Abstract
Single-cell RNA sequencing (scRNAseq) experiments provide a powerful means to identify clusters of cells that share common gene expression signatures. A major challenge in scRNAseq studies is to map the clusters to specific anatomical regions along the body and within tissues. Existing data, such as information obtained from large-scale in situ RNA hybridization studies, cell type specific transcriptomics, gene expression reporters, antibody stainings, and fluorescent tagged proteins, can help to map clusters to anatomy. However, in many cases, additional validation is needed to precisely map the spatial location of cells in clusters. Several approaches are available for spatial resolution in Drosophila, including mining of existing datasets, and use of existing or new tools for direct or indirect detection of RNA, or direct detection of proteins. Here, we review available resources and emerging technologies that will facilitate spatial mapping of scRNAseq clusters at high resolution in Drosophila. Importantly, we discuss the need, available approaches, and reagents for multiplexing gene expression detection in situ, as in most cases scRNAseq clusters are defined by the unique coexpression of sets of genes.
Keywords: Drosophila, single-cell RNAseq, spatial mapping, in situ hybridization, RNA detection, protein detection, GAL4-UAS, gene expression
Introduction
Cells within an organism or tissue have traditionally been categorized as specific cell types based on their morphology, function, and expression of a few marker genes (Trapnell 2015; 2017). In recent years, advances in single-cell and single-nucleus RNA sequencing (hereafter, scRNAseq) have provided powerful new tools for identifying cell types and precisely defining the molecular signatures of cell types, subtypes, and states (Trapnell 2015). In addition to technological advances, corresponding platforms for data analysis, visualization, and mining of scRNAseq datasets have also emerged, including the Seurat analysis platform (http://satijalab.org/seurat/) (Stuart et al. 2019), automated single-cell analysis pipeline (ASAP) (https://asap.epfl.ch) (Gardeux et al. 2017; David et al. 2020), SCope visualization platform (http://scope.aertslab.org) (Davie et al. 2018), DRscDB data mining resource (https://www.flyrnai.org/tools/single_cell/) (Hu et al. 2021), Broad Institute Single Cell Portal (https://singlecell.broadinstitute.org/single_cell), and EBI-EMBL Single Cell Expression Atlas meta-database of scRNAseq datasets (https://www.ebi.ac.uk/gxa/sc/home). Through analysis of scRNAseq data, distinct cell types found in a given sample can be identified based on the expression of a unique set of genes or a combination of coexpressed genes.
For Drosophila, efforts to generate high-quality scRNAseq data include studies by individual laboratories and by the Fly Cell Atlas (FCA; https://flycellatlas.org), a consortium of Drosophila researchers and informatics experts (for a recent review see Li 2020). The outcomes of these efforts include unprecedented new models of cell types present in adult flies, in specific dissected tissues, and at specific developmental stages. The typical output from scRNAseq data analysis is a set of defined clusters visualized as a Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2018; McInnes et al. 2018) or t-distributed stochastic neighbor embedding (t-SNE) plot (van der Maaten and Hinton 2008). The cell clusters in UMAP or t-SNE plots are generated computationally, based on the pattern of expression (or lack thereof) of “marker genes” that together define a given cluster and distinguish it from others.
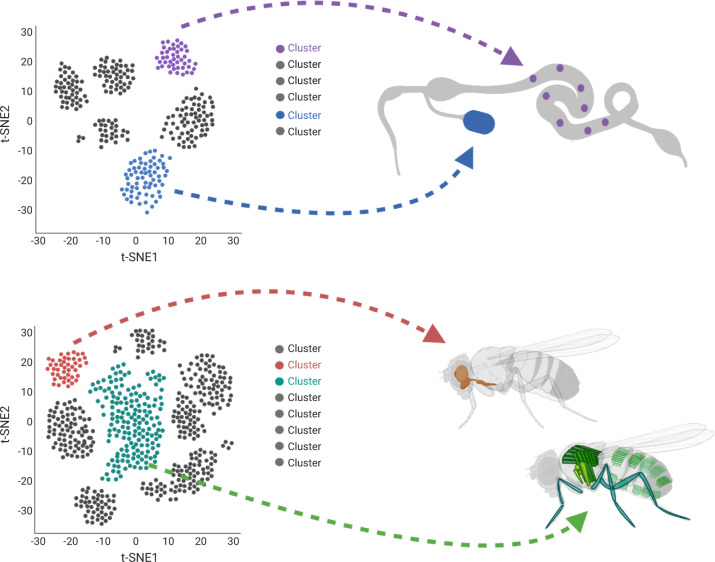
The cells in a single cluster can have any distribution within the sample that was analyzed (Figure 1). For example, they might be adjacent to one another within the tissue, enriched in a specific subregion of the tissue, or scattered throughout the tissue. Moreover, in cases where more than one tissue is analyzed, as for analysis of embryos or the entire adult, clusters might correspond to a specific stripe (e.g., along the anterior-posterior axis of the embryo) or to a cell type distributed over the entire body (e.g., hemocytes). Mapping clusters to the anatomy is necessary for creating a complete map of the body and, importantly, can inform our understanding of developmental processes, physiology, and homeostasis. For example, learning that a given specialized cell type is localized to a specific niche has different implications, and leads to different experimental hypotheses regarding function, as compared with learning that a cell type is distributed across a tissue or the whole organism.
Figure 1.
Spatial resolution of cells associated with scRNAseq clusters. Cell clusters defined by differential gene expression can be visualized in two-dimensional space using UMAP or t-SNE plots but these clusters have no defined relationship to the tissue or organisms from which the cell sample was obtained. Top: spatial mapping of clusters derived from scRNAseq of a tissue. Cells in a given cluster can have any distribution in the tissue: adjacent, dispersed, and so on. Bottom: mapping of clusters derived from scRNAseq of whole adults. Cells in a given cluster can have any distribution in the body. This figure was created using BioRender.com.
Mapping of scRNAseq clusters to the anatomy is necessarily based on the expression of the differentially expressed genes (DEGs) that define the clusters. While this can be relatively easily done if a cluster can be defined by the expression of a single DEG, the task is more complicated if a group of cells can only be defined using a combination of DEGs. Indeed, although major cell types can usually be identified based on the expression of a few specific genes, further subdivision in functionally different subtypes is usually based on the expression of a combination of genes. For example, within the hemocyte or blood cell repertoire of Drosophila, three distinct populations are well described—macrophage-like plasmatocytes, platelet-like crystal cells, and giant cell-like lamellocytes (Banerjee et al. 2019)—and can easily be identified based on the expression of single marker genes (Evans et al. 2014; Banerjee et al. 2019) such as NimC1 for plasmatocytes, prophenoloxidase-1 (PPO1) for crystal cells, and PPO3 for lamellocytes. However, in the adult fly gut, the co-expression pattern of a set of approximately 2–5 genes that encode peptide hormones in individual enteroendocrine cells could distinguish among different subtypes of enteroendocrine cells (Guo et al. 2019; Hung et al. 2020). Moreover, in the nervous system, a minimum of eleven genes were needed to distinguish among olfactory projection neuron subtypes (Li et al. 2017).
In this study, we first briefly discuss how DEGs are used to associate clusters with cell types. We then describe bioinformatics approaches used to associate cells with specific anatomical locations. As the predictions that result from these analyses must be experimentally validated, we next present an analysis of existing and new methods for direct detection of RNA, indirect detection of RNA, and detection of proteins. Throughout, we aim to provide both a thoughtful discussion of the current state-of-the-art and practical resources, including a list of relevant large-scale datasets and fly stock collections (Table 1), a list of relevant computational tools and protocols (Table 2), and a proposed workflow (Figure 2).
Table 1.
Existing datasets and resources relevant to spatial mapping in Drosophila
| Reagent or Resource | Gene or Protein Coverage | URL and/or reference(s) |
|---|---|---|
| Large-scale spatially resolved datasets | ||
| BDGP in situ hybridization project | 8,526 genes, embryonic stages | |
| FlyFISH | ∼10,000 genes, multiple stages, and tissues | |
| Dresden Ovary Table (DOT) | ∼7,000 genes, adult ovary (all stages of oogenesis) | |
| Enhancer traps, other reporters, and protein fusions | ||
| BDSC lacZ enhancer traps | 1,345 lines, 1,079 genes | https://bdsc.indiana.edu/stocks/in/lacZ_trap.html |
| BDSC lacZ reporters | 476 lines, 64 genes | https://bdsc.indiana.edu/stocks/misc/lacZ.html |
| BDSC and Kyoto GAL4 enhancer traps | ∼6,000 lines, ∼2,000 genes | |
| BDSC LexA and QF | ∼2,000 lines, ∼750 genes | |
| Janelia FlyLight GAL4a | ∼3,000 lines, ∼70 genes | |
| VDRC Vienna Tiles GAL4a | ∼200 lines, ∼160 genes | https://stockcenter.vdrc.at/control/library_vt (Tirian and Dickson 2017) |
| BDSC split-GAL4a | ∼4,000 lines, ∼950 genes | https://bdsc.indiana.edu/stocks/gal4/split_intro.html |
| MiMIC/CRIMIC GAL4 | ∼1,600 lines, 1,500 genes | |
| BDSC FP traps | ∼900 lines, ∼790 genes |
https://bdsc.indiana.edu/stocks/gfp/gfp_fluortrap.html (Morin et al. 2001; Kelso et al. 2004; Buszczak et al. 2007; Quinones-Coello et al. 2007; Singari et al. 2014; Nagarkar-Jaiswal et al. 2015) |
| VDRC fTRG FlyFos lines | 880 lines, 826 genes | |
| Antibody and protein tagging resources—information | ||
| FlyBase antibody information | https://wiki.flybase.org/wiki/FlyBase:Antibodies | |
| pAbmAbs antibody reviews | http://pabmabs.com | |
| Labome Validated Antibody Database | 472 mAbs, 280 pAbs | https://www.labome.com/index.html |
| Antibody and protein tagging resources—physical resources | ||
| Developmental Studies Hybridoma Bank | 278 mAbs for 266 genes | https://dshb.biology.uiowa.edu/collections/drosophila-antigens |
| Scarless gene editing | GFP, HA, FLAG (plasmid collection) | |
Note that most of these lines are not useful for scRNAseq cluster validation since they were generated as transgenes under the control of short regulatory regions (see text).
Table 2.
Computational tools and experimental protocols relevant to spatial mapping in Drosophila
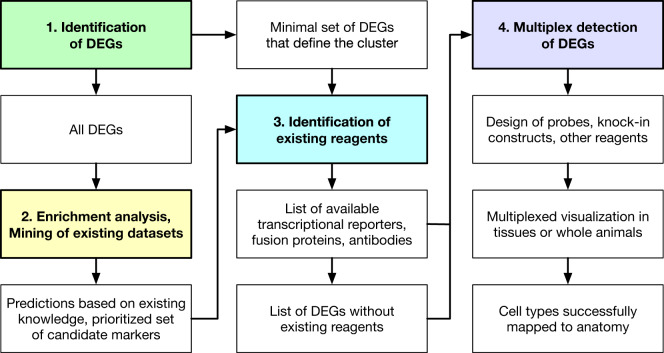
Figure 2.
Workflow for spatial resolution in Drosophila. Identification of differentially expressed (DE) genes associate with scRNAseq cell clusters provides a starting point for spatial mapping (step 1). Bioinformatics analyses (step 2) can help predict spatial mapping and prioritize candidate DEGs. Once a minimal and/or prioritized list of DEGs for experimental analysis has been defined, researchers can look for existing resources such as enhancer trap, MiMIC/CRIMIC, or GAL4 fly stocks (step 3). Finally, multiplex detection of DEGs is used to validate or newly determine spatial mapping in a given tissue or whole animal (step 4).
Use of top-enriched genes to associate clusters with cell types
Following scRNAseq, cells are grouped into clusters based on the expression in single cells of highly variable features. An immediate and major step that follows is to associate clusters with cell types or states. With Seurat, association of a cluster with a cell type is done by examining the highly significant and top enriched DEGs. This might also be done using a semi-supervised method, for example as described in (McLaughlin et al. 2020). DEGs allow users to manually annotate clusters based on prior knowledge or initiate automated annotation of clusters with the help of any of several available machine-learning algorithms (see examples from Abdelaal et al. 2019; Miao et al. 2020). From a manual scan of the top DEGs, the identity of some clusters might be readily assigned (Luecken and Theis 2019). Typically, scRNAseq analysis identifies more than one DEG per cluster and a subset of these might be helpful in annotating known cell types.
In some cases, cell clusters express a variety of top enriched genes that may not represent a specific gene signature, such that the cluster can only be reported as an “unannotated” cluster (Croset et al. 2018; Davie et al. 2018). In such cases, additional computational tools (Abdelaal et al. 2019) might be helpful in assigning cell-type identity. Automated cell-type prediction tools appear to perform best when the cell-type signatures are composed of at least three to eleven or more DEGs per cluster, and most methods tend to make “mispredictions” when clusters are defined by only one or two DEGs (Diaz-Mejia et al. 2019). This may suggest that using either manual or automated or both methods may benefit from 3 to 11 or more marker genes per cluster in accurately assigning cell identities.
The very concept of a “cell type” is being intensely debated as more dynamic “cell states” are discovered using scRNAseq (Trapnell 2015; 2017; Luecken and Theis 2019). Seurat and related algorithms cluster cells using a graph-based clustering approach and nonlinear dimensional reduction to visualize the clusters to two-dimensional space (Zheng et al. 2017; Butler et al. 2018; Korsunsky et al. 2019; Stuart et al. 2019). However, analysis of datasets can reveal large clusters with subclusters identifiable within them, suggesting heterogeneity within a given cell type. Often, these subclusters, which represent cell states or subtypes, do not possess unique DEGs, making it harder to annotate as the top enriched genes tend to overlap with other subclusters within the parent cluster. In such cases, differential expression of the top enriched marker genes that are commonly expressed across the subclusters may be used to annotate the subclusters.
Cells within a particular tissue may exist in various states or subtypes that express their own combinations of marker genes. However, when samples involve multiple tissues within a complex region, the diversity of clusters at a desired level of resolution or granularity may increase upon scRNAseq analysis (Ghosh et al. 2020). In such cases, if a marker gene that is highly specific to a cell type within a tissue is also expressed in clusters of another tissue, it loses specificity in this more complex context. Hence, although a specific marker gene represents “a cluster” in one tissue, it is highly warranted to compare its enrichment or specificity in clusters of other tissues before flagging it as a potential marker or its corresponding genetic reagent. However, such approaches are only feasible when the whole body or multiple complex tissues are investigated using scRNAseq, as has been done for example for the adult abdomen (Ghosh et al. 2020) and is being accomplished at scale for Drosophila by the FCA consortium.
Use of DEGs for spatial mapping using bioinformatics approaches
Identifying DEGs that define cell types, subtypes, and states does not necessarily lead directly to identifying the spatial location of cells. An efficient pipeline for spatial mapping would begin with mining of existing information (Figure 2). For Drosophila, we benefit from the availability of large-scale datasets and other accumulated knowledge associating gene expression with specific anatomical locations (Table 1). Among the major resources available to help associate DEGs with spatial locations are datasets from large-scale RNA in situ hybridization studies from the Berkeley Drosophila Genome Project (BDGP) in situ project, which is primarily focused on gene expression in embryos (Tomancak et al. 2002, 2007; Hammonds et al. 2013), and the Fly-FISH project, which includes analysis of gene expression in embryos and in the larval fat body, intestine, imaginal discs, nervous system, salivary gland, ovary/testis, muscle, tracheal system, and lymph/ring glands (Lecuyer et al. 2007). Both resources provide spatial mapping data in the form of images and controlled text. Additional datasets provide spatial mapping of RNA expression for specific tissues. For example, FlyTED (Zhao et al. 2010) provides mRNA in situ hybridization data for the testis, and the Dresden Ovary Table (DOT) (Jambor et al. 2015) provides mRNA in situ hybridization data for the ovary. In addition, in situ data generated for the wing disc (Butler et al. 2003) proved useful for analysis of scRNAseq data of myoblasts from wing discs (Zappia et al. 2020).
Despite the wealth of in situ data that exist, the number of genes and tissues covered is limited. One potential solution to spatial mapping is to combine existing knowledge with computational approaches. Seurat includes a computational strategy useful for inferring cellular localization by combining scRNAseq data with a spatial reference map built from in situ hybridization data for a smaller set of “landmark” genes that guide spatial assignment. This strategy has been used successfully, e.g., in zebrafish embryos, but might be challenging for some tissues, such as those in which cells with similar expression patterns are dispersed within the tissue (Satija et al. 2015). An alternative approach, DistMap, was developed and applied to Drosophila embryo scRNAseq data (Karaiskos et al. 2017). This study resulted in generation of the Drosophila Virtual Expression eXplorer (https://shiny.mdc-berlin.de/DVEX/) and became a launchpad for a DREAM challenge (Saez-Rodriguez et al. 2016) aimed at development of additional computational methods for spatial reconstruction. Outcomes from the challenge include the SCTCwhatateam R package for spatial mapping based on “location-marker genes” (Pham et al. 2020). Prior RNA in situ spatial mapping data was additionally used to compute “expression maps” of scRNAseq data from wing imaginal discs (Bageritz et al. 2019). The computed image profiles correlated well with experimentally observed distributions of mRNA. Moreover, the authors were able to compute 65 de novo “marker genes” for which new in situ data were predicted to have the highest value for spatial mapping. Altogether, the application of automated computational approaches shows promise but is limited at least in part by a lack of in situ data for many Drosophila tissues.
In addition to making use of existing RNA in situ data, bioinformatics approaches to mapping DEGs to spatial regions might also make use of bulk RNAseq data. There are numerous bulk RNA-seq datasets available for Drosophila, including datasets based on dissected fly tissues. Comprehensive datasets include RNAseq datasets for larval and adult tissues generated by the modENCODE consortium, which are available for mining at FlyBase (Thurmond et al. 2019) and DGET (Hu et al. 2017), as well as RNAseq datasets for larval and adult tissues that can be mined at the FlyAtlas2 portal (Leader et al. 2018). Additional bulk RNAseq data for specific tissues include the Flygut dataset (Marianes and Spradling 2013) and Flygut-seq dataset (Dutta et al. 2015), which provide transcription profiles from dissected subregions of the fly midgut. In the study by Dutta et al. (2015), tissue dissection was combined with fluorescence-activated cell sorting (FACS) of cells based on cell type-specific expression of GFP, which allows the bulk data to be mapped to specific cell types in defined regions. As such, this resource served as a major reference for cell-type assignment in an analysis of gut scRNAseq data (Hung et al. 2020); top DEGs for each cluster were compared with markers for different cell types in different dissected gut regions as defined in the Dutta et al. study. Finally, a combination of cell type-specific genetic manipulation with other profiling technology, such as the translating ribosome affinity purification (TRAP) method (Thomas et al. 2012) and targeted DamID (Southall et al. 2013), can be used to profile cell-specific gene expression. For example, these approaches have been used to interrogate gene expression in specific cell types in the Drosophila gut (Doupe et al. 2018; Xu et al. 2019) and as such, could have value for scRNAseq data analyses.
Bioinformatics-guided approaches to identifying or predicting spatial locations are not limited to analyses based on RNA expression data. Another approach is to look for enrichment of defined categories such as gene ontology (GO) terms that might help reveal anatomical location. For example, ASAP (Gardeux et al. 2017; David et al. 2020) can be used for enrichment analysis based on GO terms and KEGG pathways, as part of an overall scRNAseq data analysis workflow. In addition, starting with a list of DEGs, FlyMine (Lyne et al. 2007) can be used to look for enrichment of GO terms, KEGG or Reactome pathways, expression patterns in the BDGP in situ dataset, or other categories; and BioLitMine (Hu et al. 2020b) can be used to look for enrichment of medical subject heading (MeSH) terms associated with genes in the published literature, including MeSH anatomical terms. Moreover, as mutant phenotypes often manifest in the tissue(s) that normally express the mutant gene, the association of phenotype annotations with anatomical terms in FlyBase (Grumbling and Strelets 2006) might in theory be used to assist in spatial mapping. We also note that one group has developed an approach called CellAssign that facilitates annotation of scRNAseq data based on reference or de novo cell types, and the user-defined reference information inputted at CellAssign could be a set of well-established cell markers or markers from a relevant dataset (Zhang et al. 2019). Another group attempted de novo reconstruction of spatial mapping based on the assumption that proximal cells will have more similar expression profiles (Nitzan et al. 2019). The approach, novoSpaRc, was applied to several scRNAseq datasets, including for Drosophila embryos, and can be used either in the absence of prior knowledge or together with existing spatial information. In addition, the results of cross-species analyses might also help reveal information relevant to spatial mapping (Tarashansky et al. 2020).
Altogether, bioinformatics approaches can be used to associate clusters with spatial locations based on existing data or annotations, predict spatial locations either de novo or based on existing knowledge, and prioritize subsets of DEGs as potential marker genes. For both existing and new cell types or states, a next step will be to experimentally validate or determine spatial locations in whole mount samples. As discussed earlier, the results of Drosophila scRNAseq studies performed to date suggest that in most or all cases, the expression of multiple DEGs will be needed to differentiate within and among specific cell types, subtypes, and states. Thus, methods allowing for multiplexed detection of gene expression are needed in order to define the spatial locations of the cells in a given cluster. As discussed below, available methods include direct and indirect visualization of RNAs, and direct visualization of proteins.
Experimental methods for direct visualization of RNA expression
In the absence of existing data for a given set of DEGs, an obvious next step would be to use in situ hybridization to develop probes and experimentally determine the expression of potential marker genes in the stage(s) and tissue(s) of interest. A classical and powerful experimental method for in situ hybridization in Drosophila is the use of digoxigenin-labeled RNA probes to visualize RNA in fixed and permeabilized whole-mount samples using a colorimetric assay, most commonly alkaline phosphatase-based staining, as is used by the BDGP in situ project (Tomancak et al. 2002, 2007; Hammonds et al. 2013). The technique is well-established (see protocols in Table 2) and has been widely applied. A significant limit of this approach, however, is that it is not easily multiplexed. Two-color assays can be performed by combining alkaline phosphatase-based staining with the use of 3,3'-diaminobenzidine (DAB) and peroxide-based staining, which generates a precipitate that is different in color from that generated by alkaline phosphatase staining.
An attractive alternative to colorimetric approaches is the application of fluorescent in situ hybridization (FISH). As noted above, this approach was applied at large scale in the Fly-FISH project (Lecuyer et al. 2007; Wilk et al. 2016). Development of FISH technology over the years has led to new approaches to signal amplification, methods for concurrent and sequential labeling, and other improvements. For a detailed review of methods for tissue preparation, hybridization, and other technical aspects of RNA-FISH, we refer readers to (Young et al. 2020). A main focus for development of FISH technology has been on improved signal detection through amplification, which can be achieved through recruitment of multiple reporter molecules to a probe and/or the use of multiple probes per target. One of these improved approaches, hybridization chain reaction (HCR), was successfully used for validation of spatial mapping in an scRNAseq study of the Drosophila ovary (Slaidina et al. 2020). Another, RNAscope (Wang et al. 2012), has been used for detection of RNA in a number of Drosophila tissues, including cardiac tissue (Cannon et al. 2017), the brain (Yin et al. 2018), and photoreceptors (Alejevski et al. 2019). A third, the signal amplification by exchange reaction (SABER) approach (Kishi et al. 2019), has been used for detection of RNA in the Drosophila gut (Amamoto et al. 2019). SABER-FISH and RNAscope show particular promise for visualization of cellular heterogeneity in complex tissues at the single-cell level, as these methods result in bright signals and at least in some sample types, reportedly allow for detection of up to 10 (SABER-FISH) or 12 (RNAscope) targets. Another area of advancement has been in probe design. Design resources such as those associated with the Oligopaints suite of resources (Beliveau et al. 2012) can aid in the design of effective and target-specific RNA-FISH probes for Drosophila studies.
Experimental methods for indirect visualization of RNA expression
There are two major types of in vivo gene expression reporters in Drosophila: (1) those that express a reporter gene under the control of regulatory sequences contained within the transgene, independent of the site of genome integration; and (2) those that express a reporter gene under the control of native regulatory sequences near the site of genome integration. In addition, different transcriptional reporters make use of different approaches to signal detection. The first generation of gene reporters typically used the Escherichia coli lacZ gene, which encodes β-galactosidase (O'Kane and Gehring, 1987; Bellen et al. 1989). β-galactosidase expression can be detected by antibody staining or by the colorful product of cleavage of X-Gal. In either case, the approach requires development of a transgenic fly stock followed by dissection, fixation, and other manipulation of the tissue of interest. Since the discovery of GFP, fluorescent protein (FP) gene reporters have superseded lacZ, as they allow for observation of expression in both fixed and live cells. Finally, binary systems such as GAL4/UAS, LexA/lexAop, and QF/QUAS offer increased flexibility compared to single lacZ or FP reporters (Brand and Perrimon 1993; Lai and Lee, 2006 ; Potter and Luo 2011). With binary systems, an enhancer trap or promoter transcriptional activator (GAL4/LexA/QF) can be used to drive the expression of any reporter gene placed downstream of the corresponding cis-regulatory element (UAS/lexAop/QUAS). Moreover, with binary systems, signal detection is not generally an issue. That said, however, careful analyses can reveal expression patterns not noted or missed in previous analyses, for example as reported by Weaver et al. (2020).
There have been several large-scale efforts to generate gene reporters in Drosophila using an “enhancer-trapping” approach, in which a vector containing a minimal promoter and a reporter gene is randomly integrated into the genome (reviewed in Kvon 2015). The Bloomington Drosophila Stock Center (BDSC) currently houses ∼1,340 lacZ and a handful of FP enhancer trap lines. The vast majority of extant enhancer trap stocks contain insertions of binary system transcriptional activators into the 5’ regulatory region of genes, including ∼6,000 GAL4 lines and ∼180 LexA lines. As a tool for validating gene expression these lines are limited by the fact that they are untargeted insertions of transposable elements and thus may not accurately reflect the native gene expression pattern.
One of the most versatile set of reporters is the MiMIC (Minos-Mediated Integration Cassette) gene trap collection (Venken et al. 2011; Lee et al. 2018). The synthetic MiMIC transposable element contains a gene trap within a swappable insertion cassette (SIC) that can be exchanged using φC31 site-specific integrase. Multiple gene reporter reagents can therefore be derived from a single MiMIC insertion in a coding intron by swapping the SIC with different cassettes to generate reporters, mutants, protein tags, and so on. Currently, the collection includes MiMIC insertions in coding introns of ∼1,800 genes. Approximately 800 of these have been converted into T2A-GAL4 gene trap stocks and an additional 800 have been generated by CRISPR integration of a T2A-GAL4 SIC gene trap (CRIMIC). Unlike enhancer traps, these intronic GAL4 insertions are expressed under the control of undisrupted native promoter and enhancers, and are therefore a more reliable reporter of gene expression. A major limitation of the MiMIC/CRIMIC strategies is that many fly genes do not have a sufficiently large intron to integrate the SIC. We note that the GAL4-UAS system in the context of T2A-GAL4 can introduce a delay in GFP expression relative to endogenous gene expression (Ewen-Campen et al. 2020), and as such, the absence of a GFP signal in some cases may not reflect absence of gene expression.
Examples of cloned sequences used to drive the expression of a gene of interest include the pan-neuronal elav promoter (Yao and White 1994) and the ubiquitous Actin5C promoters (Fyrberg et al. 1983). While valuable, this approach has typically been limited to a relatively small number of cases where defined promoters and enhancers have been identified. However, as part of the Janelia FlyLight Project, ∼3,000 GAL4 lines and ∼1,500 LexA lines were produced using short enhancer fragments from 1,200 genes with predicted expression in the adult brain (Pfeiffer et al. 2010; Jenett et al. 2012) and are now housed at BDSC. An additional set of 200 lines, available from the Vienna Drosophila Research Center, were made using the same approach (Kvon 2015; Tirian and Dickson 2017). These are particularly well-suited for analysis of small subpopulations of neurons, but it is important to note that given the small fragments used to drive GAL4 and LexA, these lines do not provide a full picture of gene expression patterns.
An even greater refinement of cell type-specific gene expression is made possible by the split-GAL4 system, in which the GAL4 DNA-binding domain and a transcriptional activation domain are separately expressed under the control of different regulatory sequences (Luan et al. 2006). Expression of a UAS reporter gene will only occur in cells where the two domains intersect, and GAL4 is reconstituted by a leucine zipper attached to each domain. Using a similar method to their GAL4 lines (Pfeiffer et al. 2010), the FlyLight Project generated a large collection of neuronal split-GAL4 lines, of which ∼4,000 are housed at the BDSC (Dionne et al. 2018). The vast majority of split-GAL4 lines have been characterized based on their expression in the brain and have been used for spatial mapping in that tissue (see e.g., Xie et al. 2021). In addition, a recent report examined the intestinal pattern of the BDSC collection of split-GAL4s, and identified 424 drivers expressed in the midgut (Ariyapala et al. 2020).
The approaches described above provide a powerful means for identifying gene expression patterns and cell or tissue-specific reporters. Indeed, detection of expression using lacZ enhancer traps and/or binary expression systems has been reported for several scRNAseq studies (see e.g., Ariss et al. 2018; Guo et al. 2019; Tattikota et al. 2020). However, each has drawbacks. Enhancer and gene traps caused by insertion of a large element within or upstream of a gene may disrupt regulatory elements, thereby altering expression patterns. Intronic MiMIC insertions are more reliable reporters of true gene expression patterns, but only a subset of genes have suitably large introns for this approach. Although split-GAL4 lines provide exquisite information on co-expression and are uniquely suited for validation of marker genes from scRNAseq studies, almost all existing split-GAL4 lines are not useful for scRNAseq cluster validation since they have been generated as transgenes under the control of short regulatory regions (Jenett et al. 2012) (FlyLight and VDRC GAL4 lines, Table 1) and were not designed to reflect the endogenous expression pattern of the corresponding gene. Indeed, these lines almost never provide faithful representation of the pattern of gene expression because the small fragments inserted are out of their genomic context and problematically many of these lines are expressed in cell types that do not express the respective gene. Furthermore, unlike FISH, none of these approaches offer a means to reliably multiplex more than two gene expression patterns.
Methods for direct visualization of proteins
Thus far, our focus has been on detection of RNA. Given that most RNAs encode proteins, an alternative approach to spatial mapping via detection of RNA expression is direct detection of protein expression. Past experience suggests that reagents for protein-based visualization will be useful for spatial mapping information as well as for other types of experimental applications. A common approach for detection of proteins in whole mount tissues is immunofluorescence and this approach has been used for spatial mapping in Drosophila following scRNAseq analysis (see e.g., Brunet Avalos et al. 2019; Genovese et al. 2019; Allen et al. 2020). Another approach is to detect epitope- or fluorescence protein-tagged versions of the proteins that are expressed under the control of an endogenous promoter, and likewise, detection of FP fusions has been applied already to follow-up of scRNAseq studies (see e.g., Jevitt et al. 2020).
The standard method for isolating antibodies has been to immunize mammals such as rabbits or mice, then isolate polyclonal or monoclonal antibodies that detect the protein of interest. This approach is relatively complex and the quality of the reagents highly variable, and as a result, there are few antibodies available for detection of Drosophila proteins. FlyBase lists 2,414 antibodies targeting 2,028 protein targets. Most of these are polyclonal antibodies and thus are not a renewable resource. Renewable monoclonal antibody reagents are annotated in the literature for fewer than 400 Drosophila proteins (Table 1). When we compare this number with the ∼14,000 protein-coding genes annotated in the fly genome, it becomes clear that development of additional antibody resources and/or alternative methods will be needed in order to use direct detection of proteins for detection of DEGs.
Molecular genetic approaches to fusing the open reading frames (ORFs) of FPs or epitope tags to endogenous gene ORFs provide an alternative, cost-effective approach to detecting proteins. Well-established ORFs useful for tagging include a variety of FPs that can be detected either directly or using anti-FP antibodies, and short peptides (“epitope tags”) recognized by readily available and specific antibodies. A number of different strategies can be used to generate tagged proteins under endogenous expression control. Early large-scale collections of FP-tagged proteins were generated using P-element mobilization of an element containing GFP as an artificial exon (Morin et al. 2001; Kelso et al. 2004). More contemporary molecular genetic strategies have followed. The fly-TransgeneOme (fTRG) fly stocks represent a set of 880 transgenic fly lines covering 826 different genes tagged with GFP and FLAG tags using a two-step strategy to insert tags into transgenes from the FlyFos fosmid library (Sarov et al. 2016). In addition, in the case of the MiMIC approach introduced above, insertions of the SIC into coding introns can be exchanged with protein-tag cassettes such as GFP, RFP, or mCherry FPs, or with FLAG, HA, or Myc epitope tags, to create fusion proteins that can then be detected using whole mount fluorescence or immunofluorescence imaging (Venken et al. 2011). The “scarless editing” approach, which is based on CRISPR-Cas9 editing, also facilitates tagging with GFP, HA, or FLAG (Lamb et al. 2017; Li-Kroeger et al. 2018), and application of the CRISPaint approach in flies is useful for C-terminal FP tagging (Schmid-Burgk et al. 2016; Bosch et al. 2020). Notably, the MiMIC and scarless editing approaches are based on modification of the endogenous locus, such that they are most likely to preserve the normal pattern of the gene. With regards to multiplexing, although the spectrum of available FPs has increased, green and red FPs are used most often. Using antibodies to detect the FPs can further increase the potential for multiplexing, i.e., in combination with detection other types of signals. In addition, protein detection-based approaches have an advantage in that they can be combined with detection of signals generated using binary systems, i.e., detection of a protein tagged with GFP or an epitope tag can be combined with detection of another FP in a fly stock in which a GAL4 reporter drives expression of UAS-xFP.
New approaches to isolation of effective antibodies, such as phage (Moutel et al. 2016) or yeast (McMahon et al. 2018) display-based systems for isolation of nanobodies, as well as identification of new epitope tags, could be used to expand the number of Drosophila proteins that can be used as markers for spatial mapping. Nanobodies have particular appeal because of their relatively small size (12–15 kD), which makes them relatively easy to generate, e.g., using an E. coli expression system, and facilitates favorable binding properties to the target. Isolation of a nanobody that specifically recognizes 13 amino acid peptide, the ALFA nanobody, has opened the doors to detection of epitope-tagged proteins using a nanobody (Gotzke et al. 2019). Although use of the system in Drosophila has not yet been reported, the ALFA system has been shown to work for western blotting, protein purification, and imaging of live or fixed cells (Gotzke et al. 2019; Jin et al. 2020; Makhija et al. 2020), suggesting that application of the ALFA system or another nanobody-based epitope tag is likely to be useful for detection of proteins in Drosophila.
Future directions
In this study, we have described existing resources and common approaches available to the Drosophila research community for mapping cell clusters to anatomy. Cell types that can be uniquely identified based on expression of a single gene are likely to be rare, such that effective experimental approaches will require visualization of more than one RNA and/or protein. For this reason, the application in Drosophila of improved RNA-FISH techniques seem particularly promising, as RNA-FISH allows for direct detection of multiple RNAs at high resolution. We also note that a number of emerging technologies not yet implemented in Drosophila might facilitate or change dramatically the overall approach to spatial mapping. These include spatial transcriptomics technologies such as fluorescent in situ sequencing (FISSEQ) (Lee et al. 2015), multiplexed error-robust fluorescence in situ hybridization (MERFISH) (Chen et al. 2015), spatially-resolved transcript amplicon readout mapping (STARmap) (Wang et al. 2018b), sequential FISH (seqFISH) (Eng et al. 2019), and commercial technologies. These methods couple sensitive, quantitative, and highly multiplexed detection of RNA with simultaneous capture of cell and tissue morphology. As such, they might in the future have a transformative impact. Indeed, their application might be particularly useful in tissues with a high degree of transcriptional diversity across various regions, such as the fly gut (Dutta et al. 2015).
A main barrier to the application of new technologies for spatially resolved detection of RNA expression in Drosophila is that these approaches are typically performed on cell monolayers or tissue sections, whereas spatial resolution in Drosophila is typically performed in intact whole-mount animals or tissues. The relatively small size of Drosophila as compared with mammalian tissue sections might also be a barrier. This could require specific optimization of some platforms, or combining approaches such as MERFISH with expansion microscopy, as has been reported for mammalian cells (Wang et al. 2018a). The design of primers for highly multiplexed applications also presents added challenges. These are being overcome, however, through development of probe design resources including PaintSHOP (Hershberg et al. 2020) and ProbeDealer (Hu et al. 2020a), which support design of probes for Drosophila in addition to other species.
Protein-based reagents, and nanobodies in particular, also hold promise and, notably, would not only provide a potential means for spatial mapping but could also facilitate a wealth of other experimental applications. Development of a large-scale nanobody resource targeting the protein products of hundreds of DEGs is one potential approach. Another would be the development of a set of nanobody-recognized epitope tags, expanding on success of the ALFA tagging approach, and a corresponding fly stock collection in which endogenous genes are tagged with the epitopes. In either case, detection of multiple proteins could be done in a multiplexed manner using different fluorescent tags for secondary detection of the primary nanobody reagents.
Finally, we note that relevant to both RNA and protein detection technologies, new developments in fluorescence imaging, including the recent application in Drosophila of expansion microscopy (Jiang et al. 2018; Karagiannis and Boyden 2018) and improved fluorescence imaging approaches (reviewed in Dunst and Tomancak 2019), are likely to further contribute to effective spatial mapping based on the detection of either RNAs or proteins.
Altogether, we are encouraged that the available and emerging technologies are well-positioned to facilitate spatial mapping in Drosophila, allowing researchers to create anatomical maps of whole animals and specific tissues at unprecedented detail.
Acknowledgments
We thank Yifang Liu and Hongjie Li for helpful comments on the manuscript. We also thank Siyuan Wang and Mengwei Hu (Yale School of Medicine) for expanding the ProbeDealer resource to include a Drosophila database.
Funding
This work was supported by NIH NIGMS P41 GM132087. N.P. is an investigator of Howard Hughes Medical Institute.
Literature cited
- 2017. What is your conceptual definition of "cell type" in the context of a mature organism? Cell Syst. 4:255–259. [DOI] [PubMed] [Google Scholar]
- Abdelaal T, Michielsen L, Cats D, Hoogduin D, Mei H, et al. 2019. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejevski F, Saint-Charles A, Michard-Vanhee C, Martin B, Galant S, et al. 2019. The HisCl1 histamine receptor acts in photoreceptors to synchronize Drosophila behavioral rhythms with light-dark cycles. Nat Commun. 10:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AM, Neville MC, Birtles S, Croset V, Treiber CD, et al. 2020. A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. Elife. 9:e54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amamoto R, Garcia MD, West ER, Choi J, Lapan SW, et al. 2019. Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation. Elife. 8:e51452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariss MM, Islam A, Critcher M, Zappia MP, Frolov MV.. 2018. Single cell RNA-sequencing identifies a metabolic aspect of apoptosis in Rbf mutant. Nat Commun. 9:5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyapala IS, Holsopple JM, Popodi EM, Hartwick DG, Kahsai L, et al. 2020. Identification of split-GAL4 drivers and enhancers that allow regional cell type manipulations of the Drosophila melanogaster intestine. Genetics. 216:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bageritz J, Willnow P, Valentini E, Leible S, Boutros M, et al. 2019. Gene expression atlas of a developing tissue by single cell expression correlation analysis. Nat Methods. 16:750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee U, Girard JR, Goins LM, Spratford CM.. 2019. Drosophila as a genetic model for hematopoiesis. Genetics. 211:367–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, et al. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 37:38–44. [DOI] [PubMed] [Google Scholar]
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, et al. 2012. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci USA. 109:21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Kishi JY, Nir G, Sasaki HM, Saka SK, et al. 2018. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc Natl Acad Sci USA. 115:E2183–E2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, et al. 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3:1288–1300. [DOI] [PubMed] [Google Scholar]
- Bosch JA, Colbeth R, Zirin J, Perrimon N.. 2020. Gene knock-Ins in Drosophila using homology-independent insertion of universal donor plasmids. Genetics. 214:75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N.. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Brunet Avalos C, Maier GL, Bruggmann R, Sprecher SG.. 2019. Single cell transcriptome atlas of the Drosophila larval brain. Elife. 8:e50354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, et al. 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 175:1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MJ, Jacobsen TL, Cain DM, Jarman MG, Hubank M, et al. 2003. Discovery of genes with highly restricted expression patterns in the Drosophila wing disc using DNA oligonucleotide microarrays. Development. 130:659–670. [DOI] [PubMed] [Google Scholar]
- Cannon L, Zambon AC, Cammarato A, Zhang Z, Vogler G, et al. 2017. Expression patterns of cardiac aging in Drosophila. Aging Cell. 16:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X.. 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Treiber CD, Waddell S.. 2018. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife. 7:e34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David FPA, Litovchenko M, Deplancke B, Gardeux V.. 2020. ASAP 2020 update: an open, scalable and interactive web-based portal for (single-cell) omics analyses. Nucleic Acids Res. 48:W403–W414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, et al. 2018. A single-cell transcriptome atlas of the aging Drosophila brain. Cell. 174:982–998.e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Mejia JJ, Meng EC, Pico AR, MacParland SA, Ketela T, et al. 2019. Evaluation of methods to assign cell type labels to cell clusters from single-cell RNA-sequencing data. F1000Res. 8:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne H, Hibbard KL, Cavallaro A, Kao JC, Rubin GM.. 2018. Genetic reagents for making split-GAL4 lines in Drosophila. Genetics. 209:31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe DP, Marshall OJ, Dayton H, Brand AH, Perrimon N.. 2018. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Proc Natl Acad Sci USA. 115:12218–12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunst S, Tomancak P.. 2019. Imaging flies by fluorescence microscopy: principles, technologies, and applications. Genetics. 211:15–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, et al. 2015. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 12:346–358. [DOI] [PubMed] [Google Scholar]
- Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, et al. 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature. 568:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Liu T, Banerjee U.. 2014. Drosophila hematopoiesis: markers and methods for molecular genetic analysis. Methods. 68:242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B, Comyn T, Vogt E, Perrimon N.. 2020. No evidence that Wnt ligands are required for planar cell polarity in Drosophila. Cell Rep. 32:108121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg EA, Mahaffey JW, Bond BJ, Davidson N.. 1983. Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell. 33:115–123. [DOI] [PubMed] [Google Scholar]
- Gardeux V, David FPA, Shajkofci A, Schwalie PC, Deplancke B.. 2017. ASAP: a web-based platform for the analysis and interactive visualization of single-cell RNA-seq data. Bioinformatics. 33:3123–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese S, Clement R, Gaultier C, Besse F, Narbonne-Reveau K, et al. 2019. Coopted temporal patterning governs cellular hierarchy, heterogeneity and metabolism in Drosophila neuroblast tumors. Elife. 8:e50375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AC, Tattikota SG, Liu Y, Comjean A, Hu Y, et al. 2020. Drosophila PDGF/VEGF signaling from muscles to hepatocyte-like cells protects against obesity. Elife. 9:e56969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotzke H, Kilisch M, Martinez-Carranza M, Sograte-Idrissi S, Rajavel A, et al. 2019. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat Commun. 10:4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumbling G, Strelets V.. 2006. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34:D484–D488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Yin C, Yang F, Zhang Y, Huang H, et al. 2019. The cellular diversity and transcription factor code of Drosophila enteroendocrine cells. Cell Rep. 29:4172–4185.e4175. [DOI] [PubMed] [Google Scholar]
- Hammonds AS, Bristow CA, Fisher WW, Weiszmann R, Wu S, et al. 2013. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14:R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg EA, Close JL, Camplisson CK, Attar S, Chern R, et al. 2020. PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. bioRxiv: 2020.07.05.188797. [DOI] [PMC free article] [PubMed]
- Hu M, Yang B, Cheng Y, Radda JSD, Chen Y, et al. 2020a. ProbeDealer is a convenient tool for designing probes for highly multiplexed fluorescence in situ hybridization. Sci Rep. 10:22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chung V, Comjean A, Rodiger J, Nipun F, et al. 2020b. BioLitMine: advanced mining of biomedical and biological literature about human genes and genes from major model organisms. G3 (Bethesda). 10:4531–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Comjean A, Perrimon N, Mohr SE.. 2017. The Drosophila gene expression tool (DGET) for expression analyses. BMC Bioinformatics. 18:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Tattikota SG, Liu Y, Comjean A, Gao Y, et al. 2021. DRscDB: a single-cell RNA-seq resource for data mining and data comparison across species. bioRxiv: 10.1101/2021.01.29.428862. [DOI] [PMC free article] [PubMed]
- Hung RJ, Hu Y, Kirchner R, Liu Y, Xu C, et al. 2020. A cell atlas of the adult Drosophila midgut. Proc Natl Acad Sci USA. 117:1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, et al. 2015. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife. 4:e05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, et al. 2012. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevitt A, Chatterjee D, Xie G, Wang XF, Otwell T, et al. 2020. A single-cell atlas of adult Drosophila ovary identifies transcriptional programs and somatic cell lineage regulating oogenesis. PLoS Biol. 18:e3000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Kim HJ, Chozinski TJ, Azpurua JE, Eaton BA, et al. 2018. Superresolution imaging of Drosophila tissues using expansion microscopy. Mol Biol Cell. 29:1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Wang Y, Wang M, Sun M, Hattori M.. 2020. Fluorescence-detection size-exclusion chromatography utilizing nanobody technology for expression screening of membrane proteins. bioRxiv: 10.1101/2020.09.28.316307. [DOI] [PMC free article] [PubMed]
- Jory A, Estella C, Giorgianni MW, Slattery M, Laverty TR, et al. 2012. A survey of 6,300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Rep. 2:1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanca O, Bellen HJ, Schnorrer F.. 2017. Gene tagging strategies to assess protein expression, localization, and function in Drosophila. Genetics. 207:389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis ED, Boyden ES.. 2018. Expansion microscopy: development and neuroscience applications. Curr Opin Neurobiol. 50:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, et al. 2017. The Drosophila embryo at single-cell transcriptome resolution. Science. 358:194–199. [DOI] [PubMed] [Google Scholar]
- Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, et al. 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32:D418–D420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi JY, Lapan SW, Beliveau BJ, West ER, Zhu A, et al. 2019. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods. 16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, et al. 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 16:1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ. 2015. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 106:185–192. [DOI] [PubMed] [Google Scholar]
- Lai SL, Lee T.. 2006. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 9:703–709. [DOI] [PubMed] [Google Scholar]
- Lamb AM, Walker EA, Wittkopp PJ.. 2017. Tools and strategies for scarless allele replacement in Drosophila using CRISPR/Cas9. Fly (Austin). 11:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader DP, Krause SA, Pandit A, Davies SA, Dow JAT.. 2018. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46:D809–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E. 2011. High resolution fluorescent in situ hybridization in Drosophila. Methods Mol Biol. 714:31–47. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Parthasarathy N, Krause HM.. 2008. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol. 420:289–302. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, et al. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 131:174–187. [DOI] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, et al. 2015. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 10:442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Zirin J, Kanca O, Lin WW, Schulze KL, et al. 2018. A gene-specific T2A-GAL4 library for Drosophila. Elife. 7:e35574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre F, Cody N, Iampietro C, Bergalet J, Lefebvre FA, et al. 2013. Whole mount RNA fluorescent in situ hybridization of Drosophila embryos. J Vis Exp. e50057. [DOI] [PMC free article] [PubMed]
- Li H. 2020. Single-cell RNA sequencing in Drosophila: Technologies and applications. Wiley Interdiscip Rev Dev Biol. e396. [DOI] [PMC free article] [PubMed]
- Li H, Horns F, Wu B, Xie Q, Li J, et al. 2017. Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell. 171:1206–1220. e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Kroeger D, Kanca O, Lee PT, Cowan S, Lee MT, et al. 2018. An expanded toolkit for gene tagging based on MiMIC and scarless CRISPR tagging in Drosophila. Elife. 7:e38709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH.. 2006. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 52:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken MD, Theis FJ.. 2019. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, et al. 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8:R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhija S, Brown D, Bourke S, Wang Y, Zhou S, et al. 2020. Versatile labeling and detection of endogenous proteins using tag-assisted split enzyme complementation. bioRxiv: 2020.12.01.407072. [DOI] [PMC free article] [PubMed]
- Marianes A, Spradling AC.. 2013. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife. 2:e00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J.. 2018. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 1802.03426.
- McLaughlin CN, Brbić M, Xie Q, Li T, Horns F, et al. 2020. Single-cell transcriptomes of developing and adult olfactory receptor neurons in Drosophila. bioRxiv: 10.1101/2020.10.08.332130. [DOI] [PMC free article] [PubMed]
- McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, et al. 2018. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol. 25:289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner GW, Nern A, Singer RH, Wong AM, Malkesman O, et al. 2019. Mapping neurotransmitter identity in the whole-mount Drosophila brain using multiplex high-throughput fluorescence in situ hybridization. Genetics. 211:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Moreno P, Huang N, Papatheodorou I, Brazma A, et al. 2020. Putative cell type discovery from single-cell gene expression data. Nat Methods. 17:621–628. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W.. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci USA. 98:15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutel S, Bery N, Bernard V, Keller L, Lemesre E, et al. 2016. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife. 5:e16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar-Jaiswal S, Lee PT, Campbell ME, Chen K, Anguiano-Zarate S, et al. 2015. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife. 4:e05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzan M, Karaiskos N, Friedman N, Rajewsky N.. 2019. Gene expression cartography. Nature. 576:132–137. [DOI] [PubMed] [Google Scholar]
- O'Kane CJ, Gehring WJ.. 1987. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA. 84:9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro M, Martinovic M, Bevilacqua V, Hershberg EA, Rossetti G, et al. 2020. OligoMinerApp: a web-server application for the design of genome-scale oligonucleotide in situ hybridization probes through the flexible OligoMiner environment. Nucleic Acids Res. 48:W332–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, et al. 2010. Refinement of tools for targeted gene expression in Drosophila. Genetics. 186:735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VVH, Li X, Truong B, Nguyen T, Liu L, et al. 2020. The winning methods for predicting cellular position in the DREAM single-cell transcriptomics challenge. Brief Bioinform, doi: 10.1093/bib/bbaa181. [DOI] [PubMed]
- Potter CJ, Luo L.. 2011. Using the Q system in Drosophila melanogaster. Nat Protoc. 6:1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Coello AT, Petrella LN, Ayers K, Melillo A, Mazzalupo S, et al. 2007. Exploring strategies for protein trapping in Drosophila. Genetics. 175:1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Costello JC, Friend SH, Kellen MR, Mangravite L, et al. 2016. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat Rev Genet. 17:470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, Barz C, Jambor H, Hein MY, Schmied C, et al. 2016. A genome-wide resource for the analysis of protein localisation in Drosophila. Elife. 5:e12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Honing K, Ebert TS, Hornung V.. 2016. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun. 7:12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singari S, Javeed N, Tardi NJ, Marada S, Carlson JC, et al. 2014. Inducible protein traps with dominant phenotypes for functional analysis of the Drosophila genome. Genetics. 196:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaidina M, Banisch TU, Gupta S, Lehmann R.. 2020. A single-cell atlas of the developing Drosophila ovary identifies follicle stem cell progenitors. Genes Dev. 34:239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, et al. 2013. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev Cell. 26:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, et al. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarashansky AJ, Musser JM, Khariton M, Li P, Arendt D, et al. 2020. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. bioRxiv: 10.1101/2020.09.28.317784. [DOI] [PMC free article] [PubMed]
- Tattikota SG, Cho B, Liu Y, Hu Y, Barrera V, et al. 2020. A single-cell survey of Drosophila blood. Elife. 9:e54818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Lee PJ, Dalton JE, Nomie KJ, Stoica L, et al. 2012. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS ONE. 7:e40276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47:D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirian L, Dickson BJ.. 2017. The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. bioRxiv: 10.1101/198648. [DOI]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3:research0088.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, et al. 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8:R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. 2015. Defining cell types and states with single-cell genomics. Genome Res. 25:1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G.. 2008. Visualizing data using t-SNE. J Machine Learn Res. 9:2579–2605. [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, et al. 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 8:737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, et al. 2012. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 14:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Moffitt JR, Zhuang X.. 2018a. Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Sci Rep. 8:4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, et al. 2018b. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 361:eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ma T, Drummond-Barbosa D.. 2020. Analysis of Gal4 expression patterns in adult Drosophila females. G3 (Bethesda). 10:4147–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiszmann R, Hammonds AS, Celniker SE.. 2009. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nat Protoc. 4:605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R, Hu J, Blotsky D, Krause HM.. 2016. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 30:594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Brbic M, Horns F, Kolluru SS, Jones RC, et al. 2021. Temporal evolution of single-cell transcriptomes of Drosophila olfactory projection neurons. Elife. 10:e63450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Tang HW, Hung RJ, Hu Y, Ni X, et al. 2019. The septate junction protein Tsp2A restricts intestinal stem cell activity via endocytic regulation of aPKC and hippo signaling. Cell Rep. 26:670–688.e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao KM, White K.. 1994. Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J Neurochem. 63:41–51. [DOI] [PubMed] [Google Scholar]
- Yin J, Gibbs M, Long C, Rosenthal J, Kim HS, et al. 2018. Transcriptional regulation of lipophorin receptors supports neuronal adaptation to chronic elevations of activity. Cell Rep. 25:1181–1192.e1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AP, Jackson DJ, Wyeth RC.. 2020. A technical review and guide to RNA fluorescence in situ hybridization. PeerJ. 8:e8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia MP, Castro L, Ariss MM, Jefferson H, Islam AB, et al. 2020. A cell atlas of adult muscle precursors uncovers early events in fibre-type divergence in Drosophila. EMBO Rep. 21:e49555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AW, O’Flanagan C, Chavez EA, Lim JLP, Ceglia N, et al. 2019. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat Methods. 16:1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Klyne G, Benson E, Gudmannsdottir E, White-Cooper H, et al. 2010. FlyTED: the Drosophila testis gene expression database. Nucleic Acids Res. 38:D710–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]