine target lysines (mono-methyl-K190, -K290, -K317, and -K353; di-methyl K267, -K290, -K317, and -K353; and acetyl-K274) may be PTMs whose abundance genuinely differs between mice and humans.
Previous studies typically reported two or three types of modification in endogenous tau. We detected and site-assigned seven modification types in an unbiased manner—revealing a higher level of regulation of endogenous tau than previously appreciated. Although it failed to detect eight mouse or rat tau modifications identified by modification enrichment, our unbiased detection method identified 22 novel tau modifications. We also confirmed seven putative modifications for which in vitro evidence was limited, including six lysine acetylations (Supplementary Table 9)21,24,26,29. We further identified as physiological three lysine modifications (GlyGly-K311 and -K317 and methyl-K163) previously found only in pathological tissues16,23. Our findings effectively double the number of known distinct lysine modifications of physiological tau (Supplementary Table 9) and highlight the potential significance of lysine-mediated regulation.
The robust targeting of acetylation, ubiquitination, and methylation to the same set of lysines in endogenous tau suggests cross-talk between these modifications39. Findings in cultures of primary neurons and HEK293 cells suggested a competition between acetylation and ubiquitination of specific lysines in tau26. Our findings suggest that acetylation and ubiquitination also compete to modify lysines in tau in vivo. Nearly all acetylation sites on tau were alternately modified by ubiquitin, including three of the four KXGS motifs, which regulate neurite extension and the binding of tau to microtubules40,41. A cell culture study showed competitive regulation of acetylation and phosphorylation of KXGS motifs in tau25. Consistent with this finding, we did not detect any peptides simultaneously modified by both phosphorylation and acetylation. However, it is possible that any such doubly modified peptides in the MBRD would be too low in abundance to detect on tau even without competitive modification.
We found that methylation targets two critical lysines in endogenous tau that are also modified by acetylation and ubiquitination. These highly regulated, triply targeted lysines reside in the first KXGS motif and in the PHF6 sequence (306VQIVYK311), two regions that regulate the microtubule-binding and aggregation of tau.42 Lysine mono-methylation of the first KXGS motif (K259–S262) may prevent tau from binding to microtubules, similar to the effect of KXGS phosphorylation41. Mutation of K311 in the PHF6 sequence inhibits both the microtubule binding and aggregation of tau24. Di-methylation of K311 may have similar effects. K281, which is adjacent to PHF6* (275VQIINK280) and targeted by acetylation and ubiquitination in wildtype mice, may also control tau aggregation43. We only found this residue to be mono-methylated in hAPP mice, albeit at low levels. The alternate modification of lysines by at least four different processes suggests that these residues have a particularly strategic role in the regulation of tau.
It is tempting to speculate that different types of lysine modifications at triply targeted residues have different effects on tau function. At physiological pH, acetylation neutralizes, whereas methylation preserves, the positive charge on lysines. Because the high density of positively charged residues in the MBRD contributes to microtubule binding40, neutralization of charged lysines may explain the decreased microtubule binding of in vitro acetylated tau24, as in vitro methylated tau binds normally to microtubules14. In addition, mono- versus di-methylation of lysine is recognized by different methyl-binding domain proteins31. Consequently, these alternative modifications may direct the binding of tau to different proteins.
Ubiquitination often leads to protein degradation but can also modulate signaling pathways by inhibiting protein–protein interactions through a steric bulk effect. Given the short half-life of endogenous proteins targeted for degradation, many of the ubiquitination sites we found may represent longer-lived modifications serving regulatory functions. We did not find lysine ubiquitination in the proline-rich region of tau, which may be associated with tau degradation26.
The complex acetylation pattern of tau by different factors may help explain some of the divergent effects on tau aggregation. Acetylation by CREB-binding protein enhances tau aggregation24, whereas acetylation by p300 inhibits aggregation25, possibly reflecting the acetylation of different lysines. Indeed, the contributions of different lysine modifications to tau function likely vary by modification type and location and deserve to be further explored in both health and disease.
Another modification that has been proposed to have a role in tau regulation is O-GlcNAc modification. Through a suite of rigorous experiments, we confirmed the presence of a single O-GlcNAc modification site at S400 at low stoichiometry. This site was previously identified by mass spectrometry of endogenous tau isolated from rat brain20 and subsequently detected by western blotting with an antibody specific to O-GlcNAc-modification of S400 on tau35. O-GlcNAc modification has been proposed to have an inverse relationship with hyper-phosphorylation of tau on the basis of in vitro and in vivo experiments testing pharmacological blockade of β-N-acetylglucosaminidase (O-GlcNAcase, OGAse), the O-GlcNAc-removing enzyme17,18,34. However, chronic treatment of mice with OGAse inhibitors increased O-GlcNAc on S400 of tau without changing tau phosphorylation—the first indication that these modifications are not necessarily reciprocal32. Our findings do not support the notion that OGlcNAc modification blocks pathologic tau phosphorylation by competitively occupying many potential phosphorylation sites19. In vitro and in vivo studies suggest that O-GlcNAc modification of tau may prevent tau aggregation or toxicity by an unknown mechanism without altering tau phosphorylation32,33.
Arginine methylation is the most recently described modification of tau15. The MBRD arginine methylation site on tau (Table 2) was found in a broad survey of arginine methylation in mouse brain15. We found only arginine mono-methylation of tau, the functional impact of which is unknown. Because the two novel arginine methylation sites we identified were just N-terminal to the proline-rich region, arginine methylation might affect the binding of tau to membrane proteins, which is mediated by the N-terminal region44, or to SH3 domain-containing proteins, which is mediated by the proline-rich region45. Arginine methylation could also alter nucleo-cytoplasmic shuttling of tau46, the regulatory mechanism of which is unclear.
Most of the tau phosphorylation we identified was at sites adjacent to the MBRD, consistent with previous reports14,29,47. The number of tau phosphorylation sites we found on endogenous mouse tau was similar to the number identified on tau from postmortem brain tissue of cognitively normal people; more than half of the site identifications overlapped with ours14. Of the 31 phosphorylation sites identified in that study, only six (four known, two novel) were found in all samples14. The four known sites (T181, S202, T231, and S404) were consistent with four of the singly modified phosphopeptides we most commonly detected in whole lysate, suggesting that the phosphorylation state of endogenous tau is similar in wildtype mouse brain and normal human brain tissue. We also found that tau is phosphorylated differently in the PSD fraction versus unfractionated lysate, suggesting a specialized regulation or function of physiological tau in the PSD.
Notably, we did not detect any difference in tau modifications between hAPP-J20 mice and wildtype controls at an age when hAPP mice show synaptic, network, and behavioral abnormalities, consistent with our earlier findings3,4. We previously detected comparable levels of hippocampal tau phosphorylation in young hAPP mice by western blot analysis and demonstrated that increases in tau phosphorylation in older hAPP mice are restricted to neuritic plaques3. Other studies found increased tau phosphorylation in hAPP mice by western blotting48, including in the hippocampus of young hAPP-J20 mice49. Using quantitative mass spectrometry, we assessed many more tau modifications than we could assess by western blot and were able to quantify about half of the modifications found. The few modifications detected only in hAPP mice were mostly in the MBRD and included two lysine methylation sites, suggesting increased lysine methylation of tau in hAPP mice. However, these modifications were too rare to be quantified, so we could not confirm that they were specific to hAPP mice. Indeed, we found no quantitative difference in tau modification in unfractionated whole lysates or PSD preparations from hAPP-J20 mice versus wildtype controls.
These findings support the hypothesis that normally modified endogenous tau has a physiological function that allows Aβ and other epileptogenic factors to elicit aberrant network activity2–5. However, we cannot exclude the possibility that mislocalization of tau or modifications below our limit of quantification contribute to an adverse gain-of-function mechanism in hAPP mice. Nor can we exclude the possibility that tau modifications are altered at other ages, in other brain regions, or in other cellular compartments in this model, including the cytosolic compartment of dendritic spines.
The extensive modification of tau in normal brains suggests that tau regulation is important, although the exact functions of tau in the adult brain are unclear1. Many, if not all, of these modifications were substoichiometric. Phosphorylation of tau was the most abundant modification we identified; however, normal tau isolated from human brain tissue has been reported to carry 2–4 phosphorylations per molecule by molar phosphate analysis50. Lysine and arginine modifications of physiological tau were even less abundant, and O-GlcNAc was not detectable without targeted enrichment. Because several tau modifications, particularly those in the MBRD, prevent tau from binding to microtubules24,41, we speculate that many of the modifications we found belong to a small, highly regulated pool of tau that is not bound to microtubules and is free to interact with diverse molecules in different neuronal compartments. We provide these data as a resource for additional studies, which are needed to further elucidate the regulation and function of the many PTMs of tau that occur in health and disease.
Methods
Mice and hypothermia induction
All mice were on the C57BL/6J background. Heterozygous transgenic mice from line J20 carry an alternatively spliced hAPP minigene bearing the Swedish and Indiana mutations51. Mice lacking endogenous tau served as negative controls for western blotting experiments52. Food (Picolab Rodent Diet 20) and water were given ad libitum, and mice were kept on a standard 12-h light/dark cycle. All mouse experiments were done during the light cycle. Mice were group housed with up to five mice per cage. Only naive mice were used for this study. Mice were assigned to groups randomly based on mouse number and genotype. For hypothermia induction53, mice were injected intraperitoneally with ketamine HCl (75 mg/kg) and medetomidine (1 mg/kg) or with saline (negative control), and returned to their home cage for 1 h before analysis. Mice were killed by cervical dislocation, and brain tissues were immediately frozen on dry ice. All experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
Chemicals and reagents
Phosphatase inhibitor cocktails 1 or 3, and 2 (100x, Sigma) and protease inhibitor tablets (cOmplete, EDTA-free and cOmplete-mini, EDTA-free; Roche) were used interchangeably with Halt protease and phosphatase inhibitor cocktails (Thermo Scientific); all were used at 1x according to the manufacturers’ instructions. Thiamet G (4390, Tocris) was used in a range of concentrations, as specified. Trichostatin A ([R-(E,E)]-7-[4-(dimethylamino)phenyl]-N-hydroxy-4,6-dimethyl-7-oxo-2,4-heptadienamide, Sigma) was used at 3 μM and niacinamide (pyridine-3-carboxylic acid amide, Sigma) at 10 mM. Other reagents were ketamine HCl (Bionichepharma), medetomidine (Dexdomitor, Henry Schein), perchloric acid (Fisher), methanol (Fisher), 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES, Sigma), sodium chloride (5M, Cellgrow), sodium dodecyl sulfate (from 10% solution, Teknova), TritonX-100 (Sigma), and sodium deoxycholate (Sigma). All reagents for peptide digestions were from Sigma, except proteomic-grade trypsin (Promega) and AspN (Roche).
Perchloric acid enrichment
Mouse hippocampus and cortex were isolated over ice and homogenized in 1% perchloric acid54. The suspension was incubated for 20 min on ice and spun at 20,000g at 4 °C for 30 min. The supernatant was neutralized with NaOH; 1 M Tris (pH 7.4), phosphatase inhibitors and 25 μM Thiamet G were added, and the solution was concentrated over a 10-kDa MW filter (Vivaspin 20, Vivaproducts; Ultrafree, Millipore). For large-scale lectin weak-affinity chromatography preparations, 10–14 hemibrains were combined from 8–10 mice. For the first quantitative experiment (Supplementary Tables 4 and 5), wildtype mice were selected randomly; the three most hyperactive hAPP mice were selected from a total of seven hAPP mice based on their behavior in the open field test, performed as described55.
Whole-lysate preparation for immunoprecipitation
Mouse cortex and hippocampus were isolated on ice and homogenized in Thermo IP Lysis Buffer containing 25 μM Thiamet G and protease and phosphatase inhibitors. For quantitative experiments, trichostatin A and niacinamide were added to the buffer (Supplementary Tables 4 and 6). The lysates were sonicated (Episonic Multi-functional Bioprocessor 1000, Epigentek Group) at an amplitude of 40% on ice twice for 5 min and spun for 10 min at 10,000g at 4 °C. The supernatant was collected, and the protein concentration was measured by Bradford (Bio-Rad) assay according to the manufacturer's instructions.
Whole-lysate preparation for O-GlcNAc western blot
Mouse cortex and hippocampus were isolated on ice and homogenized in PBS containing 500 mM NaCl, 25 μM Thiamet G, 3 μM trichostatin A, 10 mM niacinamide, and protease/phosphatase inhibitors35. The lysates were centrifuged at 18,000g for 15 min. The supernatants were incubated at 95 °C for 7 min and centrifuged at 18,000g for 15 min to produce tau-enriched supernatants35.
Postsynaptic density fractionation
PSD fractionation was done as described, with modifications56,57. All solutions contained 1 μM Thiamet G and phosphatase inhibitors. Trichostatin A (3 μM) and niacinamide (10 mM) were used in all buffers for PSD fractionation in the second and third PSD experiments (Supplementary Tables 7 and 8). Combined hippocampus and cortex from one whole mouse brain were homogenized with a Dounce homogenizer in 1 ml of buffer A (40 mM Tris-acetate, pH 7.5., 0.3 M sucrose, and protease inhibitors). The suspension was transferred to a centrifuge tube, washed with 1 ml of buffer A, and spun at 850g (IEC Centra GP&R, rotor 216; Thermo Scientific) at 4 °C for 20 min. The supernatant was collected and stored on ice; the pellet was re-suspended in 2 ml of buffer A and spun again. The resulting supernatant was combined with the first; 50 μl was retained as the cytosolic/membrane fraction. Then, 8 ml of 40 mM Tris-acetate buffer was layered on the combined supernatants and centrifuged at 17,000g (SW41 Ti rotor) at 4 °C for 25 min. The resulting supernatant was saved as the cytosolic fraction. The pellet was re-suspended in 1.2 ml of buffer A; 50 μl was set aside as the membrane fraction, and the remainder was loaded onto a sucrose density gradient consisting of 2.2 ml each of buffer A containing 1.2 M, 1.0 M, or 0.8 M sucrose. Then, 4 ml of 40 mM Tris-acetate buffer was added to the top, and the samples were spun at 85,000g (SW41 Ti rotor) at 4 °C for 2 h. The white layer at the 1.0 M/1.2 M sucrose interface was collected, re-suspended in 10–11 ml of buffer A and spun at 85,000g (SW41 Ti rotor) at 4 °C for 30 min. The pellet was kept at –80 °C overnight, thawed, and re-suspended in buffer B (10 mM Bicine-Tris, pH 7.5, 5% N-octylglucoside, and protease inhibitors) with a 26-gauge needle; 50 μl was set aside as the synaptosomal fraction. The remaining suspension was loaded onto a sucrose gradient consisting of 3 ml each of buffer B containing 1% N-octylglucoside and 2.2 M, 1.4 M, or 1.0 M sucrose. Samples were spun at 85,000g (SW41 Ti rotor) at 4 °C for 2 h, and the top layer was collected as the non-PSD fraction. The translucent white layer at the 1.4 M/2.2 M sucrose interface was collected, resuspended in 12 ml of 10 mM Bicine-Tris, pH 7.5, and spun at 85,000g (SW41 Ti rotor) at 4 °C for 2 h. The pellet was stored at –80 °C until immunoprecipitation. The pellet was resuspended in 35 μl of PSD buffer (20 mM HEPES, 5 mM sodium chloride, 1% TritonX-100, 1% sodium dodecyl sulfate, and 1% sodium deoxycholate) and used for western blotting or tau immunoprecipitation. Samples for immunoprecipitation were diluted with 565 μl of IP lysis buffer (Pierce Crosslink IP kit) containing 25 μM Thiamet G, 3 μM trichostatin A, 10 mM niacinamide, and protease and phosphatase inhibitors. In the final PSD experiment (Supplementary Table 8), tau was enriched before immunoprecipitation by heating the 600 μl solution at 95 °C for 10 min, incubating on ice for 5–10 min, spinning the sample at 20,817g at 4 °C for 30 min, and collecting the supernatant.
Tau immunoprecipitation
Tau was immunoprecipitated from whole lysates of cortex and hippocampus or from PSD preparations with tau5 antibody free of bovine serum albumin (BSA) and azide (ab80579, Abcam), and a cross-link immunoprecipitation kit (Thermo-Pierce) following the manufacturer's instructions. Elution buffer (30 μl) was used for the second elution step of the cross-link immunoprecipitation protocol. Columns were used up to twice without noticeable loss of immunoprecipitation efficiency. For experiments using in-gel trypsin digestion of proteins before mass spectrometry, the immunoprecipitated proteins were run on a 4–12% Bis-Tris gel and stained with SafeStain (Life Technologies). A broad band around 50 kDa was then cut out for mass spectrometry analysis. For experiments using in-solution trypsin digestion before mass spectrometry, immunoprecipitated samples were neutralized with 1 M Tris, pH 9.6, containing 25 μM Thiamet G and phosphatase inhibitors. For quantitative experiments, 3 μM trichostatin A and 10 mM niacinamide were added to the neutralization solution. For the initial assignment of tau modifications, we used 1 mg of whole lysate of cortex and hippocampus and 30 μg of tau5. For quantitative immunoprecipitation experiments, we used 215 μg of whole lysate of cortex and hippocampus or the entire PSD fraction from one mouse (approximately 215 μg), and 10–20 μg of tau5 antibody.
Western blotting
Western blotting was done as described58. Signals were quantified with an Odyssey Standard or CLx infrared fluorescence imager and ImageStudio software (both from LI-COR Biosciences). For western analysis of PSD fractions, blots were incubated overnight at 4 °C with mouse anti-PSD95 (MABN68, Millipore; 1:2000), washed in TBST, and incubated overnight at 4 °C with mouse anti-α-synuclein (610787, BD Transduction Laboratories; 1:2000) and rabbit anti-tau (EP2456Y, MAB10417, Millipore; 1:2000). Blots were washed in TBST and co-incubated with the secondary antibodies 680LT donkey anti-rabbit (926-68023, LI-COR Biosciences; 1:20,000) and 800CW donkey anti-mouse (926-32212, LI-COR Biosciences; 1:20,000), for 1 h at room temperature.
To detect O-GlcNAc at S400 of tau, blots were blocked with 2% BSA for 1 h, incubated overnight at 4 °C with Ab3925 antibody35 (1:500 in 2% BSA, a gift from Dr. David Vocadlo, Simon Fraser University) and then with 800CW goat anti-rabbit secondary antibody (925-32211, LI-COR Biosciences; 1:20,000). Recombinant (nonmodified) tau protein (T-1007-1, rPeptide) included on the same blot as a negative control yielded no signal (data not shown). After signals were quantified, membranes were stripped for 15 min with NewBlot nitrocellulose stripping buffer (928-40030, LI-COR Biosciences). Blots were incubated with tau5 antibody (MAB361, Millipore; 1:2000) and then with 800CW donkey anti-mouse (926-32212, LI-COR Biosciences; 1:20,000).
To detect PHF1, PSD fractions were dissolved in 2% BSA and loaded in equal volumes onto the gel. Membranes were co-incubated with anti-PHF1 (1:2,000, a gift from Dr. Peter Davies, Feinstein Institute of Medical Research) and anti-actin (A2066, Millipore; 1:2,000) and then with 680CW goat anti-rabbit (926-32221, LI-COR Biosciences; 1:5000) and 800CW goat anti-mouse (926-32210, LI-COR Biosciences; 1:20,000) secondary antibodies.
Peptide identification and label-free quantitation by mass spectrometry
Peptides in tau-enriched samples were identified by peptide sequencing with mass spectrometry. In-gel digestion was done according to the UCSF Mass Spectrometry Facility protocol (https://msf.ucsf.edu/ingel.html). For in-solution digestion, tau-enriched samples were incubated with urea (5 M final) and 10 mM tris(2-carboxyethyl)phosphine for 10 min at 56 °C. After reduction, the samples were alkylated with 20 mM iodoacetamide at room temperature for 30 min in the dark, and quenched with 10 mM tris(2-carboxyethyl)phosphine, and the final volume was brought up to 250 μl with 100 mM ammonium bicarbonate buffer. Trypsin and AspN digestions were done overnight at 37 °C with a 1:50 ratio of enzyme to total protein. Samples were acidified with 10% formic acid to pH 2–3 and desalted with C18 Omix tips, and the extracted peptides were dried in a Speedvac concentrator (Savant) before analysis.
To target O-GlcNAcylated peptides, lectin weak-affinity chromatography enrichment was done before liquid chromatography-MS/MS analysis with electron-transfer dissociation (ETD) mass spectrometry (described below)59–61. For these experiments, tau-enriched protein samples (1.2–1.8 mg) were generated by large-scale perchloric-acid enrichment of pooled cortical lysates of 8–10 mice.
Peptides were sequenced with an LTQ-Orbitrap Velos or an LTQ-Orbitrap XL (Thermo) mass spectrometer, each equipped with a 10,000 psi system nanoACUITY (Waters) UPLC instrument for reversed-phase chromatography with a C18 column (BEH130, 1.7 μm bead size, 100 μm x 100 mm). For liquid chromatography, the flow rate was 600 nl/min, and peptides were separated over 60 or 90 min with a linear gradient of 2% to 30% acetonitrile in 0.1% formic acid.
Tau-enriched samples were analyzed by several MS/MS techniques. For collision-induced dissociation (CID) and ETD experiments, survey scans were recorded over a 350–1500 m/z range and MS/MS was done with CID or ETD activation on the three most intense precursor ions, with a minimum of 10,000 counts and 35% normalized collision energy. ETD isolation width was 3.0 Th (Thomson units) with 200-μs activation time; CID isolation width was 2.0 Th with 30-μs activation time. For the higher-energy collisional dissociation (HCD) experiments, survey scans were recorded over a 350–1400 m/z range, and MS/MS was done with HCD activation on the 10 most intense precursor ions, with minimum signal of 4000 counts, isolation width 2.5 Th, 0.1-μs activation time, and 30% normalized collision energy over a mass range of 350–1500 m/z. Internal recalibration to a polydimethylcyclosiloxane ion with m/z = 445.120025 was used for both MS and MS/MS scans62.
Mass spectrometry centroid peak lists were generated with in-house software (PAVA), and data were searched with Protein Prospector v. 5.10.163. For protein identification, database searches were performed against the Mus musculus subset of the UniProtKB database (downloaded March 21, 2012) totaling 77,771 entries. This database was concatenated with a fully randomized set of 77,771 entries to estimate the false-discovery rate64. Data were also searched against a targeted library of tau isoform sequences (P10637, P10637-2, P10637-3, P10637-4, P10637-5, and P10637-6). Peptide sequences were matched as tryptic peptides with 0, 1, or 2 missed cleavages and carbamidomethylated cysteines as a fixed modification. Variable modifications included oxidation of methionine, N-terminal pyroglutamate from glutamine, start methionine processing, protein N-terminal acetylation, phosphorylation on serine, threonine, or tyrosine, O-GlcNAcylated serine and threonine, acetylated lysine, mono-, di-, or trimethylation on lysine or arginine, and diglycyl-modified lysine. Mass accuracy tolerance was set to 20 ppm for parent and 30 ppm for fragment masses.
For reporting of protein identifications from this database search, score thresholds were selected that resulted in a protein false-discovery rate of ~1%. The Protein Prospector parameters were minimum protein score of 22, minimum peptide score of 15, and maximum expectation values of 0.02 for protein and 0.05 for peptide matches. PTM assignments on tau were scored by “SLIP” scoring within Protein Prospector27; unambiguous site assignments had a score ≥6. Site assignments were manually confirmed.
For label-free quantitation in a targeted method with HCD fragmentation, a list of parent masses with an exclusion window of ±10 ppm was developed (Supplementary Table 2). The parent mass list included experimentally observed and predicted masses for tryptic peptides containing up to three phosphorylations on serine/threonine/tyrosine, O-GlcNAcylation on serine/threonine, diglycyl-modified lysine, acetylation on lysine or arginine, and mono- or di-methylation on lysine or arginine, with up to two missed cleavages, as well as unmodified peptides that uniquely identify the known mouse isoforms of tau (SwissProt P10637, P10637-1, P10637-2, P10637-3, P10637-4, P10637-5, P10637-6). Dynamic exclusion was applied for 10 s after a single repeat count, with an exclusion list size of 25 masses. All peptides used for site assignments were manually checked with alternate site-assignment hypotheses. Data were searched as described above for information-dependent acquisition mode experiments. Quantitative analysis was done with a label-free quantitation method that integrates the base peak intensity for each identified species65. Identified peptides are reported in Supplementary Tables 5–8.
Spectra and results for all identified tau peptides may be viewed with the freely available software MS Viewer, which can be accessed through the Protein Prospector suite of software at the following URL: http://prospector2.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer. Search keys for the PTM site assignment data are listed for each set of experiments: Velos HCD: jtuvjpmrvc; OrbitrapXL CID: kfjt07okcs; and OrbitrapXL ETD: m5jt1eadj7. Search keys for the peptides identified in the label free quantitation experiments are: Cohort A: v5rszcgkcv, Cohort B: 59gqdxjiwa, Cohort D: ge5lvgitnp, and Cohort E: v6uklmtkyj. Raw mass spectrometry data files are available at the ProteoSAFE resource http://massive.ucsd.edu, and may be accessed with the accession number MSV000078796.
Quantitative comparison of tau modifications
The value of modified tau peptides in each experiment was normalized to the median peak area of unmodified tau peptides from that sample, excluding unmodified tau peptides with a value of zero for more than two samples. Any modified peptide with a peak width of zero was assigned a value of zero for the quantitative analysis. Modified peptides with the same mass to charge ratio (m/z) represent different possible peptide species within the same quantified peak and were averaged to obtain the value for that tau modification. Peptides that had the same tau modification but different m/z values (because of charge difference, missed trypsin cleavage, or oxidation) were quantified using the sum of the values for each peptide containing that tau modification. For each cohort, quantified values were normalized to the average value of that modification in wildtype mice, and the normalized values of each modification, excluding zero values, were compared in wildtype versus hAPP mice. To determine whether a modification appeared more frequently in one genotype, we also compared the number of zero values across genotypes for each tau modification.
Statistical analysis
For quantitative mass spectrometry experiments, investigators were blinded to genotype and cellular fractionation information during data collection and analysis. Data supporting quantitative results shown in figures were embedded as linked files in figure legends. Sample sizes were comparable to those used in previous PTM studies of human subjects24,66,67, although we did not predetermine them by statistical methods. The normality of data was confirmed by visual inspection but not formally tested. Variance was assessed by F-test, and unequal variance between groups was determined by an F-test with P < 0.05. A two-tailed t test was used to compare two groups and a Welch's correction was applied when comparing groups with unequal variance. Contingency analysis was done in Prizm (GraphPad) to determine significant enrichment in the MBRD; a Holm correction was used for multiple comparisons. A chi-square test was used when all expected counts were more than 5 (acetylation P = 0.21, ubiquitination P = 0.17); otherwise, we used Fisher's exact test (arginine methylation P = 1.0, lysine methylation P = 1.0, phosphorylation P = 0.44). Pearson's chi-squared test and the likelihood ratio test to determine lysine targeting above chance was done in R (R-project).
For each modification found in more than three mice per genotype per cohort with a value greater than zero, values in hAPP and wildtype mice were compared by a two-tailed t test in Excel. P values were adjusted using a Benjamini-Hochberg correction in R (R-project).
A supplementary methods checklist is available.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mariel Finucane for help with the statistical analysis, Sharon Lee, Weikun Guo, Jing Kang, Xin Wang, Daniel Kim, and Gui-Qiu Yu for technical assistance, Stephen Ordway for editorial assistance, and Monica Dela Cruz and Amy Cheung for administrative assistance. The study was supported by NIH grants NS041787 to L.M. and GM103481 to A.L.B.
Footnotes
AUTHOR CONTRIBUTIONS
M.M. designed and conducted the mouse experiments, the statistical analyses, and wrote the manuscript. G.M.K. analyzed and curated the PTM mapping data, conducted the quantitative mass spectrometry experiments, and wrote the manuscript. S.M. conducted PSD fractionation and western blots, and wrote the manuscript. J.C.T. assisted in mass spectrometry method development and wrote the manuscript. A.I. conducted PTM mapping experiments by mass spectrometry. A.L.B. and L.M. supervised the project and wrote the manuscript.
REFERENCES
- 1.Morris M, Maeda S, Vossel K, Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberson ED, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 4.Roberson ED, et al. Amyloid-β/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer's disease. J. Neurosci. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ittner LM, et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Seward ME, et al. Amyloid-β signals through tau to drive ectopic neuronal cell cycle re-entry in Alzheimer's disease. J Cell Sci. 2013;126:1278–1286. doi: 10.1242/jcs.1125880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suberbielle E, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeVos SL, et al. Antisense reduction of tau in adult mice protects against seizures. J. Neurosci. 2013;33:12887–12897. doi: 10.1523/JNEUROSCI.2107-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gheyara AL, et al. Tau reduction prevents disease in a mouse model of Dravet syndrome. Ann. Neurol. 2014;76:443–456. doi: 10.1002/ana.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holth JK, et al. Tau loss attenuates neuronal network hyperexcitability in mouse and Drosophila genetic models of epilepsy. J. Neurosci. 2013;33:1651–1659. doi: 10.1523/JNEUROSCI.3191-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Hall AM, Kelinske M, Roberson ED. Seizure resistance without parkinsonism in aged mice after tau reduction. Neurobiol. Aging. 2014;35:2617–2624. doi: 10.1016/j.neurobiolaging.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zempel H, et al. Amyloid-β oligomers induce synaptic damage via Tau-dependent microtubule severing by TTLL6 and spastin. EMBO J. 2013;32:2920–2937. doi: 10.1038/emboj.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zempel H, Thies E, Mandelkow E, Mandelkow E-M. Abeta oligomers cause localized Ca2+ elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J. Neurosci. 2010;30:11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funk KE, et al. Lysine methylation is an endogenous post-translational modification of tau protein in human brain and a modulator of aggregation propensity. Biochem. J. 2014;462:77–88. doi: 10.1042/BJ20140372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo A, et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteomics. 2014;13:372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas SN, et al. Dual modification of Alzheimer's disease PHF-tau protein by lysine methylation and ubiquitylation: a mass spectrometry approach. Acta Neuropathol. 2012;123:105–117. doi: 10.1007/s00401-011-0893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong C-X. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat. Chem. Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 19.Arnold CS, et al. The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J. Biol. Chem. 1996;271:28741–28744. doi: 10.1074/jbc.271.46.28741. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, et al. Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell Proteomics. 2010;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanger DP, et al. Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J. Biol. Chem. 2007;282:23645–23654. doi: 10.1074/jbc.M703269200. [DOI] [PubMed] [Google Scholar]
- 22.Cripps D, et al. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 23.Morishima-Kawashima M, et al. Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron. 1993;10:1151–1160. doi: 10.1016/0896-6273(93)90063-w. [DOI] [PubMed] [Google Scholar]
- 24.Cohen TJ, et al. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cook C, et al. Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 2014;23:104–116. doi: 10.1093/hmg/ddt402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min S-W, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker PR, Trinidad JC, Chalkley RJ. Modification site localization scoring integrated into a search engine. Mol. Cell Proteomics. 2011;10:M111.008078. doi: 10.1074/mcp.M111.008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 29.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradshaw RA, Medzihradszky KF, Chalkley RJ. Protein PTMs: post-translational modifications or pesky trouble makers? J. Mass Spectrom. 2010;45:1095–1097. doi: 10.1002/jms.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biggar KK, Li SS-C. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 32.Yuzwa SA, et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 33.Borghgraef P, et al. Increasing brain protein O-GlcNAc-ylation mitigates breathing defects and mortality of Tau.P301L mice. PLoS One. 2013;8:e84442. doi: 10.1371/journal.pone.0084442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F, et al. Reduced O-GlcNAcylation links lower brain glucose metabolism and tau pathology in Alzheimer's disease. Brain. 2009;132:1820–1832. doi: 10.1093/brain/awp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuzwa SA, et al. Mapping O-GlcNAc modification sites on tau and generation of a site-specific OGlcNAc tau antibody. Amino Acids. 2011;40:857–868. doi: 10.1007/s00726-010-0705-1. [DOI] [PubMed] [Google Scholar]
- 36.Hoover BR, et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68:1067–1081. doi: 10.1016/j.neuron.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goedert M, Jakes R. Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990;9:4225–4230. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMillan P, et al. Tau isoform regulation is region- and cell-specific in mouse brain. J. Comp. Neurol. 2008;511:788–803. doi: 10.1002/cne.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X-J, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biernat J, Mandelkow EM. The development of cell processes induced by tau protein requires phosphorylation of serine 262 and 356 in the repeat domain and is inhibited by phosphorylation in the proline-rich domains. Mol. Biol. Cell. 1999;10:727–740. doi: 10.1091/mbc.10.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. doi: 10.1016/0896-6273(93)90279-z. [DOI] [PubMed] [Google Scholar]
- 42.Von Bergen M, et al. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. USA. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Von Bergen M, et al. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J. Biol. Chem. 2001;276:48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- 44.Brandt R, Léger J, Lee G. Interaction of tau with the neural plasma membrane mediated by tau's amino-terminal projection domain. J. Cell Biol. 1995;131:1327–1340. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynolds CH, et al. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cgamma1, Grb2, and Src family kinases. J. Biol. Chem. 2008;283:18177–18186. doi: 10.1074/jbc.M709715200. [DOI] [PubMed] [Google Scholar]
- 46.Sultan A, et al. Nuclear tau, a key player in neuronal DNA protection. J. Biol. Chem. 2011;286:4566–4575. doi: 10.1074/jbc.M110.199976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandelkow EM, et al. Tau domains, phosphorylation, and interactions with microtubules. Neurobiol. Aging. 1995;16:355–362. doi: 10.1016/0197-4580(95)00025-a. [DOI] [PubMed] [Google Scholar]
- 48.Sturchler-Pierrat C, et al. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc. Natl. Acad. Sci. USA. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simón A-M, et al. Overexpression of wild-type human APP in mice causes cognitive deficits and pathological features unrelated to Abeta levels. Neurobiol. Dis. 2009;33:369–378. doi: 10.1016/j.nbd.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Köpke E, et al. Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J. Biol. Chem. 1993;268:24374–24384. [PubMed] [Google Scholar]
- 51.Mucke L, et al. High-level neuronal expression of Aβ1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J. Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dawson HN, et al. Inhibition of neuronal maturation in primary hippocampal neurons from tau deficient mice. J. Cell. Sci. 2001;114:1179–1187. doi: 10.1242/jcs.114.6.1179. [DOI] [PubMed] [Google Scholar]
- 53.Planel E, et al. Anesthesia leads to tau hyperphosphorylation through inhibition of phosphatase activity by hypothermia. J. Neurosci. 2007;27:3090–3097. doi: 10.1523/JNEUROSCI.4854-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanovova N, Handzusova M, Hanes J, Kontsekova E, Novak M. High-yield purification of fetal tau preserving its structure and phosphorylation pattern. J. Immunol. Methods. 2008;339:17–22. doi: 10.1016/j.jim.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Morris M, et al. Age-appropriate cognition and subtle dopamine-independent motor deficits in aged tau knockout mice. Neurobiol. Aging. 2013;34:1523–1529. doi: 10.1016/j.neurobiolaging.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trinidad JC, Thalhammer A, Specht CG, Schoepfer R, Burlingame AL. Phosphorylation state of postsynaptic density proteins. J. Neurochem. 2005;92:1306–1316. doi: 10.1111/j.1471-4159.2004.02943.x. [DOI] [PubMed] [Google Scholar]
- 57.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive Identification of Phosphorylation Sites in Postsynaptic Density Preparations. Mol. Cell Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Vossel KA, et al. Tau reduction prevents Aβ-induced axonal transport deficits by blocking activation of GSK3β. J. Cell Biol. 2015;209:419–433. doi: 10.1083/jcb.201407065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein OGlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. USA. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trinidad JC, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vosseller K, et al. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Chalkley RJ, Baker PR, Medzihradszky KF, Lynn AJ, Burlingame AL. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell Proteomics. 2008;7:2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 65.Guan S, Price JC, Prusiner SB, Ghaemmaghami S, Burlingame AL. A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell Proteomics. 2011;10:M111.010728. doi: 10.1074/mcp.M111.010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rudrabhatla P, Jaffe H, Pant HC. Direct evidence of phosphorylated neuronal intermediate filament proteins in neurofibrillary tangles (NFTs): phosphoproteomics of Alzheimer's NFTs. FASEB J. 2011;25:3896–3905. doi: 10.1096/fj.11-181297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tavares IA, et al. Prostate-derived sterile 20-like kinases (PSKs/TAOKs) phosphorylate tau protein and are activated in tangle-bearing neurons in Alzheimer disease. J. Biol. Chem. 2013;288:15418–15429. doi: 10.1074/jbc.M112.448183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.