Abstract
Cohesin is a ring-shaped protein complex that controls dynamic chromosome structure. Cohesin activity is important for a variety of biological processes, including formation of DNA loops that regulate gene expression. The precise mechanisms by which cohesin shapes local chromosome structure and gene expression are not fully understood. Recurrent mutations in cohesin complex members have been reported in various cancers, though it is not clear whether many cohesin sequence variants have phenotypes and contribute to disease. Here, we utilized CRISPR/Cas9 genome editing to introduce a variety of cohesin sequence variants into murine embryonic stem cells and investigate their molecular and cellular consequences. Some of the cohesin variants tested caused changes to transcription, including altered expression of gene encoding lineage-specifying developmental regulators. Altered gene expression was also observed at insulated neighborhoods, where cohesin-mediated DNA loops constrain potential interactions between genes and enhancers. Furthermore, some cohesin variants altered the proliferation rate and differentiation potential of murine embryonic stem cells. This study provides a functional comparison of cohesin variants found in cancer within an isogenic system, revealing the relative roles of various cohesin perturbations on gene expression and maintenance of cellular identity.
Keywords: Cohesin, cancer, variant, mutation, gene expression, cell identity, differentiation
Introduction
Proper gene expression is essential for maintaining cellular identity and dysregulation of gene control is associated with many human diseases, including cancer (Roy and Hebrok 2015). During eukaryotic gene regulation, long-range DNA interactions often form between gene promoters and distal cis-regulatory elements (Ong and Corces 2011; Bonev and Cavalli 2016). In order for a promoter to physically contact a distant cis-regulatory element, the intervening DNA must be looped out as the two elements are brought into close spatial proximity (Eagen 2018; Rowley and Corces 2018). Enhancer–promoter contacts are developmentally dynamic, and their dysregulation is associated with developmental disorders and tumorigenesis (Hill et al. 2016; Hnisz et al. 2016a; Long et al. 2016). There is limited understanding of the mechanisms through which these contacts form and subsequently function to control gene expression.
Formation of DNA loop structures that regulate transcription is dependent on the cohesin complex. Cohesin is a ring-shaped protein complex composed of three core subunits (SMC1A, SMC3, and RAD21), along with various accessory and regulatory factors including STAG1, STAG2, PDS5A, PDS5B, NIPBL, WAPL, and Sororin (Remeseiro and Losada 2013). The accessory factors associate with cohesin in combinatorial fashion and are thought to modulate and regulate its activity on the genome, although many open questions remain (Rudra and Skibbens 2013). Upon loading onto the genome at sites of transcriptional activity, cohesin appears to translocate or extrude chromatin thus generating a loop of DNA (Dowen and Young 2014; Eagen 2018; Rowley and Corces 2018). Extrusion is arrested when cohesin encounters specific chromatin-bound roadblocks, in particular the transcriptional apparatus or the 11-zinc-finger transcription factor CTCF (Hansen 2020). These sites with arrested cohesin serve as anchors of DNA loop structures that regulate local transcription in at least two ways. First, the formation of a cohesin-mediated DNA loop between an enhancer and promoter can increase expression of the gene, while loss of the interaction can decrease gene expression. Secondly, cohesin-mediated DNA loops anchored by pairs of distal CTCF sites can restrict enhancer activity to the region inside of the loop. DNA loops of this type often surround genes encoding master lineage regulators and have been termed insulated neighborhoods for their ability to constrain an enhancer’s activity to a promoter within the DNA loop defined by CTCF sites (Dowen et al. 2014; Ji et al. 2016). Disruption of an insulated neighborhood boundary can cause aberrant enhancer targeting to genes normally located outside the neighborhood and cause inappropriate gene expression (Dowen et al. 2014; Ji et al. 2016; Hnisz et al. 2016a). In addition to its role in organizing interphase chromatin, cohesin is also required for sister chromatid cohesion, DNA replication, DNA repair and dynamic restructuring of chromosomes during cell division. Because of its participation in these varied biological processes, the cohesin complex is essential for organismal and cellular function. While complete loss of cohesin function is not viable, less severe defects such as mutations or loss of non-essential subunits are often tolerated and cause diverse cellular consequences (Peters et al. 2008). Molecular insight into how cohesin organizes genome structure and impacts genome function is still lacking.
Cancer genome and exome sequencing has revealed that cohesin subunits undergo a wide spectrum of mutations in cancer (Losada 2014; Hill et al. 2016; Waldman 2020). Somatic mutations, insertions, and deletions in cohesin complex components are observed in a wide range of tumor types, whereas germline mutations in cohesin and its regulators are found in a group of developmental disorders known as cohesinopathies (Horsfield et al. 2012; Kandoth et al. 2013; Ley et al. 2013; Thol et al. 2014). Several types of mutations have been associated with disease, including missense mutations, haploinsufficiency from monoallelic gene inactivation, and complete gene inactivation. Recent studies have begun to investigate the wide variety of cellular consequences that result from distinct cohesin defects. Cohesin subunits STAG2 and SMC1A acquire somatic mutations in diverse tumor types, including bladder cancer and myeloid neoplasia (Kon et al. 2013; Ley et al. 2013; Kim et al. 2016). The STAG2 gene is X-encoded and is a frequent target of inactivating mutations, which are only partially compensated for by its paralogue, STAG1 (Hill et al. 2016; Arruda et al. 2020). Some cancers with inactivated STAG2 exhibit decreased cell viability and disrupted cell cycle, but conflicting data exist on whether these defects are attributable to cohesin’s role in mediating sister chromatid cohesion (Solomon et al. 2011, 2013; Balbás-Martínez et al. 2013; Lawrence et al. 2014; Taylor et al. 2014; Mullenders et al. 2015; Kim et al. 2016; Viny and Levine 2018; Benedict et al. 2020). An alternative hypothesis suggests that, rather than interfering with the cell cycle, cohesin mutants may contribute to disease pathology by altering genome structure and gene expression. Aberrant DNA looping could cause misregulation of oncogenes or tumor suppressor genes during tumorigenesis or alter expression of developmental regulators during cell state maintenance or differentiation (Horsfield et al. 2012; Hnisz et al. 2016a; Norton and Phillips-Cremins 2017; Bompadre and Andrey 2019). Further studies are needed to determine the diverse molecular and cellular phenotypes that arise from distinct cohesin perturbations, in order to understand both normal cohesin function in the cell and defects observed in human disease. In particular, comprehensive studies investigating multiple distinct alleles, from missense to loss of function mutations, in an isogenic system are essential for determining the importance of individual sequence variants to a pathogenic state and for further understanding of normal cohesin biology.
To investigate the model that oncogenic cohesin mutations may disrupt cohesin’s role as a spatial organizer of gene control, we utilized the CRISPR-Cas9 system to introduce disease-associated deletions and amino acid variants into cohesin subunits in murine embryonic stem cells (mESCs). Introduction of cohesin sequence variants did not alter overall levels of cohesin complex members in cells, indicating that the resulting phenotypes were not simply due to reduction or absence of the cohesin complex. Instead, most of the mESCs harboring cohesin sequence variants exhibited altered transcriptional regulation, including misexpression of genes at insulated neighborhoods and cell identity factors. Cellular differentiation and proliferation were impaired by some cohesin sequence variants but not others. The results indicate that individual cancer-associated cohesin variants are sufficient to cause misregulation of gene expression and suggest that aberrant cohesin function may contribute to human disease by altering expression of genes important for proper cellular identity and function.
Materials and methods
Cell culture
Murine embryonic stem cells (V6.5, male, derived from a C57BL/6(F) × 129/sv(M) cross) were grown under standard ESC culture conditions (Arruda et al. 2020; Justice et al. 2020). mESC media contained DMEM KO (Thermo Fisher Scientific, 10829-018), 15% fetal bovine serum (VWR, 97068-085), homemade leukemia inducing factor (LIF), 100 U/ml penicillin, 100 µg/ml streptomycin (Thermo Fisher Scientific, 15140-122), 100 µM beta-mercaptoethanol (Thermo Fisher Scientific, 21-985-023), 1× Non-essential amino acids (Thermo Fisher Scientific, 11140-050), and 1× Glutamax (Thermo Fisher Scientific, 35050-061). mESCs were grown on gelatinized tissue culture dishes and were passaged using TrypLE (Thermo Fisher Scientific, 12-604-039).
Genome editing
Genome-edited mESC lines were generated as previously described, with modifications (Arruda et al. 2020; Justice et al. 2020). mESCs were transfected with a single-stranded donor oligonucleotide (ssODN) repair template, and plasmids encoding a synthetic guide RNA (sgRNA), Cas9 and a fluorescent gene (eGFP or mCherry) using Lipofectamine 2000 (Thermo Fisher, 11-668-027). After 1–4 days, single cells that were GFP positive and/or mCherry positive were sorted by either UNC Flow Cytometry Core Facility staff using a FACSAria II (BD Biosciences, San Jose, CA, USA) or on a CytoSort Array using a CellRaft AIR System (Cell Microsystems). Fluorescent cells were either sparsely plated on irradiated murine embryonic fibroblast monolayers to form colonies or sorted into individual wells of a tissue culture dish. Individual colonies were expanded into clonal cell lines, screened for genome edits by PCR and Sanger sequencing, and cryogenically stored. Individual allele sequences were determined by PCR of the region surrounding the edit site, followed by TOPO-TA cloning (Thermo Fisher, K4575J10) and Sanger sequencing. sgRNA and ssODN sequences are provided in Supplementary Table S1 and were designed using the CRISPR tool (crispr.mit.edu) (Cong and Zhang 2015). Two independent clones were obtained for each genotype, except for Stag2V181M which only had a single clone obtained.
RT-qPCR
All mESC lines were handled side-by-side during culturing, RNA extraction, and quantitative PCR measurements. Briefly, cells were resuspended in 1 ml Trizol (Thermo Fisher, 15596018), incubated for 5 min at room temperature and either stored at −80 °C or further processed. 200 µl chloroform (Sigma Aldrich, C2432) was added and mixed before centrifugation to separate organic and aqueous phases. The aqueous phase was recovered, mixed with 200 µl additional chloroform, and centrifuged. The aqueous phase was collected and RNA was precipitated by addition of isopropanol. Total RNA was quantified using a NanoDrop instrument (Thermo Fisher). cDNA was prepared with Superscript IV and oligo-d(T) primers (Thermo Fisher, 18091050) according to the manufacturer’s instructions. Quantitative PCR was performed using SYBRgreen Master Mix on an Applied Biosystems QuantStudio 6 qPCR instrument using primers found in Supplementary Table S2. For each clone, measurements were performed in triplicate for each of four biological replicates and normalized to Tbp.
Western blotting
Adherent cells were collected by washing with PBS, scraping and transferring to a tube for centrifugation. Cell pellets were either frozen on dry ice for storage or nuclear extracts were prepared. Cell pellets were resuspended in 10 ml Lysis Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, and 0.1 mM EGTA) containing 1× protease inhibitor cocktail (PIC) (Sigma Aldrich, 11697498001) and incubated at 4 °C while rocking for 15 min. 1 ml 10% NP-40 was added, samples were immediately vortexed, and pelleted at 1350×g for 5 min at 4 °C. The pellet was resuspended in 1 ml of cold Buffer TEN250/0.1 (50 mM Tris-HCl pH 7.5, 250 mM NaCl, 5 mM EDTA, and 0.1 mM NP-40) containing 1× PIC and incubated for a minimum of 30 min while rotating at 4 °C. After spinning at max speed at 4 °C for 10 min, the nuclear fraction (supernatant) was collected. Precision Plus Protein Dual Color Standard (Bio-Rad, 1610374) and samples were run in 4–20% Tris-Glycine gels (BioRad, 4568094) and transferred to PVDF membranes (VWR 29301-856). Membranes were blocked for 1 h with 5% blocking grade buffer (BioRad, 170-6404) and incubated overnight with primary antibodies while rocking at 4 °C. Membranes were washed 3 × 10 min with TBS-T at room temperature and incubated with secondary antibodies for 1 h while rocking at 4 °C. Antibodies used: STAG1 (Bethyl Laboratories, A300-0157A), STAG2 (Bethyl Laboratories, A300-0158A), SMC1A (Bethyl Laboratories, A300-055A), H3 (Abcam, ab1791), anti-Rabbit secondary (GE Healthcare, NA934), anti-Goat secondary (Abcam, ab97100). Membranes were imaged using either Thermo SuperSignal West Pico PLUS or Thermo SuperSignal West Femto chemiluminescent substrates with an Amersham Imager 600 (GE Healthcare). ImageJ was used to quantify protein abundance.
Embryoid body differentiation
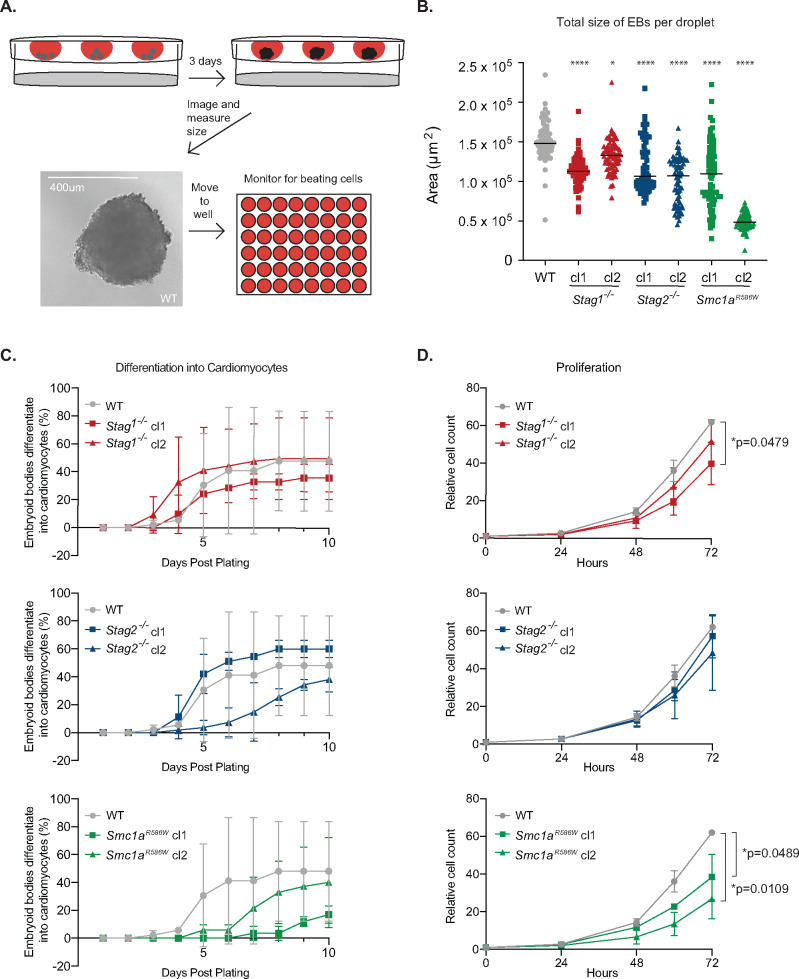
mESCs were differentiated into EBs using hanging droplet cultures as described (Behringer et al. 2016), with modifications. Briefly, mESCs were dissociated into a single-cell suspension and 1,000 cells were placed into 30 µl droplets hanging from the lid of a 10 cm dish, in media lacking LIF, and PBS was placed in the dish. After three days, images were acquired using an EVOS FL light microscope (Thermo Fisher). ImageJ was used to determine the area of the EB. If multiple EBs were found in a single droplet, the areas of each EB were summed and reported. For further differentiation, individual droplets were transferred to gelatinized wells of a six-well plate and maintained in media lacking LIF. Cultures were monitored daily for rhythmic contractions using an EVOS FL light microscope (Thermo Fisher). Significance was examined using Dunnet’s multiple comparisons test.
Proliferation assay
5 × 104 cells were plated into wells of a gelatinized six-well tissue culture plate. At the indicated timepoints (24, 48, 60, and 72 h) cells were resuspended with TrypLE (Thermo Fisher Scientific, 12-604-039) and resuspended in PBS. Cells were then mixed with trypan blue and counted on a Countess II FL instrument (Life Technologies). Measurements were performed in triplicate, with three identical wells for each sample at each timepoint. The experiment was completed four times. All calculations are represented as a fraction of initial plating density and plotted using GraphPad Prism (GraphPad). Significance was determined using Dunnet’s multiple comparisons test.
Data availability
Cell lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental Material available at figshare: https://doi.org/10.25386/genetics.13962011.
Results
Sequence variants in cohesin complex members
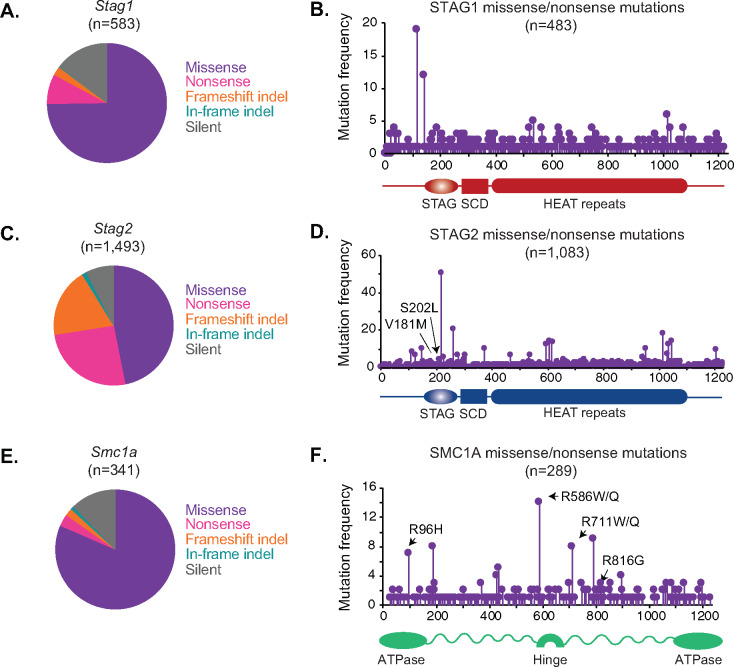
Large sequencing projects that aim to catalog disease-associated human variation have identified mutations in genes that encode members of the cohesin complex. We examined publicly available databases of disease-associated sequence variation (COSMIC, TCGA, and ENSEMBLE) to identify mutations that may impact cohesin activity (cancer.sanger.ac.uk) (Weinstein et al. 2013; Forbes et al. 2017). We observed a similar distribution of sequence variant types across these databases and report here the proportions from the COSMIC database in Figure 1. The Stag1 gene frequently acquired missense mutations, with some additional nonsense and silent mutations observed (Figure 1A). The location of missense mutations along the length of the STAG1 protein sequence reveals a broad distribution with only a few hotspots detected near the N-terminal end (Figure 1B). The Stag2 gene is encoded on the X chromosome and displays a distinct mutational spectrum from Stag1. While Stag2 frequently acquires missense mutations, it also shows an increased proportion of nonsense and frameshift insertions and deletions (indels) relative to Stag1, that are predicted to disrupt stable STAG2 protein levels (Figure 1C). The missense mutations in Stag1 and Stag2 may cause subtle defects by disrupting sites of post-translational modifications or a protein–protein interaction interface, preventing a conformational change, or may cause no defect at all. The location of missense mutations along the length of the STAG2 protein sequence is similar to STAG1, with a broad distribution punctuated by a few hotspots (Figure 1D). STAG1 and STAG2 appear to act redundantly in cohesin localization on the genome, yet loss of individual STAG proteins causes both distinct and overlapping gene expression changes (Arruda et al. 2020; Casa et al. 2020). It is interesting to note that the single copy of the Stag2 gene on the X chromosome is frequently inactivated in cancers by nonsense and frameshift insertions and deletions, whereas the two copies of the Stag1 gene tend to acquire missense mutations. While the STAG proteins are mutually exclusive members of the cohesin complex, SMC1A is a core member. We found that the X-encoded Smc1a gene rarely undergoes inactivating mutations, consistent with its essential functin in cohesin-mediated dynamic chromosome restructuring during the cell cycle (Forbes et al. 2017) (Figure 1E). Rather, Smc1a tends to acquire missense mutations predicted to be hypomorphic and not cause complete loss of function (Figure 1F). These results indicate that the Stag1, Stag2, and Smc1a genes acquire somewhat distinct mutation types at various frequencies in human cancers, which could in turn influence their contribution to disease pathology.
Figure 1.
Cancer-associated sequence variation in cohesin components. (A) Proportion of Stag1 mutations that are missense, in-frame insertion/deletion, nonsense, frameshift insertion/deletion or silent. Mutations are derived from the COSMIC database of somatic mutations in cancer (cancer.sanger.ac.uk) (Tate et al. 2019). (B) Lollipop plot showing frequency of missense and nonsense mutations in STAG1. STAG, stromal antigen domain; SCD, stromal in conserved domain; HEAT repeats, which mediate the interaction with RAD21 are indicated (Zhang et al. 2013; Hara et al. 2014; Kim et al. 2016). (C) Proportion of Stag2 mutations that are missense, in-frame insertion/deletion, nonsense, frameshift insertion/deletion or silent. Mutations are derived from the COSMIC database of somatic mutations in cancer (cancer.sanger.ac.uk) (Tate et al. 2019). (D) Lollipop plot showing frequency of missense and nonsense mutations in STAG2. STAG, stromal antigen domain; SCD, stromal in conserved domain; HEAT repeats, which mediate interaction with RAD21 are indicated (Hara et al. 2014; Kim et al. 2016). Two specific mutations are indicated and annotated with their one letter amino acid abbreviations (for example, V181M refers to p. Val118 changed to Met). (E) Proportion of Smc1a mutations that are missense, in-frame insertion/deletion, nonsense, frameshift insertion/deletion or silent. Mutations are derived from the COSMIC database of somatic mutations in cancer (cancer.sanger.ac.uk) (Tate et al. 2019). (F) Lollipop plot showing frequency of missense and nonsense mutations in SMC1A. ATPase domains and the hinge domain are indicated. Four specific mutations are indicated and annotated with their one letter amino acid abbreviations (for example, R96H refers to p. Arg96 changed to His).
Engineering isogenic cohesin variant mESCs
To investigate whether specific cohesin sequence variants cause defects in cohesin function, we created isogenic mESC lines harboring various individual cohesin sequence variants for direct comparison of the cellular and transcriptional consequences of impaired cohesin activity. The use of stable embryonic stem cell lines allows for interrogation of individual cohesin perturbations independent of other coexisting sequence variation in cancer cells and separate from the natural human variation of individual patients. mESCs also provide a well-characterized model system for investigating mechanisms of transcriptional control of cell state that are broadly applicable to other cell types. Therefore, CRISPR/Cas9 genome editing was used to engineer individual cohesin sequence variants found in human disease into their endogenous loci in the murine embryonic stem cell genome. Of particular interest are recurrent missense mutations of cohesin subunits, that are observed in multiple patients with distinct cancer types, as these variants could represent alleles of cohesin with partial loss of function that could provide insight into cohesin biology. In addition to frequent disease-associated variants, we were also interested in sequence variation within the N-terminal region of STAG2, a reported protein–protein interaction interface with CTCF (Xiao et al. 2011). Two particular disease-associated sequence variants, STAG2-V181M and STAG2-S202L, are located within the N-terminus of STAG2 and were, therefore, selected for further study. The cohesin variant mESCs generated include: frameshift indels in Stag1 and Stag2 (Stag1−/− and Stag2−/−), in-frame deletion in Stag2 (Stag2Δ164-196), and amino acid variations in Stag2 (Stag2V181M and Stag2S202L) and Smc1a (Smc1aR96H, Smc1aR586W, Smc1aR711W, and Smc1aR816G) (Table 1). The vast majority of these mutations did not co-occur with other mutations to cohesin encoding genes (Table 1). In one instance, the SMC1A-R711W and STAG2-V181M mutations co-occurred in a highly mutated colorectal adenocarcinoma. Importantly, the genes that encode cohesin subunits show strong conservation between human and mouse, with the six amino acid variants engineered in this study being 100% conserved between the two species. The sgRNA sequences and repair templates used to generate these cell lines are indicated in Supplementary Table S1. Also, included in the table are the number of potential off-target locations when allowing for mismatches between the sgRNA sequence and the genomic sequence, as deteremined using the Cas-OFFinder tool from RGEN Tools (Bae et al. 2014). The genomic edits were identified by Sanger sequencing.
Table 1.
Cohesin sequence variants found in human disease
| mESC line genotype | Sequence variant type | Disease relevance | Reported cases | Co-occurring cohesin mutationsa |
|---|---|---|---|---|
| Stag1−/− | Nonsense or frameshift indel | diverse cancer types | 42 | NDb |
| Stag2−/− | Nonsense or frameshift indel | diverse cancer types | 391 | NDb |
| Stag2V181M | V181L | lung adenocarcinoma | 1 | 0 |
| V181M | hematopoietic neoplasm | 2 | 0 | |
| V181M | large intestine adenocarcinoma | 1 | 4 (inc. SMC1A-R711W) | |
| Stag2S202L | S202L | bladder carcinoma | 1 | 0 |
| S202 nonsense | bladder carcinoma | 1 | 0 | |
| Stag2Δ164-196 | In-frame indel | none reported | 0 | ND |
| Smc1aR96H | R96H | acute myeloid leukemia | 6 | 0 |
| R96H | endometroid carcinoma | 1 | 1 | |
| Smc1aR586W | R586W | endometroid carcinoma | 1 | 0 |
| R586W | acute myeloid leukemia | 6 | 1c | |
| R586Q | acute myeloid leukemia | 6 | 0 | |
| R586Q | stomach carcinoma | 1 | 0 | |
| R586Q | colon adenocarcinoma | 1 | 0 | |
| R586Q | kidney Wilm’s tumor | 1 | 0 | |
| Smc1aR711W | R711W | large intestine adenocarcinoma | 2 | 6d (inc. STAG2-V181M) |
| R711W | endometroid carcinoma | 1 | 0 | |
| R711W | acute myeloid leukemia | 1 | 0 | |
| R711W | adult T cell lymphoma/leukemia | 1 | 0 | |
| R711W | Congenital muscular hypertrophy- cerebral syndrome | 1 | 0 | |
| R711Q | chronic myelomonocytic leukemia | 2 | 0 | |
| R711Q | endometroid carcinoma | 2 | 0 | |
| R711L | large intestine adenocarcinoma | 1 | 0 | |
| Smc1aR816G | R816H | acute myeloid leukemia | 1 | 0 |
| R816G | adenoid cystic carcinoma | 1 | 0 | |
| R816G | Cornelia de Lange syndrome | 1 | N/A |
Co-occurrence of indicated mutation with a mutation in Smc1a, Smc3, Rad21, Stag1, Stag2, Pds5a or Pds5b.
Not determined due to the large number of cases.
One instance of SMC1A-R586W co-occurred with a STAG2-R110* nonsense mutation.
Five of these occurred in one case, the sixth in a second case.
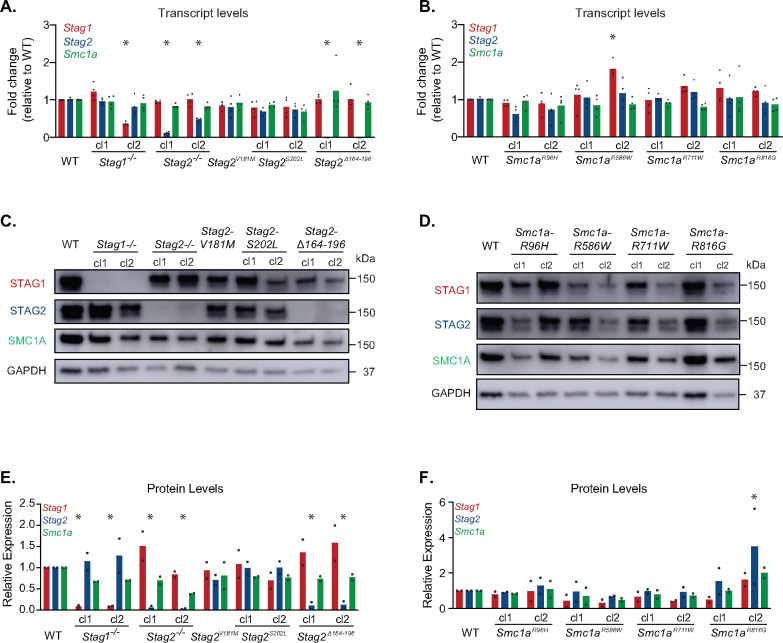
One simple mechanism by which cohesin sequence variation may impair function is by reducing the transcript and/or protein levels of cohesin complex components. We investigated how cohesin sequence variants impact the expression or stability of the variant proteins, as well as other subunits of the cohesin complex. For each sequence variant, two independent CRISPR clones were analyzed, except for Stag2V181M for which only a single clone was successfully generated. We examined Stag1, Stag2, and Smc1a transcript levels by reverse transcription-quantitative PCR (RT-qPCR), and found that Stag1−/− cl2, but not Stag1−/− cl1, showed greatly reduced levels of Stag1 transcripts, and normal levels of Stag2 and Smc1a transcripts (Figure 2A). Both Stag2−/− and Stag2Δ164-196 mESCs showed greatly reduced levels of Stag2 transcripts, but had relatively normal levels of Stag1 and Smc1a transcripts. Stag2V181M and Stag2S202L mESCS showed normal levels of Stag2, Stag1, and Smc1a transcripts. The mESCs with amino acid variants in Smc1a showed nearly normal levels of Smc1a, Stag1 and Stag2 transcripts, with less than a 2-fold change detected in any cell line (Figure 2B). These results indicate that the amino acid variants in Stag2 and Smc1a do not dramatically alter expression of the gene in which they are encoded, or expression of other cohesin complex members. Nearly all of the frameshift indels in Stag1 and Stag2, as well as the in-frame deletion in Stag2, lead to decreased transcript levels of the edited gene but did not alter levels of the other cohesin transcripts tested. We next examined the levels of SMC1A, STAG1, and STAG2 proteins in cohesin variant mESCs. Whereas the proteins containing amino acid variants were expressed at similar levels as in wild-type mESCs, the proteins that contained frameshift indels or a large in-frame deletion were not stably expressed (Figure 2C–F, Supplementary Figure 1). We note that Stag1−/− cl1 contains an in-frame deletion within an exon which lead to normal levels of Stag1 transcripts but STAG1 protein was not detected. Both Stag2−/− and Stag2Δ164-196 mESCs showed loss of Stag2 transcripts and STAG2 protein, yet Stag2 amino acid variants, Stag2V181M and Stag2S202L, showed near wild-type transcript and protein levels. Some Smc1a variant mESCs displayed an up to 2-fold increase in cohesin transcripts, though this increase was not observed at the protein level. In contrast, the Smc1aR586W mESCs showed slightly reduced STAG1 levels yet normal STAG2 protein levels. Overall, mESCs harboring cancer-derived amino acid variants exhibited near normal levels of cohesin subunits, while large deletions resulted in a dramatic reduction in protein levels. Importantly, the introduction of an amino acid variant or a large sequence deletion in one cohesin subunit did not disrupt levels of other cohesin complex members.
Figure 2.
Expression of cohesin components in isogenic mESCs harboring various cohesin sequence variants. (A) RT-qPCR analysis of Stag1, Stag2, and Smc1a transcript levels in Stag1 and Stag2 variant mESCs. n = 4 biological replicates. Cl1 refers to clone 1 and cl2 refers to clone 2. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05. (B) RT-qPCR analysis of Stag1, Stag2, and Smc1a transcript levels in Smc1a variant mESCs. Two independent clones were analyzed for each genotype. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05. (C) Levels of STAG1, STAG2, and SMC1A proteins in Stag1 and Stag2 variant mESCs. Nuclear extracts were immunoblotted for the indicated proteins and GAPDH as a control. Blots are representative of two experiments. (D) Levels of STAG1, STAG2, and SMC1A proteins in Smc1a variant mESCs. Nuclear extracts were immunoblotted for the indicated proteins and GAPDH as a control. Blots are representatitive of two experiments. (E) Quantification of 2 biological replicates of the blots presented in C. (F) Quantification of 2 biological replicates of the blots presented in D.
Aberrant transcriptional insulation in cohesin variant mESCs
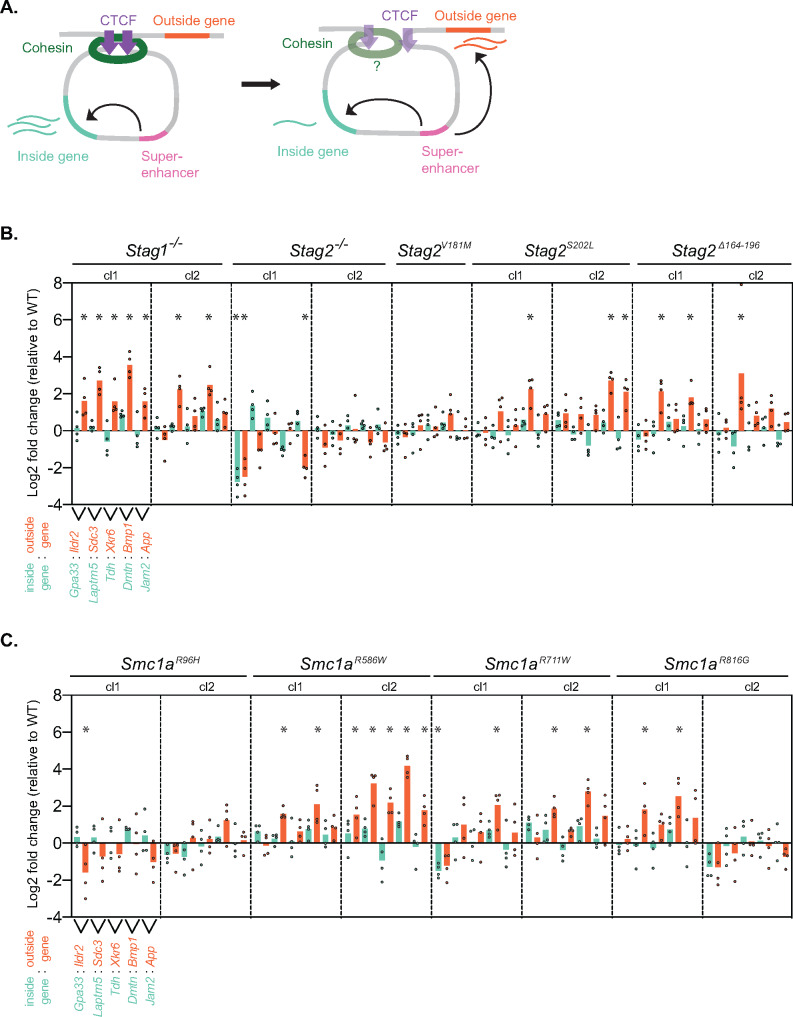
To understand if cohesin perturbations impact gene expression, we first investigated the activity of genes sensitive to transcriptional insulation. Insulated neighborhoods are DNA loop structures mediated by cohesin and CTCF, that constrain the activity of enhancers to specific target genes and prevent their activation of other nearby genes (Figure 3A). Insulated neighborhoods that contain super-enhancers and their highly expressed target genes are known as Super-enhancer Domains (SDs). SDs focus the activity of an enhancer on the highly expressed gene inside and prevent inappropriate activation of genes outside, which can occur if integrity of the insulating CTCF- and cohesin-mediated loop is compromised (Figure 3A). To test whether cohesin variants display altered gene expression at SDs, we performed RT-qPCR to measure transcript levels of gene pairs, where one gene is located inside and the other gene is outside of the SD: Gpa33/Ildr2, Laptm/Sdc3, Tdh/Xkr6, Dmtn/Bmp1, and Jam2/App. These insulated neighborhoods were previously shown to be sensitive to removal of CTCF binding sites or altered recruitment of cohesin or CTCF (Dowen et al. 2014; Arruda et al. 2020; Justice et al. 2020). Overall, mESCs lacking STAG1 showed moderate changes in gene expression at SDs with the outside gene often showing a significant increase in expression (Figure 3B). The Stag2−/−, Stag2Δ164-196 and Stag2S202 mESCs showed mild changes to expression of genes inside and outside of SDs (Figure 3B). Stag2V181M did not alter expression of the genes examined. Smc1aR586W mESCs showed the strongest misregulation of gene expression at the SDs tested, with up to 16-fold changes in gene expression detected (Figure 3C). Smc1aR711W, and Smc1aR816G mESCs showed mild gene expression changes at SDs, while Smc1aR96H did not alter expression of the genes examined (Figure 3C). Occasionally, two clones of the same mutation displayed some differences from one another. To address this, we first repeated the analysis of two such mutant cell lines, Stag2−/− and Smc1aR816G mESCs, and found that clonal differences were generally recapitulated (Supplementary Figure 2A). We next investigated whether off-target edits may underlie differences between clones. Our in silico analysis of potential off-target activity of these sgRNAs suggests that such events would be exceedingly rare, with Stag2−/−, Smc1aR816G, and Smc1aR586W having no potential off-target locations when allowing for one mismatch with the sgRNA, and only 3, 0, and 1 potential off-target locations respectively, when allowing for 2 mismatches with the sgRNA (Supplementary Table S1). Importantly, the presence of two mismatches between a sgRNA and genomic sequence significantly reduces the chance of Cas9 cutting a site and, therefore, reduces the likelihood of generating an off-target edit (Anderson et al. 2015). We performed an experimental analysis of two clones of wild-type mESCs, termed WTSmc1a-R586W exp, that experienced the sgRNA and clonal isolation process alongside two clones of Smc1aR586W mESCs. The results show no significant gene expression changes in WTSmc1a-R586W exp versus wildtype mESCs that were not exposed to the genome editing procedure (Supplementary Figure 2B). Overall, these results suggest that loss of STAG1 or STAG2, or introduction of recurring amino acid variants in cohesin observed in cancer, are sufficient to alter gene expression at Super-enhancer Domains, consistent with aberrant transcriptional insulation at cohesin-mediated insulated neighborhoods.
Figure 3.
Cohesin variant mESCs display altered gene expression at insulated neighborhoods. (A) Model of an insulated neighborhood where a cohesin and CTCF-mediated DNA loop focuses the activity of a Super-enhancer (pink) on a target gene inside the DNA loop (teal). This prevents the Super-enhancer from acting on the gene outside the DNA loop (orange). When transcriptional insulation is lost or impaired, the Super-enhancer can act on the orange gene outside the DNA loop and decrease activity on the teal gene inside the DNA loop. (B) RT-qPCR analysis of genes at five insulated neighborhoods in Stag1 and Stag2 variant mESCs. The inside gene (teal) and outside gene (orange) for each SD are adjacent to each other, and the log2 fold change value is indicated for the five insulated neighborhoods in each cell line. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05. (C) RT-qPCR analysis of genes at five insulated neighborhoods in Smc1a variant mESCs. The inside gene (teal) and outside gene (orange) for each SD are adjacent to each other, and the log2 fold change value is indicated for the five insulated neighborhoods in each cell line. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05.
Loss of pluripotency in cohesin variant mESCs
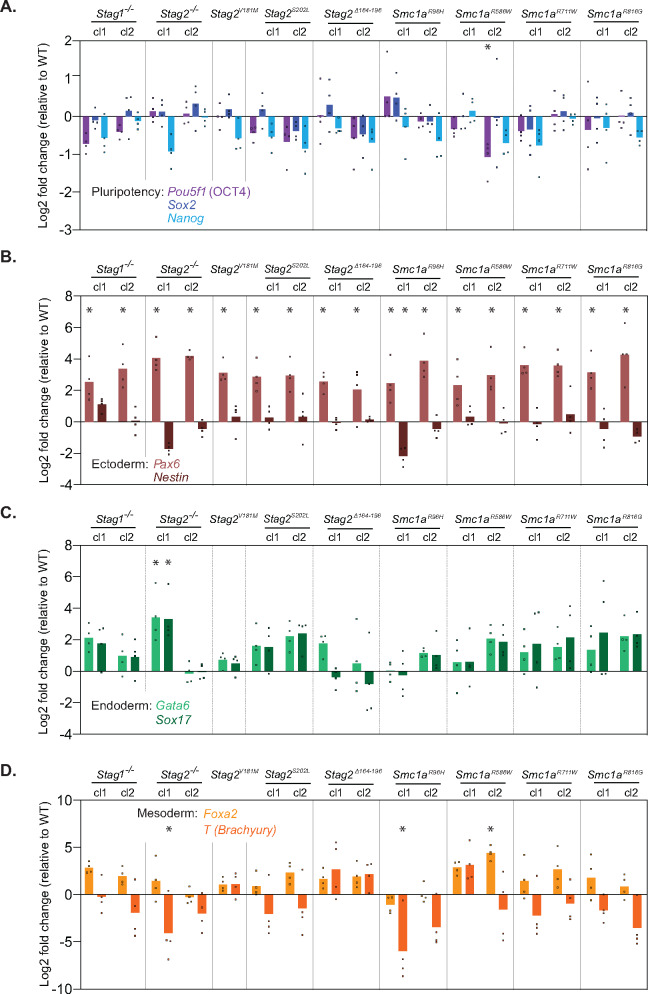
To further investigate the impact of cohesin variants in gene regulation, we examined expression of genes involved in cell identity. Embryonic stem cells maintain the pluripotent state through expression of the master transcription factors OCT4, SOX2, and NANOG. Decreased expression of these regulators in ESCs results in altered cell identity and differentiation (Nichols et al. 1998; Niwa et al. 2000; Chambers et al. 2003; Boyer et al. 2005; Loh et al. 2006). ESC differentiation, either upon receiving developmental cues or experimental disruption of pluripotency transcription factor expression, can result in entry into the endodermal, mesodermal, or ectodermal cell lineages (Keller 2005). In order to assess the cellular identity of mESCs harboring cohesin sequence variants, we performed RT-qPCR to measure transcript levels of pluripotency master transcription factors OCT4 (Pou5f1), SOX2 (Sox2), and NANOG (Nanog). Levels of pluripotency factors were generally decreased in all cohesin variant mESCs compared to wild-type, with Nanog transcripts being the most consistently reduced (Figure 4A). Expression of the ectodermal regulator PAX6 (Pax6) was significantly increased in all cohesin variant mESCs, whereas NESTIN (Nes) was mostly unchanged (Figure 4B). The endodermal regulators GATA6 (Gata6) and SOX17 (Sox17) were generally increased in expression in most cohesin variant mESCs compared to wild-type (Figure 4C). The mesodermal lineage-specifying factors FOXA2 (Foxa2) and Brachyury (T) showed inconsistent changes across the cohesin variant mESCs (Figure 4D). Many of the cohesin variants examined displayed some degree of altered stem cell identity via decreased expression of pluripotency factors and/or increased expression of ectodermal or endodermal lineage-specifying factors. To investigate whether the genome editing procedure contributes to changes in cellular identity, we performed additional analyses on two clones of wild-type mESCs that experienced the sgRNA and clonal isolation process alongside two clones of Smc1aR586W mESCs. The results show some increased expression of the ectodermal marker PAX6 and no significant increases in pluripotency, endodermal, or mesodermal markers in WTSmc1a-R586W exp mESCs versus wildtype mESCs that were not exposed to the genome editing procedure (Supplementary Figure 3). The uniform increase in Pax6 expression in all cell lines that experienced the genome editing process and clonal isolation process suggests that it may be a consequence of the procedure. While the mesodermal marker T and ectodermal marker Nestin were decreased in WTSmc1a-R586W exp mESCs relative to wildtype, the decreased expression of a gene poised for upregulation upon lineage commitment is not a clear indicator of differentiation.
Figure 4.
Altered cellular identity of cohesin variant mESCs. (A) RT-qPCR analysis of pluripotency factors. Levels of Pou5f1 (OCT4), Sox2, and Nanog transcripts were measured in Stag1, Stag2, and Smc1a variant mESCs. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05. (B) RT-qPCR analysis of ectodermal regulators. Levels of Pax6 and Nestin transcripts were measured in Stag1, Stag2, and Smc1a variant mESCs. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05. (C) RT-qPCR analysis of mesodermal regulators. Levels of Foxa2 and T (Brachyury) transcripts were measured in Stag1, Stag2, and Smc1a variant mESCs. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05. (D) RT-qPCR analysis of endodermal regulators. Levels of Gata6 and Sox17 transcripts were measured in Stag1, Stag2, and Smc1a variant mESC lines. n = 4 biological replicates. Significance was determined by Dunnet’s multiple comparisons test; *P < 0.05.
Differentiation potential is compromised in cohesin variant cells
To investigate the ability of cohesin variant mESCs to differentiate, we performed the embryoid body (EB) differentiation assay (Behringer et al. 2016). In this assay, 1,000 cells were cultured in a droplet of media lacking leukemia inhibitory factor (LIF), to allow for differentiation. The droplet was suspended from the lid of a sterile petri plate, which prevented cells from adhering to the surface of the dish, and cells were allowed to grow for three days (Figure 5A). During this process, cells differentiate into various lineages and form spherical EB structures in a process that mimics in vivo gastrulation (Sene et al. 2007). Cell lines that showed the strongest gene expression changes, were investigated for their ability to differentiate: Stag1−/−, Stag2−/−, and Smc1aR586W mESCs. The overall size of EBs was significantly decreased in all Stag1−/−, Stag2−/−, and Smc1aR586W mESC clones, as measured by their total area per droplet (Figure 5B). The two Smc1aR586W variant clones displayed some heterogeneity, with one of the two clones showing a greater reduction in size than the other (Figure 5B). To further evaluate the differentiation potential of these EBs, we transferred each droplet to an individual well of a tissue culture plate and monitored the cells as they continued to grow in media lacking LIF. During subsequent days, the cells in the EB spread out, adhered to the surface of the plate and some patches of cells began rhythmically contracting, consistent with development of ES-derived beating cardiomyocytes (Boheler et al. 2002). In this un-directed differentiation assay, Stag1−/−, Stag2−/−, and Smc1R586W cells have slightly reduced capacity to differentiate into cardiomyocytes compared to wild-type cells (Figure 5C). To test whether the decrease in EB size and potential defect in differentiation into cardiomyocytes were due to reduced cellular proliferation, we monitored cell growth rate over three days. Indeed, Smc1aR586W mESCs show a reduced proliferation rate relative to wild-type, whereas Stag1−/− and Stag2−/− mESC clones showed modest or no defects (Figure 5D). These results demonstrate that Smc1aR586W cells have a strong reduction in proliferation and possibly differentiation potential, whereas Stag1−/− and Stag2−/− mESCs have mild to no defects in cellular growth and differentiation.
Figure 5.
Impact of cohesin variants on cellular differentiation. (A) Schematic of the hanging droplet assay. 1,000 mESCs were plated in a 30 µl droplet of media hanging from the lid of a tissue culture dish. After 3 days of growth in media lacking LIF, embryoid bodies were imaged and their size was measured. Droplets were collected, dissociated and moved to individual wells of a 48-well tissue culture dish. Wells were monitored for 10 days for the presence of rhythmically contracting cells. (B) Total size of embryoid bodies after 3 days of differentiation in hanging droplet cultures lacking LIF. Within each droplet the total area of EBs was measured. If more than one EB was present in a droplet, the areas were summed and presented. WT n = 70, Stag1−/− cl1 n = 70, Stag1−/− cl2 n = 71, Stag2−/− cl1 n = 69, Stag2−/− cl2 n = 71, Smc1aR586W cl1 n = 72, Smc1aR586W cl2 n = 71. Asterisks indicate significant differences from wild-type mESCs. ****P < 0.0001, *P = 0.0124 as measured by Kruskal–Wallis test. Data merged from two biological replicates for each clone. (C) Propensity of Stag1 (top), Stag2 (middle), and Smc1a (bottom) variant mESCs to differentiate into cardiomyocytes. The proportion of wells with EB differentiation into at least one patch of rhythmically contracting cells. Error bars represent one standard deviation. No significant differences were detected as measured by two-way ANOVA. n = 2 biological replicates for each clone. (D) Proliferation rate of various Stag1 (top), Stag2 (middle), and Smc1a (bottom) variant mESCs, relative to the 50,000 cells plated at time 0. *P = 0.0479 at 72 h between wild-type and Stag1−/− cl1; *P = 0.0489 at 72 h between wild-type and Smc1AR586W cl1; *P = 0.0109 at 72 h between wild-type and Smc1aR586W cl2 mESCs as measured by two-way ANOVA. n = 4 biological replicates.
Discussion
Here, we report the functional impacts of various oncogenic cohesin sequence variants on the regulation of gene expression and cellular identity. By engineering cohesin variant cell lines, we were able to directly compare the phenotypes of individual variants in an isogenic system, free of confounding effects from use of different cancer cell lines from distinct lineages and with varied mutational burdens. Additionally, our generation and independent analysis of multiple clonal cell lines limits the possibility of off-target effects of CRISPR/Cas9 editing contributing to the phenotypes observed in the variant mESCs. Several cohesin variants were found to be sufficient to alter gene expression and the maintenance of embryonic stem cell identity, implicating aberrant cohesin activity as a potential contributor to disease via misregulation of gene expression. Generally, the cohesin variants examined did not disrupt expression or steady state protein levels of the other cohesin complex members, indicating that the phenotypes observed resulted from altered cohesin function instead of loss of the complex. The Smc1aR586W variant exhibited the strongest phenotypes with regard to altered expression of genes in insulated neighborhoods, cell identity genes and defects in proliferation and possibly differentiation. The results also indicate that the cohesin accessory subunits STAG1 and STAG2 are important regulators of gene expression and cellular identity. This work reveals that a subset of cohesin sequence variants found in cancer are sufficient to cause misregulation of gene expression and cellular identity, thus providing evidence that altered cohesin activity may contribute to disease through processes other than genome instability, aneuploidy, and altered DNA replication.
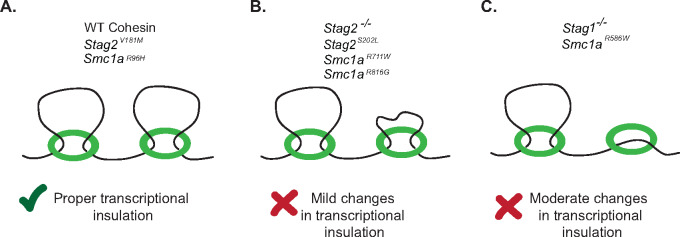
Many cohesin sequence variants tested in this study caused altered gene expression (Figure 6). Changes at insulated neighborhoods in Stag1−/−, Stag2−/−, and Smc1aR586W mESCs were consistent with potential re-wiring of enhancer-gene pairs, with up-regulation of genes located outside of the DNA loop structures observed. These changes are consistent with those observed when individual insulated neighborhood boundary elements are deleted (Dowen et al. 2014; Ji et al. 2016; Hnisz et al. 2016a). Interestingly, loss of STAG1 or STAG2 showed gene expression changes in the same direction as the presence of SMC1AR586W variant complexes, indicating that they might disrupt the function of cohesin at the anchors of DNA loop structures through a shared mechanism. SMC1AR586W causes more pronounced changes in gene expression than STAG1 or STAG2 loss, which is consistent with previous findings indicating that disruption of core cohesin subunits causes stronger changes in genome architecture than disruption of accessory subunits. Whereas inactivation of RAD21 or NIPBL was shown to cause a strong breakdown of TADs, depletion of STAG2 caused modest reductions in the number and strength of TADs (Haarhuis et al. 2017; Rao et al. 2017; Schwarzer et al. 2017; Wutz et al. 2017; Kojic et al. 2018; Cuadrado et al. 2019). Other cohesin amino acid variants analyzed in our study, including the R96H, R711W, and R816G variants in SMC1A and the STAG2 amino acid variants V181M and S202L, caused moderate to minimal impacts on SD function and pluripotency. Therefore, these variants may be only weakly hypomorphic, but we do not rule out impacts at specific loci or subsets of cohesin sites and DNA loop structures in the genome. Occasionally, two independent clones of the same mutation displayed some differences from one another. We found that these differences were repeatable and were unlikely to arise from distinct off-target mutations since relatively few off-target locations were detected when allowing for 1, 2, or 3 mismatches with the sgRNA (Supplementary Table S1). To experimentally investigate potential off-target events, we analyzed wildtype cells that experienced the genome editing process and could potentially have off-target edits, but do not have on-target edits. The results show that wild-type cells that were exposed to the sgRNA and clonal isolation process did not display gene expression changes that resolve the differences between mutant clones. Rather, in the case where two mutant clones display some differences, this may be due to on-target effects from the genome editing process followed by downstream stochastic changes in gene expression and cellular differentiation that caused independent clones to take distinct paths of aberrant cellular identity.
Figure 6.
Summary of the impact of Cohesin variants on transcriptional insulation. (A) Proper transcriptional insulation is retained in wildtype, Stag2V181M,and Smc1aR96H mESCs, with proper gene expression largely retained. (B) Mild gene expression changes are observed in Stag2−/−,Stag2S202L and Smc1aR711W, and Smc1aR816G mESCs, consistent with altered transcriptional insulation. (C) Moderate gene expression changes are observed in Stag1−/− and Smc1aR586W mESCs, consistent with altered transcriptional insulation.
While cohesin and CTCF are recognized as important structural regulators of the genome, the molecular mechanisms by which the spatial organization of DNA impacts gene expression are poorly understood. Several studies have shown that deletions or alterations of individual DNA loop boundaries disrupt transcriptional insulation causing changes in gene expression (Lupiáñez et al. 2015; Guo et al. 2015, 2018; Flavahan et al. 2016; Hnisz et al. 2016b). In this study, we show that cancer-associated cohesin mutations are also capable of disrupting transcriptional insulation causing altered gene expression. These results are consistent with other work in the field showing that altered enhancer specificity and activity can lead to pathogenic transcriptional programs in disease contexts (Sur and Taipale 2016; He et al. 2019). How the various biological functions of cohesin are spatially and temporally directed on the genome is not well understood. In particular, the recruitment of cohesin to specific cis-regulatory elements and their subsequent spatial organization into DNA loops, hubs, or domains that influence transcription requires additional investigation. Future studies addressing extrusion speed and directionality, residence time of cohesin molecules at specific genomic sites, and the multi-way interactions that bring together combinations of enhancers and promoters will provide important molecular insights.
Introduction of cancer-associated cohesin sequence variants into mESCs caused misexpression of lineage-specifying factors. Ectoderm- and endoderm-promoting factors were upregulated, while pluripotency-defining factors were downregulated in nearly all of the cohesin variant mESCs. In addition, the differentiation potential may be reduced or delayed in Smc1aR586W variant mESCs compared to wild-type. While a decreased proliferation rate may contribute to reduced differentiation, it does not fully explain the defects in Smc1aR586W mESCs. Since cardiomyocytes develop from mesodermal precursors, it is possible that the cohesin variant mESCs have impaired differentiation due to the simultaneous expression of ectodermal and endodermal lineage determining factors. Co-expression of potentially opposing transcriptional programs could disrupt a coordinated differentiation process by allowing for cell fate plasticity rather than cell fate restriction into cardiomyocytes. Together, these data suggest that oncogenic cohesin variants alter gene expression and disrupt maintenance of the pluripotent state.
Our results are consistent with and extend previous work investigating specific STAG2 alleles. Previously, STAG2-V181M was identified in a myeloid neoplasm (Kon et al. 2013), and STAG2-S202L was identified in a bladder cancer (Solomon et al. 2013). Prior studies showed that both the expression of STAG2-V181M and STAG2-S202L and their incorporation into cohesin complexes occurs at wild-type levels (Kim et al. 2016). Furthermore, STAG2-V181M and STAG2-S202L did not impact chromosome stability, cellular proliferation rate, or sister chromatid cohesion when introduced into HCT-116 cells (Kim et al. 2016). Our results indicate that STAG2-V181M and S202L cause modest changes in expression of cell identity genes, which may indicate altered cohesin activity in the context of regulating gene expression. In combination with other mutations, these point mutations may therefore contribute to tumorigenesis by impairing maintenance of cell identity-controlling transcriptional programs.
Our findings of transcriptional dysregulation in SMC1A variant or STAG-null mESCs are consistent with results in other model systems. In previous work, SMC1A-R711 was mutated to glycine in murine hematopoietic stem and progenitor cells (HSPCs) and shown to alter expression of hematopoiesis-controlling transcription factors, accessibility at transcriptional regulatory elements, and impair differentiation (Mazumdar et al. 2015). Knockdown or knockout of Stag2 had similar effects on gene expression and differentiation in HSPCs (Mullenders et al. 2015). A direct comparison of the impacts of Stag1 and Stag2 ablation on HSPC function revealed that Stag2, but not Stag1, loss resulted in altered transcription and differentiation (Viny et al. 2019). In those cells, Stag2 loss was further associated with reduced transcriptional insulation and a loss of intra-TAD contacts around master regulator genes (Viny et al. 2019). STAG2 amino acid variants have also been implicated in cancer, as STAG2-D193N reduces STAG2 incorporation into cohesin and may participate in drug resistance in melanoma (Shen et al. 2016).
This study provides insight into how cohesin sequence variants may contribute to cancer. We find that specific cohesin variants disrupt transcriptional regulation and may act in disease contexts to destabilize cell identity. Indeed, in a murine model of AML, Smc3 haploinsufficiency caused transcriptional changes to the HSC cell identity program, but tumorigenesis only occurred once the proliferative driver FLT3-internal tandem duplication (ITD) was introduced (Viny et al. 2015). Similarly, Stag2 inactivation is a frequent second hit in Ewing sarcoma, and is associated with disease recurrence and poor clinical outcomes (Brohl et al. 2014; Crompton et al. 2014; Tirode et al. 2014). Our data further support a role for cohesin variants in destabilizing cellular identity as part of multiple genetic hits that ultimately result in tumorigenesis. Indeed, a current hypothesis of tumorigenesis involves epigenetic priming of cancer-initiating cells before additional oncogenic hits establish the cancer cell state (Vicente-Dueñas et al. 2018); we propose that our data are consistent with a similar role for cohesin sequence variants in cancer. Finally, the study of cancer-derived cohesin variants provides an avenue for identifying potential hypomorphic and separation-of-function alleles for study, rather than the total loss-of function conditions mostly studied to date. Further studies are needed to elucidate the roles of individual cohesin cancer variants in cohesin-mediated gene control on chromatin.
Acknowledgements
We thank members of the Dowen lab and the lab of Dr. Dan McKay for helpful discussions and comments. We thank the staff of the UNC Flow Cytometry Core Facility for assistance with FACS to create genome edited cell lines.
Funding
This work was supported by the National Institute of General Medical Sciences under award R35GM124764 to J.M.D. N.L.R. was supported in part by the National Institute of General Medical Sciences under award 1T32GM135128. N.L.A. was supported in part by the National Institute of Health grant T32GM007092. Z.M.C. was supported in part by a grant from the National Cancer Institute under award T32CA217824. N.L.A. was supported in part by the National Science Foundation Graduate Research Fellowship Program. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650116. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- Anderson EM, Haupt A, Schiel JA, Chou E, Machado HB, et al. 2015. Systematic analysis of CRISPR-Cas9 mismatch tolerance reveals low levels of off-target activity. J Biotechnol. 211:56–65. [DOI] [PubMed] [Google Scholar]
- Arruda NL, Carico ZM, Justice M, Liu YF, Zhou J, et al. 2020. Distinct and overlapping roles of STAG1 and STAG2 in cohesin localization and gene expression in embryonic stem cells. Epigenetics Chromatin. 13:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim J-S.. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbás-Martínez C, Sagrera A, Carrillo-De-Santa-Pau E, Earl J, Márquez M, et al. 2013. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 45:1464–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer R, Gertsenstein M, Nagy KV, Nagy A.. 2016. Differentiating mouse embryonic stem cells into embryoid bodies by hanging-drop cultures. Cold Spring Harb Protoc. 2016:pdb.prot092429. [DOI] [PubMed] [Google Scholar]
- Benedict B, van Schie JJM, Oostra AB, Balk JA, Wolthuis RMF, et al. 2020. WAPL-dependent repair of damaged DNA replication forks underlies oncogene-induced loss of sister chromatid cohesion. Dev Cell. 52:683–698.e7. [DOI] [PubMed] [Google Scholar]
- Boheler KR, Czyz J, Tweedie D, Yang H-T, Anisimov SV, et al. 2002. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 91:189–201. [DOI] [PubMed] [Google Scholar]
- Bompadre O, Andrey G.. 2019. Chromatin topology in development and disease. Curr Opin Genet Dev. 55:32–38. [DOI] [PubMed] [Google Scholar]
- Bonev B, Cavalli G.. 2016. Organization and function of the 3D genome. Nat Rev Genet. 17:661–678. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Tong IL, Cole MF, Johnstone SE, Levine SS, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohl AS, Solomon DA, Chang W, Wang J, Song Y, et al. 2014. The genomic landscape of the Ewing sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. e1004475.10: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa V, Gines MM, Gusmao EG, Slotman JA, Zirkel A, et al. 2020. Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcriptional control. Genome Res. 30:515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 113:643–655. [DOI] [PubMed] [Google Scholar]
- Cong L, Zhang F.. 2015. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol. 1239:197–217. [DOI] [PubMed] [Google Scholar]
- Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, et al. 2014. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 4:1326–1341. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Gimé Nez-Llorente D, Kojic A, Gó Mez-Ló Pez G, Marti-Renom MA, et al. 2019. Specific contributions of cohesin-SA1 and cohesin-SA2 to TADs and polycomb domains in embryonic stem cells. Cell Rep. 27:3500–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Young RA.. 2014. SMC complexes link gene expression and genome architecture. Curr Opin Genet Dev. 25:131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagen KP. 2018. Principles of chromosome architecture revealed by Hi-C. Trends Biochem Sci. 43:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, et al. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 529:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, et al. 2017. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 45:D777–D783., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Perez AA, Hazelett DJ, Coetzee GA, Rhie SK, et al. 2018. CRISPR-mediated deletion of prostate cancer risk-associated CTCF loop anchors identifies repressive chromatin loops. Genome Biol. 19:160., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 162:900–910., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yáñez-Cuna JO, Amendola M, et al. 2017. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 169:693–707.e14., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS. 2020. CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism. Nucleus. 11:132–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Zheng G, Qu Q, Liu H, Ouyang Z, et al. 2014. Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion. Nat Struct Mol Biol. 21:864–870., [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Long W, Liu Q.. 2019. Targeting super-enhancers as a therapeutic strategy for cancer treatment. Front Pharmacol. 10:361–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill VK, Kim JS, Waldman T.. 2016. Cohesin mutations in human cancer. Biochim Biophys Acta Rev Cancer. 1866:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Day DS, Young RA.. 2016a. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintrau AS, Day DS, Valton AL, Bak RO, et al. 2016b. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 351:1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield JA, Print CG, Mönnich M.. 2012. Diverse developmental disorders fromthe one ring: distinct molecular pathways underlie the cohesinopathies. Front Genet. 3:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, et al. 2016. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 18:262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice M, Carico ZM, Stefan HC, Dowen JM.. 2020. A WIZ/cohesin/CTCF complex anchors DNA loops to define gene expression and cell identity. Cell Rep. 31:107503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature. 502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. 2005. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 19:1129–1155. [DOI] [PubMed] [Google Scholar]
- Kim JS, He X, Orr B, Wutz G, Hill V, et al. 2016. Intact cohesion, anaphase, and chromosome segregation in human cells harboring tumor-derived mutations in STAG2 (B. A. Sullivan, Ed). PLoS Genet. 12:e1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic A, Cuadrado A, De Koninck M, Giménez-Llorente D, Rodríguez-Corsino M, et al. 2018. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol. 25:496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, et al. 2013. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 45:1232–1237. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Cancer Genome Atlas Research Network, et al. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 38:431–440. [DOI] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J.. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 167:1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. 2014. Cohesin in cancer: Chromosome segregation and beyond. Nat Rev Cancer. 14:389–393. [DOI] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 161:1012–1025., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar C, Shen Y, Xavy S, Zhao F, Reinisch A, et al. 2015. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell. 17:675–688., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, et al. 2015. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 212:1833–1850., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, et al. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 95:379–391., [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki JI, Smith AG.. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 24:372–376. [DOI] [PubMed] [Google Scholar]
- Norton HK, Phillips-Cremins JE.. 2017. Crossed wires: 3D genome misfolding in human disease. J Cell Biol. 216:3441–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG.. 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 12:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J.. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev. 22:3089–3114. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, St Hilaire BG, Engreitz JM, Perez EM, et al. 2017. Cohesin loss eliminates all loop domains. Cell. 171:305–320.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Losada A.. 2013. Cohesin, a chromatin engagement ring. Curr Opin Cell Biol. 25:63–71. [DOI] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG.. 2018. Organizational principles of 3D genome architecture. Nat Rev Genet. 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N, Hebrok M.. 2015. Regulation of cellular identity in cancer. Dev Cell. 35:674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra S, Skibbens RV.. 2013. Cohesin codes - Interpreting chromatin architecture and the many facets of cohesin function. J Cell Sci. 126:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 551:51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, et al. 2007. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CH, Kim SH, Trousil S, Frederick DT, Piris A, et al. 2016. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nat Med. 22:1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim JS, Bondaruk J, Shariat SF, Wang ZF, et al. 2013. Frequent truncating mutations of STAG2 in bladder cancer. Nat Genet. 45:1428–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, et al. 2011. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science (80). 333:1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Taipale J.. 2016. The role of enhancers in cancer. Nat Rev Cancer. 16:483–493. [DOI] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, et al. 2019. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Platt FM, Hurst CD, Thygesen HH, Knowles MA.. 2014. Frequent inactivating mutations of STAG2 in bladder cancer are associated with low tumour grade and stage and inversely related to chromosomal copy number changes. Hum Mol Genet. 23:1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Bollin R, Gehlhaar M, Walter C, Dugas M, et al. 2014. Mutations in the cohesin complex in acute myeloid leukemia: Clinical and prognostic implications. Blood. 123:914–920., [DOI] [PubMed] [Google Scholar]
- Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, et al. 2014. Genomic landscape of ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4:1342–1353., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Dueñas C, Hauer J, Cobaleda C, Borkhardt A, Sánchez-García I.. 2018. Epigenetic priming in cancer initiation. Trends Cancer. 4:408–417. [DOI] [PubMed] [Google Scholar]
- Viny AD, Bowman RL, Liu Y, Lavallée VP, Eisman SE, et al. 2019. Cohesin members Stag1 and Stag2 display distinct roles in chromatin accessibility and topological control of HSC self-renewal and differentiation. Cell Stem Cell. 25:682–696.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, Levine RL.. 2018. Cohesin mutations in myeloid malignancies made simple. Curr Opin Hematol. 25:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viny AD, Ott CJ, Spitzer B, Rivas M, Meydan C, et al. 2015. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med. 212:1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman T. 2020. Emerging themes in cohesin cancer biology. Nat Rev Cancer. 20:504–515. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, The Cancer Genome Atlas Research Network, et al. 2013. The cancer genome atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, et al. 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. Embo J. 36:3573–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Wallace J, Felsenfeld G.. 2011. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 31:2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Jiang Y, Mao Q, Demeler B, Tao YJ, et al. 2013. Characterization of the interaction between the cohesin subunits Rad21 and SA1/2 (D. Cimini, Ed). PLoS One. 8:e69458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Cell lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental Material available at figshare: https://doi.org/10.25386/genetics.13962011.