Abstract
Exciting therapeutic targets are emerging from CRISPR-based screens of high mutational burden adult cancers. A key question, however, is whether functional genomic approaches will yield new targets in pediatric cancers, known for remarkably few mutations which often encode proteins considered challenging drug targets. To address this, we created a first-generation Pediatric Cancer Dependency Map representing 13 pediatric solid and brain tumor types. Eighty-two pediatric cancer cell lines were subjected to genome-scale CRISPR-Cas9 loss-of-function screening to identify genes required for cell survival. In contrast to the finding that pediatric cancers harbor fewer somatic mutations, we found a similar complexity of genetic dependencies in pediatric cancer cell lines compared to adult models. Findings from the Pediatric Cancer Dependency Map provide pre-clinical support for ongoing precision medicine clinical trials. The vulnerabilities seen in pediatric cancers were often distinct from adult, indicating that repurposing adult oncology drugs will be insufficient to address childhood cancers.
Outcomes for children with advanced cancers remain poor, and long-term side effects can be devastating for patients cured with chemotherapy1–7. While CRISPR-based dependency maps have focused on adult cancers8,9, it is unknown whether large-scale functional genomic approaches will yield new therapeutic targets in pediatric cancers. Pediatric cancers are known to have low mutational burdens or “quiet” genomes compared to adult tumors, with pediatric cancers having 1,000-fold fewer somatic mutations than many adult cancers10,11. In many cases, the tumors appear to be driven primarily by a single genetic aberration, such as SMARCB1 loss in rhabdoid tumors or the EWS-FLI1 fusion in Ewing sarcoma12,13. Indeed, multiple precision medicine efforts in pediatric oncology have found actionable events in only 25–30% of tumors14–16. The focus on mutation-driven dependencies in such studies highlights the hypothesis that oncogene activation drives tumor vulnerabilities. Therefore, it could be postulated that the genetic simplicity of childhood cancers might similarly translate into a limited number of genetic vulnerabilities (or dependencies) compared to adult cancers, which we show to be untrue.
Pediatric cancer cell line models
Whereas a Dependency Map of adult cancers has been established using genome-scale CRISPR-Cas9 screening across hundreds of adult cancer cell lines9,17, no such resource exists for pediatric cancer. We therefore sought to create a first-generation Pediatric Cancer Dependency Map. We assembled a collection of 178 pediatric cancer cell lines and subjected them to comprehensive genomic characterization including, to date, whole exome sequencing (n=90), SNP genotyping to facilitate copy number estimation (n=49), and RNA sequencing (n=124) (Fig. 1a, Supplementary Data Table 1, data available at depmap.org). For this first-generation map, we have focused on pediatric solid and brain tumors; however, it must be noted that we have relatively few of the diverse pediatric brain tumor types represented, a limitation of the current data set.
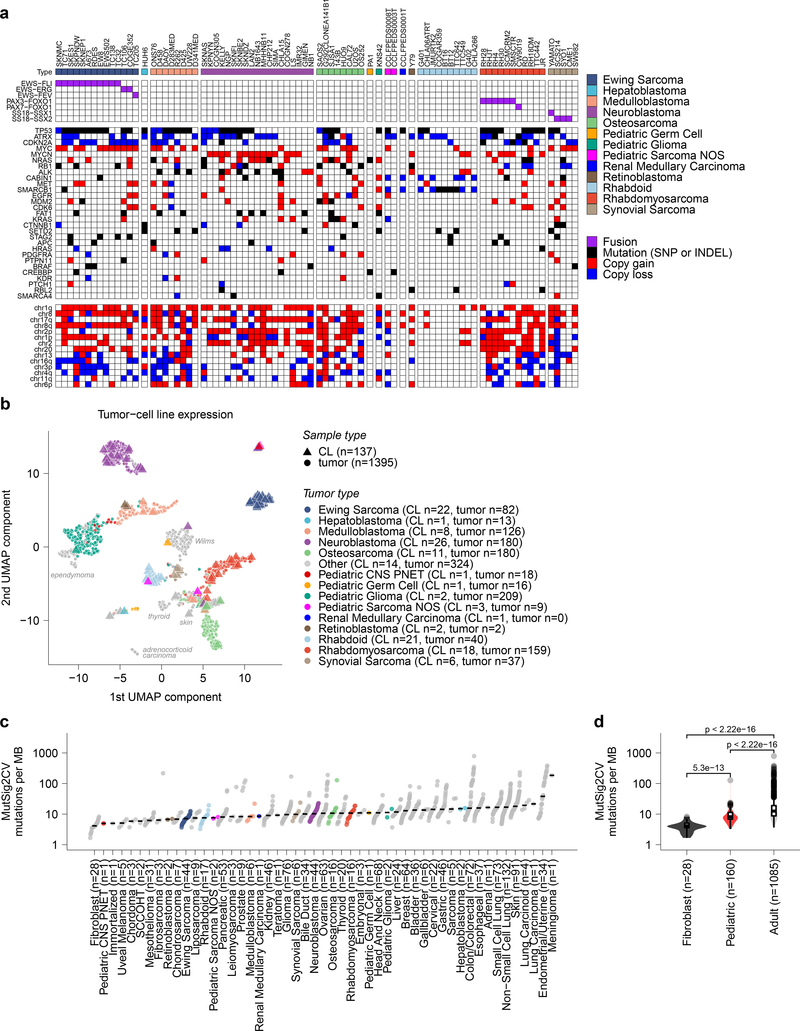
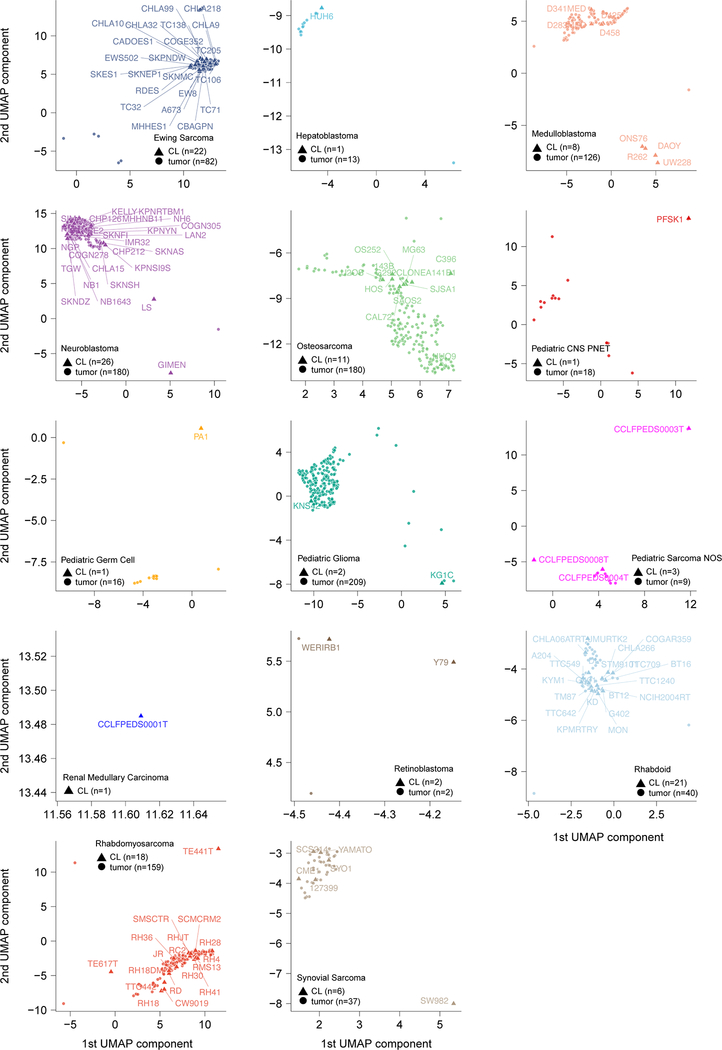
Fig 1. Pediatric solid tumor cancer models represent high-risk disease.
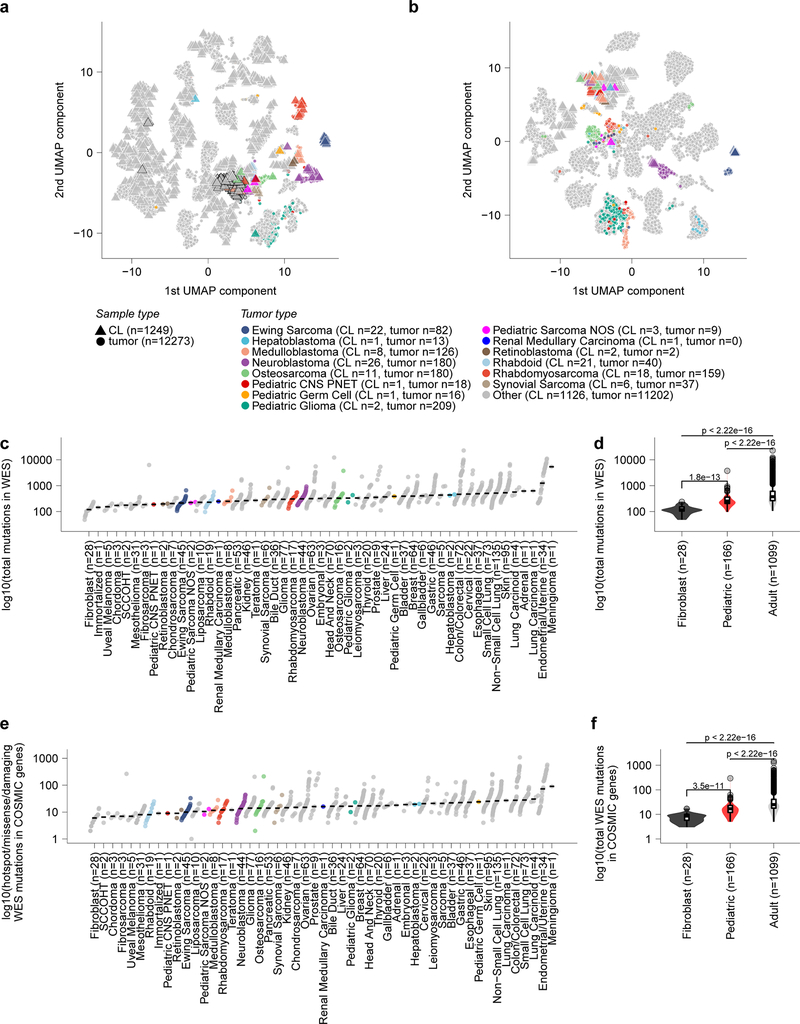
a, Overview of pediatric cancer cell line models screened in the Dependency Map project with genome-scale CRISPR-Cas9 with genomic characterization derived from whole exome and RNA sequencing. Genes and chromosomal arms highlighted are those that have been reported as commonly mutated or copy number altered in the pediatric tumor types shown13,50–59. b, Two-dimensional representation of RNA-sequencing data (after removing systematic tumor/cell line differences using the Celligner method18) using uniform manifold approximation and projection (UMAP) demonstrates high concordance between primary tumors (triangles) and cancer cell lines (circles) for pediatric tumor types. An interactive version of this plot is available at depmap.org/peddep. c, Mutational rates as mutations per megabase (MB) in whole exome sequencing as calculated using MutSig2CV (y-axis) grouped by solid tumor type (x-axis) with diseases ordered by median burden. Each circle represents an individual cell line with pediatric tumors colored by type; the black line represents the median mutation rate per tumor type. d, Pediatric solid tumor cell lines, including brain tumors (red, n=160 biologically independent cell lines), had significantly lower mutation rates (y-axis) as a whole compared to adult solid tumor lines (gray, n=1085 biologically independent cell lines) (p<2.22e−16) by two-sided Wilcoxon test, while fibroblast cell lines (black, n=28 biologically independent cell lines) had the lowest mutation rates compared to pediatric (p = 5.3e−13) or adult solid tumors (p<2.22e−16). Horizontal lines demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) represent the 10 and 90th percentiles respectively.
A key question was whether these cell lines reasonably represented the tumor types from which they were derived, or rather, have evolved in tissue culture to no longer reflect their developmental origin. Such cell line-tumor comparisons are challenging because tumors contain a diversity of cell types (tumor, stromal, and immune cells), whereas cell lines are pure tumor cells but may contain transcriptional changes due to in vitro culture. To address these challenges, we created an integrated two-dimensional map of cell line and tumor transcriptomes, using the Celligner method,18 to computationally remove systematic tumor/cell line differences in order to jointly represent the gene expression profiles of 1,249 cell lines19 and 12,273 patient tumors20. This approach, which did not use any disease type information as input, showed that the pediatric cell lines clustered closely with patient tumors of the same type (Fig. 1b, Extended Data Fig. 1), indicating that the developmental state of the cell lines was reasonably preserved despite all the caveats of extensive passaging in tissue culture. The majority of pediatric cancer cell lines express gene expression programs that align with their primary tumor counterparts; 74.0% of 123 pediatric cell lines match the respective primary tumor expression patterns (Supplementary Data Table 2). Recent pan-cancer analysis of cell line and tumor similarity using Celligner identified a cluster of 251 cell lines spanning multiple tumor types with a more undifferentiated and mesenchymal expression pattern18. Nineteen of 123 pediatric cell lines clustered in this group (Extended Data Fig. 2a, Supplementary Data Table 2); however, several of these cell lines may represent a subset of pediatric tumor biology as 9 of 11 osteosarcoma cell lines appeared in this undifferentiated group along with 23% (42 of 180) of primary osteosarcoma samples. Of note, performing a comparison of cell line to tumor expression patterns without first applying such a computational alignment procedure to remove systematic differences leads to the misperception that cell lines do not reflect the distinct transcriptional states of each tumor (Extended Data Fig. 2b) and worse performance in assigning cell lines to the correct tumor type (Supplementary Data Table 2, depmap.org/peddep).
Similarly, the mutational profiles of the cell lines largely reflected what is observed in pediatric tumors. In particular, the median mutation burden of pediatric lines was significantly lower than adult cancer cell lines (Fig. 1c-d, Supplementary Data Table 3), consistent with the lower mutation burdens seen in primary childhood tumors10,11. The magnitude of difference in mutation burden in pediatric versus adult solid tumor cell lines is not as large as that reported in primary tumors; however, we note that calling somatic mutations in cancer cell lines in the absence of matched normal tissue from the same patients is imperfect (Extended Data Fig. 2c-f). Copy number alterations in pediatric lines largely reflected patterns observed in primary tumors. For example, there were very few changes in rhabdoid tumor cell lines compared to many events in osteosarcoma cell lines (Extended Data Fig. 3a-c). As whole-genome sequencing for the majority of pediatric cancer cell lines is not available, it is difficult to systematically compare more complex non-coding events or structural variations between cell lines and primary tumors. We quantified gene fusion calls from RNA sequencing as a surrogate for translocation events and identified that cell lines from pediatric cancer types with higher numbers of structural variants in primary samples, such as osteosarcoma or rhabdomyosarcoma10, have larger median numbers of gene fusions (Extended Data Fig. 3d-e). Mutations in TP53 were seen in 50% of the pediatric solid tumor cell lines (Supplementary Data Table 4), whereas the reported frequency of TP53 mutations in pediatric cancers is only ~4%10. This discrepancy is consistent with cell lines tending to represent more advanced, aggressive cancers, and with the reported phenomenon of positive selection for TP53 mutation in vitro21. Nevertheless, the data collectively suggest that the cell lines, with their known caveats22, are reasonable models of pediatric cancers on the whole, since they capture the most common genomic alterations (Supplementary Data Table 4). However, caution must be used when focusing on single cell line models.
Mutation burden is not indicative of abundance of genetic dependencies
We next sought to quantify and characterize genetic vulnerabilities in pediatric cancer cell lines. Hence, we performed genome-scale CRISPR-Cas9 loss-of-function screens on the cell models. To date, of the 178 cell lines, we have successfully established 114 Cas9-expressing cell lines and screened 82 lines, representing 13 tumor types (Ewing sarcoma n=14, hepatoblastoma n=1, medulloblastoma n=8, neuroblastoma n=19, osteosarcoma n=8, pediatric germ cell tumor n=1, pediatric glioma n=1, pediatric sarcoma n=2, renal medullary carcinoma n=1, retinoblastoma n=1, rhabdoid tumor n=10, rhabdomyosarcoma n=11, and synovial sarcoma n=5), as previously described17. The resulting Cas9-expressing lines were subjected to pooled screening using the Avana lentiviral library of 74,378 gRNAs targeting 18,333 human genes17,23. We compared the abundance of each gRNA at the time of infection to its abundance after 21 days of cell culture to create gene dependency scores24. An important caveat of this approach is that it requires cancer models to be cultured for several weeks and is not currently amenable to short-term cell cultures.
The essentiality of each gene was scored relative to negative controls (score = 0, representing non-essential genes) and positive controls (score = −1, reflecting the median score of common essential genes) (Fig. 2a). For each gene effect score, we estimate the dependency probability as the likelihood that the gene represents a phenotype similar to positive controls in each cell line24. We focused on selective dependencies, that is, genes required for growth of a subset of cell lines (defined as a normality likelihood ratio test (normLRT) > 10025 and excluding genes that scored as common essential or non-essential in the screen) (Supplementary Data Table 5). We estimated the false positive rate for called dependencies across all genes or selective dependencies in our screen at 1.9% or 0.046%, respectively, by determining the rate at which non-expressed genes were called dependencies. The complete Pediatric Cancer Dependency Map dataset is available at depmap.org.
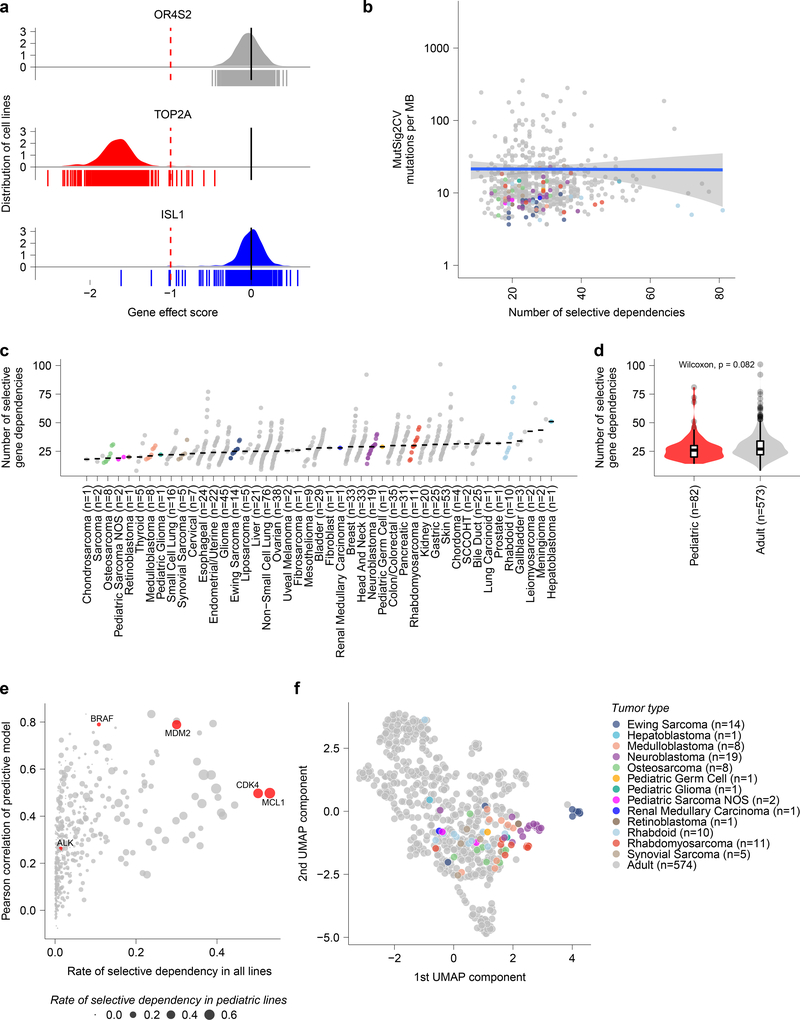
Fig 2. Cancer cell line selective dependencies are not correlated with mutation burden.
a, Example score distributions of genes that were non-essential (OR6S1), common essential (TOP2A), and a selective dependency (ISL1, with normLRT of 290) in the genome-scale CRISPR-Cas9 screen. The y-axis represents the cell line distribution with the x-axis representing CRISPR gene effect scores. Individual scores for cell lines screened (n=612) are indicated by the symbols depicted below the x-axis. b, Mutational burden as detected by whole exome sequencing by MutSig2CV in mutations per megabase (MB) compared to the number of selective dependencies per cell line in the screen. The y-axis depicts mutation rate per cell line and the x-axis the number of selective dependencies per cell line. The circles represent individual cell lines with type colored as in panel (c). The blue line represents a linear model fit to this data with Pearson correlation 0.01 with the gray shaded area representing the 95% confidence interval around the fit. c, Number of selective gene dependencies per cell line (on y-axis) grouped by tumor type ordered by median (x-axis). Each circle represents an individual cell line with pediatric tumors colored by type; the black line represents the median number of dependencies per tumor type. d, Pediatric solid and brain tumor cell lines (red, n=82 biologically independent cell lines) did not have a statistically different distribution of selective dependencies (y-axis) as a whole compared to adult solid tumor lines (gray, n=573 biologically independent cell lines) by two-sided Wilcoxon test. Horizontal lines demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) represent the 10 and 90th percentiles respectively. e, Predictive modeling of selective dependencies across all solid and brain tumor cell lines. The y-axis depicts the Pearson correlation of the predictive model for dependency on a gene, and the x-axis shows fraction of cancer cell lines that are dependent on a gene. The size of the points corresponds to the fraction of pediatric cancer cell lines that are dependent on a gene. The red color highlights examples of genes that pediatric cancers are frequently dependent on (MCL1, CDK4), genes with strong predictive models (BRAF, MDM2), or genes with low rates of dependency and poor predictive models (ALK). f, Two-dimensional representation of selective dependencies (removing genes that did not have dependency scores for all cell lines screened) using uniform manifold approximation and projection (UMAP) demonstrates strong clustering of Ewing sarcoma, neuroblastoma and rhabdomyosarcoma by tumor type. Each circle represents a cell line with pediatric tumors colored by type.
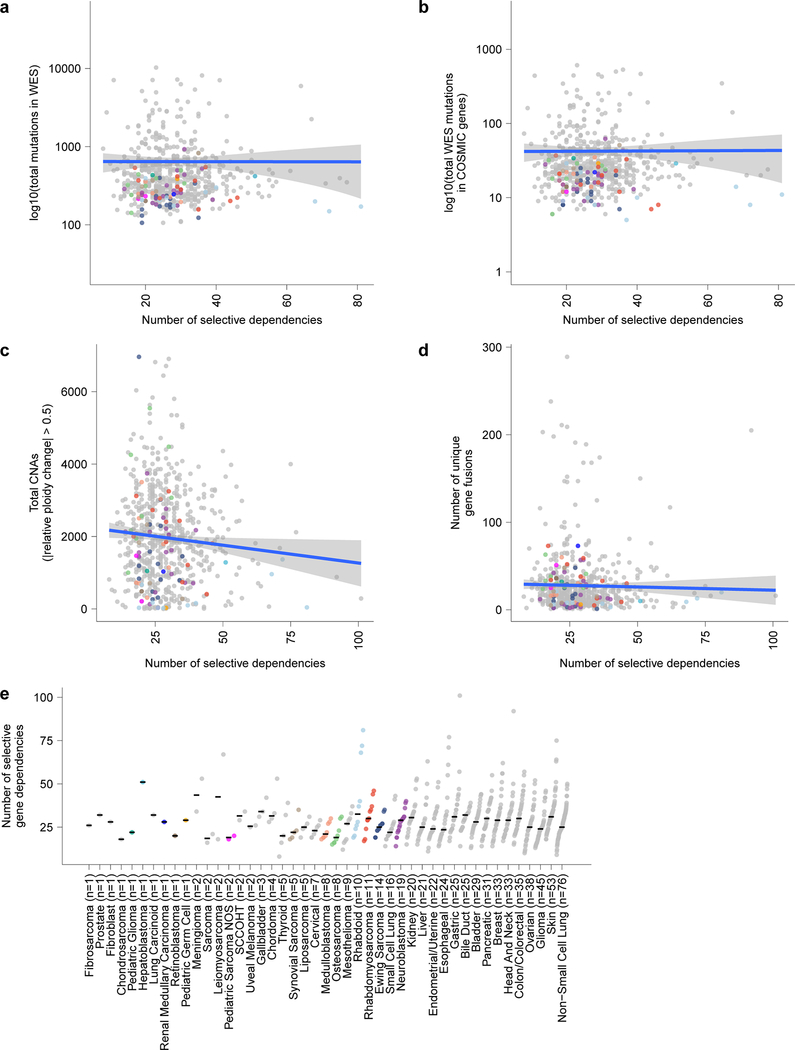
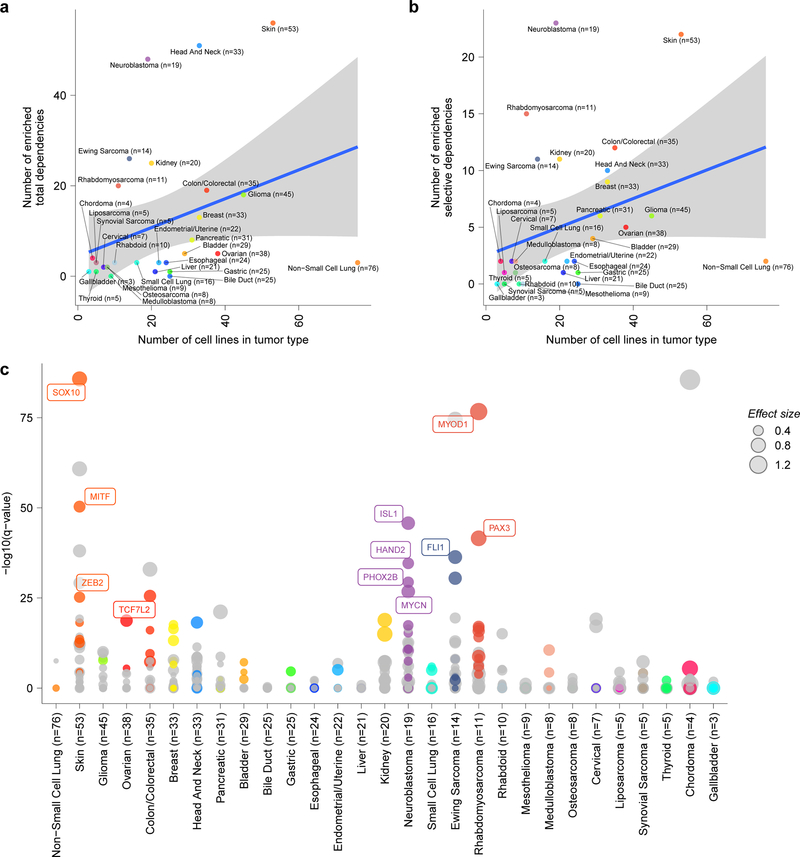
We compared the landscape of pediatric cancer dependencies to those observed in genome-wide screens of 573 adult solid tumor cell lines17,24. Surprisingly, mutational burden is not a predictor of the number of dependencies. Even within adult tumors, this observation was true: there was little correlation between the number of mutations or copy number alterations and number of dependencies (Fig. 2b, Extended Fig. 4a-d). Indeed, the numbers of selective dependencies observed in pediatric cancer cell lines were similar to that observed in adult cancers -- contrary to the expectation that genetically simpler pediatric cancers would have smaller numbers of selective dependencies (Fig. 2c-d, Extended Fig. 4e). Additionally, there was little correlation between measures of screen quality or other confounders and the number of dependencies (Extended Fig. 5a-g).
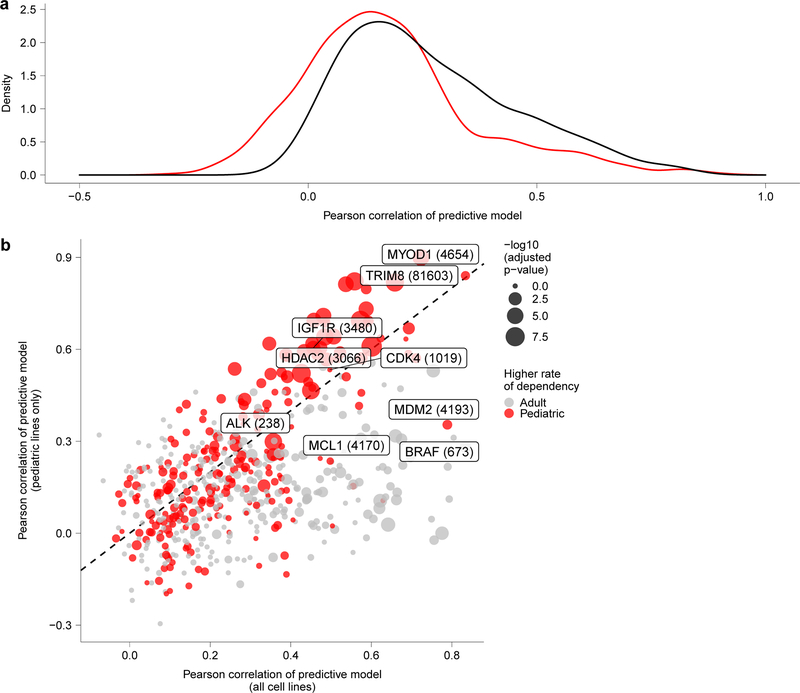
In order to identify potential biomarkers for individual genetic dependencies, we applied machine learning models (random forests) to predict gene effect scores using cell line features, including RNA expression, copy number, mutations, fusions, proteomics, metabolomics, methylation, tumor type, as well as confounders such as screen quality26,27. When examining the gene effect predictions for selective dependencies across all solid and brain tumor cell lines, 38 were found to have a Pearson score for the predictive model >0.6 (Fig. 2e). Repeating this analysis with only pediatric solid and brain tumor cell line features and gene effect scores led to overall decreased performance with 22 of the selective dependencies with Pearson score >0.6 highlighting the utility of combining the pediatric and adult data to increase the power for predicting dependencies spanning both tumor types (Extended Data Fig. 6a-b, Supplementary Data Table 6). In contrast, several pediatric tumor-specific dependencies discussed below had improved predictive modeling Pearson scores when considering pediatric cancer cell lines only (Supplementary Data Table 6).
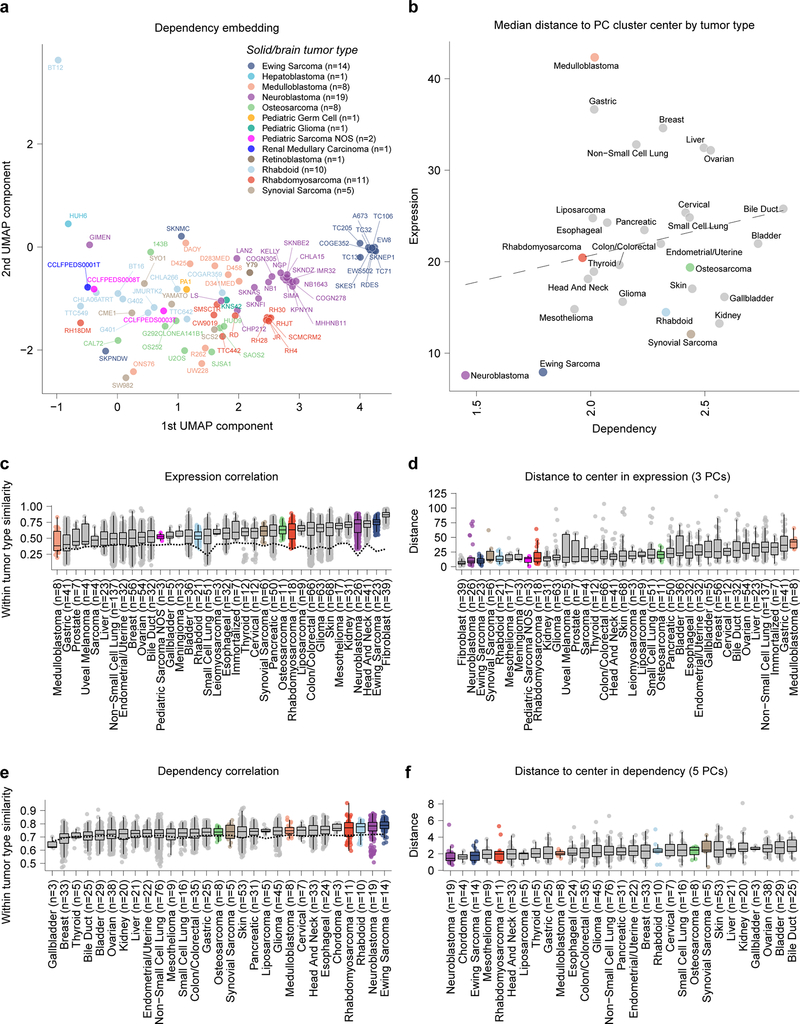
In order to understand how the patterns of dependencies exhibited by different cell lines related to each other, we created a two-dimensional projection of the cell lines’ dependency profiles. This analysis revealed tight clusters of several pediatric tumor types, suggesting that each of these tumors has a distinct set of genetic vulnerabilities enriched within a tumor type (Fig. 2f, Extended Fig. 7a-f). Therefore, we went on to identify the unique dependencies seen in pediatric cancer cell lines.
Identifying unique pediatric tumor dependencies
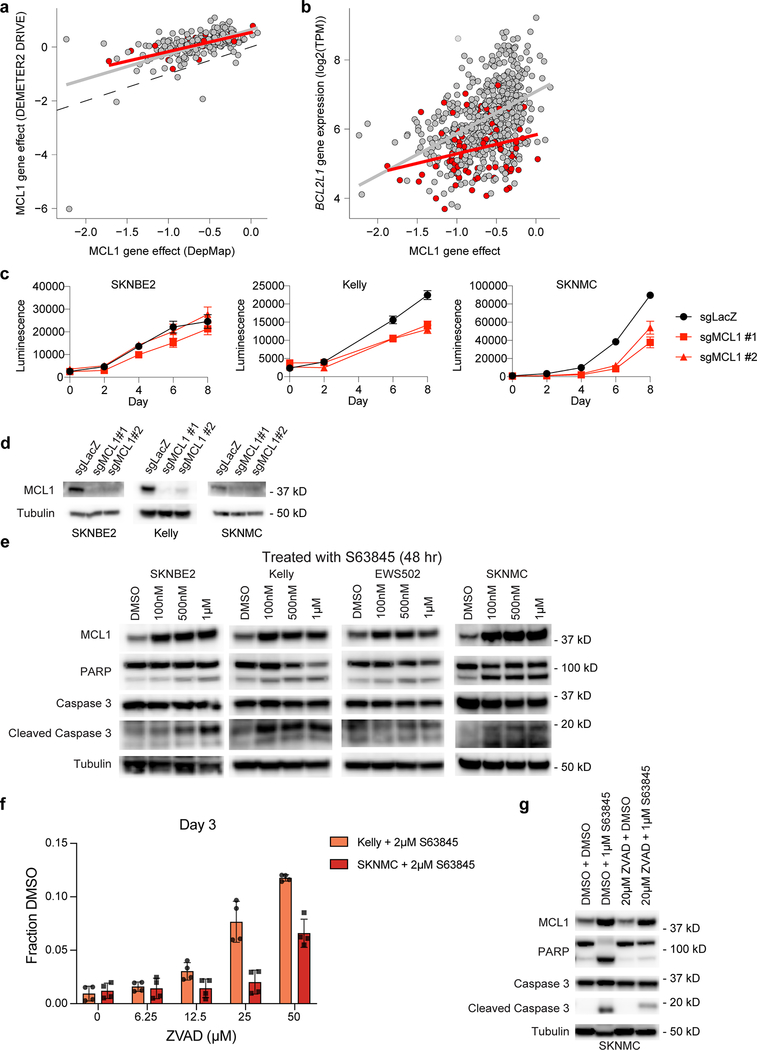
Further examination of the pediatric selective dependencies revealed that 64% were shared between adult and pediatric tumor types (of the 235 selective dependencies present in at least 2% of pediatric cancer cell lines, 151 are selective dependencies in at least 2% of adult cell lines) (Supplementary Data Table 7). For example, as in adult cancer lines, activating mutations of the kinases ALK and BRAF were associated with ALK and BRAF dependency, respectively (Fig. 3a-b), providing further support for the testing of inhibitors of these kinases in pediatric precision medicine trials28. ALK dependency did not have a strong predictive model in the random forest search above due to it being a rare dependency overall; however, BRAF dependency was predicted, as expected, by BRAF hotspot mutations. TP53-wild-type pediatric cancer cell lines were selectively dependent upon MDM2 for survival (Fig. 3c), providing further support for the clinical testing of MDM2 inhibitors in such patients29–31. Indeed, the top predictive feature for MDM2 dependency was RNA expression of EDA2R, a known direct target of p5326, as a surrogate marker for functional p53. Likewise, RB1 loss-of-function mutations were associated with lack of dependence on CDK4 or CDK6 (Fig. 3d). We note that a large proportion of pediatric cancers appear to depend on either CDK4 or CDK6 in a largely mutually exclusive fashion. These findings support the future clinical testing of CDK4/6 inhibitors in pediatric cancers, as has been recently proposed32–35. A key limitation of our screen, however, is the inability to distinguish cytostatic versus cytotoxic guide depletion and thus further studies are required. We also found that a surprisingly large proportion of pediatric cancers were dependent upon the anti-apoptotic protein MCL1 (Fig. 3e) with supporting evidence from orthogonal RNAi and CRISPR-Cas9 screens with alternative approaches and reagents (Fig. 3f, Extended Data Fig. 8a). Our modeling showed that BCL2L1 expression was the most important feature in predicting MCL1 dependency when the pediatric and adult data were combined (Extended Data Fig. 6b, Extended Data Fig. 8b). Follow-up with individual CRISPR-Cas9 MCL1 disruption and the selective MCL1-inhibiting small molecule S63845, with IC50s similar to moderately sensitive lymphoma cell lines36, confirmed this observation (Fig. 3g, Extended Data Fig. 8c-g) recapitulating reported correlations between MCL1 inhibitors and loss-of-function genetic screens37. A number of MCL1 inhibitors have recently entered clinical trials; our findings suggest that additional preclinical testing in pediatric tumors should be considered, including testing in relevant in vivo models. In comparison, signal for BCL2 dependency in pediatric solid and brain cancers was seen mainly in neuroblastoma, supporting the ongoing clinical trials evaluating BCL2 inhibition in neuroblastoma.
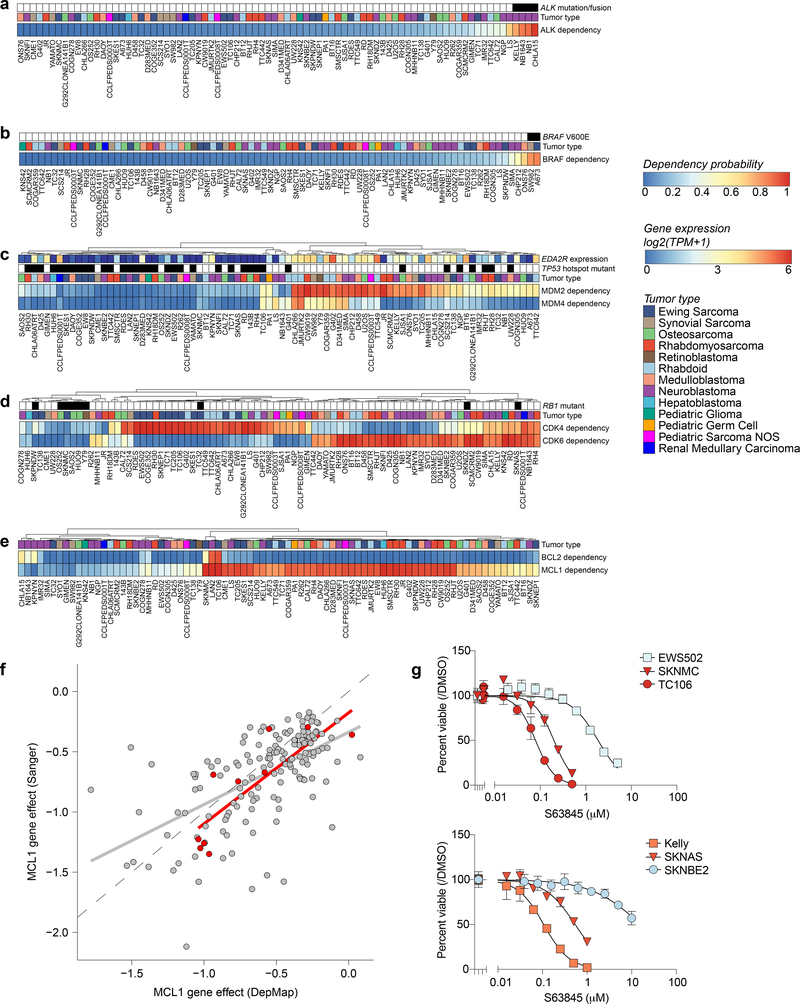
Fig 3. Genetic dependencies and potential therapeutic targeting.
For each genetic dependency, the heatmap indicates the probability of dependency of each cell line with a probability greater than 0.5 considered dependent. When multiple genes are plotted per heatmap, hierarchical clustering was performed. a, Three pediatric solid tumor cell lines demonstrate mutations or fusions in ALK and these cell lines are among the most dependent on ALK. Of note, the neuroblastoma cell line NB1 is also dependent on ALK and harbors an amplification of the gene. b, Two pediatric solid tumor cell lines demonstrate BRAF V600E mutations and these cell lines are BRAF dependent. c, Correlation between MDM2 and/or MDM4 dependency and TP53 hotspot mutations as well as EDA2R expression. d, RB1 mutation status is predictive of CDK4 or CDK6 dependency. Depicted here are all RB1 mutations. TC32 is known to have a heterozygous mutation in RB1 and thus has functional RB1. e, Neuroblastoma cell lines demonstrate dependency on BCL2 while the majority of pediatric solid tumor cell lines are dependent on MCL1. f, Correlation of MCL1 gene effect scores for overlapping cell lines DepMap 20Q1 and Sanger genome-scale CRISPR-Cas9 screen with an independent guide library. Adult cancer cell lines are colored gray while pediatric cancer cell lines are red. The gray and red line represent a linear model fit to the adult or pediatric data. g, Treatment with S63845, a selective MCL1 inhibitor, for four days in Ewing sarcoma (top) and neuroblastoma (bottom) cell lines demonstrates relative sensitivity consistent with dependency scores. The y-axis represents percent viable cells as compared to controls treated with DMSO for each experiment. The x-axis represents concentrations of inhibitor (μM). The data points for each cell line are colored by the probability of dependency on MCL1 with the same colors as the heatmap in (e). One representative experiment is shown for each cell line; each was performed in triplicate (n=3). Data are presented as mean values +/− standard error of mean (SEM).
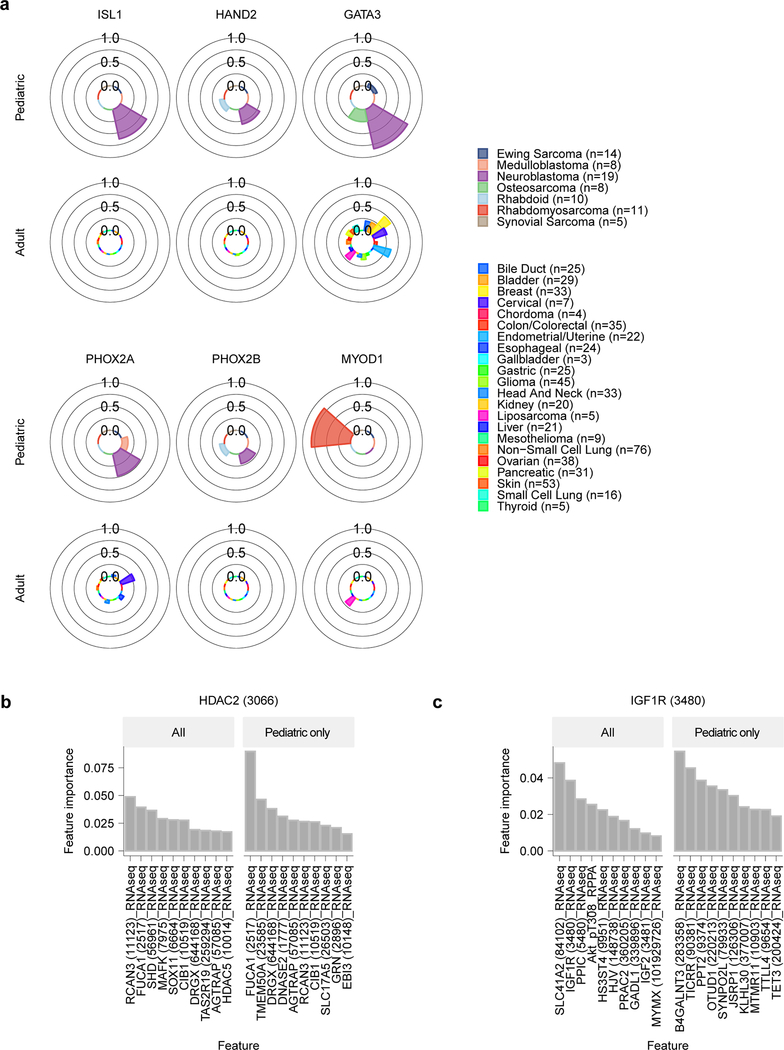
Importantly, genetic dependencies in pediatric cancer cell lines were often distinct from the adult cell lines. Of the 235 selective dependencies seen in at least 2% of pediatric cell lines, 34 (14%) were significantly more common in pediatric cancers compared to 573 adult cancer lines examined (Fig. 4a-b, Supplementary Data Table 7). For example, a potential targetable dependency on the E3 ubiquitin ligase TRIM8 was uniquely associated with Ewing sarcoma tumors (Fig. 4c). Similarly, core regulatory transcription factor dependencies were associated with neuroblastoma (ISL1, HAND2, GATA3, PHOX2A, and PHOX2B) and rhabdomyosarcoma (SOX8, MYOG, and MYOD1) (Extended Data Fig. 9a)38,39. Interestingly, HDAC2 dependency was uniquely seen in pediatric tumor types (Fig. 4c) supported by preclinical data40, and IGF1R dependency was enriched in pediatric lines (Fig. 4c) as would be predicted by the clinical signal seen for IGF1R inhibitors. For example, multiple early phase studies of IGF1R inhibitors have demonstrated that approximately 10% of patients with relapsed Ewing sarcoma respond to these agents as monotherapy41–43. Predictive feature modeling for HDAC2 highlighted RNA expression of its known target FUCA1. This suggests that lower expression of FUCA1 indicates pediatric cell lines with high HDAC activity (Extended Data Fig. 9b). Our predictive modeling for IGF1R dependency did not identify strongly predictive individual features (Extended Data Fig. 9c), reflecting the difficulties in the field in determining significant biomarkers of IGF1R inhibitor response44.
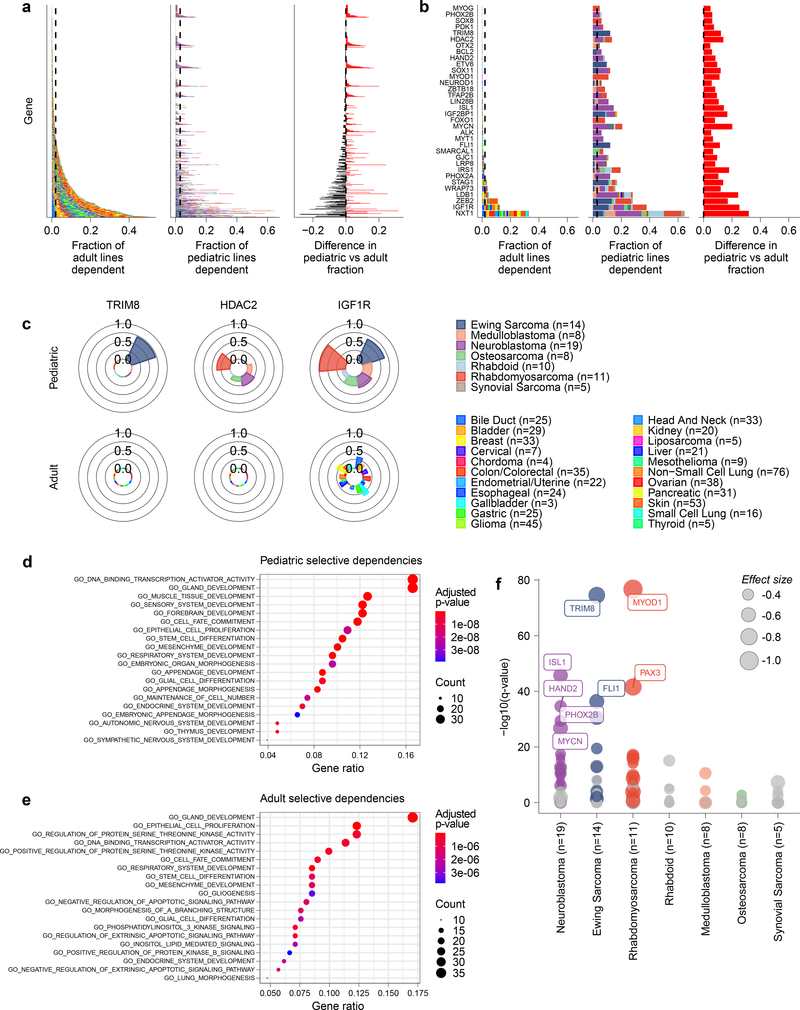
Fig 4. Selective dependencies in pediatric and adult solid tumor lines.
a, Selective dependency genes demonstrate subsets that are more common in pediatric compared to adult cancer cell lines. Each row on the y-axis represents one of the selective dependencies (removing common essential and non-essential genes) ordered across the three subpanels by rate of dependency seen in adult cancer cell lines. The left subpanel shows the rate at which adult cell lines are dependent (x-axis) and the center subpanel shows the rate at which pediatric cancer cell lines are dependent (x-axis). The right subpanel demonstrates the difference in rate of dependency in pediatric versus adult cancer cell lines (x-axis) with dependencies seen at greater frequency in pediatric cell lines colored red and those seen more frequently in adult cell lines as black. The bars in the center and right panels are colored by the contribution of each tumor type as shown in the legend. b, Thirty-four selective dependencies were significantly more common in the pediatric cell lines compared to adult cell lines (adjusted p-value <0.05 by two-sided Fisher’s exact test with Benjamini-Hochberg correction). The subpanels are arranged and colored as in panel (a). Notably, several selective dependencies were not seen in adult solid tumor cell lines and were unique to pediatric solid tumors. c, The frequency of dependency on TRIM8, HDAC2 and IGF1R are depicted in pediatric and adult solid tumor types with at least 3 cell lines screened per type in polar bar graphs. The heights of the bars correspond to the fraction of cell lines of a particular tumor type that are dependent on the gene. The tumor types are colored as in the legend. TRIM8 dependency was seen uniquely in Ewing sarcoma and no other tumor types screened. HDAC2 dependency was seen only in pediatric cell lines but across several tumor types in contrast to none of the adult solid tumor lines. IGF1R dependency was seen across adult and pediatric solid tumors but with greater frequency in pediatric tumors. d, Gene set enrichment analysis (GSEA) of selective dependencies present in >2% of pediatric cell lines using the gene ontology C5 collection from MSigDB identifies enrichment of developmental pathways. On the y-axis are the 20 gene sets with the highest overlap with the query set, plotted on the x-axis. e, GSEA of selective dependencies present in >2% of adult cancer cell lines using the C5 collection demonstrates enrichment of several signaling pathways. On the y-axis are the 20 gene sets with the highest overlap with the query set, plotted on the x-axis. f, Tumor type-enriched dependencies in pediatric tumor types with more than 2 cell lines. Plotted on the y-axis is -log10 of the q-value of enrichment as calculated by performing a two-class comparison between gene effect scores in each tumor type compared to all other cell lines screened using two-sided t-tests with Benjamini-Hochberg correction. Pediatric tumor types are plotted along the x-axis. The size of the circles reflects the mean difference in dependency score between the tumor type and all other cell lines screened. Gray circles are enriched dependencies in a tumor type that are not classified as selective and colored circles are selective dependencies in the screen.
Next, we sought to evaluate if the pediatric dependencies were enriched for specific pathways or functions. We performed gene set enrichment analyses (GSEA) of the 235 or 214 selective dependencies seen in at least 2% of pediatric or adult cell lines, respectively. Using the gene ontology collection (C5) from the Molecular Signatures Database 7.1 (MSigDB) 45, we identified that pediatric selective genetic vulnerabilities were enriched for several developmental gene sets as well as the DNA-binding transcription activator set (Fig. 4d). In contrast, adult dependencies were enriched more strongly for the epithelial cell proliferation gene set as well as several signaling pathways (Fig. 4e). These findings highlight the unique nature of pediatric solid and brain tumors as arising from the dysregulation of normal development compared to the epithelial origin and multiple mutational hits of adult tumors46.
Several selective dependencies were also identified as enriched in particular pediatric tumor types (Fig. 4f, Supplementary Data Table 7), with the caveat that several pediatric tumor types and well-defined subtypes, for example in medulloblastoma, do not yet have sufficient representation for such an analysis. However, we note that the ability to identify tumor type-enriched dependencies is a function of the number of available models representing each tumor type but lacking a clear saturation effect (Extended Data Fig. 10a-b). Therefore, a future, larger scale Pediatric Cancer Dependency Map is needed to identify additional high confidence pediatric-restricted dependencies. Moreover, tumor types with specific lineage or oncogenic transcription factor dependencies, for example neuroblastoma, rhabdomyosarcoma, Ewing sarcoma or skin cancer, appear to often have a large number of tumor-type specific dependencies possibly driven by these proteins (Extended Data 10c). A caveat of our data, however, is that it is difficult to ascertain which of these dependencies are truly cancer-specific versus lineage-specific as “normal” cells cannot be propagated in vitro sufficiently to be screened without transformation or adaptation such that the cells are not truly normal. Of note, we have excluded pediatric leukemias and lymphomas from our first-generation Pediatric Cancer Dependency Map analysis. We focused on solid tumors, including brain tumors, given the relative lack of progress in treating many of these high-risk subsets of childhood cancer. A future direction will be to expand the representation of childhood leukemias in the second-generation Pediatric Cancer Dependency Map as well as less well represented brain tumors and rare pediatric solid tumors.
Discussion
In summary, we describe here a first-generation Pediatric Cancer Dependency Map that will serve as a community resource for those studying the pathogenesis of childhood cancers and those searching for new therapeutic strategies for these diseases. Using early data from this map, vulnerabilities have been deeply characterized and validated with in vivo models in several pediatric tumor types, for example EZH2 dependency in neuroblastoma47, MDM2/4 dependency in Ewing sarcoma31 and rhabdoid tumors30, receptor tyrosine kinase dependencies in rhabdoid tumors48 and proteasome dependency in SMARCB1 deficient cancers49, highlighting the potential impact of these efforts. Raw data and data visualization tools are available at the Cancer Dependency Map Portal (depmap.org and depmap.org/peddep).
Importantly, the Pediatric Cancer Dependency Map allowed us to answer two key questions. First, do the simpler genomes of childhood cancers translate into a simpler landscape of genetic vulnerabilities? The answer here is, clearly, no. This result is significant because it indicates that a broader spectrum of therapeutic targets for pediatric cancers exists than had previously been suspected. Second, will drugs being developed against adult cancer vulnerabilities be sufficient to address pediatric cancers? Again, the answer is, clearly, no. While there are examples of dependencies that span all cancer cell lines, there indeed are new opportunities to target pediatric tumors beyond the familiar approaches in adult cancers. A substantial number of pediatric dependencies are unique to these tumors, mirroring the finding that the majority of driver genes identified in pan-pediatric tumor studies are unique to pediatric cancer11.
This finding has important societal implications because the small commercial market for pediatric cancer-restricted drugs results in limited industry investment in such diseases. The dependency landscape of childhood cancer described here highlights the need for new efforts to ensure the future development of therapeutics for children suffering from cancer.
Data availability
CRISPR-Cas9 screening results for DepMap version 20Q1 (including raw data) and the genomic characterization of cancer cell lines (whole-exome sequencing and RNA sequencing) used in this study are publicly available at https://depmap.org and also on figshare (https://figshare.com/articles/dataset/DepMap_20Q1_Public/11791698). Subsets of the raw sequencing data from whole exome sequencing and RNA sequencing used in this study are available at Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) and European Genome-phenome Archive (EGA, https://www.ebi.ac.uk/ega/) accession numbers: SRA PRJNA523380 (CCLE), SRA PRJNA261990 (Ewing sarcoma), and EGAS00001000978 (Sanger) (Supplementary Data Table 1). The remainder of the raw sequencing data is in the process of being deposited in SRA via dbGaP (https://dbgap.ncbi.nlm.nih.gov/), delayed in part as these are legacy cell lines. In the interim, we will work with specific requests to expedite the process (contact depmap@broadinstitute.org). Additionally, the pediatric-specific subsets of the processed DepMap version 20Q1 data presented in this study (dependency, mutations, copy number, expression, fusions) are available at our companion website at https://depmap.org/peddep.
Code availability
Code to complete the analyses presented in this manuscript and generate corresponding figure panels and tables is publicly available on GitHub at https://github.com/ndharia-broad/peddep.
Online Methods
Cell lines
The cell lines used for the genome-scale CRISPR-Cas9 screen were collected and validated as previously described17 with details available at depmap.org. All cell lines were short tandem repeat (STR) tested for identity and validated to be free of Mycoplasma species.
Classification of tumor cell lines
In order to limit the present study to solid and brain tumors, we performed the following for each of the cell line datasets: RNA-sequencing, whole exome sequencing, mutation calls, copy number calls, and genome-scale CRISPR-Cas9 screening results. The sample information file available for the DepMap 20Q1 dataset was used (available at depmap.org and figshare60) and contains annotations for 1,775 cell lines in total. The source and fingerprinting of the Dependency Map cell lines was as previously described17,19.
In order to concentrate on solid and brain tumor cell lines, we removed all cell lines from hematopoietic and lymphoid tissue malignancies by removing all lines that were annotated as such in their CCLE names or cancer type classification. We designated pediatric cell lines as those that represented pediatric tumor types, regardless of the age of the patient from whom the cell line was derived. These pediatric tumor types included Ewing sarcoma, hepatoblastoma, medulloblastoma, neuroblastoma, osteosarcoma, retinoblastoma, rhabdoid, rhabdomyosarcoma, synovial sarcoma and Wilms tumor. In addition, we included cell lines as pediatric for tumors that occur commonly in children as well as adults (brain and germ cell tumors) which were derived from patients less than or equal to 21 years of age. Other tumors were considered adult cancers, including those that represent common adult solid tumors but were derived from patients less than 21 years of age. For example, HEPG2 was considered an adult cancer cell line as it represents hepatocellular carcinoma even though it was initially isolated from a child. Similarly, melanoma cell lines from patients less than 21 years of age were considered adult for the purposes of this study. Of note, the cell line CHLA57 was censored from all of the analyses presented here as this line is annotated as Ewing sarcoma but does not express the hallmark EWS-ETS fusion or cluster with Ewing cell line or tumor expression. A portion of this data processing was performed using Microsoft Excel version 16. The classification of each cell line is indicated in Supplementary Data Table 1.
A literature search was performed for each of the cell lines classified as pediatric to determine if the sample was obtained prior to a patient receiving anti-tumor therapy (Supplementary Data Table 1). The reported doubling times of selected cell lines are also reported in Supplementary Data Table 1.
Cancer cell line genomics and transcriptomics
Whole exome sequencing (WES) for mutations and copy number, RNA-sequencing (RNA-seq) and fusion calling for pediatric cell lines was performed as previously described19. These data are available in the DepMap 20Q1 dataset (available at depmap.org and figshare60). Briefly, we used a modified version of the Getz Lab CGA WES Characterization pipeline (https://github.com/broadinstitute/CGA_Production_Analysis_Pipeline) developed at the Broad Institute to call, filter and annotate somatic mutations and copy number variation from WES. The pipeline employs the following tools: MuTect61, ContEst62, Strelka63, Orientation Bias Filter64, DeTiN65, AllelicCapSeg66, MAFPoNFilter67, RealignmentFilter, ABSOLUTE68, GATK69, PicardTools, Variant Effect Predictor70, Oncotator71. Copy number variants were detected in WES data using the GATK4 copy number pipeline (https://github.com/broadinstitute/gatk/)72. RNA-seq data is aligned to hg38 and expression TPM data is produced using the GTEx pipeline (https://github.com/broadinstitute/gtex-pipeline/)73. Fusion calls are produced with STAR-Fusion (https://github.com/STAR-Fusion/STAR-Fusion/)74.
Tumor to cell line expression mapping
Celligner18 combines RNA-seq gene expression datasets from primary tumor samples and cell lines to perform a joint dimensionality reduction analysis in two stages. For the analyses presented here, we used expression values from 1,249 cell lines from the DepMap 20Q1 dataset (available at depmap.org and figshare60). We used primary tumor expression values from 1,646 pediatric tumor samples from Treehouse, 821 pediatric tumor samples from TARGET, and 9,806 TCGA tumor samples20. Briefly, in the first stage contrastive principal component analysis was used to identify gene expression signatures that had increased variance in the tumor samples compared to the cell lines which represented tumor-specific signatures. The 4 top tumor-specific gene expression signatures were removed from both tumor and cell line datasets. Next in the second stage, mutual nearest neighbors batch effect correction was used to remove systematic differences between tumor and cell line expression data which was agnostic of tumor type. After correction, a two-dimensional representation of the data was produced using uniform manifold approximation and projection on the first 70 principal components using Euclidean distance, an “n.neighbors” parameter of 10 and a “min.dist” parameter of 0.5 with the Seurat version 3 R package.
To evaluate the similarity of cell lines to tumor samples we took the Pearson correlation distance between each cell line and tumor in the gene expression data, using a set of 19,188 protein-coding genes. We calculated this using both the uncorrected tumor and cell line expression data and the Celligner-aligned data. Using each data type, we classified each cell lines’ tumor type by identifying the most frequently occurring cancer type within each cell line’s 25 highest correlated tumor neighbors. To evaluate the agreement between the classifications and the annotated cancer type of the cell lines we only considered cell lines (n = 1,169) where the annotated type was also present in the tumor samples. To assess the confidence of these classifications we calculated the proportion of tumor samples within the 25 nearest neighbors that came from the most frequent cancer type.
Additionally, we classified cell lines’ tumor type using a random forest model, implemented using the R package ‘ranger’75, trained on tumor gene expression and applied to cell line gene expression, to get tumor type classifications for cell lines. The model was trained on a set of 12,301 tumor samples, across 39 cancer types (we only included cancer types with at least 5 tumor samples), and used a subset of 5,000 genes that were identified as high variance within the cell line or tumor data (Supplementary Data Table 8). This model was then applied to the 1,249 cell line samples, with cell lines classified as the tumor type with the maximum probability output by the model. To calculate the accuracy of the classifications we compared the classifications output by the model to the annotated cancer type of the cell line, using only cell lines (n = 1,132) for which the annotated cancer type of the cell line was included in the possible outputs of the model. To assess the confidence of these classifications we used the probabilities output by the random forest model.
Mutation burden and copy number analysis
Mutational burden in cancer cell lines was calculated to test the hypotheses that mutation burden would be lower in pediatric cancer cell lines compared to adult cancer cell lines. Mutation annotation format (MAF) data from the DepMap 20Q1 dataset was used (available at depmap.org and figshare60) and contains mutation calls for 18,802 genes in 1,697 cell lines called from whole exome sequencing, whole genome sequencing, targeted sequencing, and RNA-sequencing with filtering of likely germline variants19. These data were filtered as above to only include pediatric and adult solid and brain tumor cell lines of interest in this study. It should be noted that established cancer cell lines do not have paired normal samples to properly filter germline variants from somatic mutations. Therefore, we used multiple methods to assess mutation burden as follows.
MutSig2CV version 3.1167,76 (https://software.broadinstitute.org/cancer/cga) was installed along with MATLAB runtime environment R2013a. In order to run MutSig2CV to calculate mutational burden of cancer cell lines, we first filtered the MAF data to only include mutations called by whole exome sequencing performed at the Broad Institute or Wellcome Trust Sanger Institute by using the columns labeled “CGA_WES_AC” or “SangerRecalibWES_AC”. MutSig2CV was executed on each of these datasets separately with separate runs for Broad and Sanger data. The mutation rates per cell line from each run of MutSig2CV were combined by taking all cell line mutation rates from Broad whole exome sequencing and adding mutation rates for any cell lines not in the Broad dataset that were in the Sanger dataset. These rates were reported as MutSig2CV mutations per megabase per cell line.
Additionally, mutation rates were calculated by enumerating the mutations detected in either Broad or Sanger whole exome sequencing of cell lines. First, the DepMap 20Q1 MAF file was filtered to include mutations detected in either dataset by using the columns labeled “CGA_WES_AC” and “SangerRecalibWES_AC”. Next, the number of mutations was calculated for each cell line in DepMap. This was reported as total mutations in whole exome sequencing. These mutation counts were further filtered to only include mutations that were missense, predicted to be damaging, or occurred in TCGA or COSMIC hotspots within cancer-associated genes from COSMIC. The list of COSMIC genes was downloaded from https://cancer.sanger.ac.uk/cosmic/census on 1/11/2020 selecting ‘both tiers’. The following fields were used from the MAF file to perform this filtering: “isDeleterious”, “Variant_Classification”, “isCOSMIChotspot”, and “isTCGAhotspot”. These mutation rates were reported as hotspot/missense/damaging WES mutations in COSMIC genes.
The number of copy number alterations (CNAs) per cancer cell line were calculated by using the DepMap 20Q1 gene copy number data (available at depmap.org and figshare60 for 27,639 genes in 1,713 cell lines). These data were filtered as above to only include pediatric and adult solid and brain tumor cell lines of interest in this study. For each cell line with copy number data, the number of genes that were amplified (as indicated by a gene copy number >/= 1.32 which corresponds to a relative ploidy of 1.5, i.e. 3 copies of a gene in a diploid cell) or deleted (as indicated by a gene copy number </= 0.585 which corresponds to a relative ploidy of 0.5, i.e. 1 copy of a gene in a diploid cell). In order to plot the copy number across the chromosomes of each individual pediatric cell line, the DepMap 20Q1 segment level copy number data were used (available at depmap.org and figshare60).
Mutation and copy number rates from all of the above methods were compared across pediatric and adult solid and brain tumor cell lines, as well as fibroblast cell lines, with two-sided Wilcoxon tests.
Genome-scale CRISPR-Cas9 screen
Genome-scale CRISPR-Cas9 screening was conducted across human cancer cell lines with gene effect scores and gene dependency probabilities calculated as previously described17,27. For this study, DepMap 20Q1 dependency data were used (available at depmap.org and figshare60 for 18,333 genes in 739 cell lines). These data were filtered to only include pediatric and adult solid and brain tumors of interest as indicated above resulting in data for 82 pediatric cancer cell lines and 573 adult cancer cell lines. Data from the Sanger genome-scale CRISPR-Cas9 screen9 and Novartis DRIVE RNAi screen25 were used as processed by CERES17 and DEMETER277, respectively, in DepMap 20Q1.
False positive rates for individual cell lines were estimated by the rate at which non-expressed genes (TPM=0) were called dependencies (probability of dependency > 0.5) per cell line. The false positive rates for the entire screen were obtained by averaging across all cell lines.
Selective gene dependencies
With the dependency data filtered as above to include data for 656 solid or brain tumor cell lines across 18,333 genes, the normality likelihood ratio test (normLRT) was calculated to identify genetic dependencies that have skewed distributions across the cell lines screened25. The log likelihood ratio of fitting to a skewed distribution was calculated using the selm function implemented in the sn version 1.6–1 R package for the dependency scores of each gene with the skew-t parametric family of skew-elliptically contoured distribution for the error term. The log likelihood ratio of fitting to a normal distribution was calculated using the fitdistr function implemented in the MASS version 7.3–51.5 R package for the dependency scores of each gene. The normLRT score is twice the difference of the log of the likelihood ratio of fitting to a skewed distribution and the log of the likelihood ratio of fitting to a normal distribution. Selective gene dependencies were defined as those with normLRT score greater than or equal to 100, left-sided skew as indicated by mean gene effect score less than the median gene effect score, and not defined as common essential or non-essential genes in the CRISPR screen. The common essential genes in the solid and brain tumors subset of DepMap 20Q1 used in this manuscript were identified by those genes where 90% of cell lines rank the gene above a cutoff determined from the central minimum in the histogram of gene ranks in their 90th percentile least dependent line. Non-essential genes were identified by those that did not have probability of dependency greater than 0.5 in any cell lines screened.
Predictive feature modeling
A matrix of molecular and cell line annotation features was assembled from the DepMap 20Q1 dataset60. Continuous features (RNAseq, relative copy number, RPPA, total proteomics, metabolomics, RRBS) were individually z-scored per feature and joined with one-hot encodings of categorical features (damaging mutation, missense mutation, hotspot mutation, fusion, cell line tissue/disease type). Cell lines without RNAseq data were dropped and any remaining missing values were assigned a zero. Confounder variables were also included to represent technical aspects of the CRISPR-Cas9 screens (SSMD, NNMD, Cas9 activity, media type, and culture type).
The CERES gene effect for each perturbation in the CRISPR dataset is modeled using two sets of features. The first is the related model where features are only selected if there is a prior known relationship between the perturbation target and the measured molecular feature suggested by PPI, CORUM, or paralogs based on DNA sequence similarity (exception of confounders and tissue/disease annotations that are always included). The second model is the unbiased model where all features are included, but filtered by Pearson correlation to use the top 1,000.
Random forest regression models (100 trees, max-depth of 8, and a minimum of 5 cell lines per leaf) from the Python scikit-learn package were trained using stratified 5-fold cross-validation. Once predictions were made for each held-out set, the correlation between predicted and observed CERES gene effects was used as the accuracy per model. To get a final score per gene, we took the maximum of the accuracies for the related and unbiased models.
Dependency clustering
Clustering on genetic dependencies was performed by first performing principal component analysis on the dependency gene effect scores for the selective dependencies. As principal component analysis implemented in the prcomp function of the stats version 3.6.2 R package does not handle NAs, selective dependencies that contained NA values for any of the 612 cell lines analyzed in this manuscript were removed prior to principal component analysis. Subsequently, uMAP was performed on the first 50 principal components with default parameters using the umap function of the umap version 0.2.5.0 R package to produce a two-dimensional representation of dependency data.
Homogeneity in gene expression and dependencies by tumor type
The pairwise Pearson correlations were calculated for all solid and brain tumor types that had at least 3 cell lines with data across the 2,000 most variable genes in expression as evaluated by the standard deviation of expression. The same was done across all solid and brain tumor types with at least 3 cell lines with data across the 500 most variable dependencies as evaluated by the standard deviation of gene effect score. For each tumor type, the median Pearson correlation was calculated between cell lines within that tumor type and compared to the median Pearson correlation between cell lines of the tumor type compared to other cell lines screened.
For all solid and brain tumor types with at least 3 cell lines with expression data, principal component analysis was performed (prcomp function of the stats version 3.6.2 R package) on the 2000 most variable genes in expression as evaluated by the standard deviation of expression. The top 3 principal components captured 33.8% of the variance with the next components capturing <3.5% of the variance. The center of each tumor type expression cluster was calculated as the median of each of the top 3 principal components for cell lines of that tumor type. Then the average distance of each cell line to the median for its tumor type across the top 3 principal components was calculated. Similarly, for all solid and brain tumor types with at least 3 cell lines with dependency data, principal component analysis was performed (prcomp function of the stats version 3.6.2 R package) on the 500 most variable dependencies as evaluated by the standard deviation of gene effect score. The top 5 principal components captured 20.5% of the variance with the next components capturing <2.5% of the variance. The center of each tumor type dependency cluster was calculated as the median of each of the top 5 principal components for cell lines of that tumor type. Then the average distance of each cell line to the median for its tumor type across the top 5 principal components was calculated.
Dependencies and drug targets
Cell lines with ALK mutations or fusions were identified by filtering DepMap 20Q1 MAF data mentioned above for COSMIC hotspot mutations in ALK. Additionally, ALK fusions were identified by filtering DepMap 20Q1 fusion data for fusions that contained ALK. Cell lines with BRAF V600E mutations were identified by filtering the DepMap 20Q1 MAF data for this particular mutation. Lines with TP53 mutations were identified by filtering all TP53 hotspot mutations. Cell lines with RB1 mutations were likewise identified by filtering all RB1 mutations except silent mutations, including cell lines without complete RB1 loss like TC32, which has a heterozygous mutation in RB1. When more than one genetic dependency was considered, hierarchical clustering was performed on dependency scores and heatmaps were generated using the pheatmap function in the pheatmap version 1.0.12 R package.
Comparing pediatric and adult selective dependencies
Selective dependencies were identified as above to include 573 genetic dependencies. The rate of dependency for pediatric or adult solid and brain tumor cell lines was calculated as the percent of cell lines in either category that had probability of dependency greater than or equal to 0.5. For each selective dependency, a two-sided Fisher’s exact test with Benjamini-Hochberg correction was performed. Genetic dependencies with p-value of less than 0.05 and a higher rate of dependency in pediatric cell lines compared to adult cell lines were identified. Gene set enrichment analyses (GSEA) were performed using the enricher function implemented in the clusterProfiler version 3.14.3 R package using the NCBI Entrez GeneID78 from the C5 gene sets version 7.1 downloaded from MSigDB45.
Dependency enrichment analysis
For each solid or brain tumor type in the screen with at least two cell lines screened, a two-class comparison was performed between the gene effect scores for cell lines of each tumor type (in-group) and the remainder of all other cell lines in the screen (out-group). The two-class comparison was performed using the lmFit and eBayes functions implemented in the limma version 3.42.2 R package. Briefly, lmFit was used to fit a linear model to the gene effect scores divided in the in-group and out-group. Then, eBayes was used to compute t-statistics and log-odds ratios of differential gene effect. Effect size was calculated as difference in the mean gene effect dependency score in the in-group compared to the out-group. In addition to two-sided p-values, one-sided “left” p-values were calculated to identify gene dependency effects that were more negative (more dependent) in the in-group compared to the out-group, and one-sided “right” p-values were calculated to identify those that were less dependent in the in-group compared to the out-group. All p-values were corrected for multiple hypothesis testing using the Benjamini-Hochberg correction and these adjusted p-values were reported as q-values. Enriched genetic dependencies were identified in each tumor type as those with q-value less than 0.05 with a negative effect size (mean of dependency gene effect score more negative in in-group than out-group).
Figure creation
Figure panels relating to DepMap data were created using RStudio version 1.2.5033 with R version 3.6.2 (2019–12-12). Data from validation of MCL1 dependency were plotted with GraphPad Prism version 8. All manuscript figures were compiled using Adobe Illustrator version 24.
Extended Data
Extended Data Fig 1. Pediatric solid tumor cancer models represent primary tumors.
Two-dimensional representation of RNA-sequencing data using uniform manifold approximation and projection (UMAP) following alignment by Celligner for all primary tumors (triangles) and cancer cell lines (circles) with each cancer type separated for clarity. Cell line names are labelled.
Extended Data Fig 2. Pediatric solid tumor cancer models represent high-risk disease.
a, Two-dimensional representation of RNA-sequencing data using uniform manifold approximation and projection (UMAP) after alignment by Celligner for primary tumors (triangles) and cancer cell lines (circles). Cell lines and primary tumors that were classified as belonging to the undifferentiated cluster are outlined by a black border. b, Two-dimensional representation of RNA-sequencing data using UMAP prior to alignment by Celligner for primary tumors (triangles) and cancer cell lines (circles). c, The total count of mutations in whole exome sequencing (WES) (y-axis) grouped by solid tumor type (x-axis) with diseases ordered by median burden. d, Number of mutations in WES (y-axis) of pediatric solid tumor cell lines (red, n=166 biologically independent cell lines) compared to adult solid tumor (gray, n=1099 biologically independent cell lines) (p<2.22e−16 by two-sided Wilcoxon test) and fibroblast cell lines (black, n=28 biologically independent cell lines) (p=1.8e−13). e, The count of mutations in WES filtered to only include hotspot, missense or damaging mutations in COSMIC genes (y-axis) grouped by solid tumor type (x-axis) with diseases ordered by median burden. Each circle in panels (c, e) represents an individual cell line with pediatric tumors colored by type; the black line represents the median mutation burden per tumor type. f, Mutations in WES filtered to only include hotspot, missense or damaging mutations in COSMIC genes (y-axis) of pediatric solid tumor cell lines (red, n=166 biologically independent cell lines) compared to adult solid tumor (gray, n=1099 biologically independent cell lines) (p<2.22e−16 by two-sided Wilcoxon test) and fibroblast cell lines (black, n=28 biologically independent cell lines) (p=3.5e−11). Horizontal lines in panels (d, f) demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) in panels (d, f) represent the 10 and 90th percentiles respectively.
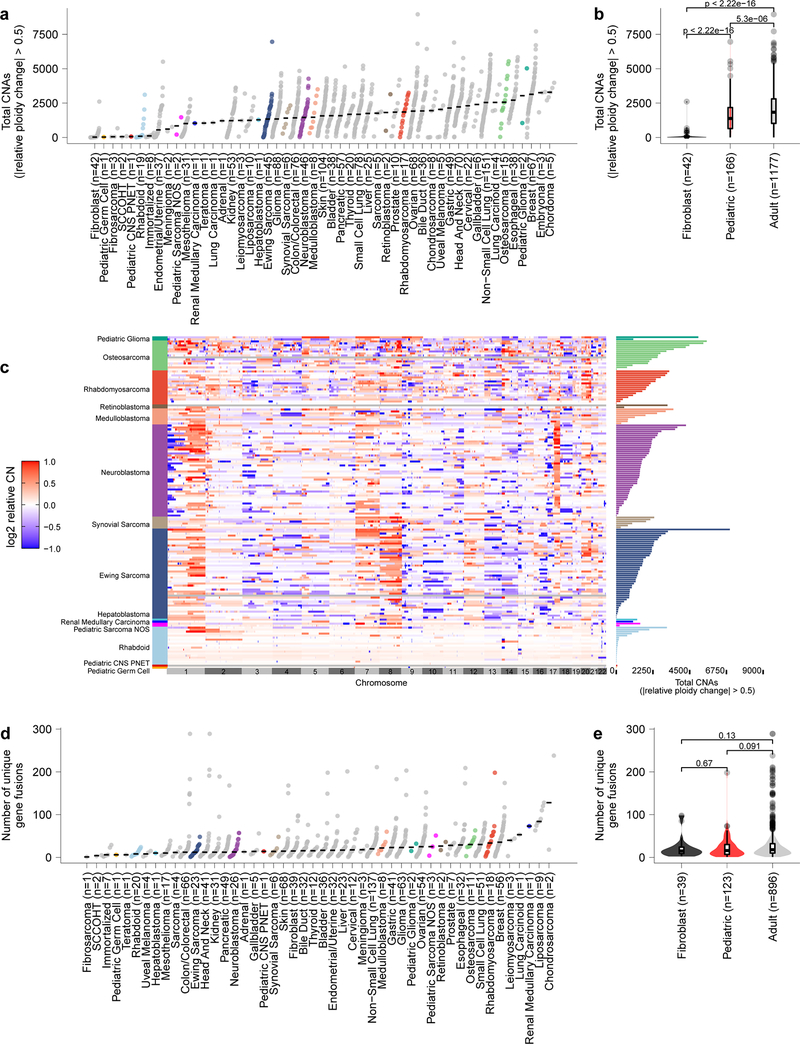
Extended Data Fig. 3. Pediatric solid tumors have fewer total copy number events and gene fusions than adult tumor cell lines with expected profiles for disease subtypes.
a, Total number of genes with copy number alterations (CNA) as identified by genes that had a relative change in ploidy of 0.5 is plotted on the y-axis with tumor types along the x-axis. Each circle represents an individual cell line with pediatric tumors colored by type; the black line represents the median number of CNAs per tumor type. Of note, rhabdoid tumors have very few CNAs, consistent with primary patient tumors. b, CNAs (y-axis) in pediatric solid tumor cell lines (red, n=166 biologically independent cell lines) compared to adult solid tumor (gray, n=1177 biologically independent cell lines) (p=5.3e−06 by two-sided Wilcoxon test) and fibroblast cell lines (black, n=42 biologically independent cell lines) (p<2.22e−16). c, Copy number heatmap across the genome for pediatric cancer cell lines demonstrates multiple CNAs in osteosarcoma as expected with few events in rhabdoid tumors. d, Total number of genes fusions per cell line from RNA sequencing is plotted on the y-axis with tumor types along the x-axis. Each circle represents an individual cell line with pediatric tumors colored by type; the black line represents the median number of gene fusions per tumor type. Of note, osteosarcoma cell lines have high numbers of gene fusions, consistent with primary patient tumors. e, Gene fusion calls from RNA sequencing (y-axis) in pediatric solid tumor cell lines (red, n=123 biologically independent cell lines) compared to adult solid and brain tumor (gray, n=896 biologically independent cell lines) and fibroblast cell lines (black, n=39 biologically independent cell lines) by two-sided Wilcoxon test. Horizontal lines in panels (b, e) demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) in panels (b, e) represent the 10 and 90th percentiles respectively.
Extended Data Fig. 4. Selective dependencies in pediatric cell lines and the relationship to mutation burden.
a, Mutational burden count of mutations in whole exome sequencing (WES) (y-axis) compared to the number of selective dependencies per cell line (x-axis) in the screen. b, Mutational burden count of mutations in WES filtered to only include hotspot, missense or damaging mutations in COSMIC genes (y-axis) compared to the number of selective dependencies per cell line (x-axis) in the screen. c, Total number of genes with copy number alterations (CNA) (y-axis) compared to the number of selective dependencies per cell line (x-axis) in the screen. d, Total number of unique gene fusions (y-axis) compared to the number of selective dependencies per cell line (x-axis) in the screen. The circles in panels (a-d) represent individual cell lines with tumor types colored as in panel (e). The blue lines in panels (a-d) represent a linear model fit to this data with the gray shaded area representing the 95% confidence interval around the fit. e, Number of selective dependencies per cell line (y-axis) grouped by tumor type ordered by number of cell lines (x-axis). Each circle represents an individual cell line with pediatric tumors colored by type; the black line represents the median number of selective dependencies per tumor type.
Extended Data Fig. 5. Selective dependencies in pediatric cell lines and the relationship to confounders.
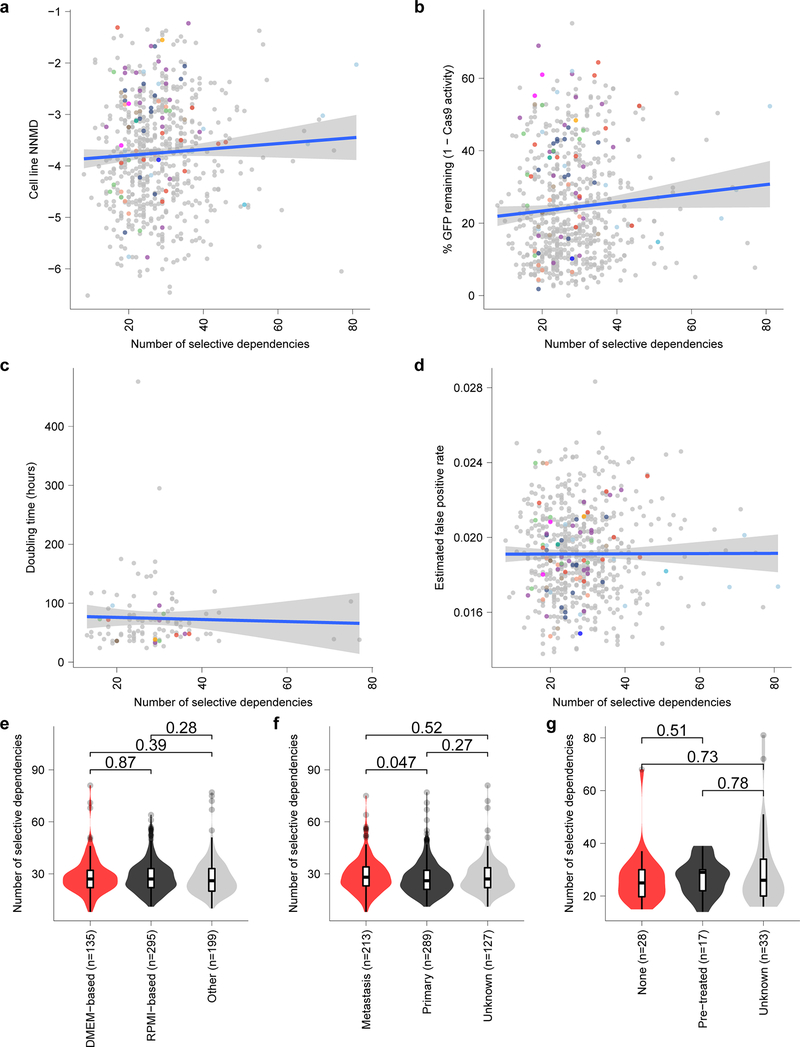
a, Screen quality measured by null-normalized mean difference (NNMD) between positive and negative controls (y-axis) compared to number of selective dependencies per cell line (x-axis). b, Cas9 activity expressed as percent of GFP remaining after CRISPR-Cas9-mediated disruption of exogenous GFP (y-axis) compared to number of selective dependencies per cell line (x-axis). c, Cell line doubling time (y-axis) compared to number of selective dependencies per cell line (x-axis). d, Estimated false positive rate calculated as the fraction of genetic dependencies in a cell line that are not expressed in RNA sequencing data (y-axis) compared to the number of selective dependencies per cell line (x-axis). Circles in panels (a-d) represent individual cell lines with tumor types colored as in panel (Extended Data Fig. 4e). Blue lines in panels (a-d) represent a linear model fit to this data with gray shaded area representing the 95% confidence interval around the fit. e, Number of selective dependencies in cell lines cultured in DMEM-based media (red, n=135 biologically independent cell lines), RPMI-based media (black, n=295 biologically independent cell lines), or other media (gray, n=199 biologically independent cell lines). f, Number of selective dependencies per cell line annotated as derived from metastatic samples (red, n=213 biologically independent cell lines), primary tumors (black, n=289 biologically independent cell lines), or unknown (gray, n=127 biologically independent cell lines). g, Number of selective dependencies per pediatric cancer cell line annotated by literature search as derived from a patient with no pre-treatment (“none”, red, n=28 biologically independent cell lines), after treatment (“pre-treated”, black, n=17 biologically independent cell lines), or unknown (gray, n=33 biologically independent cell lines). Horizontal lines in panels (e-g) demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) in panels (e-g) represent 10 and 90th percentiles respectively.
Extended Data Fig. 6. Predictive modeling of dependencies.
a, Distribution of Pearson correlations of predictive modeling of all dependencies in the screen when using all solid or brain cancer cell lines (black) versus using only the pediatric solid or brain tumor cell lines (red) demonstrates better overall performance when considering all cell lines. b, Predictive modeling of selective dependencies across all solid and brain tumor cell lines versus pediatric solid and brain cancer cell lines. The y-axis depicts the Pearson correlation of the predictive model for dependency on a gene when only considering pediatric cancer cell lines, and the x-axis depicts the Pearson correlation of the predictive model for dependency on a gene when only considering all solid or brain cancer cell lines. The size of the points corresponds to the -log10(adjusted p-value) comparing the rates of dependency in pediatric versus adult cancer cell lines with the points colored by whether the rate is higher in pediatric or adult cancer cell lines for a particular genetic dependency.
Extended Data Fig. 7. Homogeneity of tumor type in expression space is correlated to homogeneity in dependency space.
a, Two-dimensional representation of selective dependencies using uniform manifold approximation and projection (UMAP) demonstrates clustering of cell lines by tumor type. Each circle represents a cell line with pediatric tumors colored by type and adult tumors not depicted for clarity. b, Median distance from panel (d) (y-axis) compared to median distance from panel (f) (x-axis) demonstrating a trend that tumor types with more homogeneity in expression tend toward more homogeneity in dependency. c, Pairwise Pearson correlation of gene expression of the top 2000 most variable genes across cell line pairs from the same tumor type (y-axis) versus tumor types ordered by median (x-axis). Dotted line represents the median correlation to cell lines not of the same tumor type. d, Distance between each cell line in a tumor type and the center of the tumor type cluster in the first 3 principle components of gene expression of the top 2000 most variable genes (y-axis) versus tumor types ordered by median (x-axis). e, Pairwise Pearson correlation of gene dependency of top 500 most variable dependencies across cell line pairs from the same tumor type (y-axis) versus tumor types ordered by median (x-axis). Dotted line represents median correlation to cell lines not of the same tumor type. f, Distance between each cell line in a tumor type and the center of the tumor type cluster in the first 5 principle components of gene dependency of the top 500 most variable dependencies (y-axis) versus tumor types ordered by median (x-axis). Horizontal lines in panels (c-f) demonstrate the median (center) with minima and maxima box boundaries demonstrating the 25 and 75th percentiles. Upper and lower bounds (whiskers) in panels (c-f) represent the 10 and 90th percentiles respectively.
Extended Data Fig. 8. Validation of MCL1 dependency in pediatric cell lines.
a, MCL1 gene effect scores for overlapping cell lines in DepMap 20Q1 (x-axis) and DRIVE RNAi (y-axis) for adult (gray) and pediatric cancer cell lines (red). b, MCL1 gene effect scores (x-axis) versus gene expression of BCL2L1 (y-axis) for adult (gray) and pediatric cancer cell lines (red). Gray and red lines in panels (a-b) represent linear model fits to adult or pediatric data, respectively. c, CRISPR-Cas9 mediated disruption of MCL1 by two independent sgRNAs reveals decreased cell growth in vitro as demonstrated by CellTiter-Glo luminescence (y-axis) versus time (x-axis), correlated with the larger screen. One representative experiment shown for each cell line; each time-point measured in replicate (n=8). Data presented as mean values +/− SEM. d, Western blotting after MCL1 disruption by CRISPR-Cas9 2 days post-selection (SKNBE2, SKNMC) or 3 days post-selection (Kelly). e, Western blotting after MCL1 inhibition with S63845 at 48 hours demonstrates increased protein expression of MCL1 after inhibition with S63845 at 48 hours with less induction of cleaved PARP or Caspase 3 at lower concentrations in SKNBE2 or EWS503 compared to the more sensitive neuroblastoma or Ewing cell lines, Kelly and SKNMC, respectively. f, Treatment with increasing concentrations of ZVAD, a pan-caspase inhibitor, reveals a concentration-dependent rescue of 2 μM S63845 treatment in Kelly and SKNMC at day 3 as demonstrated by the fraction of CellTiter-Glo luminescence compared to DMSO control (y-axis). One representative experiment shown for each cell line; each time-point measured in replicate (n=4). Data presented as mean values +/− SEM. g, Western blotting after one hour of pre-treatment with either DMSO or 20 μM ZVAD followed by either DMSO or 1 μM S63845 treatment at 48 hours show increased protein expression of MCL1 after inhibition with S63845 at 48 hours with decreased induction of cleaved PARP or Caspase 3 following pre-treatment with ZVAD in SKNMC. Experiments shown in panels (c-g) were performed independently at least in duplicate, with one representative experiment shown.
Extended Data Fig. 9. Selective and enriched dependencies in pediatric and adult solid tumor lines.
a, The frequency of dependency on the neuroblastoma core regulatory transcription factors (ISL1, HAND2, GATA3, PHOX2A, PHOX2B) and rhabdomyosarcoma regulatory transcription factors (MYOD1) are depicted in pediatric and adult solid tumor types with at least 3 cell lines screened per type in polar bar graphs. The tumor types are colored as in the legend. The neuroblastoma transcription factor dependencies were seen uniquely in neuroblastoma and MYOD1 dependency was seen in rhabdomyosarcoma. b, Feature importance for the predictive models of HDAC2 dependency using data from all solid and brain tumor cell lines (left) or pediatric solid and brain cancer cell lines only (right). The y-axis shows the feature importance as calculated by the predictive model with features listed on the x-axis. c, Feature importance for the predictive models of HDAC2 dependency using data from all solid and brain tumor cell lines (left) or pediatric solid and brain cancer cell lines only (right). The y-axis shows the feature importance as calculated by the predictive model with features listed on the x-axis.
Extended Data Fig. 10. Selective and enriched dependencies in pediatric and adult solid tumor lines.
a, Quantification of tumor type-enriched dependencies per tumor-type (y-axis) compared to number of cell lines screened per tumor type (x-axis). The number of enriched dependencies per tumor type with a q-value <0.05 was calculated by performing a two-class comparison between gene effect scores in each tumor type compared to all other cell lines screened using two-sided t-tests with Benjamini-Hochberg correction. b, Quantification of tumor type-enriched dependencies that are also classified as selective dependencies in the screen per tumor-type (y-axis) compared to number of cell lines screened per tumor type (x-axis). The number of enriched dependencies per tumor type with a q-value <0.05 was calculated by performing a two-class comparison between gene effect scores in each tumor type compared to all other cell lines screened using two-sided t-tests with Benjamini-Hochberg correction. Each circle in panels (a-b) represents a tumor type colored as in the legend. The blue lines in panels (a-b) represent a linear model fit to this data with the gray shaded area representing the 95% confidence interval around the fit. c, Tumor type-enriched dependencies in all solid and brain tumor types with more than 2 cell lines. Plotted on the y-axis is -log10 of the q-value of enrichment as calculated by performing a two-class comparison between gene effect scores in each tumor type compared to all other cell lines screened using two-sided t-tests with Benjamini-Hochberg correction. Tumor types are plotted along the x-axis. The size of the circles reflects the mean difference in dependency score between the tumor type and all other cell lines screened. Gray circles are enriched dependencies in a tumor type that are not classified as transcription factors and colored circles are transcription factor dependencies in the screen.
Supplementary Material
Supplementary Data Table 1. Characteristics of pediatric and adult cancer cell lines used in this study. Columns indicate various identifications for cell lines including the DepMap ID, CCLE name, COSMIC ID and Sanger Model ID. The tumor type information is contained in columns labeled lineage, lineage_subtype, lineage_sub_subtype, lineage_molecular_subtype, disease and disease_subtype. In addition, when available, the sex and age of the patient from whom the cell line was originally derived is indicated. The source, culture type and media of each cell line is indicated. The column Achilles_n_replicates indicates the number of replicates used in the genome-scale CRISPR-Cas9 screen that passed quality control. Cell_line_NNMD is a measure of CRISPR-Cas9 screen quality quantified as the difference in the means of positive and negative controls normalized by the standard deviation of the negative control distribution. Cas9_activity indicates the percentage of cells remaining GFP positive on days 12–14 of Cas9 activity assay as measured by flow cytometry. Estimated_Doubling_Time_Hours indicates a doubling time estimate in hours when available. The columns with information from a literature search for established pediatric cell lines include whether the DepMap annotation of age and sex is matched by the literature (LiteratureSearch_Age_Sex_Match_DepMap_Annotation), whether the cell line was derived after a tumor was treated (LiteratureSearch_Previous_Treatment) and any available data on the treatment (LiteratureSearch_Previous_Treatment_Details), the earliest citation year for the cell line (LiteratureSearch_Year_Earliest_Citation), any other annotations from the literature search (LiteratureSearch_Other_Annotations), and the PubMed Central (PMC) IDs for the literature source (LiteratureSearch_Paper_PMC_ID). The columns labeled RNAseq, WES, SangerWES and SNParray indicate which lines have the respective data in the DepMap 20Q1 dataset used for this study and the source of the data with previously published data indicated with CCLE or Sanger and newly generated data with DepMap. The column labeled RNAseq_SRA_accession contains the SRA accession for the raw RNA sequencing data when available or indicates in process for legacy cell lines being deposited by DepMap. The column labeled WES_SRA_accession contains the SRA accession for the raw WES data when available or indicates in process for legacy cell lines being deposited by DepMap. The column labeled WES_EGA_accession contains the EGA accession for the raw WES data from Sanger.
Supplementary Data Table 2. Celligner tumor type assignments for cell lines. Columns indicate the various identifications for cell lines and primary tumors in the sampleID and DepMap_ID. The coordinates from the UMAP projection of all tumor and cell line samples are contained in columns UMAP_1 and UMAP_2. The tumor type is contained in lineagesubtype with primary or metastasis annotated in the column Primary/Metastatis if known. The column type contains whether the sample is a cell line (CL) or primary tumor. The cluster column contains the cluster for the sample as defined by Warren et al18. The cancer_type column contains a more specific tumor type than lineagesubtype. RF_class, RF_probability, and RF_probability_margin contain the primary tumor expression classification of cell line expression and the associated probability of that classification. Celligner_class and Celligner_class_confidence contain the cell line classification which is the most frequently occurring cancer type from the 25 highest correlated primary tumor neighbors for each cell line after Celligner correction and the confidence is the proportion of tumor samples within the 25 nearest neighbors that came from the most frequent cancer type. Uncorrected_tumor_class and uncorrected_tumor_prop contain the cell line classification which is the most frequently occurring cancer type from the 25 highest correlated primary tumor neighbors for each cell line without Celligner correction and the confidence is the proportion of tumor samples within the 25 nearest neighbors that came from the most frequent cancer type. The undifferentiated_cluster column indicates those cell lines and primary tumors that were classified as belonging to the undifferentiated cluster by Warren et al.
Supplementary Data Table 3. Mutation rates of COSMIC genes as detected by MutSig2CV in pediatric cancer cell lines. The columns indicate the gene name, number of pediatric cell lines with a mutation in that gene called by MutSig2CV and the total number of pediatric cell lines analyzed with MutSig2CV.
Supplementary Data Table 4. Common genomic alterations in pediatric cancer cell lines. A survey of reported recurrent genomic alterations in primary pediatric tumors and the rates of the same alterations in the corresponding pediatric cancer cell lines in this study.
Supplementary Data Table 5. NormLRT scores for all genes. The columns indicate the gene names with Entrez ID in parenthesis, the normLRT score across all solid and brain tumor cell lines in DepMap 20Q1, the median and mean gene effect score for each gene across the same subset of cell lines.
Supplementary Data Table 6. Predictive modeling of dependencies. The results of the random forest regression models are presented with the gene column indicating the genetic dependency being modeled. The model column indicates whether the method was executed using only related or all (top 1000 correlated unbiased) cell line features. The pearson column contains the correlation between the predicted and observed CERES gene effects as an accuracy measure per model. The top model for each gene is indicated in the column labeled best. The remaining columns indicate the top features used to build the model and the feature importance of each.
Supplementary Data Table 7. Selective and enriched gene dependencies. For selective dependencies, the gene names with Entrez ID in parenthesis are provided, as are the corrected p-value of the two-sided Fisher’s exact test with Benjamini-Hochberg correction comparing the number of pediatric and adult cell lines dependent on each gene, the fraction of pediatric cell lines dependent on each gene (dependency probability >0.5), the fraction of adult cell lines dependent on each gene (dependency probability >0.5), and the difference in the fractions. For enriched dependencies, the gene names with Entrez ID in parenthesis are provided in the first column followed by the enrichment statistics for each tumor type including the effect size (ES) or difference in mean gene effect score for the tumor type compared to all other cell lines, the p-value that the gene has more negative gene effect scores in the tumor type compared to all other cell lines, and the q-value for each tumor type which is adjusted for multiple hypothesis testing with Benjamini-Hochberg correction.
Supplementary Data Table 8. High variance genes used for random forest tumor-cell line expression alignment. The primary tumor expression standard deviations (from TCGA, Treehouse and TARGET data) and cell line expression standard deviations (from CCLE) for the top 5,000 most variable genes that were used for the random forest classification of cell line to primary tumor expression.
Acknowledgements
This work was supported by the National Cancer Institute R35 CA210030, R01 CA204915, P01 CA217959, a St. Baldrick’s Foundation Robert J. Arceci Innovation Award, the Four C’s Fund, and PMC Team Eradicate (KS). This work was funded in part by the Slim Initiative in Genomic Medicine for the Americas (SIGMA), a joint U.S-Mexico project funded by the Carlos Slim Foundation (TRG). This work was supported in part by Walter and Marina Bornhorst (TRG). This work was supported by Team Sciarappa Strong (Jimmy Fund Walk) (KS, ADD). This work was funded in part by the Alexandra Simpson Pediatric Research Fund (CWMR, KS). This work was supported by the NBTII Foundation (JSB). This work was supported by the NCI U01 CA176058 (WCH).
NVD was a Julia’s Legacy of Hope St. Baldrick’s Foundation Fellow and received support from the Rally Foundation for Childhood Cancer Research. LMG is a William Raveis Charitable Fund Physician-Scientist of the Damon Runyon Cancer Research Foundation (PST-20-18) and receives support from the Rally Foundation for Childhood Cancer Research, as well as received support from Boston Children’s Hospital Office of Faculty Development. CFM was supported by a Helen Gurley Brown Presidential Initiative Fellowship and by the National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (F32CA243266). ADD was supported by a Damon Runyon Sohn Fellowship from the Damon Runyon Cancer Research Foundation (DRSG-24-18), the Alex’s Lemonade Stand Foundation, Rally Foundation for Childhood Cancer Research, CureSearch for Children’s Cancer and American Society for Clinical Oncology. ALH was supported by grants from the American Cancer Society MRSG-18-202-01 and Department of Defense CDMRP W81XWH-19-1-0281. TPH was supported by National Institutes of Health grants T32GM007753 and T32GM007226. PB was supported by the Pediatric Brain Tumor Foundation, Jared Branfman Sunflowers for Life Fund, The Isabel V Marxuach Fund for Medulloblastoma Research and NCI R00CA201592.
Conflicts of interest
NVD is a current employee of Genentech, Inc., a member of the Roche Group. PB receives funding from Novartis Institute of Biomedical Research for an unrelated project and serves as a consultant for QED Therapeutics. WCH is a consultant for ThermoFisher, Solvasta Ventures, MPM Capital, KSQ Therapeutics, iTeos, Tyra Biosciences, Frontier Medicine, Paraxel, and Jubilant Therapeutics. AT is a consultant for Tango Therapeutics. TRG receives research funding unrelated to this project from Bayer HealthCare, Calico Life Sciences, and Novo Ventures. TRG was formerly a consultant and equity holder in Foundation Medicine, which was acquired by Roche. TRG is a consultant to GlaxoSmithKline and is a founder and equity holder of Sherlock Biosciences and FORMA Therapeutics. FV and BRP receive research support from Novo Ventures unrelated to this project. KS has funding from Novartis Institute of Biomedical Research, consults for and has stock options in Auron Therapeutics and served as an advisor for Kronos Bio. The remaining authors declare no competing interests.
References
- 1.Park JR et al. A phase III randomized clinical trial (RCT) of tandem myeloablative autologous stem cell transplant (ASCT) using peripheral blood stem cell (PBSC) as consolidation therapy for high-risk neuroblastoma (HR-NB): A Children’s Oncology Group (COG) study. JCO 34, LBA3–LBA3 (2016). [Google Scholar]
- 2.Northcott PA et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol 29, 1408–1414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho Y-J et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol 29, 1424–1430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dome JS et al. Children’s Oncology Group’s 2013 blueprint for research: renal tumors. Pediatr Blood Cancer 60, 994–1000 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigel BJ et al. Intensive Multiagent Therapy, Including Dose-Compressed Cycles of Ifosfamide/Etoposide and Vincristine/Doxorubicin/Cyclophosphamide, Irinotecan, and Radiation, in Patients With High-Risk Rhabdomyosarcoma: A Report From the Children’s Oncology Group. J. Clin. Oncol 34, 117–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grier HE et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. N. Engl. J. Med 348, 694–701 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Yeh JM et al. Life Expectancy of Adult Survivors of Childhood Cancer Over 3 Decades. JAMA Oncol (2020). doi: 10.1001/jamaoncol.2019.5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan EM et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behan FM et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568, 511–516 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Gröbner SN et al. The landscape of genomic alterations across childhood cancers. Nature 555, 321–327 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Ma X et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts CWM & Biegel JA The role of SMARCB1/INI1 in development of rhabdoid tumor. Cancer Biol. Ther 8, 412–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crompton BD et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 4, 1326–1341 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Harris MH et al. Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol 2, 608–615 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Mody RJ et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA 314, 913–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons DW et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol 2, 616–624 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nature genetics 350, 1096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren A et al. Global computational alignment of tumor and cell line transcriptional profiles. Nat Commun 12, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghandi M et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morozova O, Newton Y, Cline M, Zhu J & Learned K Abstract lb-212: Treehouse childhood cancer project: a resource for sharing and multiple cohort analysis of pediatric cancer genomics data. (2015).
- 21.Drexler HG et al. p53 alterations in human leukemia-lymphoma cell lines: in vitroartifact or prerequisite for cell immortalization? Leukemia 14, 198–206 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Ben-David U, Beroukhim R & Golub TR Genomic evolution of cancer models: perils and opportunities. Nat. Rev. Cancer 19, 97–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossen J & Pan J Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv 20, 720243 (2019). [Google Scholar]
- 25.McDonald ER et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Tsherniak A et al. Defining a Cancer Dependency Map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dempster JM et al. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv 20, 720243 (2019). [Google Scholar]
- 28.Children Successfully MATCHed to Therapies. Cancer Discov 9, OF3–OF3 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Tisato V, Voltan R, Gonelli A, Secchiero P & Zauli G MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J Hematol Oncol 10, 133–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard TP et al. MDM2 and MDM4 are Therapeutic Vulnerabilities in Malignant Rhabdoid Tumors. Cancer Res canres.3066.2018 (2019). doi: 10.1158/0008-5472.CAN-18-3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolte B et al. Genome-scale CRISPR-Cas9 screen identifies druggable dependencies in TP53 wild-type Ewing sarcoma. J. Exp. Med 215, 2137–2155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther LM et al. A Combination CDK4/6 and IGF1R Inhibitor Strategy for Ewing Sarcoma. Clin. Cancer Res 25, 1343–1357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood AC et al. Dual ALK and CDK4/6 Inhibition Demonstrates Synergy against Neuroblastoma. Clin. Cancer Res. 23, 2856–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mills CC, Kolb EA & Sampson VB Recent Advances of Cell-Cycle Inhibitor Therapies for Pediatric Cancer. Cancer Res 77, 6489–6498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olanich ME et al. CDK4 Amplification Reduces Sensitivity to CDK4/6 Inhibition in Fusion-Positive Rhabdomyosarcoma. Clin. Cancer Res 21, 4947–4959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotschy A et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. 538, 477–482 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Gonçalves E et al. Drug mechanism-of-action discovery through the integration of pharmacological and CRISPR screens. bioRxiv 20, 2020.01.14.905729 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durbin AD et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nature genetics 50, 1240–1246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gryder BE et al. Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nature genetics 51, 1714–1722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frumm SM et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem. Biol 20, 713–725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pappo AS et al. R1507, a monoclonal antibody to the insulin-like growth factor 1 receptor, in patients with recurrent or refractory Ewing sarcoma family of tumors: results of a phase II Sarcoma Alliance for Research through Collaboration study. J. Clin. Oncol 29, 4541–4547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juergens H et al. Preliminary efficacy of the anti-insulin-like growth factor type 1 receptor antibody figitumumab in patients with refractory Ewing sarcoma. J. Clin. Oncol 29, 4534–4540 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tap WD et al. Phase II study of ganitumab, a fully human anti-type-1 insulin-like growth factor receptor antibody, in patients with metastatic Ewing family tumors or desmoplastic small round cell tumors. J. Clin. Oncol 30, 1849–1856 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Beckwith H & Yee D Minireview: Were the IGF Signaling Inhibitors All Bad? Mol. Endocrinol 29, 1549–1557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filbin M & Monje M Developmental origins and emerging therapeutic opportunities for childhood cancer. Nat. Med 25, 367–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L et al. CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J. Clin. Invest 128, 446–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberlick EM et al. Small-Molecule and CRISPR Screening Converge to Reveal Receptor Tyrosine Kinase Dependencies in Pediatric Rhabdoid Tumors. Cell Rep 28, 2331–2344.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong AL et al. Renal medullary carcinomas depend upon SMARCB1 loss and are sensitive to proteasome inhibition. Elife 8, 818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eichenmüller M et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J. Hepatol 61, 1312–1320 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Thériault BL, Dimaras H, Gallie BL & Corson TW The genomic landscape of retinoblastoma: a review. Clin. Experiment. Ophthalmol 42, 33–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shern JF et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 4, 216–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johann PD et al. Atypical Teratoid/Rhabdoid Tumors Are Comprised of Three Epigenetic Subgroups with Distinct Enhancer Landscapes. Cancer Cell 29, 379–393 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Chun H-JE et al. Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer Cell 29, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Northcott PA et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pugh TJ et al. The genetic landscape of high-risk neuroblastoma. Nature genetics 45, 279–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kovac M et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun 6, 8940 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braunstein S, Raleigh D, Bindra R, Mueller S & Haas-Kogan D Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J. Neurooncol 134, 541–549 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Lafin JT, Bagrodia A, Woldu S & Amatruda JF New insights into germ cell tumor genomics. Andrology 7, 507–515 (2019). [DOI] [PubMed] [Google Scholar]
Online Methods References
- 60.DepMap B DepMap 20Q1 Public. (2020). doi: 10.6084/m9.figshare.11791698.v3 [DOI] [Google Scholar]
- 61.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature biotechnology 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cibulskis K et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27, 2601–2602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Costello M et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res 41, e67–e67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor-Weiner A et al. DeTiN: overcoming tumor-in-normal contamination. Nat. Methods 15, 531–534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landau DA et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152, 714–726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lawrence MS et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramos AH et al. Oncotator: cancer variant annotation tool. Hum. Mutat 36, E2423–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van der Auwera GA et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Consortium GTEx et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haas BJ et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 20, 213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright MN & Ziegler A ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. Journal of Statistical Software 77, (2015). [Google Scholar]
- 76.Lawrence MS et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McFarland JM et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun 9, 4610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maglott D, Ostell J, Pruitt KD & Tatusova T Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 39, D52–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data Table 1. Characteristics of pediatric and adult cancer cell lines used in this study. Columns indicate various identifications for cell lines including the DepMap ID, CCLE name, COSMIC ID and Sanger Model ID. The tumor type information is contained in columns labeled lineage, lineage_subtype, lineage_sub_subtype, lineage_molecular_subtype, disease and disease_subtype. In addition, when available, the sex and age of the patient from whom the cell line was originally derived is indicated. The source, culture type and media of each cell line is indicated. The column Achilles_n_replicates indicates the number of replicates used in the genome-scale CRISPR-Cas9 screen that passed quality control. Cell_line_NNMD is a measure of CRISPR-Cas9 screen quality quantified as the difference in the means of positive and negative controls normalized by the standard deviation of the negative control distribution. Cas9_activity indicates the percentage of cells remaining GFP positive on days 12–14 of Cas9 activity assay as measured by flow cytometry. Estimated_Doubling_Time_Hours indicates a doubling time estimate in hours when available. The columns with information from a literature search for established pediatric cell lines include whether the DepMap annotation of age and sex is matched by the literature (LiteratureSearch_Age_Sex_Match_DepMap_Annotation), whether the cell line was derived after a tumor was treated (LiteratureSearch_Previous_Treatment) and any available data on the treatment (LiteratureSearch_Previous_Treatment_Details), the earliest citation year for the cell line (LiteratureSearch_Year_Earliest_Citation), any other annotations from the literature search (LiteratureSearch_Other_Annotations), and the PubMed Central (PMC) IDs for the literature source (LiteratureSearch_Paper_PMC_ID). The columns labeled RNAseq, WES, SangerWES and SNParray indicate which lines have the respective data in the DepMap 20Q1 dataset used for this study and the source of the data with previously published data indicated with CCLE or Sanger and newly generated data with DepMap. The column labeled RNAseq_SRA_accession contains the SRA accession for the raw RNA sequencing data when available or indicates in process for legacy cell lines being deposited by DepMap. The column labeled WES_SRA_accession contains the SRA accession for the raw WES data when available or indicates in process for legacy cell lines being deposited by DepMap. The column labeled WES_EGA_accession contains the EGA accession for the raw WES data from Sanger.
Supplementary Data Table 2. Celligner tumor type assignments for cell lines. Columns indicate the various identifications for cell lines and primary tumors in the sampleID and DepMap_ID. The coordinates from the UMAP projection of all tumor and cell line samples are contained in columns UMAP_1 and UMAP_2. The tumor type is contained in lineagesubtype with primary or metastasis annotated in the column Primary/Metastatis if known. The column type contains whether the sample is a cell line (CL) or primary tumor. The cluster column contains the cluster for the sample as defined by Warren et al18. The cancer_type column contains a more specific tumor type than lineagesubtype. RF_class, RF_probability, and RF_probability_margin contain the primary tumor expression classification of cell line expression and the associated probability of that classification. Celligner_class and Celligner_class_confidence contain the cell line classification which is the most frequently occurring cancer type from the 25 highest correlated primary tumor neighbors for each cell line after Celligner correction and the confidence is the proportion of tumor samples within the 25 nearest neighbors that came from the most frequent cancer type. Uncorrected_tumor_class and uncorrected_tumor_prop contain the cell line classification which is the most frequently occurring cancer type from the 25 highest correlated primary tumor neighbors for each cell line without Celligner correction and the confidence is the proportion of tumor samples within the 25 nearest neighbors that came from the most frequent cancer type. The undifferentiated_cluster column indicates those cell lines and primary tumors that were classified as belonging to the undifferentiated cluster by Warren et al.
Supplementary Data Table 3. Mutation rates of COSMIC genes as detected by MutSig2CV in pediatric cancer cell lines. The columns indicate the gene name, number of pediatric cell lines with a mutation in that gene called by MutSig2CV and the total number of pediatric cell lines analyzed with MutSig2CV.
Supplementary Data Table 4. Common genomic alterations in pediatric cancer cell lines. A survey of reported recurrent genomic alterations in primary pediatric tumors and the rates of the same alterations in the corresponding pediatric cancer cell lines in this study.
Supplementary Data Table 5. NormLRT scores for all genes. The columns indicate the gene names with Entrez ID in parenthesis, the normLRT score across all solid and brain tumor cell lines in DepMap 20Q1, the median and mean gene effect score for each gene across the same subset of cell lines.
Supplementary Data Table 6. Predictive modeling of dependencies. The results of the random forest regression models are presented with the gene column indicating the genetic dependency being modeled. The model column indicates whether the method was executed using only related or all (top 1000 correlated unbiased) cell line features. The pearson column contains the correlation between the predicted and observed CERES gene effects as an accuracy measure per model. The top model for each gene is indicated in the column labeled best. The remaining columns indicate the top features used to build the model and the feature importance of each.
Supplementary Data Table 7. Selective and enriched gene dependencies. For selective dependencies, the gene names with Entrez ID in parenthesis are provided, as are the corrected p-value of the two-sided Fisher’s exact test with Benjamini-Hochberg correction comparing the number of pediatric and adult cell lines dependent on each gene, the fraction of pediatric cell lines dependent on each gene (dependency probability >0.5), the fraction of adult cell lines dependent on each gene (dependency probability >0.5), and the difference in the fractions. For enriched dependencies, the gene names with Entrez ID in parenthesis are provided in the first column followed by the enrichment statistics for each tumor type including the effect size (ES) or difference in mean gene effect score for the tumor type compared to all other cell lines, the p-value that the gene has more negative gene effect scores in the tumor type compared to all other cell lines, and the q-value for each tumor type which is adjusted for multiple hypothesis testing with Benjamini-Hochberg correction.
Supplementary Data Table 8. High variance genes used for random forest tumor-cell line expression alignment. The primary tumor expression standard deviations (from TCGA, Treehouse and TARGET data) and cell line expression standard deviations (from CCLE) for the top 5,000 most variable genes that were used for the random forest classification of cell line to primary tumor expression.
Data Availability Statement
CRISPR-Cas9 screening results for DepMap version 20Q1 (including raw data) and the genomic characterization of cancer cell lines (whole-exome sequencing and RNA sequencing) used in this study are publicly available at https://depmap.org and also on figshare (https://figshare.com/articles/dataset/DepMap_20Q1_Public/11791698). Subsets of the raw sequencing data from whole exome sequencing and RNA sequencing used in this study are available at Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) and European Genome-phenome Archive (EGA, https://www.ebi.ac.uk/ega/) accession numbers: SRA PRJNA523380 (CCLE), SRA PRJNA261990 (Ewing sarcoma), and EGAS00001000978 (Sanger) (Supplementary Data Table 1). The remainder of the raw sequencing data is in the process of being deposited in SRA via dbGaP (https://dbgap.ncbi.nlm.nih.gov/), delayed in part as these are legacy cell lines. In the interim, we will work with specific requests to expedite the process (contact depmap@broadinstitute.org). Additionally, the pediatric-specific subsets of the processed DepMap version 20Q1 data presented in this study (dependency, mutations, copy number, expression, fusions) are available at our companion website at https://depmap.org/peddep.