Abstract
TMEM106B, encoding a lysosome membrane protein, has been recently associated with brain aging, hypomyelinating leukodystrophy and multiple neurodegenerative diseases, such as frontotemporal lobar degeneration (FTLD) and limbic-predominant age-related TDP-43 encephalopathy (LATE). During the past decade, considerable progress has been made towards our understanding of the cellular and physiological functions of TMEM106B. TMEM106B regulates many aspects of lysosomal function, including lysosomal pH, lysosome movement and lysosome exocytosis. Both an increase and decrease in TMEM106B levels results in lysosomal abnormalities. In vivo, TMEM106B deficiency leads to lysosome trafficking and myelination defects and potentiates FTLD pathology in mouse models. In humans, alterations in TMEM106B levels caused by TMEM106B polymorphisms are intimately linked to neuronal proportions, brain aging and brain disorders. Further elucidation of the physiological function of TMEM106B and alternation of TMEM106B in the disease states will yield insights into the role of TMEM106B in brain health and therapeutic development to treat brain disorders associated with TMEM106B.
Keywords: FTLD, myelination, lysosome, TMEM106B, progranulin, TDP-43, aging
Introduction
TMEM106B is associated with brain disorders and brain aging
Frontotemporal lobar degeneration (FTLD) is a devastating neurodegenerative disease that results in personality and behavioral changes and language disorders [50,60]. FTLD overlaps in pathology and genetic causes with amyotrophic lateral sclerosis (ALS), and these two are often considered part of a continuous disease spectrum [45]. A large subset of FTLD patients accumulates ubiquitin positive aggregates containing the RNA binding protein TDP-43. In an effort to identify genetic risk factors for FTLD with TDP-43 inclusions (FTLD-TDP), TMEM106B was discovered as a main risk factor for FTLD with Granulin (GRN) mutations [78]. The rs1990622 single nucleotide polymorphism (SNP), in the 3’ untranslated region (UTR), is most commonly associated with modulating the disease risk; almost all other disease associated TMEM106B SNPs are in linkage disequilibrium with rs1990622, making it a sentinel SNP [14,20] [21,42,57,78,79]. While the major TMEM106B allele (or the most common allele, rs1990622T) acts as a risk factor, the minor allele (rs1990622C) reduces the odds of patients with GRN mutations developing FTLD [78]. TMEM106B has also been shown to affect brain size [29], neuronal proportion [43], and cognitive decline in FTLD patients [76] (Table 1). Further research has also found that TMEM106B might modify disease progression in ALS/FTLD patients carrying mutations in another gene, C9ORF72[17,22,77]. Despite this, surprisingly no strong link has been found between TMEM106B and ALS risk [32,80], although TMEM106B appears to affect cognitive impairment in ALS [80] (Table 1).
Table 1:
Association of TMEM106B with brain disorders
| Disease | TMEM106B SNPs/Mutations | Risk allele/ Major allele | Protective allele/ Minor allele | Phenotypes associated with SNPs/Mutations | References |
|---|---|---|---|---|---|
| FTLD-GRN | rs1990622 | T | C | Increased risk for FTLD-GRN (all risk alleles). | [14, 20, 21, 29, |
| rs1990621 | C | G | Altered chromatin structure (rs1990620). | 42, 43, 57, 76, 78, | |
| rs1990620 | A | G | Reduced brain size (rs1990620). | 79] | |
| rs1020004 | A | G | Decreased neuron proportion (rs1990621). | ||
| rs6966915 | C | T | Worse cognitive decline (rs1990622). | ||
| rs3173615 (T185S) | C | G | |||
| FTLD-C9ORF72 | rs1990622 | T | C | Major allele associated with higher risk. | [17, 22, 77] |
| rs3173615 | C | G | Minor allele associated with reduced risk. | ||
| ALS | rs1990622 | T | C | Worse cognitive decline in ALS patients (rs1990622) | [80] |
| AD | rs1990622 | T | C | Worse TDP-43 pathology (rs1990622). | [35, 46, 49, 64] |
| rs1990620 | A | G | Increased AD risk in APOE4 mutation carriers (rs1990622; rs1596014). | ||
| rs1595014 | T | A | Reduced inflammation in LOAD (rs1990620) | ||
| PD | rs1990622 | T | C | Worse cognitive decline (rs1990622). | [76] |
| CTE | rs3173615 | C | G | Less inflammation, ante-mortem dementia, and Tau pathology in CTE patients (rs3173615). | [5, 11] |
| HS-Aging | rs1990622 | T | C | Increased risk and more advanced TDP-43 pathology (rs1990622). | [52, 87] |
| LATE | rs1990622 | T | C | Increased risk (rs1990622) | [30, 51] |
| HLD | c.754G>A (D252N) | Hypomyelination in the brain. Delayed motor | [33, 70, 84] | ||
Abbreviations FTLD: Frontotemporal lobar degeneration; ALS: Amyotrophic lateral sclerosis; AD: Alzheimer’s disease; PD: Parkinson’s disease; CTE: Chronic traumatic encephalopathy; HS-aging: Hippocampal sclerosis of aging; LATE: Limbic-predominant age-related TDP-43 encephalopathy; HLD: Hypomyelinating leukodystrophy.
In addition to ALS/FTLD, recent genome-wide association (GWAS) studies and gene expression studies have found that TMEM106B modulates several other neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease (PD), chronic traumatic encephalopathy (CTE), hippocampal sclerosis in aging (HS-Aging) and limbic-predominant age-related TDP-43 encephalopathy (LATE) (Table 1). In Alzheimer’s disease, the major allele has been associated with enhanced TDP-43 pathology [64]. Additionally, TMEM106B variants interact with APOE polymorphisms to confer risks in AD [35,46]. In a recent study using an iterative weighted gene co-expression network analysis (WGCNA), a TMEM106B variant (rs1990620G) was shown to confer protection from the inflammatory subtype of Late-Onset Alzheimer’s disease (LOAD) [49]. In Parkinson’s disease, the TMEM106B major allele (rs1990622T) is associated with faster cognitive decline [76]. In chronic traumatic encephalopathy (CTE), the minor TMEM106B allele is associated with reduced ante-mortem dementia, reduced phosphorylated Tau pathology, reduced inflammation and increased synaptic protein density, but has not been significantly associated with disease risk [5,11]. The major allele of TMEM106B was also found to be a risk factor for hippocampal sclerosis in aging (HS-Aging) [52] and associated with more advanced TDP-43 pathology in old persons without FTLD [87]. Recently researchers have discovered that many people of advanced age, especially those over 80, have TDP-43 pathology and dementia; they named this disease “limbic-predominant age-related TDP-43 encephalopathy (LATE)” [51]. This new disease criteria contains a significant portion of patients previously considered as having hippocampal sclerosis in aging (HS-Aging) with TDP-43 pathology [51]. TMEM106B has been shown to be one of the main risk factors for TDP-43 proteiopathy in LATE [30,51].
More intriguingly, a novel dominant SNP in TMEM106B causing an amino acid substitution D252N has been recently associated with several cases of hypo-myelinating leukodystrophy (HLD)[33,70,84]. Unlike the previously mentioned rs1990622 sentinel SNP, D252N has a very strong disease presentation and alters the molecular function of TMEM106B, providing novel insight into the role of TMEM106B in the lysosome [19].
Additionally, a genome wide transcriptome study associated TMEM106B with brain aging [63]. The protective variant of the TMEM106B gene was also found to confer a neuronal protection effect against general aging, independent of disease status [43]. In culmination, the last decade worth of genetic studies have found TMEM106B to be closely linked to overall brain health and implicated TMEM106B as a central player in brain aging, myelination disorders and neurodegenerative diseases.
TMEM106B encodes a lysosomal membrane protein expressed by many cell types
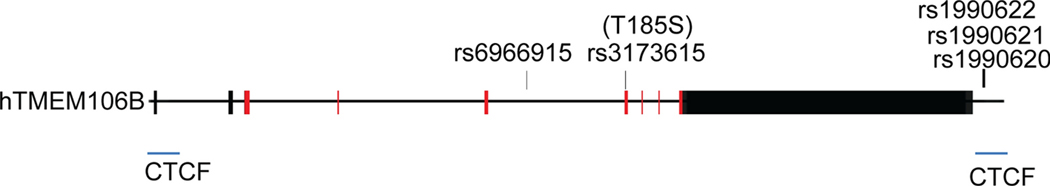
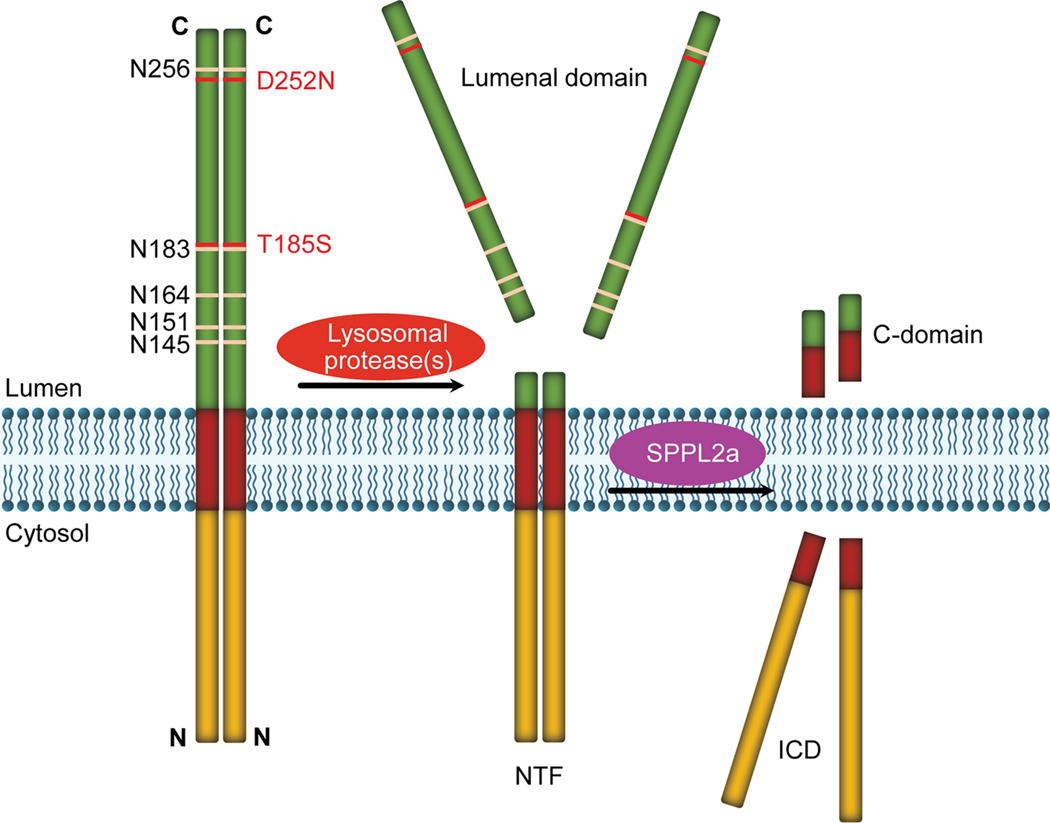
The human TMEM106B gene is comprised of nine exons located on chromosome 7p21 (Fig. 1). Alternative splicing gives rise to two TMEM106B mRNA isoforms, but they encode the same protein. TMEM106B is a type II transmembrane protein containing 274 amino acid residues, which can be divided into three domains: an N-terminal cytosolic domain, a transmembrane domain and a C-terminal lumenal domain containing five N-glycosylation sites [41] (Fig. 2). With a predicted molecular weight of 31 kDa, TMEM106B runs at ~43kDa due to glycosylation [7,10,41]. In the central nervous system (CNS), TMEM106B is expressed highest in neurons and oligodendrocytes [7,10,19,41]. At the subcellular level, TMEM106B is localized in late endosome/lysosome compartments [7,10,41].
Figure 1. TMEM106B genomic structure and variants.
The human TMEM106B gene is comprised of 9 exons. The coding regions are labelled in red. The major SNPs associated with neurodegenerative diseases are indicated. The CTCF binding sites are labelled in blue.
Figure 2. Domain structure and proteolytic processing of TMEM106B protein.
TMEM106B is a type II transmembrane protein localized on lysosome membrane, with its N-terminus facing the cytosol and C-terminus facing lysosomal lumen. The lumenal domain contains five predicted N-glycosylation sites. Heterozygous D252N mutation in TMEM106B is associated with HLD. The T185S variant has been to be protective towards several neurodegenerative diseases. TMEM106B forms homodimers and heterodimers with its homolog TMEM106C. TMEM106B is processed into an N-terminal fragment (NTF) by lysosomal proteases, which is further cleaved by SPPL2a through intramembrane proteolysis to generate intracellular cytosolic domain (ICD).
Lysosomal function of TMEM106B
Lysosomes as the main degradative organelle in the cell contain many acid hydrolases that digest a variety of substrates, including proteins, lipids, glycans and nucleic acids, that they receive from multiple pathways, including endocytosis, phagocytosis and autophagy [65]. In addition, lysosomes have emerged as the a central signaling hub within the cell that senses and integrates multiple signals to respond to changes in the environment [65]. Lysosomes are very dynamic that traffic along microtubules within the cell and constantly undergoes fusion and fission [6,59]. Lysosomal dysfunction is closely linked to neurodegeneration and mutations in the lysosome genes are frequently associated with neurodegenerative diseases[44]. During the past decade, studies by multiple groups have shown a role of TMEM106B in regulating several aspects of lysosomal function.
TMEM106B, lysosomal morphology and lysosome movement
Several studies in cell lines show that overexpression of TMEM106B results in lysosome enlargement [7,10,41], induces cell death and enhances oxidative stress-induced cytotoxicity [74] [21]. This phenotype is also observed in primary neurons [72]. On the other hand, reduced TMEM106B expression does not appear to have an obvious effect on lysosomal size or morphology in in vitro studies [7,72]. Surprisingly, loss of TMEM106B in mice leads to the formation of lysosomal vacuoles at the distal end of the axon initial segment (AIS) of motor neurons [18,47] (Fig. 3), and in the axon initial segment in aged Purkinje cells. Since lysosome size is intricately regulated by fission events and fusion with other organelles, such as endosomes and autophagosomes [16,75], it is likely that either lysosome fusion or fission is affected by TMEM106B.
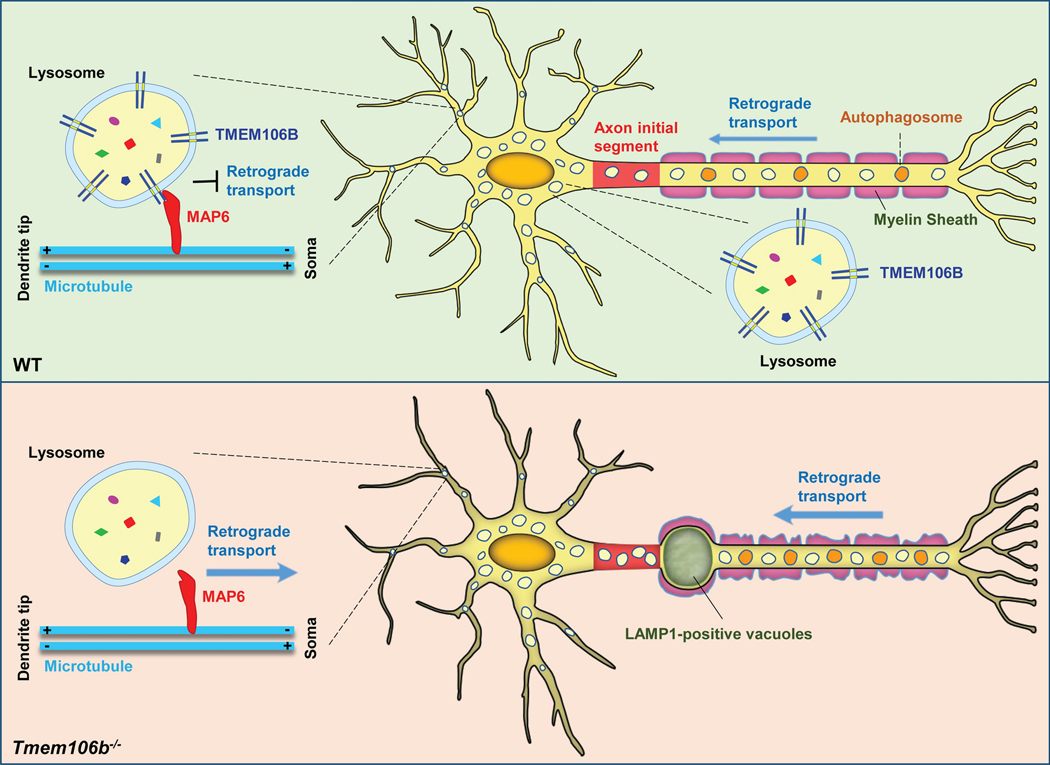
Figure 3. TMEM106B regulates lysosomal trafficking in neurons.
In dendrites, TMEM106B interacts with MAP6 to restrict retrograde transport of lysosomes. In axons, TMEM106B regulates lysosome transport across the axon initial segment (AIS) region. TMEM106Bdeficiency results in increased retrograde transport of lysosomes in dendrites and reduced dendritic branching in cultured neurons. In mouse models, loss of TMEM106B leads to the formation of LAMP1 positive lysosomal vacuoles at the AIS region in motor neurons and Purkinje neurons. This is partly due to increased retrograde transport of lysosomes along axons. TMEM106B ablation also results in alterations in the myelin sheath along axons.
In addition to lysosome size, TMEM106B has also been shown to affect lysosome movement within the cell [18,47]. In mammalian cells, lysosome movement is regulated by its tracking along microtubules mediated by kinesin and dynein motors [6]. The positioning of lysosomes within the cell affects many aspects of lysosomal function and cell physiology [59]. Knockdown/knockout of TMEM106B induces lysosomal clustering near the nucleus, resulting in reduced lysosomal exocytosis (fusion of the lysosome with plasma membrane) in nonpolar cells [40,67] and in the oligodendroglial precursor cell line Oli-neu [19]. In cultured primary cortical neurons, knockdown of TMEM106B was shown to increase axonally transported lysosomes [72]. In motor neurons, TMEM106B ablation results in increased retrograde transport of lysosomes along axons, which could contribute to the formation of giant lysosomal vacuoles at the distal end of the AIS [47] (Fig. 3).
The mechanism by which TMEM106B regulates lysosome movement remains to be determined. It has been shown that TMEM106B physically interacts with microtubule associated protein 6 (MAP6) to inhibit retrograde transport of lysosomes in neuronal dendrites [67] (Table 2, Fig. 3). In primary neurons, downregulation of TMEM106B significantly increases retrograde transport of lysosomes in dendrites and reduced dendritic branching (Fig. 3). This phenotype can be rescued by MAP6 knockdown, or the expression of dominant-negative Rab7-interacting lysosomal protein (RILP), which has been shown to enhance anterograde lysosomal transport [67]. How TMEM106B regulates lysosome movement in axons is still a mystery. TMEM106B might have a specialized role in transporting lysosomes across AIS in motor neurons and Purkinje cells, given the unique lysosome vacuolation phenotypes in the AIS region in the TMEM106B deficient mice [18,47,73].
Table 2:
List of TMEM106B-interacting proteins
| Name | Methods | References |
|---|---|---|
| AP2M1 | Yeast two-hybrid | [72] |
| CLTC | Yeast two-hybrid | [72] |
| VPS11 | Yeast two-hybrid | [72] |
| VPS13D | Yeast two-hybrid | [72] |
| TMEM106C | Yeast two-hybrid | [72] |
| MAP6 | Co-Immunoprecipitation | [67] |
| CHMP2B | Co-Immunoprecipitation | [36] |
| V-ATPase accessory protein 1 (AP1) | Co-Immunoprecipitation | [19, 38] |
| Cathepsin D | Co-Immunoprecipitation | [19] |
Taken together, these studies have revealed that TMEM106B is a critical regulator of lysosome positioning within the cell body and lysosomal transport in axons and dendrites. However, general lysosomal dysfunction has also been shown to cause the clustering of lysosomes near the nucleus [59]. It remains to be determined whether the lysosome trafficking defects in Tmem106b−/− cells are due to a direct regulation of lysosome movement by TMEM106B or secondary effects caused lysosomal dysfunction because of TMEM106B loss.
TMEM106B and lysosomal pH
The lysosome is maintained at pH 4.5–5.0. Acidification of the lysosomal lumen is critical for the proper stability and activity of many lysosomal hydrolases and degradation of lysosomal substrates [75]. Lysosomal acidification is achieved by the action of vacuolar ATPase (V-ATPase) [75]. TMEM106B was shown to interact with V-ATPase accessory protein 1 (AP1) [19,38] and AP2M1 [72] (Table 2). Ablation of TMEM106B results in a reduction in the levels of AP1 and V0 subunits of V-ATPase, leading to impaired lysosomal acidification [38]. On the other hand, contradictory data has been reported with TMEM106B overexpression. In some studies, overexpression of TMEM106B was shown to induce cellular alkalization, and impair endosomal-lysosomal acidification [10,41]. However, another study found that overexpression of GFP tagged TMEM106B in murine and human lung cancer cell lines leads to increased fluorescence intensity of lysotracker compared to control cells, indicating enhanced lysosomal acidification in response to TMEM106B overexpression [40]. Similar results have been reported in HEK293T cells with the application of both pH-sensitive and pH-insensitive dyes to more accurately measure lysosomal pH [19]. Further studies are needed to resolve the discrepancies and elucidate the molecular mechanisms by which TMEM106B regulates lysosomal pH.
TMEM106B regulates the levels and activities of lysosomal proteins
TMEM106B was shown to physically interact with the lysosomal protease cathepsin D and maintain proper cathepsin D levels in oligodendrocytes [19]. In addition to cathepsin D, other lysosomal proteins have also been shown to be affected by alteration in TMEM106B levels. In vitro, TMEM106B overexpression leads to the upregulation of lysosomal gene expression, while TMEM106B knockdown results in decreased lysosomal gene expression [40]. This is likely due to the effect of TMEM106B on the cytoplasm-nuclear translocation of the transcription factor EB (TFEB), a master transcriptional regulator for lysosomal biogenesis [40,72]. However, conflicting results have been reported from in vivo studies. One study reported a decrease in the levels of several lysosomal proteins, including LAMP1, and lysosomal enzymes (CatB, CatL, and DPPII) in the brain lysates of TMEM106B deficient mice generated using the gene-trap approach [38], which still express 5–10% full‐length TMEM106B protein [88]. In another study using the same gene-trap knockout line, LAMP1 levels are not altered, but the activities of lysosomal enzymes β-Glucuronidase and β-hexosaminidase A are decreased in TMEM106B deficient mice [2]. However, in two other studies using complete TMEM106B knockout mice generated using the CRISPR/Cas9 technique, increased levels of cathepsins and elevated activities of several lysosomal enzymes, including β-hexosaminidase A, α-mannosidase and β-galactosidase have been reported [19,47]. Thus it is likely that the presence of residual full length TMEM106B protein in the gene-trap line[88]is the cause for the differences in these studies. Nevertheless, in all studies, the mRNA levels of these lysosomal genes were not affected by TMEM106B ablation [18,38,47], indicating that TMEM106B ablation does not affect TFEB activities and instead regulates the levels of lysosomal proteins through post-transcriptional mechanisms. In addition, the protein levels of LAMP1, cathepsin D and L are all decreased in TMEM106B-deficient Oli-neu cells [19], suggesting that TMEM106B might have different effects on lysosomal proteins in different cell types.
TMEM106B and autophagy
Autophagy is a conserved intracellular degradative process essential for cellular homeostasis and self-renewal, which is especially critical for the health of postmitotic neurons [86]. Autophagy dysfunction is tightly associated with neurodegeneration and mutations in several autophagy genes, including p62/SQSTM1, optineurin, TBK1 and ubiquilin 2 are linked to ALS/FTD [48]. In the last step of autophagy, autophagosomes fuse with lysosomes and autophagy substrates engulfed in the autophagosome membrane are degraded by lysosomal enzymes. Thus, lysosome activity is essential for the completion of autophagy flux. Although TMEM106B is not required for autophagy induction and does not localize on the autophagosome membrane, TMEM106B has been shown to regulate autophagy through modulating lysosomal functions. Loss of TMEM106B in mice significantly increases the protein levels of LC3, an autophagy marker; p62, an autophagy adaptor degraded in the lysosome; and ubiquitinated proteins in the brain lysates [47] [18]. This phenotype is further exacerbated during aging [18]. These observations support that TMEM106B influences lysosomal cargo clearance and autophagic turnover by modulating lysosomal function or autophagosome-lysosome fusion, although it is possible that TMEM106B might also affect other steps of autophagy.
Regulation of TMEM106B function on the lysosomal membrane
TMEM106B has been shown to form homodimers (Fig. 2) as well as heterodimers with its homolog TMEM106C [72] (Table 2). Homodimerization of TMEM106B can be detected in western blot analysis as an SDS-resistant homo-dimer at ~75 kDa in cold conditions compared to the 43 kDa monomer when boiled [7,10]. Thus at the lysosomal membrane, TMEM106B is likely to function as a homodimer or heterodimer with its homologs, TMEM106A or TMEM106C, both of which have been shown to localize on the lysosomal membrane [8,67,72] (and our unpublished observations), although TMEM106C was shown to be in the endoplasmic reticulum (ER) compartment in one of the studies [8].
In addition, TMEM106B is cleaved by lysosomal proteases to generate a N-terminal fragment (NTF) containing the transmembrane and intracellular domains [8]. The N-terminal fragment is further processed into a small, unstable intracellular domain (ICD) by the GxGD aspartyl proteases SPPL2a through the intramembrane proteolysis [8] (Fig. 2). Whether NTF and ICD process biological functions remains to be tested. It is possible that TMEM106B processing regulates its levels on lysosomal membrane, since too much or too little TMEM106B could have profound effects on lysosomal functions.
TMEM106B, brain aging and brain disorders
Alteration of TMEM106B levels in human diseases
TMEM106B was first identified as a risk factor for FTLD-TDP, especially in patients with GRN mutations [14,20,78,79]. The SNP rs1990622 in the 3’UTR region of TMEM106B was found to be the most significant [20,78] (Fig. 1). Several studies have demonstrated that the levels of TMEM106B mRNA [78] and protein [10,26] were significantly increased in FTLD patients with GRN mutations. A recent study has shown that the risk allele of a candidate causal variant (rs1990620) preferentially recruits the chromatin-organizing protein CCCTC-binding factor (CTCF) and forms a long-range chromatin-looping interaction within a sub-topologically associated domain (sub-TAD), which contains five CTCF-binding sites [21] (Fig. 1). One of the five CTCF sites is 48 bp away from SNP rs1990620, and the other one is located at the TMEM106B promoter [21] (Fig. 1). This indicates that TMEM106B upregulation associated with the rs1990620 risk allele is probably due to transcriptional activation. Besides chromatin structure alterations, another mechanism regulatingTMEM106B levels involves microRNA-132/212 cluster, which was shown to bind to the 3’UTR of TMEM106B gene and inhibits TMEM106B expression [10]. This microRNA is significantly decreased in FTLD-TDP patients’ brains, thus leading to increased TMEM106B expression [10].
Among the associated TMEM106B variants, rs3173615 (encoding p.T185S) is the only coding variant. This SNP was found to be in perfect linkage disequilibrium (LD) with SNP rs1990622 [14]. Cellular studies have shown that both risk (T185) and protective (S185) TMEM106B localize in the late endosomes [7,36]. In one study, the risk TMEM106B variant (T185) was shown to have higher expression levels than the protective allele (S185), due to a slower protein degradation rate and possible differences in glycosylation at N183 [54]. However, T185 and S185 appear to be expressed at the same levels in another study [7]. Additionally, TMEM106B has been shown to interact with CHMP2B, a component of ESCRT-III involved in multivesicular body formation and a protein mutated in FTLD, and this interaction appears to be stronger with T185 than S185 [36]. In an effort to identify gene networks associated with the risk (TT) and protective (SS) TMEM106B haplotypes using WGCNA, several gene clusters were found to be associated with TT or SS [61], indicating that TMEM106B SNPs could have a profound effect on gene expression in the brain.
A robust increase of TMEM106B protein levels was observed in several conditions with lysosomal dysfunction, such as mice deficient in the FTLD gene GRN or lysosomal protease cathepsin D [26]. In both cases, the upregulation of TMEM106B protein is not caused by increased transcription [26]. The increase in TMEM106B protein levels might involve alternations in the lysosomal activities. In line with this, bafilomycin treatment, which inhibits lysosomal activities by interfering with lysosome acidification, significantly increases the protein levels, but not mRNA levels of TMEM106B [10,41]. In addition, TFEB, a master transcriptional regulator for lysosomal biogenesis [69] does not directly regulate TMEM106B expression [40].
In summary, these studies demonstrate that TMEM106B levels can be regulated at transcriptional levels through chromatin modulation, at mRNA levels by miRNAs and at the protein levels through lysosomal activities.
TMEM106B and brain aging
Brain aging is the primary risk factor for neurodegenerative diseases. Normal brain aging and neurodegenerative disease share common pathological hallmarks, such as cognitive deficits, neuronal loss, synaptic impairment and inflammation. A recent transcriptome-wide cerebral cortex gene expression study has shown that the TMEM106B risk variant (rs1990622T) is associated with neuronal loss, inflammation and cognitive deficits and selectively affects the frontal cerebral cortex of older individuals (>65 years), even without any known brain disorder [63]. The TMEM106B rs1990622 risk allele has also been associated with worse residual cognition [82], and reduced left temporal brain size [1]. In neuropathologically normal old individuals, the TMEM106B risk haplotype (rs3173615 TT) is associated with reduced expression of neuronal markers and increased expression of inflammatory genes in the temporal cortex [61]. Consistent with this result, TMEM106B protective variant rs1990621, which is in high LD with the TMEM106B variant rs3173615 (p.T185S) was found to confer neuronal protection against aging [43].
In mouse models, TMEM106B deficiency leads to age-dependent ALS/FTLD pathology, including accumulation of ubiquitinated proteins, autophagy adaptor protein p62 and phosphorylated TDP-43 in the brain and spinal cord [47] [18].
TMEM106B and TDP-43 proteinopathy
Transactive response DNA binding protein of 43 kDa (TDP-43), a DNA and RNA binding protein [12,28], has been linked to the pathophysiology of several neurodegenerative diseases such as ALS/FTLD, HS-Aging[62] and AD [13,24]. Hyper-phosphorylation, ubiquitination and cleavage of TDP-43 serve as a pathological hallmark for a large subset of FTLD and TDP-43 often translocates from nucleus to cytosol to form ubiquitin- positive aggregates, resulting in depletion of TDP-43 in the nucleus [53]. TMEM106B was first identified as a risk factor for FTLD with TDP-43 aggregates, especially in patients with GRN mutations [14,20,78,79]. Recent genetic studies identified the TMEM106B risk allele as one of the five risk factors for neuropathological change in LATE, a disease characterized by TDP-43 proteinopathy in older adults [51]. Additionally, the TMEM106B rs1990622 risk allele is associated with severe CA1 neuronal loss in LATE-NC with hippocampal sclerosis pathology [39]. Genome-wide transcription analysis found that the TMEM106B risk allele enhances the expression of genes associated with myelination and lysosomal membrane proteins [85], which is associated with LATE TDP-43 burden [85].
In mouse models, TMEM106B deficiency does not affect the cytoplasmic translocation of TDP-43 or TDP-43 cleavage, but leads to the accumulation of phosphorylated TDP-43 in an age dependent manner [18]. Since the clearance of TDP-43 relies on the autophagy-lysosome pathway [68], TMEM106B could affect TDP-43 homeostasis through regulating lysosomal functions. However, further studies are needed to determine how TMEM106B risk variants affect TDP-43 proteiopathy.
TMEM106B and myelination
Recently, a recurrent dominant mutation, c.754G>A p.(D252N) in the lumenal domain of TMEM106B was identified in patients with hypomyelinating leukodystrophy (HLD) in several studies [33,70,84]. All patients with the TMEM106B D252N mutation present with congenital nystagmus and mild motor developmental delay [33,70,84]. Additionally, in human samples, genome-wide transcription analyses have found that the TMEM106B risk allele enhances the expression of genes associated with myelination [85]. These findings indicate that TMEM106B might play a critical role in myelination. This was directly tested in mouse models recently. TMEM106B deficiency in mice was reported to result in myelination defects with a significant reduction of protein levels of proteolipid protein (PLP) and myelin oligodendrocyte glycoprotein (MOG), two membrane proteins found in the myelin sheath [19]. Moreover, TMEM106B-deficient mice show eye movement and mild motor coordination defects [19,47]. In another study, TMEM106B deficiency was shown to alter myelination-related cellular processes including axon ensheathment and oligodendrocyte differentiation in brain transcriptomic analysis [89]. A significant loss of oligodendrocytes and a reduction in the expression levels of mature oligodendrocyte markers have been observed in the corpus callosum of Tmem106b−/− mice at different ages. In the same study, TMEM106B deficiency was shown to result in poor recovery from cuprizone-induced demyelination [89].
Several studies have demonstrated that lysosomes play an essential role in myelination, and lysosomal dysfunction caused by mutations in lysosomal genes leads to myelination defects [23,27,58,66]. Cellular studies revealed that TMEM106B is expressed and localized to the lysosome compartment in oligodendrocytes in vivo and in vitro [19]. In the Oli-neu oligodendroglial precursor cells, TMEM106B ablation results in lysosome clustering in the perinuclear region and a decrease in lysosome exocytosis [19]. Interestingly, PLP, the main transmembrane protein found in the myelin sheath, was shown to reside in the lysosome compartment and get delivered to myelin sheath (extension of plasma membrane) upon lysosome exocytosis [83]. This process is significantly altered by thelysosome positioning and exocytosis defects due to TMEM106B loss, leading to an increased accumulation of PLP in the lysosome compartment and decreased levels of PLP on the cell surface [19,89] (Fig. 4). In addition, TMEM106B physically interacts with cathepsin D (Table 2) and regulates cathepsin D levels in oligodendrocytes[19]. Thus, TMEM106B affects several aspects of lysosomal function in oligodendrocytes, which is critical for proper PLP trafficking and myelination.
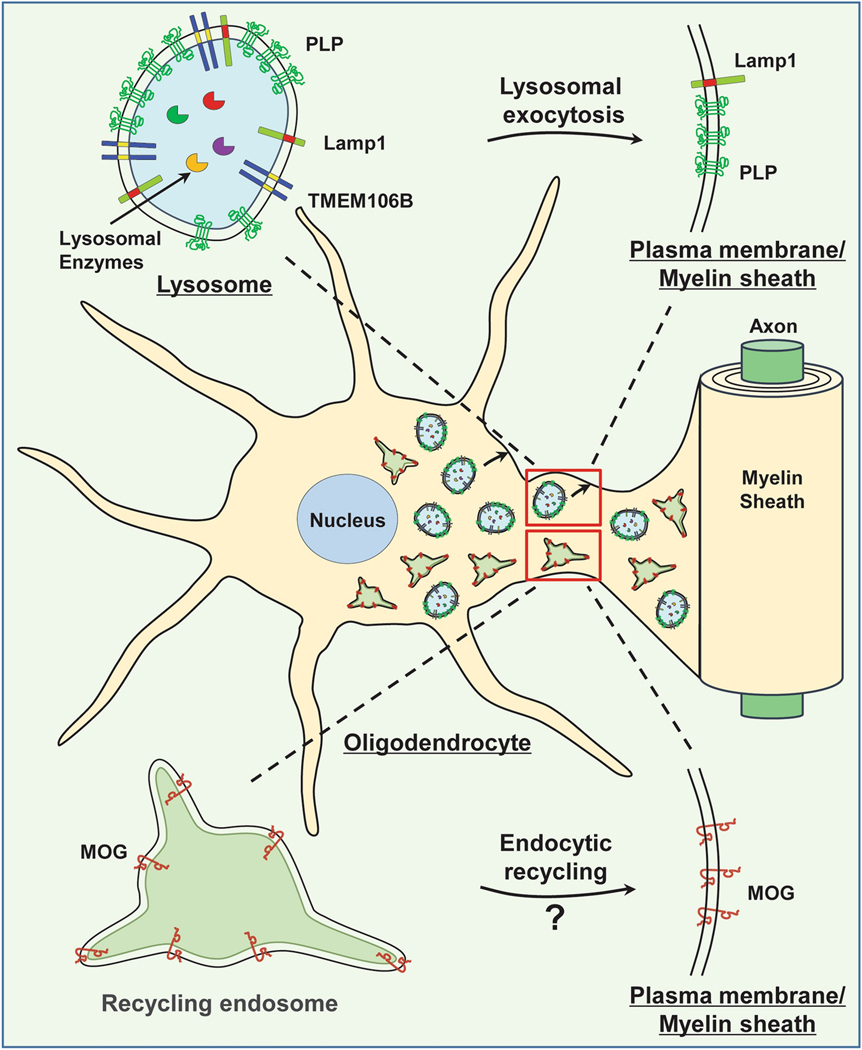
Figure 4. TMEM106B and myelination.
TMEM106B is highly expressed in oligodendrocytes and is localized in the lysosome in these cells. TMEM106B deficiency in oligodendrocytes results in lysosome clustering near the nucleus and a defect in lysosomal exocytosis, which leads to trafficking defects and decreased cell surface levels of myelin membrane protein, PLP. Additionally, TMEM106B might modulate the endocytic recycling of MOG, a type I transmembrane protein found in the myelin sheath.
Recently, the effect of the D252N mutation on TMEM106B function and the lysosome has also been analyzed. It does not affect TMEM106B messenger RNA (mRNA) and protein levels in vitro [19,34,83]. The stability and dimerization of TMEM106B is also not affected [19], but TMEM106B processing might be slightly decreased by D252N [19]. However, interesting phenotypes have been observed regarding lysosomal alteration induced by the D252N mutation. While the D252N mutation fails to induce lysosome enlargement when overexpressed, it results in lysosome clustering near the nucleus [19]. Furthermore, the D252N mutation abolishes lysosomal acidification induced by wild-type TMEM106B overexpression [19]. Lysosomal dysfunction and impaired lysosomal acidification have also been observed in patient-derived fibroblasts with the TMEM106B D252N mutation [34]. Based on these studies, it is likely that D252N is a loss-of-function mutation and it acts in a dominant negative manner by forming a heterodimer with the wild type TMEM106B protein. This mutation might also affect the interaction between TMEM106B and its binding partners. However, the D252N mutation does not affect the binding between TMEM106B and vacuolar-ATPase accessory protein 1 (AP1) or cathepsin D [19].
Taken together, these findings support that TMEM106B regulates myelination through modulation of lysosomal function in oligodendrocytes. Efforts to identify interactions and cellular processes affected by the D252N mutation might yield novel insights into how TMEM106B regulates lysosomal function and myelination. In addition to PLP, the levels of another myelin transmembrane protein, MOG, are also affected by TMEM106B deletion[19]. MOG, a type I transmembrane protein in the outer loop (abaxonal) region of myelin sheath, is transported to myelin sheath via recycling endosomes [83] (Fig. 4). It will be interesting to examine whether this process is affected by the loss of TMEM106B or the D252N mutation. Additionally, since the interaction between axons and oligodendrocytes is critical for proper myelin formation [71] and TMEM106B is highly expressed in neurons, it is possible that TMEM106B might also regulate myelination through its action in neurons.
Genetic interaction between Progranulin (PGRN) and TMEM106B
Haploinsufficiency of progranulin (PGRN) protein, caused by heterozygous mutations in the GRN gene, is a leading cause of familial FTLD-TDP [3,15,25]. PGRN, initially identified as a secreted glycoprotein, has been shown recently to play an important role in maintaining proper lysosomal function in neurons and microglia in the central nervous system (CNS), especially during aging [56] [4,9,37]. PGRN is comprised of 7.5 granulin repeats, which can be liberated in the lysosome compartment or extracellularly [56]. The granulin peptides have been shown to modulate the activities of several lysosomal enzymes [56].
TMEM106B was initially identified as a genetic risk factor of FTLD-TDP with GRN mutations [78]. The TMEM106B risk allele is associated with lower PGRN levels [14,78]. The effect of TMEM106B on PGRN levels have been subsequently tested in cell culture and in mouse models. Cellular studies have revealed that overexpression of TMEM106B leads to an increase in PGRN levels in several cell lines [7,10,31], through post transcriptional mechanisms [7] [54]. Increased levels of TMEM106B reduces the processing of PGRN into granulin peptides [31]. Although TMEM106B knockdown does not alter PGRN protein and mRNA levels in cell lines [7,31,41], elevated levels of full length PGRN have been detected in the brain lysates from TMEM106B-deficient mice [47]. Increased expression of TMEM106B might also affect PGRN trafficking by impairing the endosomal lysosomal pathway [10]. In summary, these studies support that TMEM106B affects PGRN levels and processing indirectly by altering lysosomal activities.
The TMEM106B risk allele was reported to increase the levels of TMEM106B protein [10,78] and TMEM106B protein levels are increased in aged PGRN deficient mice [26,90]. Thus, modulation of TMEM106B levels might affect disease progression of FTLD patients with GRN mutations. Consistent with this notion, transgenic mice expressing TMEM106B under the neuronal CamKII promoter have been shown to exacerbate lysosomal abnormalities and lipofuscin accumulation caused by PGRN loss [90]. Using the gene-trap mouse line which still express 5–10% of full length TMEM106B, TMEM106B deficiency was shown to cause opposite changes in the lysosomal transcriptome and proteome compared to PGRN deficiency [38]. Moreover, TMEM106B ablation normalizes changes in lysosomal enzyme and proteolysis, rescues FTLD-related behavioral deficits and retinal degeneration, but not the accumulation of lipofuscin, microglia activation and complement C1q deposition in PGRN-deficient mice [38]. However, heterozygous TMEM106B mice fails to rescue phenotypes of Grn+/− mice in another study using the same gene trap line [2]. In addition, three recent studies have shown that TMEM106B deficiency exacerbates lysosomal abnormalities and FTLD pathology in PGRN deficient mice using independently generated mouse lines [18,81,88]. Severe neuronal loss and glial activation have been detected in the retina, spinal cord and brain in Tmem106b−/− Grn−/− mice. Lysosome enlargement in both microglia and astrocytes and increased accumulation of lysosomal vacuoles in the distal end of the axon initial segment in motor neurons have been observed. FTLD phenotypes, such as the significant accumulation of ubiquitinated proteins, autophagy adaptor protein p62, and phosphorylated TDP-43 are also enhanced in the double knockout mice with a much earlier onset. In two studies, Tmem106b−/− Grn−/− mice also show severe myelination defects [81,88]. In all three studies, deficiency of both PGRN and TMEM106B leads to severe hindlimb weakness, paralysis and premature death [18,81,88]. The same gene-trap line was used in one of the studies [88] and delayed onset of phenotypes was observed compared to other two other lines with complete loss of full length TMEM106B and PGRN, indicating that residual TMEM106B in the gene trap line is able to rescue some of the early phenotypes associated with PGRN and TMEM106B loss [18,81,88]. In addition, the gene trap line might also express an N-terminal fragment of TMEM106B fused to LacZ, which might also contribute to some of phenotypic differences [55]. Nevertheless, results from these independently generated mouse lines from recent studies strongly support that the loss of TMEM106B results in lysosomal abnormalities and potentiates FTLD pathology. In mouse models, both up- and down-regulation of TMEM106B lead to lysosomal alternation and exacerbate phenotypes associated with loss of PGRN. Thus, TMEM106B protein levels need to be tightly regulated to ensure proper lysosomal function and brain health.
Conclusions and Perspectives
Since its association with FTLD was first identified in 2010, TMEM106B has been found to be intimately linked to brain health. We have learned a great deal about the cellular and physiological functions of TMEM106B during the past decade. A series of robust cellular, molecular and genetic studies have revealed that TMEM106B plays a critical role in regulating lysosome dynamics in vitro and in vivo, and TMEM106B is associated with brain aging and multiple brain disorders. However, the exact function of TMEM106B on the lysosomal membrane remains to be determined, such as its role in lysosomal pH regulation and lysosome movement within the cell body and along axons and dendrites. The role of TMEM106B in the normal physiology of different brain cell types and neuronal subtypes still needs to be elucidated. How TMEM106B variants mechanistically alter diseases-related pathology through regulating lysosomal function is still a mystery. A better understanding on the molecular and cellular function of TMEM106B under normal and diseased conditions will help develop the therapeutics necessary to treat the brain disorders associated with TMEM106B.
Acknowledgements
We thank Mohammed Ullah and Isabel I. Katz for proofreading this manuscript. This work is supported by NINDS/NIA (R01NS088448 & R01NS095954) and the Bluefield project to cure frontotemporal dementia to F.H.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Adams HH, Verhaaren BF, Vrooman HA, Uitterlinden AG, Hofman A, van Duijn CM, vander Lugt A, Niessen WJ, Vernooij MW, Ikram MA (2014) TMEM106B influences volume of left-sided temporal lobe and interhemispheric structures in the general population. Biol Psychiatry 76:503–508. doi: 10.1016/j.biopsych.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 2.Arrant AE, Nicholson AM, Zhou X, Rademakers R, Roberson ED (2018) Partial Tmem106breduction does not correct abnormalities due to progranulin haploinsufficiency. Mol Neurodegener 13:32. doi: 10.1186/s13024-018-0264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442:916–919 [DOI] [PubMed] [Google Scholar]
- 4.Bateman A, Bennett HP (2009) The granulin gene family: from cancer to dementia. Bioessays 31:1245–1254. doi: 10.1002/bies.200900086 [DOI] [PubMed] [Google Scholar]
- 5.Bieniek KF, Ross OA, Cormier KA, Walton RL, Soto-Ortolaza A, Johnston AE, DeSaro P, Boylan KB, Graff-Radford NR, Wszolek ZK, Rademakers R, Boeve BF, McKee AC, Dickson DW (2015) Chronic traumatic encephalopathy pathology in a neurodegenerative disorders brain bank. Acta Neuropathol 130:877–889. doi: 10.1007/s00401-015-1502-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino JS, Neefjes J (2017) Moving and positioning the endolysosomal system. Curr Opin Cell Biol 47:1–8. doi: 10.1016/j.ceb.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady OA, Zheng Y, Murphy K, Huang M, Hu F (2013) The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet 22:685–695. doi: 10.1093/hmg/dds475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady OA, Zhou X, Hu F (2014) Regulated intramembrane proteolysis of the frontotemporal lobar degeneration risk factor, TMEM106B, by signal peptide peptidase-like 2a (SPPL2a). J Biol Chem 289:19670–19680. doi: 10.1074/jbc.M113.515700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cenik B, Sephton CF, Kutluk Cenik B, Herz J, Yu G (2012) Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem 287:32298–32306. doi: 10.1074/jbc.R112.399170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, Trojanowski JQ, Lee VM (2012) TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci 32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherry JD, Mez J, Crary JF, Tripodis Y, Alvarez VE, Mahar I, Huber BR, Alosco ML, Nicks R, Abdolmohammadi B, Kiernan PT, Evers L, Svirsky S, Babcock K, Gardner HM, Meng G, Nowinski CJ, Martin BM, Dwyer B, Kowall NW, Cantu RC, Goldstein LE, Katz DI, Stern RA, Farrer LA, McKee AC, Stein TD (2018) Variation in TMEM106B in chronic traumatic encephalopathy. Acta Neuropathol Commun 6:115. doi: 10.1186/s40478-018-0619-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen TJ, Lee VM, Trojanowski JQ (2011) TDP-43 functions and pathogenic mechanisms implicated in TDP-43 proteinopathies. Trends Mol Med 17:659–667. doi: 10.1016/j.molmed.2011.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook C, Zhang YJ, Xu YF, Dickson DW, Petrucelli L (2008) TDP-43 in neurodegenerative disorders. Expert Opin Biol Ther 8:969–978. doi: 10.1517/14712598.8.7.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC, Goate A (2011) Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol 68:581–586. doi: 10.1001/archneurol.2010.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C (2006) Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442:920–924 [DOI] [PubMed] [Google Scholar]
- 16.de Araujo MEG, Liebscher G, Hess MW, Huber LA (2020) Lysosomal size matters. Traffic 21:60–75. doi: 10.1111/tra.12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deming Y, Cruchaga C (2014) TMEM106B: a strong FTLD disease modifier. Acta Neuropathol 127:419–422. doi: 10.1007/s00401-014-1249-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng T, Mai S, Roscoe JM, Sheng RR, Ullah M, Zhang J, Katz II, Yu H, Xiong W, Hu F (2020) Loss of TMEM106B and PGRN leads to severe lysosomal abnormalities and neurodegeneration in mice. EMBO Rep:e50219. doi: 10.15252/embr.202050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng T, Sheng RR, Sole-Domenech S, Ullah M, Zhou X, Mendoza CS, Enriquez LCM, Katz II,Paushter DH, Sullivan PM, Wu X, Maxfield FR, Hu F (2020) A role of the frontotemporal lobar degeneration risk factor TMEM106B in myelination. Brain 143:2255–2271. doi: 10.1093/brain/awaa154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, Crook J, Finger E, Hantanpaa KJ, Karydas AM, Sengdy P, Gonzalez J, Seeley WW, Johnson N, Beach TG, Mesulam M, Forloni G, Kertesz A, Knopman DS, Uitti R, White CL 3rd, Caselli R, Lippa C, Bigio EH, Wszolek ZK, Binetti G, Mackenzie IR, Miller BL, Boeve BF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Geschwind DH, Rademakers R (2011) TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL, Chesi A, Manduchi E, Wells AD, Grant SFA, Blobel GA, Brown CD, Chen-Plotkin AS (2017) A Dementia-Associated Risk Variant near TMEM106B Alters Chromatin Architecture and Gene Expression. Am J Hum Genet 101:643–663. doi: 10.1016/j.ajhg.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher MD, Suh E, Grossman M, Elman L, McCluskey L, Van Swieten JC, Al-Sarraj S, Neumann M, Gelpi E, Ghetti B, Rohrer JD, Halliday G, Van Broeckhoven C, Seilhean D, Shaw PJ, Frosch MP, Alafuzoff I, Antonell A, Bogdanovic N, Brooks W, Cairns NJ, Cooper-Knock J, Cotman C, Cras P, Cruts M, De Deyn PP, DeCarli C, Dobson-Stone C, Engelborghs S, Fox N, Galasko D, Gearing M, Gijselinck I, Grafman J, Hartikainen P, Hatanpaa KJ, Highley JR, Hodges J, Hulette C, Ince PG, Jin LW, Kirby J, Kofler J, Kril J, Kwok JB, Levey A, Lieberman A, Llado A, Martin JJ, Masliah E, McDermott CJ, McKee A, McLean C, Mead S, Miller CA, Miller J, Munoz DG, Murrell J, Paulson H, Piguet O, Rossor M, Sanchez-Valle R, Sano M, Schneider J, Silbert LC, Spina S, van der Zee J, Van Langenhove T, Warren J, Wharton SB, White CL 3rd, Woltjer RL, Trojanowski JQ, Lee VM, Van Deerlin V, Chen-Plotkin AS (2014) TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol 127:407–418. doi: 10.1007/s00401-013-1239-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamp AC, Tanaka Y, Lullmann-Rauch R, Wittke D, D’Hooge R, De Deyn PP, Moser T, Maier H, Hartmann D, Reiss K, Illert AL, von Figura K, Saftig P (2003) LIMP-2/LGP85 deficiency causes ureteric pelvic junction obstruction, deafness and peripheral neuropathy in mice. Hum Mol Genet 12:631–646 [PubMed] [Google Scholar]
- 24.Gao J, Wang L, Huntley ML, Perry G, Wang X (2018) Pathomechanisms of TDP-43 in neurodegeneration. J Neurochem. doi: 10.1111/jnc.14327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, Josephs K, Pickering-Brown SM, Graff-Radford N, Uitti R, Dickson D, Wszolek Z, Gonzalez J, Beach TG, Bigio E, Johnson N, Weintraub S, Mesulam M, White CL 3rd, Woodruff B, Caselli R, Hsiung GY, Feldman H, Knopman D, Hutton M, Rademakers R (2006) Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet 15:2988–3001 [DOI] [PubMed] [Google Scholar]
- 26.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J, Lang CM, Kremmer E, Martin JJ, Engelborghs S, Kretzschmar HA, Arzberger T, Van Broeckhoven C, Haass C, Capell A (2014) Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 127:845–860. doi: 10.1007/s00401-014-1262-6 [DOI] [PubMed] [Google Scholar]
- 27.Guo DZ, Xiao L, Liu YJ, Shen C, Lou HF, Lv Y, Pan SY (2018) Cathepsin D deficiency delays central nervous system myelination by inhibiting proteolipid protein trafficking from late endosome/lysosome to plasma membrane. Exp Mol Med 50:e457. doi: 10.1038/emm.2017.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo L, Shorter J (2017) Biology and Pathobiology of TDP-43 and Emergent Therapeutic Strategies. Cold Spring Harb Perspect Med 7. doi: 10.1101/cshperspect.a024554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harding SR, Bocchetta M, Gordon E, Cash DM, Cardoso MJ, Druyeh R, Ourselin S, Warren JD, Mead S, Rohrer JD (2017) The TMEM106B risk allele is associated with lower cortical volumes in a clinically diagnosed frontotemporal dementia cohort. Journal of Neurology, Neurosurgery & Psychiatry 88:997–998. doi: 10.1136/jnnp2017-315641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hokkanen SRK, Kero M, Kaivola K, Hunter S, Keage HAD, Kiviharju A, Raunio A, Tienari PJ, Paetau A, Matthews FE, Fleming J, Graff C, Polvikoski TM, Myllykangas L, Brayne C, Collaboration EC (2020) Putative risk alleles for LATE-NC with hippocampal sclerosis in population-representative autopsy cohorts. Brain Pathol 30:364–372. doi: 10.1111/bpa.12773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holler CJ, Taylor G, Deng Q, Kukar T (2017) Intracellular Proteolysis of Progranulin Generates Stable, Lysosomal Granulins that Are Haploinsufficient in Patients with Frontotemporal Dementia Caused by GRN Mutations. eNeuro 4. doi: 10.1523/ENEURO.0100-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu T, Chen Y, Ou R, Wei Q, Cao B, Zhao B, Wu Y, Song W, Chen X, Shang HF (2017) Association analysis of polymorphisms in VMAT2 and TMEM106B genes for Parkinson’s disease, amyotrophic lateral sclerosis and multiple system atrophy. J Neurol Sci 377:65–71. doi: 10.1016/j.jns.2017.03.028 [DOI] [PubMed] [Google Scholar]
- 33.Ikemoto S, Hamano SI, Kikuchi K, Koichihara R, Hirata Y, Matsuura R, Hiraide T, Nakashima M, Inoue K, Kurosawa K, Saitsu H (2020) A recurrent TMEM106B mutation in hypomyelinating leukodystrophy: A rapid diagnostic assay. Brain Dev 42:603–606. doi: 10.1016/j.braindev.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 34.Ito Y, Hartley T, Baird S, Venkateswaran S, Simons C, Wolf NI, Boycott KM, Dyment DA, Kernohan KD (2018) Lysosomal dysfunction in TMEM106B hypomyelinating leukodystrophy. Neurol Genet 4:e288. doi: 10.1212/NXG.0000000000000288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, Kunkle BW, Wang LS, Bis JC, Bellenguez C, Harold D, Lunetta KL, Destefano AL, Grenier-Boley B, Sims R, Beecham GW, Smith AV, Chouraki V, Hamilton-Nelson KL, Ikram MA, Fievet N, Denning N, Martin ER, Schmidt H, Kamatani Y, Dunstan ML, Valladares O, Laza AR, Zelenika D, Ramirez A, Foroud TM, Choi SH, Boland A, Becker T, Kukull WA, van der Lee SJ, Pasquier F, Cruchaga C, Beekly D, Fitzpatrick AL, Hanon O, Gill M, Barber R, Gudnason V, Campion D, Love S, Bennett DA, Amin N, Berr C, Tsolaki M, Buxbaum JD, Lopez OL, Deramecourt V, Fox NC, Cantwell LB, Tarraga L, Dufouil C, Hardy J, Crane PK, Eiriksdottir G, Hannequin D, Clarke R, Evans D, Mosley TH Jr., Letenneur L, Brayne C, Maier W, De Jager P, Emilsson V, Dartigues JF, Hampel H, Kamboh MI, de Bruijn RF, Tzourio C, Pastor P, Larson EB, Rotter JI, O’Donovan MC, Montine TJ, Nalls MA, Mead S, Reiman EM, Jonsson PV, Holmes C, St George-Hyslop PH, Boada M, Passmore P, Wendland JR, Schmidt R, Morgan K, Winslow AR, Powell JF, Carasquillo M, Younkin SG, Jakobsdottir J, Kauwe JS, Wilhelmsen KC, Rujescu D, Nothen MM, Hofman A, Jones L, Consortium I, Haines JL, Psaty BM, Van Broeckhoven C, Holmans P, Launer LJ, Mayeux R, Lathrop M, Goate AM, Escott-Price V, Seshadri S, Pericak-Vance MA, Amouyel P, Williams J, van Duijn CM, Schellenberg GD, Farrer LA (2016) A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 21:108–117. doi: 10.1038/mp.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jun MH, Han JH, Lee YK, Jang DJ, Kaang BK, Lee JA (2015) TMEM106B, a frontotemporal lobar dementia (FTLD) modifier, associates with FTD-3-linked CHMP2B, a complex of ESCRT-III. Mol Brain 8:85. doi: 10.1186/s13041-015-0177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao AW, McKay A, Singh PP, Brunet A, Huang EJ (2017) Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci 18:325–333. doi: 10.1038/nrn.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, Lam TT, Strittmatter SM (2017) Loss of TMEM106B Ameliorates Lysosomal and Frontotemporal Dementia-Related Phenotypes in Progranulin-Deficient Mice. Neuron 95:281–296 e286. doi: 10.1016/j.neuron.2017.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koga S, Lin WL, Walton RL, Ross OA, Dickson DW (2018) TDP-43 pathology in multiple-system atrophy: colocalization of TDP-43 and alpha-synuclein in glial cytoplasmic inclusions. Neuropathol Appl Neurobiol 44:707–721. doi: 10.1111/nan.12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundu ST, Grzeskowiak CL, Fradette JJ, Gibson LA, Rodriguez LB, Creighton CJ, Scott KL, Gibbons DL (2018) TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat Commun 9:2731. doi: 10.1038/s41467-018-05013-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, Capell A, Haass C (2012) Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem 287:19355–19365. doi: 10.1074/jbc.M112.365098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lattante S, Le Ber I, Galimberti D, Serpente M, Rivaud-Pechoux S, Camuzat A, Clot F, Fenoglio C, French research network on FTD, Ftd ALS, Scarpini E, Brice A, Kabashi E (2014) Defining the association of TMEM106B variants among frontotemporal lobar degeneration patients with GRN mutations and C9orf72 repeat expansions. Neurobiol Aging 35:2658 e2651–2658 e2655. doi: 10.1016/j.neurobiolaging.2014.06.023 [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Farias FHG, Dube U, Del-Aguila JL, Mihindukulasuriya KA, Fernandez MV, Ibanez L, Budde JP, Wang F, Lake AM, Deming Y, Perez J, Yang C, Bahena JA, Qin W, Bradley JL, Davenport R, Bergmann K, Morris JC, Perrin RJ, Benitez BA, Dougherty JD, Harari O, Cruchaga C (2020) The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol 139:45–61. doi: 10.1007/s00401-019-02066-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lie PPY, Nixon RA (2019) Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol Dis 122:94–105. doi: 10.1016/j.nbd.2018.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ling SC, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–438. doi: 10.1016/j.neuron.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu RC, Wang H, Tan MS, Yu JT, Tan L (2014) TMEM106B and APOE polymorphisms interact to confer risk for late-onset Alzheimer’s disease in Han Chinese. J Neural Transm (Vienna) 121:283–287. doi: 10.1007/s00702-013-1106-x [DOI] [PubMed] [Google Scholar]
- 47.Luningschror P, Werner G, Stroobants S, Kakuta S, Dombert B, Sinske D, Wanner R, Lullmann-Rauch R, Wefers B, Wurst W, D’Hooge R, Uchiyama Y, Sendtner M, Haass C, Saftig P, Knoll B, Capell A, Damme M (2020) The FTLD Risk Factor TMEM106B Regulates the Transport of Lysosomes at the Axon Initial Segment of Motoneurons. Cell Rep 30:3506–3519 e3506. doi: 10.1016/j.celrep.2020.02.060 [DOI] [PubMed] [Google Scholar]
- 48.Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Fullgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, Licitra F, Lopez Ramirez A, Pavel M, Puri C, Renna M, Ricketts T, Schlotawa L, Vicinanza M, Won H, Zhu Y, Skidmore J, Rubinsztein DC (2017) Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 93:1015–1034. doi: 10.1016/j.neuron.2017.01.022 [DOI] [PubMed] [Google Scholar]
- 49.Milind N, Preuss C, Haber A, Ananda G, Mukherjee S, John C, Shapley S, Logsdon BA, Crane PK, Carter GW (2020) Transcriptomic stratification of late-onset Alzheimer’s cases reveals novel genetic modifiers of disease pathology. PLoS Genet 16:e1008775. doi: 10.1371/journal.pgen.1008775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neary D, Snowden J, Mann D (2005) Frontotemporal dementia. Lancet Neurol 4:771–780 [DOI] [PubMed] [Google Scholar]
- 51.Nelson PT, Dickson DW, Trojanowski JQ, Jack CR, Boyle PA, Arfanakis K, Rademakers R, Alafuzoff I, Attems J, Brayne C, Coyle-Gilchrist ITS, Chui HC, Fardo DW, Flanagan ME, Halliday G, Hokkanen SRK, Hunter S, Jicha GA, Katsumata Y, Kawas CH, Keene CD, Kovacs GG, Kukull WA, Levey AI, Makkinejad N, Montine TJ, Murayama S, Murray ME, Nag S, Rissman RA, Seeley WW, Sperling RA, White Iii CL, Yu L, Schneider JA (2019) Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 142:1503–1527. doi: 10.1093/brain/awz099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson PT, Wang W-X, Partch AB, Monsell SE, Valladares O, Ellingson SR, Wilfred BR, Naj AC, Wang L- S, Kukull WA, Fardo DW (2015) Reassessment of Risk Genotypes (GRN, TMEM106B, and ABCC9 Variants) Associated With Hippocampal Sclerosis of Aging Pathology. Journal of Neuropathology & Experimental Neurology 74:75–84. doi: 10.1097/nen.0000000000000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133. doi: 10.1126/science.1134108 [DOI] [PubMed] [Google Scholar]
- 54.Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB 3rd, Castanedes-Casey M, Rousseau L, Benussi L, Binetti G, Ghidoni R, Hsiung GY, Mackenzie IR, Finger E, Boeve BF, Ertekin-Taner N, Graff-Radford NR, Dickson DW, Rademakers R (2013) TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem 126:781–791. doi: 10.1111/jnc.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholson AM, Zhou X, Perkerson RB, Parsons TM, Chew J, Brooks M, DeJesus-Hernandez M, Finch NA, Matchett BJ, Kurti A, Jansen-West KR, Perkerson E, Daughrity L, Castanedes-Casey M, Rousseau L, Phillips V, Hu F, Gendron TF, Murray ME, Dickson DW, Fryer JD, Petrucelli L, Rademakers R (2018) Loss of Tmem106b is unable to ameliorate frontotemporal dementia-like phenotypes in an AAV mouse model of C9ORF72-repeat induced toxicity. Acta Neuropathol Commun 6:42. doi: 10.1186/s40478-018-0545-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paushter DH, Du H, Feng T, Hu F (2018) The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol 136:1–17. doi: 10.1007/s00401-018-1861-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pottier C, Zhou X, Perkerson RB, Baker M, Jenkins GD, Serie DJ, Ghidoni R, Benussi L, Binetti G, López de Munain A, Zulaica M, Moreno F, Le Ber I, Pasquier F, Hannequin D, Sánchez-Valle R, Antonell A, Lladó A, Parsons TM, Finch NA, Finger EC, Lippa CF, Huey ED, Neumann M, Heutink P, Synofzik M, Wilke C, Rissman RA, Slawek J, Sitek E, Johannsen P, Nielsen JE, Ren Y, van Blitterswijk M, DeJesus-Hernandez M, Christopher E, Murray ME, Bieniek KF, Evers BM, Ferrari C, Rollinson S, Richardson A, Scarpini E, Fumagalli GG, Padovani A, Hardy J, Momeni P, Ferrari R, Frangipane F, Maletta R, Anfossi M, Gallo M, Petrucelli L, Suh E, Lopez OL, Wong TH, van Rooij JGJ, Seelaar H, Mead S, Caselli RJ, Reiman EM, Noel Sabbagh M, Kjolby M, Nykjaer A, Karydas AM, Boxer AL, Grinberg LT, Grafman J, Spina S, Oblak A, Mesulam MM, Weintraub S, Geula C, Hodges JR, Piguet O, Brooks WS, Irwin DJ, Trojanowski JQ, Lee EB, Josephs KA, Parisi JE, Ertekin-Taner N, Knopman DS, Nacmias B, Piaceri I, Bagnoli S, Sorbi S, Gearing M, Glass J, Beach TG, Black SE, Masellis M, Rogaeva E, Vonsattel J-P, Honig LS, Kofler J, Bruni AC, Snowden J, Mann D, Pickering-Brown S, Diehl-Schmid J, Winkelmann J, Galimberti D, Graff C, Öijerstedt L, Troakes C, Al-Sarraj S, Cruchaga C, Cairns NJ, Rohrer JD, Halliday GM, Kwok JB, van Swieten JC, White CL, Ghetti B, Murell JR, Mackenzie IRA, Hsiung G- YR, Borroni B, Rossi G, Tagliavini F, Wszolek ZK, Petersen RC, Bigio EH, Grossman M, Van Deerlin VM, Seeley WW, Miller BL, Graff-Radford NR, Boeve BF, Dickson DW, Biernacka JM, Rademakers R (2018) Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. The Lancet Neurology 17:548–558. doi: 10.1016/s1474-4422(18)30126-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prolo LM, Vogel H, Reimer RJ (2009) The lysosomal sialic acid transporter sialin is required for normal CNS myelination. J Neurosci 29:15355–15365. doi: 10.1523/JNEUROSCI.3005-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS (2016) Mechanisms and functions of lysosome positioning. J Cell Sci 129:4329–4339. doi: 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ratnavalli E, Brayne C, Dawson K, Hodges JR (2002) The prevalence of frontotemporal dementia. Neurology 58:1615–1621 [DOI] [PubMed] [Google Scholar]
- 61.Ren Y, van Blitterswijk M, Allen M, Carrasquillo MM, Reddy JS, Wang X, Beach TG, Dickson DW, Ertekin-Taner N, Asmann YW, Rademakers R (2018) TMEM106B haplotypes have distinct gene expression patterns in aged brain. Mol Neurodegener 13:35. doi: 10.1186/s13024-018-0268-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renton AE, Chio A, Traynor BJ (2014) State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17:17–23. doi: 10.1038/nn.3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rhinn H, Abeliovich A (2017) Differential Aging Analysis in Human Cerebral Cortex Identifies Variants in TMEM106B and GRN that Regulate Aging Phenotypes. Cell Syst 4:404–415 e405. doi: 10.1016/j.cels.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 64.Rutherford NJ, Carrasquillo MM, Li M, Bisceglio G, Menke J, Josephs KA, Parisi JE, Petersen RC, Graff-Radford NR, Younkin SG, Dickson DW, Rademakers R (2012) TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology 79:717–718. doi: 10.1212/WNL.0b013e318264e3ac [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saftig P, Puertollano R (2020) How Lysosomes Sense, Integrate, and Cope with Stress. Trends Biochem Sci. doi: 10.1016/j.tibs.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmiedt ML, Blom T, Blom T, Kopra O, Wong A, von Schantz-Fant C, Ikonen E, Kuronen M, Jauhiainen M, Cooper JD, Jalanko A (2012) Cln5-deficiency in mice leads to microglial activation, defective myelination and changes in lipid metabolism. Neurobiol Dis 46:19–29. doi: 10.1016/j.nbd.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 67.Schwenk BM, Lang CM, Hogl S, Tahirovic S, Orozco D, Rentzsch K, Lichtenthaler SF, Hoogenraad CC, Capell A, Haass C, Edbauer D (2014) The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J 33:450–467. doi: 10.1002/embj.201385857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scotter EL, Chen HJ, Shaw CE (2015) TDP-43 Proteinopathy and ALS: Insights into Disease Mechanisms and Therapeutic Targets. Neurotherapeutics 12:352–363. doi: 10.1007/s13311-015-0338-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A (2011) TFEB links autophagy to lysosomal biogenesis. Science 332:1429–1433. doi: 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simons C, Dyment D, Bent SJ, Crawford J, D’Hooghe M, Kohlschutter A, Venkateswaran S, Helman G, Poll-The BT, Makowski CC, Ito Y, Kernohan K, Hartley T, Waisfisz Q, Taft RJ, Care4Rare C, van der Knaap MS, Wolf NI (2017) A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain 140:3105–3111. doi: 10.1093/brain/awx314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simons M, Trajkovic K (2006) Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. J Cell Sci 119:4381–4389. doi: 10.1242/jcs.03242 [DOI] [PubMed] [Google Scholar]
- 72.Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM (2014) Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci 61:226–240. doi: 10.1016/j.mcn.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stroobants S, D’Hooge R, Damme M (2020) Aged Tmem106b knockout mice display gait deficits in coincidence with Purkinje cell loss and only limited signs of non-motor dysfunction. Brain Pathol. doi: 10.1111/bpa.12903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki H, Matsuoka M (2016) The Lysosomal Trafficking Transmembrane Protein 106B Is Linked to Cell Death. J Biol Chem 291:21448–21460. doi: 10.1074/jbc.M116.737171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trivedi PC, Bartlett JJ, Pulinilkunnil T (2020) Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 9. doi: 10.3390/cells9051131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tropea TF, Mak J, Guo MH, Xie SX, Suh E, Rick J, Siderowf A, Weintraub D, Grossman M, Irwin D, Wolk DA, Trojanowski JQ, Van Deerlin V, Chen-Plotkin AS (2019) TMEM106B Effect on cognition in Parkinson disease and frontotemporal dementia. Ann Neurol 85:801–811. doi: 10.1002/ana.25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Blitterswijk M, Mullen B, Nicholson AM, Bieniek KF, Heckman MG, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Murray ME, Hsiung GY, Stewart H, Karydas AM, Finger E, Kertesz A, Bigio EH, Weintraub S, Mesulam M, Hatanpaa KJ, White CL 3rd, Strong MJ, Beach TG, Wszolek ZK, Lippa C, Caselli R, Petrucelli L, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Mackenzie IR, Seeley WW, Grinberg LT, Miller BL, Boylan KB, Graff-Radford NR, Boeve BF, Dickson DW, Rademakers R (2014) TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol 127:397–406. doi: 10.1007/s00401-013-1240-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, Hodges J, Spillantini MG, Gilman S, Lieberman AP, Kaye JA, Woltjer RL, Bigio EH, Mesulam M, Al-Sarraj S, Troakes C, Rosenberg RN, White CL 3rd, Ferrer I, Llado A, Neumann M, Kretzschmar HA, Hulette CM, Welsh-Bohmer KA, Miller BL, Alzualde A, Lopez de Munain A, McKee AC, Gearing M, Levey AI, Lah JJ, Hardy J, Rohrer JD, Lashley T, Mackenzie IR, Feldman HH, Hamilton RL, Dekosky ST, van der Zee J, Kumar-Singh S, Van Broeckhoven C, Mayeux R, Vonsattel JP, Troncoso JC, Kril JJ, Kwok JB, Halliday GM, Bird TD, Ince PG, Shaw PJ, Cairns NJ, Morris JC, McLean CA, DeCarli C, Ellis WG, Freeman SH, Frosch MP, Growdon JH, Perl DP, Sano M, Bennett DA, Schneider JA, Beach TG, Reiman EM, Woodruff BK, Cummings J, Vinters HV, Miller CA, Chui HC, Alafuzoff I, Hartikainen P, Seilhean D, Galasko D, Masliah E, Cotman CW, Tunon MT, Martinez MC, Munoz DG, Carroll SL, Marson D, Riederer PF, Bogdanovic N, Schellenberg GD, Hakonarson H, Trojanowski JQ, Lee VM (2010) Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 42:234–239. doi: 10.1038/ng.536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van der Zee J, Van Langenhove T, Kleinberger G, Sleegers K, Engelborghs S, Vandenberghe R, Santens P, Van den Broeck M, Joris G, Brys J, Mattheijssens M, Peeters K, Cras P, De Deyn PP, Cruts M, Van Broeckhoven C (2011) TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain 134:808–815. doi: 10.1093/brain/awr007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vass R, Ashbridge E, Geser F, Hu WT, Grossman M, Clay-Falcone D, Elman L, McCluskey L, Lee VM, Van Deerlin VM, Trojanowski JQ, Chen-Plotkin AS (2011) Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol 121:373–380. doi: 10.1007/s00401-010-0782-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Werner G, Damme M, Schludi M, Gnörich J, Wind K, Fellerer K, Wefers B, Wurst W, Edbauer D, Brendel M, Haass C, Capell A (2020) Loss of TMEM106B potentiates lysosomal and FTD-like pathology in progranulin deficient mice. EMBO Rep:e50241. doi: 10.15252/embr.202050241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White CC, Yang HS, Yu L, Chibnik LB, Dawe RJ, Yang J, Klein HU, Felsky D, Ramos-Miguel A, Arfanakis K, Honer WG, Sperling RA, Schneider JA, Bennett DA, De Jager PL (2017) Identification of genes associated with dissociation of cognitive performance and neuropathological burden: Multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med 14:e1002287. doi: 10.1371/journal.pmed.1002287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winterstein C, Trotter J, Kramer-Albers EM (2008) Distinct endocytic recycling of myelin proteins promotes oligodendroglial membrane remodeling. J Cell Sci 121:834–842. doi: 10.1242/jcs.022731 [DOI] [PubMed] [Google Scholar]
- 84.Yan H, Kubisiak T, Ji H, Xiao J, Wang J, Burmeister M (2018) The recurrent mutation in TMEM106B also causes hypomyelinating leukodystrophy in China and is a CpG hotspot. Brain 141:e36. doi: 10.1093/brain/awy029 [DOI] [PubMed] [Google Scholar]
- 85.Yang HS, White CC, Klein HU, Yu L, Gaiteri C, Ma Y, Felsky D, Mostafavi S, Petyuk VA, Sperling RA, Ertekin-Taner N, Schneider JA, Bennett DA, De Jager PL (2020) Genetics of Gene Expression in the Aging Human Brain Reveal TDP-43 Proteinopathy Pathophysiology. Neuron 107:496–508 e496. doi: 10.1016/j.neuron.2020.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu L, Chen Y, Tooze SA (2018) Autophagy pathway: Cellular and molecular mechanisms. Autophagy 14:207–215. doi: 10.1080/15548627.2017.1378838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu L, De Jager PL, Yang J, Trojanowski JQ, Bennett DA, Schneider JA (2015) The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology 84:927–934. doi: 10.1212/WNL.0000000000001313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou X, Brooks M, Jiang P, Koga S, Zuberi AR, Baker MC, Parsons TM, Castanedes-Casey M, Phillips V, Librero AL, Kurti A, Fryer JD, Bu G, Lutz C, Dickson DW, Rademakers R (2020) Loss of Tmem106b exacerbates FTLD pathologies and causes motor deficits in progranulin-deficient mice. EMBO Rep:e50197. doi: 10.15252/embr.202050197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou X, Nicholson AM, Ren Y, Brooks M, Jiang P, Zuberi A, Phuoc HN, Perkerson RB, Matchett B, Parsons TM, Finch NA, Lin W, Qiao W, Castanedes-Casey M, Phillips V, Librero AL, Asmann Y, Bu G, Murray ME, Lutz C, Dickson DW, Rademakers R (2020) Loss of TMEM106B leads to myelination deficits: implications for frontotemporal dementia treatment strategies. Brain 143:1905–1919. doi: 10.1093/brain/awaa141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou X, Sun L, Brady OA, Murphy KA, Hu F (2017) Elevated TMEM106B levels exaggerate lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency. Acta Neuropathol Commun 5:9. doi: 10.1186/s40478-017-0412-1 [DOI] [PMC free article] [PubMed] [Google Scholar]