Abstract
Centromeres are defined by the location of Centromeric Histone H3 (CENP-A/CENH3) which interacts with DNA to define the locations and sizes of functional centromeres. An analysis of 26 maize genomes including 110 fully assembled centromeric regions revealed positive relationships between centromere size and genome size. These effects are independent of variation in the amounts of the major centromeric satellite sequence CentC. We also backcrossed known centromeres into two different lines with larger genomes and observed consistent increases in functional centromere sizes for multiple centromeres. Although changes in centromere size involve changes in bound CENH3, we could not mimic the effect by overexpressing CENH3 by threefold. Literature from other fields demonstrate that changes in genome size affect protein levels, organelle size and cell size. Our data demonstrate that centromere size is among these scalable features, and that multiple limiting factors together contribute to a stable centromere size equilibrium.
Keywords: centromere, kinetochore, cellular scaling, CENP-A, genome-site
Introduction
The histone variant known as CENP-A/CENH3 recruits a set of constitutive centromere proteins that in turn recruit the kinetochore proteins that interact with microtubules (Zhong et al. 2002; Kixmoeller et al. 2020; Mitra et al .2020). When CENH3 binds to DNA, that sequence is referred to as centromeric DNA. How and where CENH3 binds is determined by a host of factors that differ among species. In human and mouse, centromere sequences can specifically promote the deposition of overlying centromere proteins (Aldrup-MacDonald et al. 2016; Iwata-Otsubo et al. 2017) although the fact that centromeres occasionally form on regions that lack canonical centromere repeats suggests that the underlying mechanism is epigenetic (Murillo-Pineda and Jansen 2020). This contrasts with maize and other plants where centromere sequences vary dramatically and the specification mechanisms are largely or entirely epigenetic (Oliveira and Torres 2018). Whether DNA sequence helps to guide the deposition of centromere proteins or not, centromere/kinetochore domains adopt predictable sizes and are stably propagated (Bodor et al. 2014; Gent et al. 2017).
Under the epigenetic model for centromere positioning, the location and size of the existing centromere is used as a template for the location of a newly replicated centromere (Mitra et al. 2020). However, we have observed extensive plasticity in the size and locations of centromeres that presumably represents both stochastic and physiological variation (Gent et al. 2015, 2017). The observed plasticity fits well with the proposal that centromeres are highly dynamic and their average size is determined in part by the concentration of kinetochore proteins (Bodor et al. 2014). An analysis of multiple grass species demonstrated that the sum of all kinetochore sizes in a cell scales linearly with genome size (Bennett et al. 1981; Zhang and Dawe 2012), suggesting that the amount of kinetochore proteins is at least partially dependent on genome size and cell volume (Zhang and Dawe 2012). As a test, maize chromosomes were introduced into the larger oat genomic background, and centromere size measured by the amount of DNA occupied by CENH3 as interpreted by ChIP-seq. The maize centromeres increased in size by twofold in the oat background as predicted (Wang et al. 2014). These results mirror a variety of studies showing that subcellular structures frequently scale with genome and cell size (Price et al. 1973; Gregory 2001; Cavalier-Smith 2005; Gillooly et al. 2015; Robinson et al. 2018).
As outlined by Marshall (2016), cellular scaling could occur by several mechanisms. The simplest is the limiting precursor model, where the amount of a key component increases with cell size and directly contributes to the size of the structure of interest. In the case of centromeres, a likely candidate is CENH3/CENP-A. Prior data from Drosophila have shown that overexpression of CENP-A causes ectopic centromere formation in noncentromeric regions (Heun et al. 2006) whereas in human cell lines, excessive CENP-A alone is not sufficient to form ectopic kinetochores (Van Hooser et al. 2001; Lacoste et al. 2014). Other likely limiting components are those involved in CENP-A deposition. CENP-A loading involves licensing factors such as Kinetochore Null 2 (KNL2) (Lermontova et al. 2013; Sandmann et al. 2017; Boudichevskaia et al. 2019), specific chaperones (Sanchez-Pulido et al. 2009; Chen et al. 2014) and interactions with other kinetochore proteins such as Centromere Protein C (CENP-C) (French et al. 2017; Sandmann et al. 2017;). CENH3/CENP-A, its licensing factors, chaperones, and other inner kinetochore proteins may directly or indirectly regulate centromere size either alone or in combination.
In this study, we tested the idea that maize centromeres are scalable by analyzing recent genome assemblies of multiple inbreds, experimentally manipulating genome size using genetic crosses, and overexpressing CENH3. We find no consistent association between specific sequences and centromere size, and no change in centromere size after overexpressing CENH3 by threefold. However, we found evidence of centromere scaling among inbreds that naturally vary in genome size and in lines with experimentally manipulated genome sizes. The data support the conclusion that centromere size is not controlled by DNA sequence or by CENH3 alone, but by a mass-action mechanism that may be sensitive to cell volume and regulated by the concentration of multiple precursors.
Materials and methods
Plant materials and crossing
The plant materials used in this study were obtained from the Germplasm Resources Information Network (GRIN), Ames, Iowa. The lines were B73 (PI 550473), a domesticated landrace from Oaxaca, Mexico (PI 628470) and Zea luxurians (PI 462368). Crosses among lines were made over several years in the UGA Plant Biology greenhouses or an adjoining outdoor field site.
ChIP-seq
Whole seedlings including roots were collected from inbreds B73, CML103, CML277, CML333, HP301, IL14H, Ki11, Ki3, NC350, Oh7b, P39 and Tzi8, and CENH3 ChIP conducted as described previously (Gent et al. 2017) with the following modifications: During the nuclei extraction, we did not cut off pipet tips, and during micrococcal nuclease digestion, we used 2 µL micrococcal nuclease per 50 µL pelleted nuclei. For the overnight antibody incubation, we used 8.5 µg of anti-maize CENH3 antibodies (Zhong et al. 2002) and anti-rice CENH3 antibodies (Nagaki et al. 2004). During sequencing library preparation with the KAPA Hyper Prep kit (KK8502), we used a double-sided size selection with Mag-Bind® TotalPure NGS magnetic beads rather than a post-PCR gel purification. We removed large fragments with a 0.6X Mag-Bind bead cleanup (by adding 66 µL of beads to the 110 µL of ligation product and discarding the pellet), then removed small fragments with a 0.8X bead cleanup (by adding 136 µL of beads to the resulting 170 µL of supernatant from the last step and then discarding the new supernatant). All ChIP libraries were amplified with 5 cycles of PCR. Multiple adapters were used for pooling libraries (KAPA Single-Indexed Adapter Kit KK8700 and NEBNext®Multiplex Oligos for Illumina NEB #E7535S/L). The DNA samples were sequenced using the Illumina NextSeq 500 platform and 150-nucleotide single-end reads were generated. The Sequence Read Archive run IDs for all the ChIP data of this study are listed in Supplementary Table 6.
PE100 Illumina CENH3 ChIP-seq reads for lines B97, CML228, CML322, CML247, CML52, CML69, KY21, MO18W, M37W, M162W, MS71, NC358, Oh43, and Tx303 were published previously (Schneider et al. 2016). The data were obtained from Genbank (SRP067358) and converted to single-end format using seqtk (https://github.com/lh3/seqtk).
Measuring centromere size
The efficiency of CENH3 chromatin immunoprecipitation varies from day-to-day and sample to sample. The results are generally assessed after the experiment is over, when the average read depth within centromere cores is compared to the average read depth over chromosome arms. We observed previously (Gent et al. 2015), as well as in the current datasets, that measured centromere sizes vary with ChIP efficiency. This is because CENH3 ChIP-seq profiles take on the shape of bell-shaped curves. When there is low efficiency, a smaller profile of the curve exceeds the enrichment cutoff used to define edges of the centromere. To ameliorate this effect, we designed a custom workflow that amplifies ChIP-seq signals in the centromere relative to the chromosome arms, and consequently sharpens the centromere curve for samples with low efficiency (Supplementary Figure 1). The workflow involved four steps as follows:
(1) Input data normalization: PE150 genomic input reads of all NAM lines (Hufford et al. 2021) (www.maizegdb.org/NAM_project) were subsampled to 30x with seqtk (v1.2, https://github.com/lh3/seqtk) relative to assembly size. The CENH3 ChIP-seq data in the form of PE100 reads were downloaded from SRP067358, converted to single-end data, and subsampled to 5 million reads using seqtk (v1.2). The SE150 ChIP reads generated in this study were subsampled to 3.33 million. Subsampled ChIP and genomic data were subjected to adapter removal with trimglore (v0.4.5, https://github.com/FelixKrueger/TrimGalore/) and mapped to corresponding NAM genomes (Hufford et al. 2021) with bwa-mem (v0.7.17) at default parameters (Li and Durbin, 2009). PCR Duplicates were removed from bam files using piccard (v2.16) and alignments with a mapping quality higher than 20 were extracted with samtools (v1.9) (Li et al. 2009). The resulting CENH3 ChIP bam files were then normalized against input with deeptools (v3.2.1) (Ramírez et al. 2014) using the RPKM method with 5Kb nonoverlapping windows (–binSize 5,000 –normalizeUsing RPKM –scaleFactorsMethod None). Regions with an enrichment higher than 5 were extracted and merged into islands with bedtools (v2.28) (Quinlan 2014).
(2) Normalization of ChIP efficiency: Centromeres were located manually and placed into 5 Mb windows, then all remaining genomic space classified as background. The ratio between the sum of ChIP RPKM values (≥0) in the 5 Mb centromere regions (core) and background areas (all noncore areas) were used to calculate ChIP enrichment (Supplementary Figure 1A. The core/background ratios were then modified using the formula (1+ core/background) × ChIP RPKM enrichment. This scaling step amplified signal in the centromere relative to chromosome arms. The resulting ChIP bedgraph files exhibited more pronounced curves compared with that before scaling (Supplementary Figure 1C).
(3) Merging ChIP islands separated by alignment gaps: Highly repetitive regions result in either an absence of aligned reads or the alignment of far more reads than expected. Alignment gaps were defined as regions (>100bp) with lower than 2 or higher than 101 reads mapped using bedtools (v2.28). The resulting gaps resulted in isolated “ChIP islands” that we presumed would be connected if not for the intervening gap. Islands within 100 kb of each other that exceeded the fivefold enrichment cutoff were merged using bedtools merge (v2.28; -d 15000). After this initial merging step, islands larger than 15 kb with enrichments higher than 3 were merged with a 50 kb interval using bedtools merge (v2.28; -d 50000).
(4) Using replicates to remove outliers: For lines with ChIP replicates, final coordinates were determined and centromere sizes were calculated for each replicate. Centromere sizes were compared among replicates, and outliers were removed using the mean absolute deviation (MAD) method. The mean centromere sizes among replicates were calculated after outlier removal.
Using these methods we compared centromere sizes across four different IL14H biological replicates. While the ChIP enrichment ranged from 7.36 to 13 (fold increase over background in the 5 Mb core region), their measured centromere sizes after processing were similar (Supplementary Figure 1C).
CRM and CentC annotation
CRM elements were identified by aligning complete copies of CRM1, CRM2, CRM3, and CRM4 (Sharma and Presting 2008) to the assemblies using BLAST (v2.2.26). The output was filtered by applying a 150 bp alignment length cutoff and enforcing an E-value lower than 0.0001. CentC was identified by aligning a consensus sequence (Gent et al. 2017) using BLAST and applying a 30 bp alignment length cutoff and enforcing an E-value lower than 0.001. The amount of CentC and CRM in active centromeres and chromosome arms were then quantified using bedtools merge and bedtools intersect from bedtools (v2.28).
Genome size measurements of Oaxaca and Z. luxurians lines
Genome sizes were estimated by flow cytometry. Young leaf samples from single plants were sent to Plant Cytometry Services (Schijndel, the Netherlands) for flow cytometry measurements using Vinca major (2 C = 4.2 pg) as an internal standard. We also included the reference maize inbred B73 in each batch as a second internal control to reduce technical error. Genome sizes were divided by the size of the B73 genome (where B73 was assigned a value of 1.0). Genome sizes measured in this study are listed in Supplementary Table 2.
B73 X Oaxaca, B73 X Z. luxurians and CENH3-Ox-1 centromere analyses
CENH3 ChIP-seq SE150 data from the Oaxaca-B73, Z. luxurians-B73 and CENH3-Ox-1 overexpression lines described here were subsampled to 3.33 million reads. Additional ChIP and input samples from the parental Oaxaca and Z. luxurians lines were downloaded from SRP105290 (Gent et al. 2017). The subsampled reads were trimmed with TrimGalore (version 0.4.5) and mapped to Zm-B73-REFERENCE-NAM-5.0 (https://nam-genomes.org/) with BWA-mem (version 0.7.17) at default parameters (Li and Durbin 2009). Only uniquely mapped reads (defined with MAPQ scores of at least 20) were used for peak calling. Centromere sizes were determined using the same methods used for NAM centromere analysis, except that B73 30X genomic Illumina data were used as input reads for all samples. After the merging steps, small islands less than 100 kb were manually removed. The results were visualized using IGVTools (version 2.3.98) at coverage calculated on 5 kb intervals (Thorvaldsdóttir et al. 2013).
Centromere genotyping
Mapping ChIP-seq reads from Oaxaca to B73 revealed that Oaxaca centromeres 2, 3, 8, and 9 have similar locations as the centromeres in B73 (Supplementary Figure 2). Using ChIP-seq reads, we identified SNPs with GATK HaplotypeCaller (v3.8-1) at default parameters (Poplin et al. 2018). SNP2CAPS software (Thiel et al. 2004) was used to design Cleaved Amplified Polymorphic Sequence (CAPS) markers that distinguish B73 centromeres 2, 3, 8, and 9 from those in the Oaxaca line (Supplementary Table 3). Young leaf DNA was prepared (Clarke 2009) and PCR products digested with restriction enzymes (Supplemental Table 3) to identify lines homozygous for the B73 centromeres.
In Oaxaca-B73 F2 and BC1F2 progeny, we genotyped the samples directly using ChIP-seq data. ChIP-seq reads were mapped to Zm-B73-REFERENCE-NAM-5.0 with BWA-mem (version 0.7.17) at default parameters. The results were then used for SNP calling with GATK HaplotypeCaller at default parameters. If no SNPs were present in the centromeric region, the corresponding centromere was classified as homozygous for the B73 centromere.
Measuring CENH3 copy number and expression in CENH3-Ox-1 lines
We prepared the CENH3-Ox-1 line by transforming maize Hi-II with the construct gRNA-ImmuneCENH3 using Agrobacterium mediated transformation (Wang et al. 2021). The ImmuneCENH3 gene contains 6454 bp of the native CENH3 gene (coordinates Chr6:166705239-166711693 on Zm-B73-REFERENCE-NAM-5.0) with five silent codon changes to render it immune to a transacting guide RNA. The promoter includes 2184 bp of sequence upstream of the ATG. The transgene is sufficient to fully complement a cenh3 null allele.
Quantitative PCR was used to determine CenH3 gene copy number. Young leaf DNA was prepared (Clarke 2009) from three biological replicates from wild-type and CENH3-Ox-1 transgenic lines. qPCR was carried out using a BioRad CFX96 Real-Time PCR system using a SYBR Green qPCR kit (Thermo Fisher Scientific). The single copy Adh1 gene (Osterman and Dennis 1989) was used as an internal control gene. Primers are listed in Supplemental Table 3.
For RNA-seq, mRNA was prepared from young leaves of three wild-type and three overexpression lines using a plant total RNA kit (IBI Scientific IB47342). Eight-hundred nanogram of total RNA was used for library construction with a mRNA-seq kit (KAPA mRNA hyper prep kit #KK8580). RNA-seq reads were trimmed with Trimmomatic at the following parameters: LEADING : 3 TRAILING : 3 SLIDINGWINDOW : 4:15 MINLEN : 36 (Bolger et al. 2014), then the trimmed reads were mapped to Zm-B73-REFERENCE-NAM-5.0 with hisat2 at the following parameters: –min-intronlen 20, –max-intronlen 500000, –rna-strandness R (Kim et al. 2019). The alignments were converted to BAM files and sorted with SAMtools. Stringtie was used to compute gene expression levels using Transcripts Per Kilobase Million (TPM) = 1 as the cutoff (Pertea et al. 2015; Kim et al. 2019).
Nuclear protein isolation and protein blotting
Approximately 2 g of flash-frozen leaves or roots were collected and chopped into 1.5 mL pre-chilled nuclei extraction buffer (1 mM EDTA, 1x cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail, 10 mM Tris-HCl pH 7.5, 10 mM NaCl, 0.2% NP-40, 5 mM 2-mercaptoethanol, 0.1 mM PMSF). The mixture was poured through miracloth and filtered through a 40 µm cell strainer. Then, 30 µL of the filtered sample was stained with 4,6-diamidino-2-phenylindole and nuclei counted using fluorescence microscopy. Nuclei concentrations were normalized based on these measurements. The nuclei were centrifuged at 5,000 g for 5 minutes and the pellets flash-frozen and stored at −80°C until used for protein blots. Nuclei were resuspended in Laemmli buffer and loaded into 4%−20% Mini-PROTEAN® TGX™ Precast Protein Gels (Bio-Rad Cat #4561093). SDS-PAGE and protein blotting were performed according to Dawe et al. (2018). CENH3 was detected with anti-CENH3 antibodies (Zhong et al. 2002) (1:1,000 dilution) and normalized to total H4 histones revealed by anti-H4 antibodies (1:1,000 dilution, Abcam, ab7311). Primary antibodies were detected using anti-rabbit secondary antibodies (1:5000 dilution, Anti-Rabbit IgG HRP Linked Whole Ab Sigma Cat# GENA934-1ML). The band intensities were quantified with Image J (Schneider et al. 2012).
Statistical analysis
Welch’s analysis of variance (ANOVA) tests were performed to determine whether genome size and centromere size were significantly different across a variety of subgroups (Welch 1951). Pairwise comparisons among different subgroups were conducted with the available R package for Welch’s test (http://www.r-project.org; Dag et al. 2018). Significance was set at P < 0.05. To assess the relationship between centromere size and all parameters (including genome size, assembled chromosome size, CentC length, CRM length), Spearman’s rank correlation coefficient (r) was calculated with cor.test in R, with the method set as spearman (Spearman 1961).
Data availability
ChIP-seq data can be obtained from the NCBI Sequence Read Archive (ncbi.nlm.nih.gov/sra) under project PRJNA639705. RNA-seq data are available under project PRJNA688370. Scripts used in this study are available at https://github.com/dawelab/centromere_size. Strains and reagents are available upon request. Supplemental Material available at figshare: https://doi.org/10.25386/genetics.13697332.
Results
Limited impact of sequence on centromere location or size
In maize, the major centromere repeats are a tandem repeat known as CentC and an abundant class of Gypsy transposons known as Centromeric Retroelements (Wolfgruber et al. 2009). While both components are repetitive, they are diverse enough that many maize centromeres have been fully assembled, and a surprising number of short reads align to the assembled centromeres uniquely (Gent et al. 2012, 2015, 2017). For instance, seven B73 centromeres (2, 3, 4, 5, 8, 9, and 10) were assembled gaplessly in the recent B73-Ab10 assembly (Liu et al. 2020). This makes it possible to identify the sequence occupied by centromeric nucleosomes by aligning CENH3 ChIP-seq data from each inbred to the subset of assembled centromeres from that inbred. Functional centromere sizes can be estimated by identifying regions where the depth of ChIP-seq reads exceed an enrichment threshold, and enforcing a minimal peak size and maximal distance between peaks (Supplementary Figure 1 and Methods). Throughout this report, “centromere size” is defined as the length of DNA under the CENH3 ChIP-seq enrichment curve. We only analyzed centromeres that were fully scaffolded where sequence gaps (if any) were of known size.
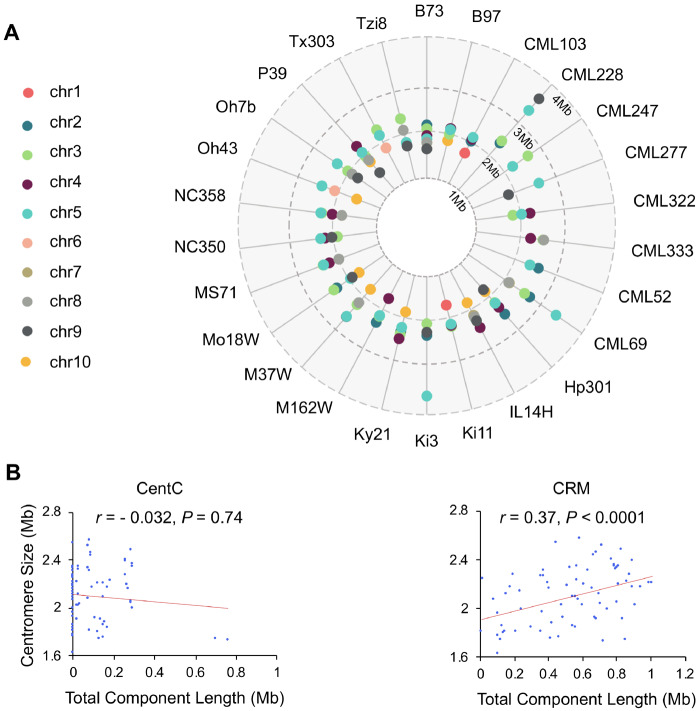
To assess natural centromere variation in a variety of maize lines, we measured centromere size in 26 inbred lines from the nested association mapping (NAM) population (McMullen et al. 2009). High-quality de novo genome assemblies of these lines have recently been completed (Hufford et al. 2021). CENH3 ChIP-seq data are available for most of the NAM inbreds (Schneider et al. 2016). We carried out CENH3 ChIP-seq for eight of the NAM lines, either because the data were absent from the prior study, showed poor ChIP efficiencies, or because we wanted biological replicates prepared under the same conditions (Supplemental Table 1). Alignment of ChIP-seq data to the assemblies revealed that of the 260 centromeres present, 110 centromeres were fully scaffolded (Figure 1A). Of these, 88 were assembled gaplessly and 22 contained one or more gaps of known size.
Figure 1.
Centromeric repeats and their relationship to centromere size in the 26 NAM inbred lines. (A) The sizes of 110 fully scaffolded centromeres in each inbred line as measured by CENH3 ChIP-seq. (B) Spearman’s rank correlation analysis between centromere size and the abundance of CentC and CRM. Only 73 centromeres without any gaps (of known or unknown size) were used for this analysis. Correlation coefficients (r) and P-values are indicated in the graph. The data show that CentC is not a primary driver of centromere size in maize, as is the case for alpha satellite in human. CRM shows a positive correlation and is commonly observed in all centromeres.
In humans the alpha satellite actively recruits CENP-A and directly contributes to centromere size (Aldrup-MacDonald et al. 2016; Iwata-Otsubo et al. 2017; Hoffmann et al. 2020). Our analysis of the CENH3-occupied regions of the 110 fully scaffolded centromeres revealed that larger centromeres do not necessarily have more CentC (Figure 1B). In contrast, the summed length of centromeric retroelements (CRM elements), which preferentially target active centromeres (Wolfgruber et al. 2009; Schneider et al. 2016), showed the expected positive correlation with CENH3-occupied area. Neither repeat is found exclusively with functional centromeres: 59.5% of the CRM elements and 30.1% of the CentC repeats in the assembled reference genomes lie in flanking pericentromeric areas. The actual percentages may be smaller given that unknown amounts of each are present in the gaps of the 150 centromeres excluded from this analysis. Nonetheless, it is clear that large expanses of CRM and CentC can exist outside of functional centromeres.
Positive correlation between centromere size and genome and chromosome sizes in NAM inbreds
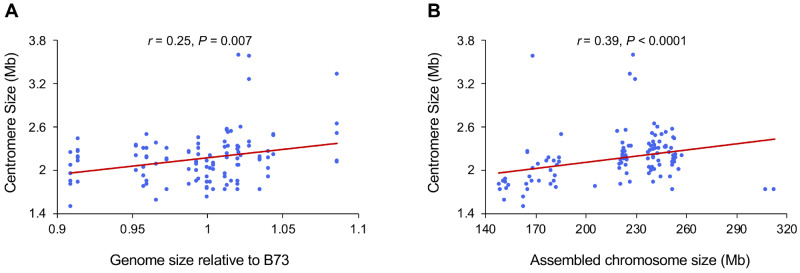
The NAM founder inbreds were drawn from a wide genetic and demographic range, including tropical and northern lines as well as popcorn and sweet corn (McMullen et al. 2009). The genome sizes among the NAM founder inbreds vary from 2.09 to 2.50 Gb (0.91 to 1.19 fold relative to B73), where most of the differences in genome size can be attributed to differences in the amount of tandem repeat arrays within heterochromatic regions known as knobs (Chia et al. 2012).
We plotted the 110 measured centromere sizes against genome size and chromosome size. The data revealed a weak but significant positive correlation between centromere size and genome size (Figure 2A), supporting a prior cross-species study that came to the same conclusion (Zhang and Dawe 2012). Although we have speculated that maize chromosomes each have similar sized centromeres (Zhang and Dawe 2012), our diverse collection of assembled centromeres revealed that larger chromosomes accommodate larger centromeres (Figure 2B). On human chromosomes, DNA sequence has a larger role in centromere specification, and this trend is not observed (Dumont et al. 2020).
Figure 2.
Correlation of centromere size with genome size and chromosome size. The graphs show data from 110 fully assembled centromeres in the 26 NAM genomes. (A) Spearman’s rank correlation analysis between genome size and centromere size. (B) Spearman’s rank correlation analysis between chromosome size and centromere size. When the > 3 Mb outliers were removed, the correlations were still significant (genome size/centromere size, r = 0.19, P < 0.05; chromosome size/centromere size, r = 0.43, P < 0.0001).
Centromeres expand when introduced into larger genomes
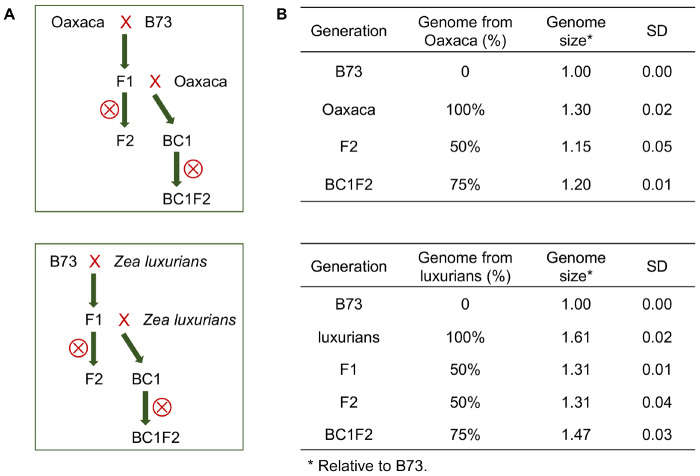
The only empirical data supporting a correlation between centromere size and genome size comes from an unnatural wide cross between maize and oat (Wang et al. 2014). We sought to confirm these results using natural crosses between the B73 inbred and two different genetic backgrounds: a maize landrace from Oaxaca Mexico with a genome about 1.3 times the size of the B73 genome, and the intercrossing species Z. luxurians with a genome size about 1.6 times the size of B73 (Oaxaca and Z. luxurians are not NAM lines). The genomes of these two accessions are larger primarily because they contain more heterochromatic knob repeats (Bilinski et al. 2018), although Z. luxurians also contains a larger proportion of retroelements (Tenaillon et al. 2011). Figure 3 shows the basic crossing schemes. We first crossed B73 with either Oaxaca or Z. luxurians to create F1s, which were self-crossed to create F2s or crossed again to the larger-genome parent to obtain BC1 lines. The BC1 lines were then self-crossed to create BC1F2 lines segregating for B73 centromeres. The genome sizes were measured for each cross using flow cytometry (Figure 3B and Supplementary Table 2). For the Oaxaca crosses, we found that the genomes of F2 progeny were 1.15 times larger than the B73 genome and that the BC1F2 progeny were 1.2 times larger. For the Z. luxurians crosses, the genomes of F2 progeny were 1.31 times larger than the B73 genome and the BC1F2 progeny were 1.47 times larger. The seven fully assembled B73 centromeres segregate in these progeny, providing the opportunity to measure changes in CENH3 area as a function of genome size in the context of identical centromere sequences.
Figure 3.
Crossing scheme for generating maize lines with different genome sizes. (A) Crossing schemes for transferring B73 centromeres into the Oaxaca and Z. luxurians backgrounds. (B) Genome sizes of B73, Oaxaca, Z. luxurians and their hybrids. Genome sizes are averages from 3 to 11 plants per family. ⊗ is the self-cross symbol. SD indicates standard deviation.
B73 X Oaxaca hybrids
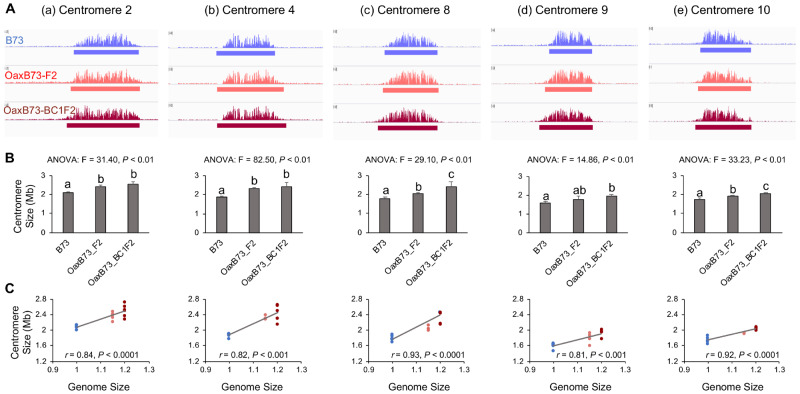
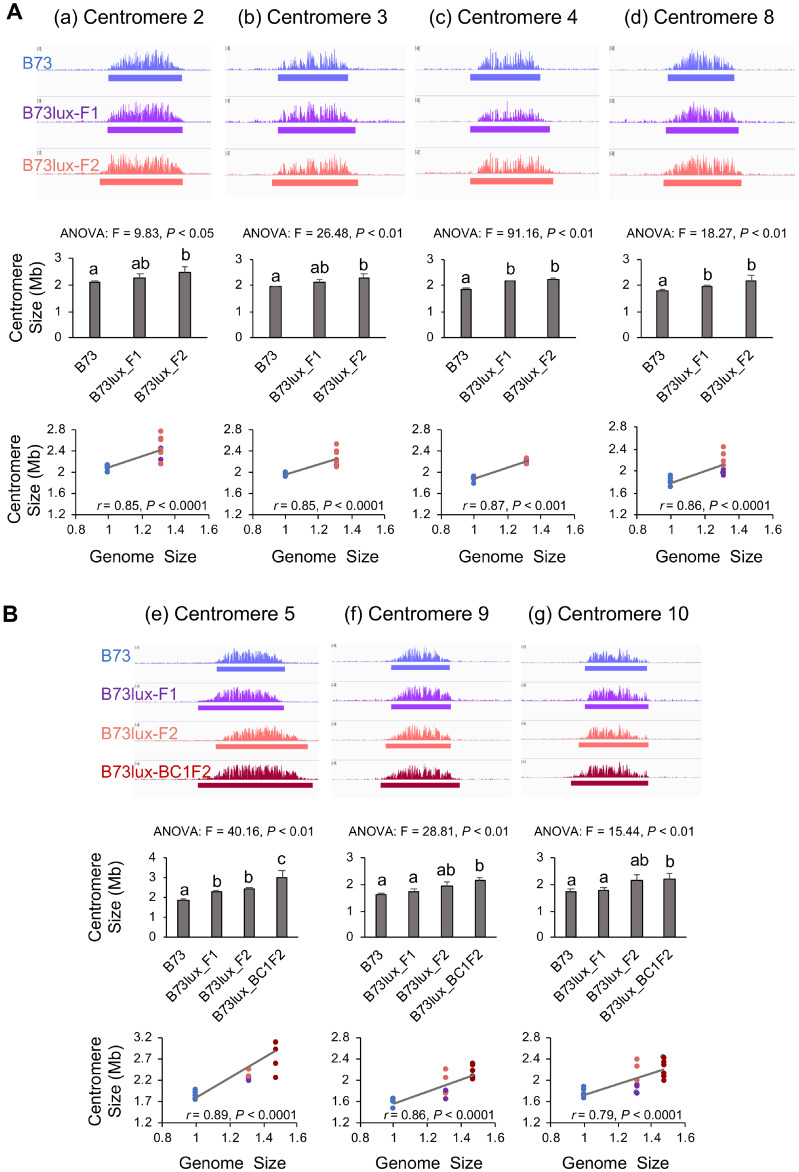
While the Oaxaca genome has not been sequenced, CENH3 ChIP-seq revealed that multiple centromeres from Oaxaca are similar to those in B73 (Supplementary Figure 2A). To avoid complexities associated with mapping two centromeres onto a single reference, we developed PCR markers to distinguish Oaxaca centromeres and focused our analysis entirely on B73 centromeres that were homozygous in the F2 or BC1F2 progeny (Supplementary Table 3). Analysis of the data revealed that four of the B73 centromeres examined (2, 4, 8, and 10) were significantly larger in the F2 and BC1F2 Oaxaca backgrounds than in their original smaller-genome context, and that the increases were correlated with genome size (Figure 4, Supplementary Table 4). The single exception was centromere 9 which had a statistically similar size in the B73 and the Oaxaca F2 progeny.
Figure 4.
Centromeres are expanded in B73 X Oaxaca F2 and BC1F2 progeny. (A) CENH3-ChIP profiles of B73, Oaxaca X B73 F2 and Oaxaca X B73 BC1F2 progeny for five centromeres. Centromere size is defined as the length of DNA under the ChIP-seq enrichment curve (see solid bars under the read depth plots). (B) ANOVA analysis of centromere sizes across different lines. Bar graphs show mean centromere size comparison among different lines. Letters represent different groups that are statistically different (P < 0.05). Centromere sizes in the F2 and BC1F2 progeny were significantly larger than in B73, with the sole exception of centromere 9 in the F2 generation. Bars represent SD. (C) Spearman’s rank correlation analyses of centromere sizes across different lines. Dots represent different individuals. Blue: B73, orange/red: F2, dark red: BC1F2. Genome sizes are expressed relative to B73, and are averages based on 3–11 individuals.
B73 X Z. luxurians hybrids
The centromeres in Z. luxurians are known to contain long CentC arrays on every centromere (Albert et al. 2010). ChIP data from Z. luxurians consistently yielded high enrichment for CentC, but when these data were mapped to the B73 reference, there were no clear peaks (Supplementary Figure 2B). This is because uniquely mapping reads generally exclude simple tandem repeats such as CentC. The absence of significant ChIP-seq read alignment from Z. luxurians centromeres allowed us to assess B73 centromere size in both the heterozygous and homozygous conditions.
The data reveal that in first generation F1 hybrids between B73 and Z. luxurians, there were little or no changes in centromeres size. We observed only minor increases in the sizes of Cen4, Cen5, and Cen8 in F1 progeny and no obvious changes in Cen2, Cen3, Cen9, and Cen10 (Figure 5). The differences were more pronounced in the F2 progeny where Cen2, Cen3, Cen4, Cen5, and Cen8 were significantly larger and Cen9 and Cen10 showed positive trends that were not statistically significant (Figure 5, Supplementary Table 5). An example is centromere 5, which is 1.86 Mb in the B73 inbred but expanded to 2.4 Mb in the B73 X Z. luxurians F2 progeny. Although there were only three B73 centromeres segregating in the BC1F2 population (Cen5, Cen9, and Cen10), all three were significantly expanded, confirming the trends observed in the F2 progeny. Taken together, the data from NAM centromere comparisons and Oaxaca and Z. luxurians crosses indicate that centromere size is positively correlated with genome size.
Figure 5.
Centromeres are expanded in the B73 X Z. luxurians F2 and BC1F2 progeny. Each panel shows ChIP-seq, ANOVA and Spearman’s rank correlation analyses as described in Figure 4. (A) Data for B73 X Z. luxurians F1 and F2 progeny from four centromeres. (B) Data for B73 X Z. luxurians F1, F2, and BC1F2 progeny from three centromeres (BC1F2 data were available for these three centromeres only). Letters represent different groups that are statistically different (P < 0.05). In the F1, the sizes of Cen4, Cen5, and Cen8 were significantly larger than in B73 (and the others were not). In the F2, the sizes of Cen2, Cen3, Cen4, Cen5, and Cen8 were significantly larger than in B73 (while Cen9 and Cen10 were not). The sizes of all centromeres in the BC1F2 were significantly larger than in B73. Blue: B73, purple: F1, orange/red: F2, dark red: BC1F2. Genome sizes are expressed relative to B73, and are averages based on 3–10 individuals.
Threefold overexpression of CENH3 does not affect centromere size
It is possible that centromere size is determined by the total amount of CENH3 that is available to bind to centromeric DNA. This hypothesis is supported by early work in Drosophila showing that extreme overexpression of CENP-A/Cid caused a spreading of centromere locations to ectopic sites (Heun et al. 2006). A recent study of maize lines overexpressing a YFP-tagged version of CENH3 described subtle shifting of centromere locations, partially supporting this view (Feng et al. 2020). However, the YFP-tagged CENH3 gene was not sufficient to complement a strong hypomorphic mutation, indicating it may not be fully functional or not expressed in all required cell types (Feng et al. 2020).
We were able to address this hypothesis using materials created for a study of how a cenh3 null mutation behaves in crosses (Wang et al. 2021). CRISPR-Cas9 was used to create a stop codon in the N-terminal tail of the cenh3 gene. As a means to propagate the null, we also introduced a complete genomic copy of the transgene that differs from wild-type by five silent nucleotide changes. RNA-seq of one of the transgenic lines (CENH3-Ox-1) showed approximately fourfold higher expression of the transgene than the wild-type copy of CENH3 (Figure 6B). Quantitative PCR analysis of genomic DNA from the CENH3-Ox-1 line suggested that the high CENH3 expression in this line is caused by multiple transgene insertions (Figure 6B), which is a frequent occurrence in Agrobacterium transformants (Shou et al. 2004; Jupe et al. 2018). These four transgenes are sufficient to fully complement the cenh3 null mutation (Wang et al. 2021).
Figure 6.
Centromere size is stable in the CENH3-overexpression lines. (A) Protein blot analysis of maize CENH3 expression levels in roots of wild-type (WT) and CENH3-Ox-1 (Ox) lines. Nuclear protein was diluted to 0.25X, 0.5X, and 1X. The same blot was incubated with antibodies to histone H4 as a loading control. (B) Quantification of CENH3 gene copy number. DNA (gene) copy number was estimated by qPCR, mRNA expression was estimated from RNA-seq, and protein levels interpreted as the relative staining intensity of CENH3 and histone H4 on protein blots. Error bars show standard deviation. WT expression was set to one in each experiment. CENH3 is a single copy gene in WT lines. (C and D) CENH3 ChIP-seq profiles for B73, WT siblings of the Ox lines, and Ox lines for centromeres 4 and 10. The window size is 5 Mb. (E and F) ANOVA analyses of centromere sizes across different lines. There were no significant differences in the sizes of Cen4 and Cen10 between WT and Ox (P < 0.05). The unit of centromere size is Mb. Bars in (B, E, and F) represent SD.
Analysis of leaf and root protein revealed that CENH3-Ox-1 lines have approximately threefold higher nuclear CENH3 levels than wild-type lines (Figure 6, A and B), providing excellent material to test whether altered CENH3 levels change centromere size. The transgenic lines have a mixed genetic background but centromeres 4 and 10 are identical to those in B73. CENH3 ChIP-seq analysis of these two centromeres revealed no significant size differences between CENH3-Ox-1 lines and wild-type siblings or the B73 inbred (Figure, 6C–F). We also did not observe any new peaks in noncentromeric regions. These data indicate that centromere size, as measured by the length of DNA occupied by CENH3, is not affected by a threefold increase in CENH3 protein levels. Our results leave open the possibility that higher levels of overexpression might alter centromere size or promote ectopic centromere formation in maize. CENP-A expression levels of up to ∼10 to 30-fold higher than wild-type were required to see cytological defects in Drosophila and human (Van Hooser et al. 2001; Heun et al. 2006; Shrestha et al. 2017).
Discussion
Here, we combine data from a large collection of centromere sequences, empirical manipulations of genome size, and a novel CENH3 overexpression line to test how maize cells determine centromere size. Data from many sources suggest that at least in plants, DNA sequence alone does not determine centromere location. Here, viewed from a total centromere size perspective, we again find that centromere sequence does not itself determine the distribution of CENH3 (Figure 2B). Comparisons among the fully assembled centromeres in NAM inbreds revealed a weak positive correlation between total genome size and centromere size (Figure 2A) as predicted from earlier work (Zhang and Dawe 2012; Wang et al. 2014). The weakness of the correlation suggests that other factors also influence centromere size, possibly including small sequence motifs that can have an effect on local positioning of CENH3 nucleosomes (Gent et al. 2011). In the oat-maize addition experiment, some centromeres expanded unidirectionally, suggesting barriers to CENH3 encoded in the DNA (Wang et al. 2014). It is also possible that our observed correlation between chromosome size and centromere size (Figure 2B) reflects inherent structural constraints encoded in the DNA.
CENP-A/CENH3 binds directly to DNA and is widely interpreted as a limiting factor for centromere establishment. Early work in Drosophila demonstrated that heavily overexpressed (∼10- to 30-fold) CENP-A was inappropriately distributed along chromosome arms, where it was sufficient to recruit all overlying kinetochore proteins and activate spurious centromeres (Heun et al. 2006). Similarly, extreme overexpression of human CENP-A resulted in mislocalization of centromeric proteins and chromosome instability (Van Hooser et al. 2001; Shrestha et al. 2017). However, we found that in maize, milder overexpression of native CENH3 protein by threefold had no discernible effect on the size or distribution of CENH3 as assayed by ChIP (Figure 6). These results suggest that other factors limit the incorporation of excess CENH3 in maize, with likely candidates being the histone chaperones that direct CENH3 to centromeric locations (Mizuguchi et al. 2007; Dunleavy et al. 2009; Foltz et al. 2009; Chen et al. 2014). Among these are KNL2, which is required for CENH3 deposition in Arabidopsis (Lermontova et al. 2013) and NASPSIM3, which modulates soluble CENH3 levels (Le Goff et al. 2020). Another potential limiting factor is CENP-C, a key inner centromere protein that has been implicated in multiple aspects of centromere specification and stability (Du et al. 2010; Mitra et al. 2020).
Prior results demonstrated that when maize centromeres were transferred into the oat genome, their sizes increased roughly twofold, in line with expectations based on the difference in genome size (Zhang and Dawe 2012; Wang et al. 2014). However, the wide oat-maize cross rarely succeeds and does not result in a stable hybrid (Kynast et al. 2001). Here we took a different approach of making natural crosses within Zea and tracking changes in centromere size over several generations. The results demonstrate that B73 centromeres increase in size when crossed into the larger Oaxaca and Z. luxurians backgrounds. Changes were clearly evident in F2 as well as BC1F2 progeny, revealing heritable increases (Figures 4 and 5). However, the size increases were less apparent in the first generation B73 X Z. luxurians individuals, consistent with prior results with the same F1 cross (Gent et al. 2017). The weak effect in the F1 argues against the possibility that the increases in centromere size observed in later generations are a result of extreme overexpresssion of CENH3. Multiple cellular generations or passage through meiosis may be required for CENH3 and associated binding partners and redistribute along chromosomes and reach a new centromere size equilibrium. Taken together, our results indicate that centromere size is scalable and responsive to changes in genome or cell size.
The impact of genome size on centromere size can be explained as a general cellular scaling process. Many forms of evidence from multiple species show strong correlations between genome size, nuclear size, and cell size (Price et al. 1973; Gregory 2001; Cavalier-Smith 2005; Gillooly et al. 2015; Robinson et al. 2018). With remarkably few exceptions, the entire cellular system scales in response to changes in genome size (Gregory 2001; Schmoller and Skotheim 2015; Amodeo and Skotheim 2016). These trends have been explained as an outcome of the fact that larger genomes are packaged into larger nuclei, and that nuclear size is at least indirectly correlated with cell size (Gregory 2001). Larger cells have more protein per cell and more and larger macromolecular structures such as mitochondria, microtubules, and ribosomes (Schmoller and Skotheim 2015). Given that the number of centromeres is constrained by the number of chromosomes, any increases in centromere size will be manifested as extensions of existing centromeres spread over larger chromosomal areas.
The scaling model not only requires scalable centromeres, but a deposition mechanism that is responsive to the amount of soluble precursors. A prior study of CENP-A dynamics in human cells provides support for the view that a mass-action mechanism regulates the number of CENP-A molecules bound to DNA (Bodor et al. 2014). The authors showed that about 4% of total CENP-A binds to centromeres over a range of natural expression variation, implying that CENP-A deposition varies with the amount of CENP-A available to bind (Bodor et al. 2014). They further observed that up to 2.5-fold increases in the quantity of CENP-A on centromeric DNA (perhaps increasing the density or size of the CENP-A domain) did not result in corresponding changes in the amounts of the conserved kinetochore proteins CENP-C or NDC80, suggest that these and/or other key kinetochore proteins are limiting and help to buffer the effects of CENP-A overexpression. The available information from both human and maize show that while centromere sizes are malleable, moderate overexpression of CENP-A/CENH3 alone does not alter the size of the functional centromere domain, consistent with the view that multiple limiting factors together contribute to a stable centromere size equilibrium.
Acknowledgments
We appreciate the support of the Georgia Genomics and Bioinformatics Core facility, the Georgia Advanced Computing Resource Center, and the UGA Plant Biology greenhouse staff.
Funding
This work was funded by grant 1444514 from the National Science Foundation.
Competing Interests
The authors declare no competing interests.
Literature cited
- Albert PS, Gao Z, Danilova TV, Birchler JA.. 2010. Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet Genome Res. 129:6–16. [DOI] [PubMed] [Google Scholar]
- Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA.. 2016. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26:1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Skotheim JM.. 2016. Cell-size control. Cold Spring HarbPerspect Biol. 8:a019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB, Ward J, Jenkins G.. 1981. The relationship between nuclear DNA content and centromere volume in higher plants. J Cell Sci. 47:91–115. [DOI] [PubMed] [Google Scholar]
- Bilinski P, Albert PS, Berg JJ, Birchler JA, Grote MN, et al. 2018. Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLoS Genet. 14:e1007162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, et al. 2014. The quantitative architecture of centromeric chromatin. Elife 3:e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudichevskaia A, Houben A, Fiebig A, Prochazkova K, Pecinka A, et al. 2019. Depletion of KNL2 results in altered expression of genes involved in regulation of the cell cycle, transcription, and development in Arabidopsis. IJMS 20:5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot. 95:147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Dechassa ML, Bettini E, Ledoux MB, Belisario C, et al. 2014. CAL1 is the Drosophila CENP-A assembly factor. J Cell Biol. 204:313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia J-M, Song C, Bradbury PJ, Costich D, de Leon N, et al. 2012. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 44:803–807. [DOI] [PubMed] [Google Scholar]
- Clarke JD. 2009. Cetyltrimethyl Ammonium Bromide (CTAB) DNA Miniprep for Plant DNA Isolation. Cold Spring Harb Protoc. 2009:pdb.prot5177. [DOI] [PubMed] [Google Scholar]
- Dag O, Dolgun A, Konar Naime M.. 2018. onewaytests: an R package for one-way tests in independent groups designs. R J. 10:175. [Google Scholar]
- Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, et al. 2018. A kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell 173:839–850.e18. [DOI] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK.. 2010. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6:e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Gamba R, Gestraud P, Klaasen S, Worrall JT, et al. 2020. Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 39:e102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, et al. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137:485–497. [DOI] [PubMed] [Google Scholar]
- Feng C, Yuan J, Bai H, Liu Y, Su H, et al. 2020. The deposition of CENH3 in maize is stringently regulated. Plant J. 102:6–17. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, et al. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137:472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French BT, Westhorpe FG, Limouse C, Straight AF.. 2017. Xenopuslaevis M18BP1 directly binds existing CENP-A nucleosomes to promote centromeric chromatin assembly. Dev Cell 42:190–199.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Dong Y, Jiang J, Dawe RK.. 2012. Strong epigenetic similarity between maize centromeric and pericentromeric regions at the level of small RNAs, DNA methylation and H3 chromatin modifications. Nucleic Acids Res. 40:1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Schneider KL, Topp CN, Rodriguez C, Presting GG, et al. 2011. Distinct influences of tandem repeats and retrotransposons on CENH3 nucleosome positioning. Epigenet Chromatin 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Wang N, Dawe RK.. 2017. Stable centromere positioning in diverse sequence contexts of complex and satellite centromeres of maize and wild relatives. Genome Biol. 18:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Wang K, Jiang J, Dawe RK.. 2015. Stable patterns of CENH3 occupancy through maize lineages containing genetically similar centromeres. Genetics 200:1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly JF, Hein A, Damiani R.. 2015. Nuclear DNA content varies with cell size across human cell types. Cold Spring Harb Perspect Biol. 7:a019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev. 76:65–101. [DOI] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, et al. 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10:303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann S, Izquierdo HM, Gamba R, Chardon F, Dumont M, et al. 2020. A genetic memory initiates the epigenetic loop necessary to preserve centromere position. EMBO J. 39:e105505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, et al. 2021.De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory 2021.01.14.426684. [DOI] [PMC free article] [PubMed]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmátal L, et al. 2017. Expanded satellite repeats amplify a discrete CENP-a nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 27:2365–2373.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe F, Rivkin AC, Michael TP, Zander M, Motley TS, et al. 2018. The complex architecture of plant transgene insertions. bioRxiv 282772.
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kixmoeller K, Allu PK, Black BE.. 2020. The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol. 10:200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira SB, et al. 2001. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 125:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, et al. 2014. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell 53:631–644. [DOI] [PubMed] [Google Scholar]
- Le Goff S, Keçeli BN, Jeřábková H, Heckmann S, Rutten T, et al. 2020. The H3 histone chaperone NASPSIM3 escorts CenH3 in Arabidopsis. Plant J. 101:71–86. [DOI] [PubMed] [Google Scholar]
- Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, et al. 2013. Arabidopsis KINETOCHORE NULL2 Is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 25:3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, , Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Seetharam AS, Chougule K, Ou S, Swentowsky KW, et al. 2020. Gapless assembly of maize chromosomes using long-read technologies. Genome Biol. 21:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. 2016. Cell geometry: how cells count and measure size. Annu Rev Biophys. 45:49–64. [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, et al. 2009. Genetic properties of the maize nested association mapping population. Science 325:737–740. [DOI] [PubMed] [Google Scholar]
- Mitra S, Srinivasan B, Jansen LET.. 2020. Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control. J Cell Biol. 219:e202005099. 10.1083/jcb.202005099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C.. 2007. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129:1153–1164. [DOI] [PubMed] [Google Scholar]
- Murillo-Pineda M, Jansen LET.. 2020. Genetics, epigenetics and back again: lessons learned from neocentromeres. Exp Cell Res. 389:111909. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, et al. 2004. Sequencing of a rice centromere uncovers active genes. Nat Genet. 36:138–145. [DOI] [PubMed] [Google Scholar]
- Oliveira LC, Torres GA.. 2018. Plant centromeres: genetics, epigenetics and evolution. MolBiol Rep. 45:1491–1497. [DOI] [PubMed] [Google Scholar]
- Osterman JC, Dennis ES.. 1989. Molecular analysis of the ADH1-Cm allele of maize. Plant Mol Biol. 13:203–212. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, et al. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory 2018.07.24.201178.
- Price HJ, Sparrow AH, Nauman AF.. 1973. Correlations between nuclear volume, cell volume and DNA content in meristematic cells of herbaceous angiosperms. Experientia 29:1028–1029. [Google Scholar]
- Quinlan AR. 2014. BEDTools: the swiss-army tool for genome feature analysis. CurrProtoc Bioinformatics 47:11.12.1–34.[10.1002/0471250953.bi1112s47] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T.. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DO, Coate JE, Singh A, Hong L, Bush M, et al. 2018. Ploidy and size at multiple scales in the Z Sepal. Plant Cell 30:2308–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC.. 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137:1173–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann M, Talbert P, Demidov D, Kuhlmann M, Rutten T, et al. 2017. Targeting of Arabidopsis KNL2 to centromeres depends on the conserved CENPC-k Motif in its C terminus. Plant Cell 29:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller KM, Skotheim JM.. 2015. The biosynthetic basis of cell size control. Trends Cell Biol. 25:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider KL, Xie Z, Wolfgruber TK, Presting GG.. 2016. Inbreeding drives maize centromere evolution. ProcNatl Acad Sci USA. 113:E987–E996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Presting GG.. 2008. Centromeric retrotransposon lineages predate the maize/rice divergence and differ in abundance and activity. Mol Genet Genomics 279:133–147. [DOI] [PubMed] [Google Scholar]
- Shou H, Frame BR, Whitham SA, Wang K.. 2004. Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breed. 13:201–208. [Google Scholar]
- Shrestha RL, Ahn GS, Staples MI, Sathyan KM, Karpova TS, et al. 2017. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8:46781–46800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman C. 1961. The proof and measurement of association between two things. In: Jenkins JJ, editor.Studies in Individual Differences: The Search for Intelligence. p. 45–58. [Google Scholar]
- Tenaillon MI, Hufford MB, Gaut BS, Ross-Ibarra J.. 2011. Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome BiolEvol. 3:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T, Kota R, Grosse I, Stein N, Graner A.. 2004. SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 32:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. BriefBioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, et al. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 114:3529–3542. [DOI] [PubMed] [Google Scholar]
- Wang K, Wu Y, Zhang W, Dawe RK, Jiang J.. 2014. Maize centromeres expand and adopt a uniform size in the genetic background of oat. Genome Res. 24:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Gent JI, Dawe RK.. 2021. Haploid induction by a maize cenh3 null mutant. Sci Adv. 7:eabe2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch BL. 1951. On the comparison of several mean values: an alternative approach. Biometrika 38:330–336. [Google Scholar]
- Wolfgruber TK, Sharma A, Schneider KL, Albert PS, Koo D-H, et al. 2009. Maize centromere structure and evolution: sequence analysis of centromeres 2 and 5 reveals dynamic Loci shaped primarily by retrotransposons. PLoSGenet. 5:e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dawe RK.. 2012. Total centromere size and genome size are strongly correlated in ten grass species. Chromosome Res. 20:403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, et al. 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ChIP-seq data can be obtained from the NCBI Sequence Read Archive (ncbi.nlm.nih.gov/sra) under project PRJNA639705. RNA-seq data are available under project PRJNA688370. Scripts used in this study are available at https://github.com/dawelab/centromere_size. Strains and reagents are available upon request. Supplemental Material available at figshare: https://doi.org/10.25386/genetics.13697332.