Abstract
Sickle cell disease (SCD) is the most common monogenic blood disorder marked by severe pain, end-organ damage, and early mortality. Treatment options for SCD remain very limited. There are only four FDA approved drugs to reduce acute complications. The only curative therapy for SCD is hematopoietic stem cell transplantation, typically from a matched, related donor. Ex vivo engineering of autologous hematopoietic stem and progenitor cells followed by transplantation of genetically modified cells potentially provides a permanent cure applicable to all patients regardless of the availability of suitable donors and graft-vs-host disease. In this review, we focus on the use of CRISPR/Cas9 gene-editing for curing SCD, including the curative correction of SCD mutation in β-globin (HBB) and the induction of fetal hemoglobin to reverse sickling. We summarize the major achievements and challenges, aiming to provide a clearer perspective on the potential of gene-editing based approaches in curing SCD.
Keywords: Sickle cell disease, Gene editing, CRISPR/Cas9
1. Introduction
Sickle cell disease (SCD) is the most common monogenic blood disorder affecting ~100,000 Americans and millions more worldwide [1,2]. SCD is caused by a single nucleotide change in the β-globin gene (HBB), replacing a hydrophilic glutamic acid with a hydrophobic valine at the sixth residue. The resulting hemoglobin S (HbS) polymerizes under hypoxic or acidic conditions [3], deforming the red blood cells (RBCs) into a rigid sickle shape with a reduced deformity and a shortened lifespan. Damaged RBCs lead to chronic hemolysis and hemolytic anemia, resulting in severe pain, end-organ damage, and early mortality in SCD patients [4,5].
Despite being the first molecular disease for which the genetic basis was known more than 60 years ago [6], treatment options for SCD remain very limited, and the average lifespan of patients with SCD has not improved over the last few decades [7]. There are only four FDA approved drugs to reduce acute complications; hydroxyurea (approved in 1998), L-glutamine (approved in 2018), crizanlizumab-tmca (approved in 2019) and voxelotor (approved in 2020). The only curative therapy for SCD is a hematopoietic stem cell transplant (HSCT), typically from a matched related donor, which is available to only ~15 % of patients [8,9]. Morbidity and mortality from HSCT increase significantly when using matched but unrelated donors [8,9] or haploidentical donors [10]. Furthermore, there are substantial treatment-related risks and complications [11,12], and without modifications to existing regimens, this therapy is not safe for widespread adoption [13].
Autologous gene therapy, whereby in patients’ own cells a copy of the “healthy” gene is added, or the mutated gene is corrected, or genes are inactivate, has the advantage of eliminating the need to find a matched donor. Ex vivo engineering of autologous hematopoietic stem and progenitor cells (HSPCs) followed by transplantation of genetically modified cells potentially provides a permanent cure applicable to patients regardless of the availability of suitable donors and without the risk of graft-vs-host disease [11,12,14]. Sickle RBCs mature inefficiently and have shorter lifespans compared to healthy RBCs, implying selective advantage of gene-corrected HSPCs over SCD HSPCs in vivo. As little as 2–5 % of donor chimerism post allogeneic transplantation is adequate to ameliorate SCD-related symptoms in patients with SCD, thus providing the rationale for a gene therapy approach [8,15]. Thus, successful gene addition or correction in relatively few HSCs could translate into a clinically meaningful level of RBC chimerism in the peripheral blood [16].
In the last two decades, gene therapy for SCD using a lentiviral-vector has proven to be curative in preclinical and clinical studies. The first patient with SCD treated with lentiviral vector–mediated addition of an antisickling HBB into autologous HSCs was successful, demonstrating a high level of therapeutic antisickling β-globin 15 months after treatment [14]. A self-inactivating (SIN) lentiviral vector encoding the human anti-sickling HBB, LentiGlobin, is currently being evaluated for safety and efficacy in clinical trials [17]. However, the use of lentiviral vectors poses potential risks such as generation of a replication-competent lentivirus (RCL) capable of infecting non-target cells, and insertional mutagenesis leading to clonal dominance and genotoxicity [18]. Although the recent results from the lentiviral gene therapy clinical trials provide the promise of ex vivo engineering of autologous HSPCs, longer follow-up is required to confirm the durability of the safety and efficacy of lentiviral-vector based gene therapy for SCD.
In contrast to conventional gene therapy approaches, gene-editing technologies offer the potential to permanently modify disease-causing genes through precise correction, deletion, addition, and disruption of specific sequences [19]. The product for therapy are gene-edited HSPCs from patients with SCD (SCD HSPCs) for autologous transplantation. Several gene editing strategies for curing SCD have shown promise in recent preclinical studies, including: (i) correction of the causative point mutation in HBB [20–24], (ii) induction of fetal hemoglobin (HbF) via gene-disruption of γ-globin (HBG) repressors [25–31], and (iii) induction of HbF via introducing beneficial hereditary persistence of fetal hemoglobin (HPFH) mutations on the β-globin locus [32–36].
The hemoglobin molecules consist of four subunits, two α polypeptide chains, and two β polypeptide chains. In a healthy adult, the overall hemoglobin composition is 97 % adult hemoglobin (HbA, α2β2), 3 % or less HbA2 (α2δ 2), and up to 1 % HbF (HbF, α2γ2). HPFH is a benign condition caused by mutations within the β-globin gene cluster, which results in elevated HbF levels in adulthood [37]. Patients with SCD who have concomitant HPFH have milder clinical consequences, and elevated levels of HbF are correlated with reduced morbidity and mortality [37]. With an improved understanding of the globin locus regulation, there is considerable interest in developing approaches to induce HbF expression for therapeutic purposes. HbF induction can be achieved by silencing transcription factors such as B-cell lymphoma/leukemia 11A (BCL11A) that mediate silencing of HBG after birth [38] or mimicking beneficial HPFH mutations [39]. In addition, the identification of novel HbF regulators is an active area of research [40].
A gene-editing strategy using engineered nucleases such as TAL-effector nucleases (TALENs), zinc finger nucleases (ZFNs) and clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 systems, creates a DNA double-strand break (DSB) at a user-defined location. The technology offers the potential to permanently repair disease-causing mutations through correction, deletion, addition, and disruption of the specific sequences mediated by targeted DSB generation followed by non-homologous end joining (NHEJ) or homology-directed repair (HDR) [19]. ZFNs and TALENs have distinct DNA binding domains, and both utilize the FokI endonuclease domain for cleaving the DNA [41]. However, the programming of these nucleases is complicated, time-consuming, and requires significant expertise. The class of programmable nuclease that has proved the most versatile and effective in recent years is the CRISPR/Cas9 system. CRISPR/Cas9 utilizes single guide RNA sequences (gRNA) that bind to a specific target site in the genome and to the Cas9 endonuclease. The Cas9 endonuclease is guided to a specific target site by homology between the gRNA and the target DNA sequences. Although the off-target effect remains a potential issue, it can be significantly reduced by rational gRNA designs or utilizing high-fidelity Cas9 protein [42]. Base editors are created by fusing a nucleotide deaminase with catalytically disabled Cas9 protein. Base editors directly convert one base into another without inducing DSBs and therefore not relying on HDR, enabling the point mutation correction in non-dividing cells. Therefore, base editors are a promising DNA editing tool and considered to be preferable to using Cas9 nuclease which may lead to the generation of unwanted small insertions/deletions (indels), translocations, or chromosomal rearrangements [43]. With the advancement of gene-editing technologies, each of the four technologies (ZFNs, TALENs, CRISPR/Cas9, base editor) have been tested in HSPCs for treating SCD. Studies showed the correction of the SCD mutation by delivering ZFNs [44] or TALENs [45] along with DNA donor template. Other groups developed ZFNs and TALEN targeting the HbF transcriptional repressors or the repressor binding site to induce HbF [26,46]. ZFN targeting the BCL11A locus has been utilized in a Phase-1/2 clinical trial (BIVV003, clinicaltrials.gov).
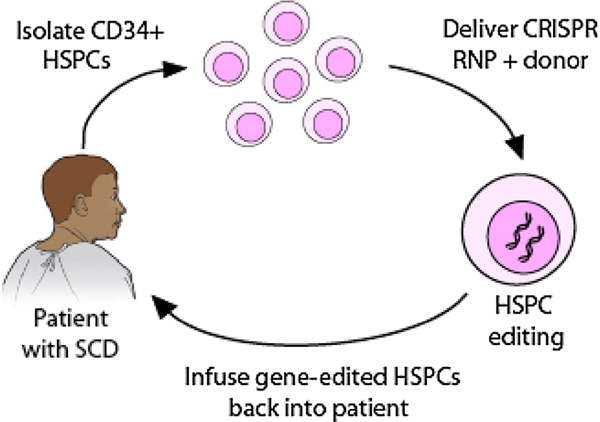
In this review, we focus on several approaches using CRISPR/Cas9 gene-editing for the treatment of SCD; specifically correcting the sickle mutation in HBB (Fig. 1), producing sufficient levels of HbF to reverse sickling by targeting the HbF transcriptional repressors, and introducing beneficial HPFH mutations. One particular example is the CRISPR/Cas9 gene-editing based Phase-1 clinical trial in a patient with severe SCD (CTX001, clinicaltrials.gov) using autologous CD34+ HSPCs in which the erythroid lineage-specific enhancer of the BCL11A gene was modified to induce HbF expression. We summarize the major achievements and challenges in order to provide a clearer perspective on the potential of gene editing strategies as a cure for SCD. To translate the gene-editing based strategy to the clinic, many challenges exist, including the potential off-target effects, the need to further increase the efficiency of gene correction, and the in vivo engraftment of gene-edited HSPCs. Optimization of the genome-editing method, including the CRISPR Cas9/gRNA and donor template as well as the delivery method, is critical in achieving high safety and efficacy. Small improvements in each step is key for clinical translation.
Fig. 1. Genome editing based strategy for treating sickle cell disease.
CD34+ HSPCs are first isolated from a patient with sickle cell disease. The RNP (ribonecleoprotein) complex of CRISPR guide RNA with Cas9 protein and DNA donor template are delivered into the nuclei of HSPCs via electroporation for gene correction. The gene-edited HSPCs are then infused back into the patient to reverse the disease phenotype. To make the gene-editing strategy a clinically viable approach, both high efficacy and adequate safety need to be achieved.
2. Preclinical studies for ex vivo HSPCs gene editing and xenotransplantation
Most preclinical studies utilized ex vivo gene-editing of human HSPCs followed by transplantation in an immune-deficient mouse model. This is to assess the long-term engraftment potential of gene-edited HSCs since the durability of an autologous HSCT depends on the ability to modify HSCs permanently.
2.1. Cell culture
Human CD34+ HSPCs can be isolated from several sources, such as cord blood, bone marrow, and mobilized peripheral blood. The majority of genome editing studies utilized peripheral blood CD34+ HSPCs. Isolated CD34+ cells were cultured in pre-stimulation media with cytokines for several days before gene-editing, as post-isolation culture with cytokine exposure has been shown to increase the gene-editing efficiency [47]. Several strategies have been employed to prime HSPCs for efficient gene-editing and stimulate the expansion of gene-edited HSCs.
Low cell density culture conditions and using a hematopoietic stem cell self-renewal agonist such as UM171 and StemRegenin 1 (SR1) have been shown to stimulate the expansion of gene-edited HSPCs as measured by higher engraftment levels in immunodeficient mice [48–50]. Edited CD34+ cells can be cultured in media supporting erythroid differentiation for globin analysis by HPLC and flow cytometry. The functional impact of gene-editing on HSPC lineage commitment is evaluated using the colony-forming unit (CFU) assay by comparing the distribution of CFU between edited and non-edited controls.
2.2. Gene editing reagent delivery
Most studies used the CRISPR/Cas9 system derived from Streptococcus pyogenes (Spy Cas9). SpyCas9’s gRNAs typically contain a 20-nt guide sequence with a 5′-NGG-3′ PAM requirement. Over the years, the development and optimization of the CRISPR/Cas9 genome editing reagent and delivery method substantially improved the safety and efficiency of gene editing in HSPCs [33]. Early attempts used a plasmid DNA based system for Cas9 and gRNA expression, which resulted in low editing efficiency and high toxicity [51]. Achieving high editing efficiency, however, needs to be balanced with potential safety concern regarding off-target mutations and immunogenicity arising from sustained or excess expression of CRISPR components. Although all editing machinery components elicited immune, stress, and apoptotic responses, delivery of gRNA and Cas9 as a pre-complexed ribonucleoprotein (RNP) is well-tolerated in CD34+ HSPCs, despite eliciting a DNA damage response (DDR) [52,53]. Electroporation using a nucleofection protocol is often the preferred method for direct delivery of RNPs to HSPCs, as this allows the RNP to enter the cell nucleus quickly, so it can immediately start cutting the genome. The majority of genome editing studies utilized RNP to achieve a high editing efficiency and specificity with lower cytotoxicity in CD34+ HSPCs. Chemical alterations to gRNAs further enhanced genome editing efficiency while reducing toxicity in CD34+ HSPCs [54]. High-fidelity variants of SpyCas9 maintain on-target activities comparable to wild-type SpyCas9 with reduced off-target activities in HSPCs [42]. For SCD mutation correction using the corrective donor template, clinical translation is hindered by a low ratio of HDR to NHEJ in long-term reconstituting HSCs. Cell cycle phase-specific regulation of DNA repair pathways through temporal regulation of Cas9 nuclease activity and transient synchronization of HSPCs in HDR-preferred phases have shown to improve HDR/NHEJ ratio in vitro [55]. The chemically modified synthetic gRNAs and high-fidelity Cas9 protein are commercially available, and optimal electroporation conditions for CD34+ cells have been established [21]. Further optimization steps, however, are still needed for each specific application concerning nuclease and donor template amount as ex vivo manipulation may negatively impact HSPCs engraftment and long-term repopulation capacity.
2.3. Transplantation study
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) and NOD,B6.SCID Il2rγ−/− KitW41/W41 (NBSGW) are commonly used xenotransplant hosts for accessing multilineage engraftment of human hematopoietic cells. NSG strain requires sublethal myeloablative irradiation to achieve a high level of human chimerism. Since NSG does not support human erythropoiesis, engrafted HSCs are subjected to in vitro erythroid differentiation post-engraftment to assess globin expression and editing frequency in the erythroid lineages. NBSGW is an NSG-derivative strain and does not require preconditioning irradiation to support the engraftment of human HSCs. NBSGW supports not only myeloid and lymphoid but also erythroid engraftment [56]. Despite the widespread use of NSG and NBSGW models, it is difficult to extrapolate from a xenotransplant model of how these cells would behave in a clinical setting. Hans-Peter Kiem and colleagues [35] used a nonhuman primate (NHP) model to closely reproduce human stem cells in terms of kinetics of hematopoietic recovery, immunophenotypic markers, and cross-reactivity between cytokines. The use of a highly clinically relevant large-animal model for ex vivo HSPCs gene editing and transplantation offers an opportunity to monitor both long-term engraftment and hemoglobin profile, facilitating the translation of gene editing therapy to the clinic [26,35].
3. HbF induction by BCL11A gene editing
The level of HbF is a key modifier of the clinical severity of SCD, and reactivation of HbF by targeting genes involved in HbF regulation is a promising approach for treating SCD. Genome-wide association studies (GWAS) investigating individuals with HPFH have identified multiple causative genetic loci, and numerous transcription factors have been indirectly implicated in HbF silencing. BCL11A is the chief regulator of HbF level and suppresses fetal hemoglobin expression by association with other DNA-bound factors at numerous positions within the β-globin locus, including direct HBG promoter repression by BCL11A [57]. Therefore, HbF reactivation through disruption of BCL11A or BCL11A binding motifs represents an attractive and discrete target for therapeutic gene editing for the treatment of SCD.
3.1. BCL11A gene disruption
An earlier study by Humbert et al. [26] validated the HbF repressor function of BCL11A and performed a proof-of-concept transplantation study in the NHP model with TALE nuclease mRNA targeting the BCL11A coding sequence. However, BCL11A plays varied roles in different hematopoietic lineages and coding variation at BCL11A is highly deleterious. Alternatively, naturally occurring genetic variation at the BCL11A enhancer is well-tolerated. Numerous studies have validated BCL11A erythroid enhancer as a target for HbF induction, offering a framework for erythroid-specific therapeutic genome editing by targeting the core sequences of BCL11A enhancer in HSPCs [25]. For example, Canver et al. [25] employed CRISPR lentiviral pooled gRNA screening to perform in situ saturating mutagenesis to study the organization and function of the BCL11A erythroid enhancer. A GATA1 motif that forms the core of an enhancer is essential for human erythroid BCL11A expression and HbF repression. Enhancer disruption by individual gRNAs in primary erythroid precursors results in substantial HbF induction while sparing BCL11A expression and function in non-erythroid contexts [25]. Chang et al. [58] and Psatha et al. [28] directly compared the functional consequences of BCL11A exonic versus enhancer disruption by using ZFN in HSPCs. BCL11A enhancer disruption showed comparable level of HbF reactivation with the BCL11A coding knockout (KO) while retaining the ability of BCL11A to support HSCs function including differentiation, reconstitution, and long-term engraftment potential [28,58]. Wu et al. [30] demonstrated highly efficient therapeutic gene-editing in HSPCs by CRISPR/Cas9 mediated disruption of GATA1 binding site at the +58 BCL11A erythroid enhancer. This resulted in a erythroid specific reduction of BCL11A expression, and therapeutic induction of fetal γ-globin in engrafting SCD HSCs [30]. The gRNA directly cleaving at the core of the +58 erythroid enhancer of BCL11A gave the highest levels HbF induction in erythroid progeny with the high rate of indels. Based on the clonal analysis of the erythroid progeny of CD34+ HSPCs edited at the BCL11A enhancer, biallelic modifications at the cleavage site resulted in robust induction of γ-globin. Edited human SCD CD34+ HSPCs were transplanted into immunodeficient NBSGW mice to study the impact of BCL11A enhancer editing on long-term engrafting HSCs. NBSGW supported similar levels of human myeloid, lymphoid, and erythroid engraftment of edited cells compared to unedited cells, validating gene editing of self-renewing HSCs. Indel frequencies at BCL11A enhancer persisted after secondary transplant demonstrating that BCL11A enhancer editing does not have a deleterious impact on stem cell function. Interestingly, the spectrum of indels in bulk HSPCs was different compared to that in the long-term engrafting HSCs, suggesting that engrafting HSCs may favor NHEJ as compared to Microhomology-mediated end joining (MMEJ) repair. Most prior studies reported a reduction of therapeutic allele levels after engraftment which raises questions about the durability of gene-editing in transplantation. The persistence of BCL11A enhancer edited cells suggests that the NHEJ-mediated gene disruption strategy could be more efficient over other gene-editing strategies relying on HDR or MMEJ. This is due to the fact that NHEJ is active in all stages of the cell cycle and HSCs preferentially undergo NHEJ [30]. This study also tested the specificity and genotoxicity of CRISPR/Cas9 editing and did not observe detectable activities at Circularization for in vitro reporting of cleavage effects by sequencing (CIRCLE-seq) and in-silico predicted off-target sites or genotoxicity in terms of TP53 variants and stem cell function. Together, this work demonstrated that editing the BCL11A enhancer by CRISPR/Cas9 is a practical therapeutic strategy to produce a durable therapeutic level of HbF induction in engrafting HSCs. Vertex Pharmaceuticals and CRISPR Therapeutics have obtained promising results in their Phase-1 clinical trial (CTX001, clinicaltrials.gov) using CRISPR/Cas9 to edit the BCL11A erythroid enhancer to induce HbF expression.
3.2. BCL11A base editing
A recent study by Zeng et al. [31] demonstrated the feasibility of producing therapeutic levels of base edits in multilineage-repopulating and self-renewing human HSCs. Base editing can potentially offer a high purity gene corrected product compared to nuclease-based editing. Base editors directly introduce base changes without inducing DSBs bypassing the low-efficiency HDR as well as DSB induced unwanted indels and off-target effects [30]. The A3A(N57Q)-BE3 base editor was delivered as an RNP targeting the BCL11A erythroid enhancer in SCD HSPCs. This base editor targets cytosine within the base editing window to disrupt the GATA1 motif. Two cycles of electroporation increased the therapeutic base editing rate but this also resulted in decreased viability and engraftment potential. Biallelic single base edits at the BCL11A enhancer within the GATA1 motif led to potent HbF induction similar to nuclease editing. Following transplantation into NBSGW mice, the base editing frequencies were reduced in engrafted HSCs compared to input HSPCs. Base edited cells showed multilineage reconstitution with similar base editing frequencies in each lineage. There was erythroid lineage specific BCL11A knockdown from erythroid enhancer disruption. For base-editing, both gRNA dependent and independent off-target editing need to be investigated. Although off-target base-editing can be minimized by reducing exposure to RNP and by utilizing the base editor with an attenuated cytosine deaminase domain, comprehensive off-target analysis needs to be performed before clinical implementation of base editing.
4. HbF induction by introducing HPFH mutations
The major fetal hemoglobin gene repressors, BCL11A and Leukemia/lymphoma-related factor (LRF), are directly bound to the HbG promoter at regions residing around 115 bp and 200 bp upstream of the transcription start site, respectively [59]. CRISPR/Cas9 mediated disruption of either the LRF- or the BCL11A-binding site in the HBG promoters induced significant HbF production. Traxler et al. [39] identified a naturally occurring 13 nucleotide (nt) HPFH deletion within the HBG promoter as a DNA target for genome editing. After CRISPR/Cas9 editing, the 13-nt deletion identical to the naturally occurring mutation predominates among other indels because the Cas9 cleavage site is flanked by 8-nt tandem repeats that facilitate MMEJ repair edited progenitors produced RBCs with increased HbF levels that were sufficient to reverse sickling in vitro [39]. This strategy has been advanced to demonstrate high-level editing in human HSCs capable of multilineage engraftment after transplantation into immunodeficient mice, and absence of detectable off-target mutations or deleterious hematopoietic effects [60]. Humbert et al. [35] used an NHP autologous transplantation model to show the curative potential of this approach. The previously validated CRISPR gRNA target sites for human cells were used as the CRISPR target sites as this region of HBG is conserved between human and rhesus macaque. The gRNA target sites are located on the promoter of the homologous HBG1 and HBG2 genes. Considerable levels of large deletions due to simultaneous cleavage have been reported which remove the entire HBG2 gene and part of the HBG1 promoter. Although the frequency of large deletions was significantly reduced post-engraftment in NHP, the underlying mechanism remains unknown and the long-term clinical effect of the large deletion had not been determined [35].
4.1. HBG base editing
A recent study by Wang et al. [61] demonstrated that base editing that induced a single nucleotide substitution at the BCL11A binding site on the HBG promoter was enough to disrupt BCL11A binding and increase HBG expression. Since the base editor mediates base conversion without inducing DSBs, the HBG copy number was not affected, demonstrating that base editing may lead to safer therapeutic applications without creating further DSB induced damage in the genome. The efficiency of this approach has not been tested in engrafting HSCs.
4.2. LRF binding site editing
The transcription factor LRF represses expression of HbF independently from BCL11A [62]. Since knockdown of LRF increases HbF expression but delays erythroid differentiation, targeting of the LRF binding site in the HBG promoter was tested. Disruption of the LRF binding site by CRISPR/Cas9 ameliorated the SCD phenotype. HBG promoter editing is maintained in repopulating HSCs that differentiate into RBCs expressing therapeutically relevant HbF levels [36]. This study by Weber et al. [36] paved the way for simultaneously blocking both LRF and BCL11A which resulted in an additive effect on HbF. Given the independent role of LRF and BCL11A [62] and the efficient multiplex base-editing demonstrated by Zeng et al. [31], base editing to simultaneously disrupt both LRF and BCL11A repressor binding sites in the HBG promoters represent a promising strategy.
4.3. Large deletional HPFH
Several investigators have reported proof-of-concept CRISPR/Cas9 mediated gene editing to recapitulate large deletional HPFH mutation in the β-globin gene cluster as a new approach to treat SCD [32–34]. These approaches rely on NHEJ of paired DSBs to yield precise large deletions to mimic the naturally occurring Sicilian HPFH encompassing the δ- and β-globin genes or the corfu deletion of the γ-δ intergenic region. The efficiency of these maneuvers in engrafting HSCs has not been reported. Introducing a large deletional HPFH mutation requires the simultaneous processing of two DSBs and the re-joining of their distant ends, which led to low rates of large deletions and higher risks of off-target effects. In addition, competing genome editing outcomes, such as small indels and inversions accompanying these deletions, may limit clinical application.
5. SCD mutation correction using DNA donor template
Correction of the disease causing sickle mutation using gene-editing represents the most straightforward therapeutic approach. As shown in Fig. 1, in this approach, the CRISPR gRNA/Cas9 RNP complex targeting HBB together with DNA donor template are delivered into HSPCs isolated from patients with SCD, resulting in the HDR mediated correction of the causative mutation. Many viral based vector options have been evaluated in HSPCs for donor template delivery including integrase-deficient lentiviral systems (IDLVs), adenovirus 5/35 serotype (Ad5/35) and adeno-associated viruses (AAVs) [44]. Compared to other viral vectors, one main advantage of AAV is the low frequency of vector integration into the host genomic DNA and the low risk of related insertional mutagenesis and genotoxicity. Several studies demonstrated efficient targeted integration at the HBB locus in CD34+ HSPCs by using RNP combined with single-stranded oligodeoxynucleotides (ssODNs) [20–23,63]. rAAV6 [24,64] and ssODNs [20–23,63] donors have been used by most studies due to safety considerations and efficient HDR mediated by these donors. Differentiated erythroblasts from gene edited cells had an increase in mean levels of HbA and reduced the sickle cell phenotype [21]. Gene edited cells from patients with SCD were able to engraft in NSG [21] or NBSGW mouse transplant models [20] with gene correction observed at standard times post-transplant.
5.1. ssODN vs. rAAV6 donors
Recent reports directly compared the efficiencies of co-delivery of RNP in association with ssODN template or rAAV6-packaged template and demonstrated that the methodology for donor template delivery impacts long-term persistence of HBB gene-modified HSPCs [44,65]. In vitro, rAAV6 outperformed ssODN by causing less acute toxicity and inducing greater HDR. The RNP and rAAV6-edited cells showed lower engraftment in the NSBGW [65] or NSG [44] mice, suggesting that rAAV6 caused a decrease in the hematopoietic capacity. These results suggest that the ssODN template is likely to be more amenable for clinical translation than the viral based approach. The use of the ssODN template for gene correction also has the advantages of being easy to produce and having a low manufacturing cost, which will facilitate the application of a gene-editing based cure for SCD.
5.2. Potential risk of inducing β-thalassemia
Clinical translation of SCD mutation correction using the corrective donor template is currently hindered by a low ratio of HDR to NHEJ in long-term reconstituting HSCs. The possibility of inducing β-thalassemia major, intermediate or minor due to Cas9 cutting of HBB has not been carefully evaluated. In addition, the in vivo effects of Cas9 cleavage of HBB and reduction in functional β-globin levels in a patient with SCD remain unclear and will need to be addressed in a clinical trial.
5.3. HbF induction
Several studies reported upregulation of HbF as a result of HBB disruption in CD34+ HSPCs [20–22]. It is possible that the increase in HbF percentage is due to Cas9 cleavage induced HBB KO or due to the increase in hemoglobin formation between α-globin and γ-globin given that β-globin chains are unavailable. However, the molecular mechanism underlying the HbF induction observed here remains elusive and warrants further investigation. Recently, Boontanrart et al. [66] reported that cellular erythroid stress caused by β-globin knockout can induce robust re-expression of γ-globin, providing mechanistic insight to the poorly understood phenomenon of stress-induced globin compensation. It is also still unclear if Cas9 HBB-cleavage induced increase in HbF percentage would have a long-lasting effect at a therapeutically relevant level, and if the resulting benefits to patients with SCD would outpace the potential harm of HBB disruption, including the possibility of inducing β-thalassemia major or minor. Addressing these issues will facilitate the safe and effective clinical translation of a gene-editing based treatment strategy for SCD.
6. In vivo gene editing
Although ex vivo gene-editing has many advantages, including a high editing efficiency and the ability to ablate the unedited HSPCs in the patient, the high cost will prevent the applicability of this therapy to patients with SCD in resource-poor regions. Attempts have been made to develop in vivo HSC transduction/selection technology using non-integrating adenovirus. In vivo HBG-promoter editing by CRISPR/Cas9 in β-YAC/CD46-transgenic mice has been performed [67]. The human CD46-targeting adenovirus vector (HDAd-HBG-CRISPR/mgmt) expresses CRISPR/Cas9 which targets the HBG promoter for γ-globin reactivation. The vector also contains a O-6-methylguanine-DNA methyltransferase (MGMTP140K) cassette for in vivo selection of transduced cells using chemotherapeutic drugs [67]. CD46 is uniformly expressed on HSPCs for hematopoietic tissue targeted viral transduction. The β-YAC/CD46 mice carrying the human β-globin gene locus express human CD46 at a level and in a pattern similar to humans which allows for direct in vivo analysis of γ-globin reactivation using the human CD46-targeting adenovirus vector. Because direct transduction of HSPCs localized in the BM is inefficient, the in vivo HSPC transduction approach involves HSPCs mobilized from the bone marrow into the peripheral blood followed by intravenous injections of the adenovirus vector (HDAd-HBG-CRISPR/mgmt). This resulted in the reactivation of human γ-globin in erythrocytes of adult animals that was maintained after secondary transplantation of HSPCs [67]. Since mobilized HSCs transduced in the peripheral blood could home to the bone marrow and renew themselves [68], this approach could generate a long-lasting effect.
While promising, in vivo gene editing for curing SCD has a lot challenges. Both high in vivo delivery efficiency and high editing efficiency in SCD HSCs are required, and off-target cell/tissue editing is a potential concern. Although viral vector based in vivo delivery of gene-editing machinery can be highly efficient, it may lead to uncontrollable expression of Cas9/gRNA, causing genotoxicity and activating an immune response [69–71]. On the other hand, non-viral based in vivo delivery vehicles may have low efficiencies and broad biodistribution, and repeated injections are often needed for a high delivery efficiency [72]. It is also necessary to compare systemic delivery and local injection to determine the best delivery strategy. The percentage of in vivo gene-edited HSCs required for a cure is currently unknown since the unedited HSCs would still produce sickle cells.
7. Potential risks of off-target mutations caused by CRISPR/Cas9
Genome editing poses new challenges since its mechanism of action is different from the conventional gene therapy. In contrast to the knock-in or knockdown gene therapy, which generally requires the continuous and long-term expression of therapeutic genes for treatment, permanent gene modification can be achieved with a single delivery of CRISPR/Cas9. Due to the tolerance for nucleotide mismatches between target DNA and gRNA, the utility of CRISPR-Cas9 systems for genome editing may be compromised by their off-target activity [73–75]. The off-target activity of Cas9 nuclease can cause disruption of normal gene function and genome instability via large chromosomal rearrangements [76], which is of serious concern in human gene therapies, potentially leading to difficult-to-predict side effects. Importantly, the long-term expression of Cas9 nuclease via plasmid and viral vectors in treated cells means there is a potential for off-target cleavages to accumulate over time [75]. While delivery of gRNA and Cas9 as RNP and utilization of high-fidelity Cas9 have shown to significantly reduce off-target editing, off-target effects are not eliminated. Therefore, better systems for detecting and quantifying these aberrant events are needed to validate potential off-target sites and to aid in optimizing strategies to minimize off-target mutations without sacrificing gene correction efficiency. In addition, a robust, rapid, high-throughput method for monitoring off-target events over time is necessary to assess the long-term toxicity or off-target effects of the system especially for clinical applications of CRISPR-Cas9. Recent advances in off-target site identification using genome-wide unbiased methods such as Chip-seq [77], GUIDE-seq [78], BLESS [79], and END-seq [80] has given rise to a new problem in off-target site validation. The current gold standard for quantifying Cas9 off-target activity is PCR amplification, followed by next-generation sequencing. This method allows for multiple sites to be assessed simultaneously with a high degree of sensitivity. However, recent advances in CRISPR/Cas9 off-target site identification has revealed many sites that cannot be identified by deep sequencing due to a detection limit of 0.1 % by deep sequencing for accurate indel identification [81,82]. Recent publications have reported frequent large deletions and insertions after CRISPR-Cas9 cleavage in mouse embryonic stem cells, mouse hematopoietic progenitors and a human differentiated cell line [83]. Although the large deletions/insertions at the on- and off-target sites and the large chromosomal rearrangements between on- and off-target sites typically have low occurrence, they pose a significant safety concern since even a very small number of HSCs harboring these detrimental events could cause hematological malignancies after HSCT. Next generation sequencing also has significant costs, long turnaround times, and requires the development of robust bioinformatics pipelines, all of which prevent quick sample screening. In addition, any gross chromosomal rearrangements between an on- and off-target DNA break or two off-target DNA breaks cannot be identified by most methods. There are currently no standard methods for quantification of large chromosomal rearrangements induced by CRISPR/Cas9. For therapeutic genome editing, potential off-target effects need to be carefully analyzed because significant challenges exist in both accurately predicting potential off-target sites and in performing genome-wide unbiased searches. As CRISPR/Cas9 moves towards clinical application, there is a need for robust patient follow up and monitoring methods. Just as drug treatments have side-effects, CRISPR/Cas9 treatments will likely have some degree of off-target edits (side effects) that will require careful monitoring over time to ensure that these events do not have a proliferative effect.
8. Future perspectives and challenges
With the advancement of CRISPR/Cas9 technology, autologous transplant of gene-edited hematopoietic stem cells could potentially provide a cure for most patients with SCD. However, to translate the gene-editing based SCD treatment strategy to the clinic, many challenges exist, including the need for high editing efficiency and low off-target effects. Quantitative understanding of the genotypic and phenotypic consequences of a diverse array of mutations in the CRISPR/Cas9 edited SCD CD34+ cells is essential for safe clinical applications. The development of the editing strategies which allow high yields of long-term repopulating HSCs that have a polyclonal and a high proportion of gene-edited cells after engraftment remains a challenge. Furthermore, there is limited knowledge of the impact of SCD pathology on HSPC viability and engraftment potential, particularly in patients exposed to years of SCD related chronic inflammation. To date, most SCD-related in vivo engraftment studies are performed with cells derived from healthy individuals, limiting our understanding of the effects of chronic systemic inflammation and ineffective erythropoiesis associated with HSPCs from patients with SCD. The source of HSPCs and the SCD pathology of the individuals, including differences in patient conditions, could have a significant effect on both the gene-editing outcomes and the engraftment potential. Genetic and environmental factors could likely influence the viability and functions of SCD HSPCs.
Current ex vivo gene-editing approaches have some shortcomings throughout the process. Only a small percentage of CD34+ cells from patients with SCD are typically HSCs. Harvesting HSCs from the bone marrow is invasive. Patients undergoing myeloablative chemotherapy also experience chemotherapy related side effects such as low blood counts and infections. In vitro culture and gene editing of HSCs lead to loss of HSC pluripotency and engraftment potential. Furthermore, providing an ex vivo gene-editing based cure to patients may be prohibitive due to the high cost which is driven by the need for highly specialized facilities and the technical expertise required. In vivo gene-editing of HSCs can potentially overcome the limitations of ex vivo gene-editing since administration of in vivo therapy could be minimally invasive and cost effective; therefore, more readily available in resource-poor regions. However, major challenges exist in developing in vivo gene-editing as a clinically viable approach, including achieving both high in vivo delivery efficiency and high editing efficiency. Although the development of in vivo gene-editing based therapies for SCD is still in its infancy, the collaborative between the NIH and the Bill and Melinda Gates Foundation to support the development of a curative in vivo gene therapy approach for SCD will greatly accelerate technological development and innovation.
Funding
This work was supported by the National Institutes of Health [R01HL152314 and OT2HL154977 to G.B.]
Footnotes
Declaration of Competing Interest
The authors have no conflict of interest and nothing to disclose
References
- [1].Hassell KL. Population estimates of sickle cell disease in the US. Am J Prev Med 2010;38:S512–21. [DOI] [PubMed] [Google Scholar]
- [2].Odame I. Perspective: We need a global solution. Nature 2014;515:S10–. [DOI] [PubMed] [Google Scholar]
- [3].Eaton WA, Hofrichter J. Hemoglobin S gelation and sickle cell disease. Blood 1987; 70:1245–66. [PubMed] [Google Scholar]
- [4].Platt OS, Thorington BD, Brambilla DJ, Milner PF, Rosse WF, Vichinsky E, et al. Pain in sickle cell disease. Rates and risk factors. N Engl J Med 1991;325:11–6. [DOI] [PubMed] [Google Scholar]
- [5].Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, et al. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med 1994;330:1639–44. [DOI] [PubMed] [Google Scholar]
- [6].Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature 1957;180:326–8. [DOI] [PubMed] [Google Scholar]
- [7].Lanzkron S, Carroll CP, Haywood C. Mortality rates and age at death from sickle cell disease: U.S., 1979–2005. Public Health Rep 2013;128:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Walters MC, Patience M, Leisenring W, Rogers ZR, Aquino VM, Buchanan GR, et al. Stable mixed hematopoietic chimerism after bone marrow transplantation for sickle cell anemia. Biol Blood Marrow Transplant 2001;7:665–73. [DOI] [PubMed] [Google Scholar]
- [9].Mentzer WC, Heller S, Pearle PR, Hackney E, Vichinsky E. Availability of related donors for bone marrow transplantation in sickle cell anemia. Am J Pediatr Hematol Oncol 1994;16:27–9. [PubMed] [Google Scholar]
- [10].Dallas MH, Triplett B, Shook DR, Hartford C, Srinivasan A, Laver J, et al. Long-term outcome and evaluation of organ function in pediatric patients undergoing haploidentical and matched related hematopoietic cell transplantation for sickle cell disease. Biol Blood Marrow Transplant 2013;19:820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Galanello R, Origa R. Beta-thalassemia. Orphanet J Rare Dis 2010;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].El-Kamah GY, Amr KS. Thalassemia — from genotype to phenotype. Inherited hemoglobin disorders. InTech; 2015. [Google Scholar]
- [13].Shenoy S, Eapen M, Panepinto JA, Logan BR, Wu J, Abraham A, et al. A BMT CTN phase II trial of unrelated donor marrow transplantation for children with severe sickle cell disease. Blood 2016;128:2361–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ja Ribeil, Hacein-Bey-Abina S, Payen E, Magnani A, Semeraro M, Magrin E, et al. Gene therapy in a patient with sickle cell disease. N Engl J Med 2017;376:848–55. [DOI] [PubMed] [Google Scholar]
- [15].Abraham A, Hsieh M, Eapen M, Fitzhugh C, Carreras J, Keesler D, et al. Relationship between mixed donor-recipient chimerism and disease recurrence after hematopoietic cell transplantation for sickle cell disease. Biol Blood Marrow Transplant 2017;23:2178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kean LS, Manci EA, Perry J, Balkan C, Coley S, Holtzclaw D, et al. Chimerism and cure: hematologic and pathologic correction of murine sickle cell disease. Blood 2003;102:4582–93. [DOI] [PubMed] [Google Scholar]
- [17].Urbinati F, Hargrove PW, Geiger S, Romero Z, Wherley J, Kaufman ML, et al. Potentially therapeutic levels of anti-sickling globin gene expression following lentivirus-mediated gene transfer in sickle cell disease bone marrow CD34+ cells. Exp Hematol 2015;43:346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Negre O, Bartholomae C, Beuzard Y, Cavazzana M, Christiansen L, Courne C, et al. Preclinical evaluation of efficacy and safety of an improved lentiviral vector for the treatment of β-thalassemia and sickle cell disease. Curr Gene Ther 2015;15:64–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cox DBT, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med 2015;21:121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Magis W, DeWitt MA, Wyman SK, Vu JT, Heo S-J, Shao SJ, et al. High-level correction of the sickle mutation amplified in vivo during erythroid differentiation. bioRxiv; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Park SH, Lee CM, Dever DP, Davis TH, Camarena J, Srifa W, et al. Highly efficient editing of the beta-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Res 2019;47:7955–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med 2016;8:360ra134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hoban MD, Lumaquin D, Kuo CY, Romero Z, Long J, Ho M, et al. CRISPR/Cas9-mediated correction of the sickle mutation in human CD34+ cells. Mol Ther 2016; 24:1561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 2016;539:384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 2015; 527:192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Humbert O, Peterson CW, Norgaard ZK, Radtke S, Kiem HP. A nonhuman primate transplantation model to evaluate hematopoietic stem cell gene editing strategies for beta-hemoglobinopathies. Mol Ther Methods Clin Dev 2018;8:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khosravi MA, Abbasalipour M, Concordet JP, Berg JV, Zeinali S, Arashkia A, et al. Targeted deletion of BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: a promising approach for gene therapy of beta thalassemia disease. Eur J Pharmacol 2019;854:398–405. [DOI] [PubMed] [Google Scholar]
- [28].Psatha N, Reik A, Phelps S, Zhou Y, Dalas D, Yannaki E, et al. Disruption of the BCL11A erythroid enhancer reactivates fetal hemoglobin in erythroid cells of patients with beta-thalassemia major. Mol Ther Methods Clin Dev 2018;10:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].The Lancet H. CRISPR-Cas9 gene editing for patients with haemoglobinopathies. Lancet Haematol 2019;6. [DOI] [PubMed] [Google Scholar]
- [30].Wu Y, Zeng J, Roscoe BP, Liu P, Yao Q, Lazzarotto CR, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med 2019;25: 776–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zeng J, Wu Y, Ren C, Bonanno J, Shen AH, Shea D, et al. Therapeutic base editing of human hematopoietic stem cells. Nat Med 2020;26:535–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chiara Antoniani GP, Wassim El Nemer VM, Hoss Sara El, Lattanzi Annalisa, Kurita Ryo, et al. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human b-globin locus. Blood 2021;131:17. [DOI] [PubMed] [Google Scholar]
- [33].Lattanzi A, Meneghini V, Pavani G, Amor F, Ramadier S, Felix T, et al. Optimization of CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells for therapeutic genomic rearrangements. Mol Ther 2019;27: 137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ye L, Wang J, Tan Y, Beyer AI, Xie F, Muench MO, et al. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: an approach for treating sickle cell disease and beta-thalassemia. Proc Natl Acad Sci U S A 2016;113: 10661–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Humbert O, Radtke S, Samuelson C, Carrillo RR, Perez AM, Reddy SS, et al. Therapeutically relevant engraftment of a CRISPR-Cas9–edited HSC-enriched population with HbF reactivation in nonhuman primates. Sci Transl Med 2019;11. eaaw3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Weber L, Frati G, Felix T, Hardouin G, Casini A, Wollenschlaeger C, et al. Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci Adv 2020;6. eaay9392-eaay. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Musallam KM, Sankaran VG, Cappellini MD, Duca L, Nathan DG, Taher AT. Fetal hemoglobin levels and morbidity in untransfused patients with β-thalassemia intermedia. Blood 2012;119:364–7. [DOI] [PubMed] [Google Scholar]
- [38].Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell APW, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 2016;351:285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Traxler EA, Yao Y, Wang YD, Woodard KJ, Kurita R, Nakamura Y, et al. A genome-editing strategy to treat beta-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med 2016;22:987–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Grevet JD, Lan X, Hamagami N, Edwards CR, Sankaranarayanan L, Ji X, et al. Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells. Science 2018;361:285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gaj T, Gersbach CA, Barbas 3rd CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med 2018;24:1216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018;19:770–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Romero Z, Lomova A, Said S, Miggelbrink A, Kuo CY, Campo-Fernandez B, et al. Editing the sickle cell disease mutation in human hematopoietic stem cells: comparison of endonucleases and homologous donor templates. Mol Ther 2019;27: 1389–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hoban MD, Cost GJ, Mendel MC, Romero Z, Kaufman ML, Joglekar AV, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 2015;125:2597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lux CT, Pattabhi S, Berger M, Nourigat C, Flowers DA, Negre O, et al. TALEN-mediated gene editing of HBG in human hematopoietic stem cells leads to therapeutic fetal hemoglobin induction. Mol Ther Methods Clin Dev 2019;12: 175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gundry MC, Brunetti L, Lin A, Mayle AE, Kitano A, Wagner D, et al. Highly efficient genome editing of murine and human hematopoietic progenitor cells by CRISPR/Cas9. Cell Rep 2016;17:1453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Charlesworth CT, Camarena J, Cromer MK, Vaidyanathan S, Bak RO, Carte JM, et al. Priming human repopulating hematopoietic stem and progenitor cells for Cas9/sgRNA gene targeting. Mol Ther Nucleic Acids 2018;12:89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Fares I, Chagraoui J, Gareau Y, Gingras S, Ruel R, Mayotte N, et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 2014;345:1509–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zonari E, Desantis G, Petrillo C, Boccalatte FE, Lidonnici MR, Kajaste-Rudnitski A, et al. Efficient ex vivo engineering and expansion of highly purified human hematopoietic stem and progenitor cell populations for gene therapy. Stem Cell Rep 2017;8:977–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mandal PK, Ferreira LMR, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 2014;15:643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cromer MK, Vaidyanathan S, Ryan DE, Curry B, Lucas AB, Camarena J, et al. Global transcriptional response to CRISPR/Cas9-AAV6-Based genome editing in CD34(+) hematopoietic stem and progenitor cells. Mol Ther 2018;26:2431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schiroli G, Conti A, Ferrari S, Della Volpe L, Jacob A, Albano L, et al. Precise gene editing preserves hematopoietic stem cell function following transient p53-Mediated DNA damage response. Cell Stem Cell 2019;24:551–65. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 2015;33:985–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lomova A, Clark DN, Campo-Fernandez B, Flores-Bjurstrom C, Kaufman ML, Fitz-Gibbon S, et al. Improving gene editing outcomes in human hematopoietic stem and progenitor cells by temporal control of DNA repair. Stem Cells 2019;37: 284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].McIntosh BE, Brown ME, Duffin BM, Maufort JP, Vereide DT, Slukvin II, et al. Nonirradiated NOD,B6.SCID Il2rgamma−/−Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Rep 2015;4: 171–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, et al. Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell 2018;173: 430–42. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chang KH, Smith SE, Sullivan T, Chen K, Zhou Q, West JA, et al. Long-term engraftment and fetal globin induction upon BCL11A gene editing in bone-marrow-derived CD34(+) hematopoietic stem and progenitor cells. Mol Ther Methods Clin Dev 2017;4:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Martyn GE, Wienert B, Yang L, Shah M, Norton LJ, Burdach J, et al. Natural regulatory mutations elevate the fetal globin gene via disruption of BCL11A or ZBTB7A binding. Nat Genet 2018;50:498–503. [DOI] [PubMed] [Google Scholar]
- [60].Metais JY, Doerfler PA, Mayuranathan T, Bauer DE, Fowler SC, Hsieh MM, et al. Genome editing of HBG1 and HBG2 to induce fetal hemoglobin. Blood Adv 2019;3: 3379–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wang L, Li L, Ma Y, Hu H, Li Q, Yang Y, et al. Reactivation of gamma-globin expression through Cas9 or base editor to treat beta-hemoglobinopathies. Cell Res 2020;30:276–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 2016;351:285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 2016;539:384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bak RO, Dever DP, Porteus MH. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat Protoc 2018;13:358–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pattabhi S, Lotti SN, Berger MP, Singh S, Lux CT, Jacoby K, et al. In vivo outcome of homology-directed repair at the HBB gene in HSC using alternative donor template delivery methods. Mol Ther Nucleic Acids 2019;17:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Boontanrart M, Stehli G, Banovic M, Schroder MS, Wyman S, Lew R, et al. ATF4¨ mediates fetal globin upregulation in response to reduced β-globin. bioRxiv; 2020. 2020.01.15.905943. [Google Scholar]
- [67].Li C, Psatha N, Sova P, Gil S, Wang H, Kim J, et al. Reactivation of gamma-globin in adult beta-YAC mice after ex vivo and in vivo hematopoietic stem cell genome editing. Blood 2018;131:2915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liesveld JL, Sharma N, Aljitawi OS. Stem cell homing: from physiology to therapeutics. Stem Cells 2020. [DOI] [PubMed] [Google Scholar]
- [69].Li A, Tanner MR, Lee CM, Hurley AE, De Giorgi M, Jarrett KE, et al. AAV-CRISPR gene editing is negated by pre-existing immunity to Cas9. Mol Ther 2020;28: 1432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 2019;25:249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wagner DL, Amini L, Wendering DJ, Burkhardt LM, Akyüz L, Reinke P, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med 2019;25:242–8. [DOI] [PubMed] [Google Scholar]
- [72].Tong S, Moyo B, Lee CM, Leong K, Bao G. Engineered materials for in vivo delivery of genome-editing machinery. Nat Rev Mater 2019;4:726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31: 827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 2013;41: 9584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013;31:822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun 2014;5:3964. [DOI] [PubMed] [Google Scholar]
- [77].Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 2014;32:677–83. [DOI] [PubMed] [Google Scholar]
- [78].Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015;520:186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Canela A, Sridharan S, Sciascia N, Tubbs A, Meltzer P, Sleckman BP, et al. DNA breaks and end resection measured genome-wide by end sequencing. Mol Cell 2016;63:898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 2015;12: 237–43. [DOI] [PubMed] [Google Scholar]
- [82].Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 2014;24:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018;36:765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]