Abstract
Human subtelomere regions are highly enriched in large segmental duplications and structural variants, leading to many gaps and misassemblies in these regions. We develop a novel method, NPGREAT (NanoPore Guided REgional Assembly Tool), which combines Nanopore ultralong read datasets and short-read assemblies derived from 10x linked-reads to efficiently assemble these subtelomere regions into a single continuous sequence. We show that with the use of ultralong Nanopore reads as a guide, the highly accurate shorter linked-read sequence contigs are correctly oriented, ordered, spaced and extended. In the rare cases where a linked-read sequence contig contains inaccurately assembled segments, the use of Nanopore reads allows for detection and correction of this error. We tested NPGREAT on four representative subtelomeres of the NA12878 human genome (10p, 16p, 19q and 20p). The results demonstrate that the final computed assembly of each subtelomere is accurate and complete.
Keywords: segmental duplications, nanopore, genome assembly, subtelomere, linked reads sequencing
I. INTRODUCTION
Telomeres are complex nucleoprotein structures that are essential for proper replication and stability of chromosomes. They consist of stretches of (TTAGGG) repeat DNA at the ends of chromosomes with their associated proteins and at least one lncRNA, TERRA; their dysfunction can contribute to disease at multiple levels [1]. Only one or a few critically short telomeres in a cell are sufficient to induce DDR-mediated senescence or apoptosis [2], resulting in the disruption of tissue microenvironments and age-related diseases including cancer [3], [4]. Subtelomere regions regulate adjacent single-telomere lengths and stabilities of human chromosomes in both telomerase-positive and telomerase-negative contexts [5], [6]; thus, accurate maps and DNA sequences for human subtelomere regions, along with detailed knowledge of subtelomere variation and long-range telomere-terminal haplotypes in individuals, are critical for understanding telomere function and its roles in human biology. However, the human subtelomere regions are highly enriched in large segmental duplications and structural variants, leading to many gaps and misassemblies in these regions, even in the large-insert clone-based finished human reference sequence [7]. Fifteen years after completion of the human genome project, there is still no robust method to fully sequence and accurately assemble subtelomeric DNA. We hypothesized that new ultralong Nanopore read technology combined with synthetic long read methods might be used to successfully solve the large segmental duplication assembly problem.
10x Linked-Reads Sequencing.
The recently developed Linked-read approach pioneered by 10X Genomics generates short-read datasets from large genomic DNA molecules first partitioned and barcoded using the “Gel Bead in Emulsion” (GEM) microfluidic method [8], [9]. Each set of linked reads is comprised of low-read coverage of a small number of large genomic DNA molecules and is associated with a unique bar code. Our group recently devised a computational approach called Regional Extension of Assemblies Using Linked-Reads (REXTAL) [10], [11] for improved region-specific assembly of segmental duplication-containing DNA, leveraging these relatively inexpensive genomic short-read datasets. We have shown that, using REXTAL, it is possible to extend assemblies corresponding to single-copy diploid DNA segments into adjacent, otherwise inaccessible subtelomere segmental duplication regions and subtelomeric gap regions [11].
Oxford Nanopore Sequencing.
Nanopore-based long-read sequencing technology has opened up the possibility of bridging large segmental duplication and structurally variant regions with ultralong single-molecule reads. Ranging up to roughly 800 kb in length [12], these ultralong reads are currently expensive to produce on large scales and are relatively error-prone – single ultralong Nanopore reads mapped to the human reference genome with only about 66% – 88% nucleotide sequence identity. Despite the enormous promise of this technology these limitations currently preclude its routine use for sequencing large genomes. Nevertheless, these ultralong reads and especially the ones that include the telomere terminal repeat tracts, could be ideal for solving the long-standing assembly problems caused by segmental duplications and structural variation by providing unprecedented long-range contiguity of assemblies that incorporate them.
Combining the technologies.
In order to accurately assemble the large segmentally duplicated, structurally variant, and biologically critical human subtelomere regions, we propose a pipeline that combines the Nanopore ultralong read datasets and the short-read assemblies derived from 10x linked-reads. For the first time, complete haplotype-specific DNA sequences would become available for these understudied regions, completing subtelomeric regions of the existing as well as future reference genomes. With our method, NPGREAT (NanoPore Guided REgional Assembly Tool), the high-quality REXTAL assemblies, are integrated with relatively low-quality but ultralong Nanopore reads from the same genome. The ultralong Nanopore reads are essentially used as sequence-level resolution scaffolds upon which the REXTAL contigs can be corrected and placed, replacing the low-quality Nanopore sequence with high-quality REXTAL sequence for matching regions while marking and retaining the parts of lower-quality Nanopore sequence (which lack REXTAL assembly coverage) as “connectors” useful for spacing, orienting and ordering multiple REXTAL contigs along the ultralong reads. Leveraging the qualities of both technologies, the ultimate outcome of NPGREAT is the production of a single DNA sequence.
Previous work.
A similar strategy has been used in the past for shorter (5 to 50 kb-sized) Nanopore reads in simpler genomes [13], but our particular challenge is to achieve contiguity across 30 – 300 kb sized segmental duplication regions of human subtelomeres. A superficially similar effort targeting the human genome with the incorporation of Nanopore reads and the 10x Supernova assembly technologies [14] does not capture large segmentally duplicated regions, which is a primary goal of our effort. On the other hand, a recent assembly strategy intended specifically for segmental duplication regions [15] utilizes the Pacific Biosciences (Pacbio) long-read technology, which is limited to small segmental duplications (10 −20 kb reads) and did not work well with error-prone Nanopore ultralong reads. This approach would be ineffective on the segmental duplications that occur in subtelomeric regions, since they are typically long and have high similarity amongst each other. Here, we develop a novel method for using Nanopore ultralong reads to guide and correct REXTAL assemblies in large subtelomeric segmental duplication regions which are currently refractory to all previously attempted assembly methods.
II. METHODOLOGY
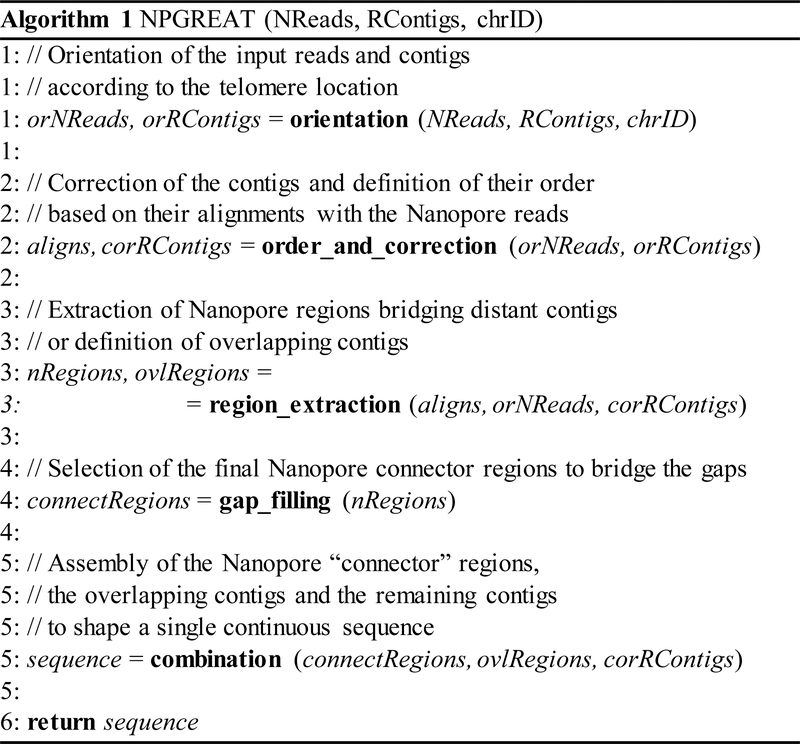
A general description of our algorithm, which assembles ultralong Nanopore reads and short REXTAL contigs of a chromosome’s telomeric region, can be seen in Fig. 1. With the use of five main operations, it generates one continuous sequence. Details of those operations are presented in subsections A – E.
Figure 1.
The assembly algorithm. Input: The selected ultralong Nanopore reads (NReads), the selected short REXTAL contigs (RContigs) and the chromosome number (chrID). Output: The assembled sequence (sequence).
A. Data and Tools
We tested NPGREAT on the genome of the human cell line NA12878, specifically on subtelomeres 10p, 16p, 19q and 20p. Those four regions have different segmental duplication sizes and complexities that are representative of subtelomeres overall.
The input data to the assembly algorithm are the REXTAL contigs and the Nanopore reads. The REXTAL procedure uses 10X Genomics Linked-Reads in paired-end format, with a mean read size of approximately 150bp. The output data from REXTAL are sequence scaffolds, which need to be converted into a set of sequence contigs in order to insert them into our algorithm. For that to be done, we had to take into consideration the assembly of scaffolds by Supernova [9]. Each scaffold may contain a number of “N”s, notably a series of 10 Ns designating a de Bruijn graph cycle, a series of 100 Ns designating a small gap, and a longer series of NN’s designating a large gap. To obtain the sequence contigs of a scaffold, we deleted the 10 N segments, and split the scaffold into discrete sequence contigs at points where 100 Ns or larger existed (simultaneously removing these stretches of NN’s).The resulting set of sequence contigs were designated REXTAL contigs.
We then obtained the Nanopore reads using the following procedure:
-
1)
Raw Data: The raw FAST5 files of the ultralong reads dataset from the NA12878 genome were downloaded from the European Nucleotide Archive (ENA) with accession number PRJEB23027 [12].
-
2)
Basecalling: The Oxford Nanopore Technologies (ONT) Guppy basecalling software version 2.3.7 was used to re-analyze the raw data and obtain the FASTQ files for these reads.
-
3)
Minimum required length: Only ultralong Nanopore reads of length 40K bases or more were retained.
-
4)
Telomere tract screen: Candidate telomeric Nanopore reads were found by searching the database of selected ultralong Nanopore reads > 40K with (TTAGGG)n using a simple pattern-matching to a TTAGGG)n 24-mer [16]. To distinguish the subtelomere to which the selected (TTAGGG)n containing reads belong, they were screened with a panel of unique single-copy sequences (10 K baits) closest to the telomere. The result was the set of mapped telomere-terminal Nanopore reads.
-
5)
1-copy region screen: For each subtelomere, the 10K, 50K and 100K bases 1-copy regions closest to the telomere were extracted from HG38, as shown on Table I. The regions were masked with the Repeat Masker open-4.0.9 and Tandem Repeat Finder 4.09 and then set as baits to find the matching Nanopore reads from the dataset of ultralong reads above 40Kb. The screen was done with the use of BLASTn 2.8.1 alignment software, requiring a percent identity (pid) of at least 80 and retained only the output alignments with expect value (evalue) less than e(−250). Subtelomeric Nanopore reads thus selected were further validated by BLASTn against the whole genome to ensure their mapping to the correct subtelomere location. The number of identified subtelomeric Nanopore reads is 14, 5, 5, 9 and the telomeric Nanopore reads 3, 2, 5, 2 for 10p, 16p, 19q and 20p respectively.
TABLE I.
COORDINATES OF SUBTELOMERE REGIONS
| Subtelomere region |
HG38 Reference | Segmental Duplication region |
1-copy region | 10K 1-copy | 50K 1-copy | 100K 1-copy |
|---|---|---|---|---|---|---|
| 10p | 10,001 – 588,571 | 10,001 – 88,570 | 88,571 – 588,571 | 88,506 – 98,505 | 88,506 – 138,505 | 88,506 – 188,505 |
| 16p | 10,000 – 240,859 | 10,000 – 40,859 | 40,860 – 240,859 | 43,549 – 58,548 | 43,549 – 93,548 | 43,549 – 143,548 |
| 19q | 58,386,558 – 58,607,616 | 58,586,558 – 58,607,616 | 58,386,558 – 58,586,557 | 58,576,001 – 58,586,000 | 58,536,001 – 58,586,000 | 58,486,001 – 58,586,000 |
| 20p | 66,335 – 266,334 | N/A | 66,335 – 266,334 | 67,230 – 77,229 | 67,230 – 117,229 | 67,230 – 167,229 |
B. Orientation
Telomeric Nanopore reads have a strong strand bias always ending in the 5’- (TTAGGG)n −3’ telomere repeat tract sequences. This is possibly due to the blocking of Nanopore sequencing linker ligation to the 5’-CCCTAA – 3” at the telomere end by a G-strand single-stranded 3’ DNA overhang at natural human telomeres [17]; this would result in only terminal strands oriented from the centromere to the telomere getting sequenced. To follow the convention of visualizing a DNA sequence from 5’ to 3’ end (plus strand), based on the chromosome’s ID, the telomeric Nanopore reads are either kept in their initial state (subtelomere 19q) or are reverse complemented (subtelomeres 10p, 16p, 20p). The result is the set of oriented telomeric Nanopore reads, which will guide the orientation of the subtelomeric Nanopore reads as well as the REXTAL contigs’.
The non-telomere containing subtelomeric Nanopore reads are oriented by determining the relative orientation of their pairwise BLAST alignments (+/+ or +/−) to telomere-anchored Nanopore reads and to each other. REXTAL contigs for specific subtelomeres are determined in similar fashion, by the orientation of their BLAST alignments with the oriented Nanopore reads. Because the Nanopore reads are repeat-masked prior to this alignment, some small REXTAL contigs are set aside during this step (in most cases due to their small size and repeat content).
C. Order and Correction
The oriented Nanopore reads are used as sequence-resolution guides to order the oriented REXTAL contigs. We align them with the repeat masked and tandem repeat masked oriented telomere Nanopore reads, requiring a pid of at least 80. Short local alignments are not included in the final aligned contig if they have noncontiguous features suggestive of very short duplications. REXTAL contigs that are totally covered by other REXTAL contigs are also set aside.
NPGREAT is able to identify problematic regions within REXTAL sequence contigs by investigating its local alignments with the Nanopore read. A local alignment consists of four coordinates: The first and last coordinates of the REXTAL contig’s region and the corresponding coordinates of the Nanopore read’s region. When two local alignments are present, there exists an unidentified segment between them whose length is defined differently by the Nanopore and by the REXTAL coordinates. Wherever this segment according to the Nanopore read’s coordinates is bigger than the segment according to the REXTAL contig’s coordinates, it’s an indication that the contig should “split”.
To detect misassemblies caused by deletions of 1 kb length or greater within the REXTAL contigs, we require that the difference between the two segments is at least 1 kb. In addition, if multiple Nanopore reads align with this portion of the REXTAL contig, the majority should confirm the split.
To confirm the need for correction and identify the exact location in the case of the “split”, we align the two segment regions plus 1 kb to their left and right to the unmasked region of the Nanopore read, requiring a pid of at least 80. Then, the contig is split into two and we align the corrected contigs with the repeat masked and tandem repeat masked oriented telomere Nanopore reads, with the aforementioned requirements.
The final alignments consist of the contigs which remained after the initial elimination and the corrected contigs. Each of the output files contains the alignments of a single Nanopore read and the REXTAL contigs that align well with it, obtaining the contigs’ order.
D. Filling the gaps
The ordered REXTAL contigs based upon alignment with repeat-masked and tandem repeat-masked Nanopore reads (red line segments in Fig. 2) provide positional information on the repeat-free parts of these contigs.
Figure 2.
Definition of a gap or an overlap between two REXTAL contigs. The red color designates the border local alignments of the neighboring REXTAL contigs and the blue color shows the Nanopore read’s alignment with the two contigs. The green color and prime letters constitute REXTAL contigs extended in repeat-masked parts of Nanopore reads. The purple color defines the bridging region. (a) A gap between the two contigs. (b) An overlap between two contigs. (c) Possible overlap, further investigation required.
The parts of these uniquely positioned contigs that extend into repeat-masked parts of the Nanopore read are added based upon the very high quality of these sequence contigs [10], [11] (green line segments, Fig. 2). The entire positioned contigs either overlap, or they have a “gap” between them. Based on the alignments of the Order and Correction step, in cases where neighbor contigs overlap, they are merged. The overlap region from the contig with the higher percentage id with the Nanopore sequence is retained in the merge region. In the case of Fig. 2c, where it is uncertain whether the adjacent contigs overlap, the DNA sequence of the Nanopore read containing the potential gap and the flanking region in question is aligned with the flanking contig ends in unmasked mode, and either a small overlap or a small gap is identified and processed as such in downstream steps. Where there is a gap between the contigs, parts of the Nanopore reads that bridge the gap (purple line segment, Fig. 2) or extend the most distal REXTAL contigs outward are extracted and analyzed for inclusion in the final sequence as described below.
The region of Nanopore reads spanning gaps in the REXTAL coverage have two key characteristics: their sequence similarity to the neighbor REXTAL contigs and the total number of sequencing errors that they might contain. Prior to such a region’s extraction for analysis the two flanking local alignments with REXTAL contigs have a pid, defining the similarity of the Nanopore read to each of the REXTAL contigs at these alignments. The % sequence identity of the Nanopore region with a REXTAL contig is a reasonable indicator of its local Nanopore sequence quality, since REXTAL contigs have very high sequence quality [10], [11].We define the average pid of a Nanopore read region as the average of the neighboring pids with the flanking REXTAL contigs according to their local alignments. The greater this number is, the higher the quality of the Nanopore read.
Nanopore reads are error-prone, with read regions rich in homopolymers of five or greater nucleotides correlating with the highest error rates [12]. In the analysis of a Nanopore read-derived bridging sequence, we define as errors the total number of 5-mer occurrences ( i.e. the occurrence of AAAAA, TTTTT, CCCCC and GGGGG) within a sequence. The greater this number is, the lower the quality of the Nanopore read.
These two variables, the average pid and errors are used by the algorithm that will compute the final connector region of a gap between two REXTAL contigs. If there exist more than one Nanopore regions that can bridge a gap, one is selected as the final connector. The user can define their preference regarding the selection approach they would like to take place.
The algorithm.
For each gap region between the REXTAL contigs, if there exists only one Nanopore region to cover it, it is selected as the connector region of that gap. Otherwise, if there are multiple Nanopore regions:
-
1)
The average pid for the Nanopore segments is calculated based on the aligned REXTAL contigs flanking the gap.
-
2)
The number of errors in the Nanopore segments is calculated.
-
3)
Selection of the final gap connector Nanopore region:
Approach 1. The region with the highest average pid.
Approach 2. The region with the lowest number of errors.
E. Combination
The DNA sequence of the Nanopore connector regions (including the gap connector, as well as the extension regions) and overlapping REXTAL contigs, are combined in the final assembly step.
According to their order, the pieces are connected one after the other. Starting from the 5’ extension region, then the first REXTAL contig, then the gap connector Nanopore region, then the next REXTAL contig, and so on. The sequence ends with the 3’ extension region. The final assembled sequence has been computed without the existence of any sequence gaps.
III. RESULTS AND DISCUSSION
We present our results with the use of QUAST and the Icarus genome viewer, as its use is outlined in the REXTAL analysis [11]. In order to evaluate our assembly, we compare it against the HG38, specifying the coordinates of the reference for every chromosome region as defined in Table I. The following figures consist of two sections, the lowest portion presenting the total data and the upper displaying a selection of the lowest part in detail. In each section, the first row is our assembly (approach 1), the second row is the REXTAL assembly and the third row is the HG38 reference sequence designated with gaps at the places where tandem repeats exist.
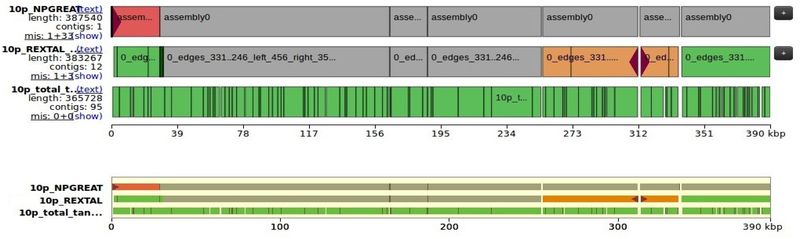
Chromosome 10p.
In Fig. 3, we present the 10p subtelomere region, which has the telomere set at its left side. Our assembly is almost complete, with a few small gaps, as well as a gap induced by QUAST and mistakenly designated with red color as misassembly.
Figure 3.
10p Subtelomere region comparison with REXTAL and HG38.
A close-up view of the 10p telomere shows that the misassembly towards the telomere is in fact artifactual. It is caused by the correct presence of the 5,000 bp telomere sequence (TTAGGG)n in our assembly and its absence in the reference sequence. This is occurring because the reference sequence, being clone-based does not include most of the telomere sequence because it is not stable in bacterial and yeast clones [16].
Correction.
The region previously designated as a misassembly within the REXTAL contig (orange region near coordinate 312), has been corrected. The particular contig was identified by the Nanopore read that aligns with it, locating the deletion within the REXTAL contig. The gap has now been bridged with the use of the connector Nanopore read.
The few small gaps shown in our final 10p assembly when compared to the reference using QUAST are artifactual; they occur at VNTR tandem repeat regions and are actually just the difference between the tandem repeats of the reference vs the tandem repeats in the Nanopore reads (as well as the REXTAL contigs).
Orientation and Ordering.
In NPGREAT, all initial REXTAL contigs have been placed at the correct location and with the correct orientation according to the reference and the telomere, which is located on the left.
Filling and Extending.
Furthermore, regarding the extension towards the Telomere’s side, our assembly extends the telomeric end by about 5 kb, corresponding to the length of (TTAGGG)n on the Nanopore read used.
Evaluation.
The bridging and extending Nanopore “connectors” of each approach are being evaluated compared to the reference coordinates they are meant to align with, i.e. bridge. The evaluation takes place with the use of BLASTn. The best approach for 10p, based on the percent identity with the reference, appears to be approach 1. In detail, the percent identity of approach 1 in areas where multiple Nanopore reads could bridge a gap, ranges from 91.14% to 96.23%, whereas approach 2, ranges from 90.04% to 96.23%.
Chromosome 20p.
The assembly is correct and complete, with no misassemblies. There exists a large extension of the assembly in the telomeric direction beyond the HG38 reference sequence used in the QUAST comparison.
Orientation and Ordering.
All contigs have been oriented and ordered correctly.
Filling and Extending.
NPGREAT has successfully filled all 8 gaps (and an additional 4 in the telomeric extension region not compared to HG38) between the REXTAL contigs. The 20p HG38 reference is not complete and does not contain the Segmental Duplication region, but our sequence has a 113 kb extension towards the telomere on the left. This extension region consists of two overlapping REXTAL contigs and a Nanopore region.
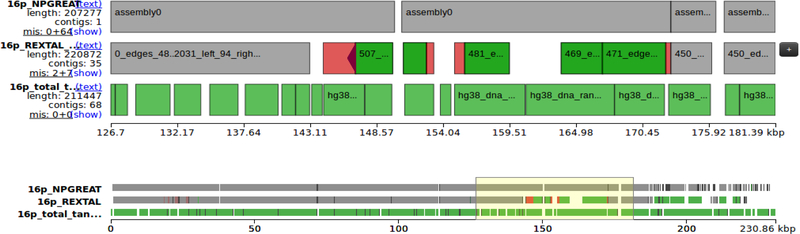
Chromosome 16p.
In Fig. 4 we present the 16p subtelomere region.
Figure 4.
16p Subtelomere region comparison with REXTAL and HG38.
Correction.
In the REXTAL assembly, there exist two misassemblies, designated with red color, where each contig should have been split into two and there exists another contig to be placed in between. In NPGREAT, the two problematic contigs have been “split”, placed at the right locations and connected accordingly.
Orientation and Ordering.
All contigs have been oriented and ordered correctly.
Filling and Extending.
As was the case for 10p, the few small gaps shown in our final sequence are artifacts caused by QUAST comparison of variable tandem repeat regions. The Nanopore-guided assembly extends telomerically about 7 kb, corresponding to the length of the telomere (TTAGGG)n tract in the Nanopore read used.
Chromosome 19q.
The telomere’s location is on the right side.
Correction.
In the REXTAL assembly, there exist two misassemblies. In the first misassembly, the contig should have been split, allowing for another contig to be placed in between the two portions. While, in the second misassembly, there was a deletion within a contig. Our enhanced method was able to detect and resolve both issues, splitting one REXTAL contig, and using the Nanopore read to effectively fill a gap corresponding to a tandem repeat region.
Orientation and Ordering.
All contigs have been oriented and ordered correctly.
Filling and Extending.
As before, the few small gaps shown in the QUAST analysis relative to the HG38 reference are artifactual and occur at tandem repeat regions.
In all four subtelomere regions, the NPGREAT assembly has percent identity ≥ 98% with the HG38 reference in the large portion close to the telomere. In the example of 20p, the total percent identity with HG38 is 99.19%.
IV. CONCLUSION
By combining two of the latest sequencing technologies, Oxford Nanopore ultralong read sequencing [12] and 10x Linked-Read sequencing [8], [9], we show here that we can obtain highly accurate and continuous sequences in segmentally duplicated subtelomere regions of human chromosomes. Specifically, the ultralong Nanopore reads were used as sequence resolution guides upon which the REXTAL contigs were placed, leveraging the contiguity of the Nanopore reads and the base-call accuracy of the REXTAL sequence contigs. When applied to subtelomeres 10p, 16p, 19q and 20p of the NA12878 genome, it is apparent that with the use of the Nanopore reads, the short contigs are oriented and ordered correctly, extended on the sides and extending through the telomere (TTAGGG)n sequence and, where necessary, split and corrected. The Nanopore reads guide the assembly process successfully and have the capability of producing long-sequence assemblies in regions that would otherwise consist of gaps and remain unobserved. Notable is the case of the 20p subtelomere region, where the HG38 reference sequence lacks the segmental duplication region, and yet our assembly is capable of detecting ~100 Kb of it.
ACKNOWLEDGMENT
Supported by NIH RO1CA140652 (HR, Paul Lieberman PI).
Contributor Information
Eleni Adam, Department of Computer Science, Old Dominion University, Norfolk, VA, USA.
Tunazzina Islam, Department of Computer Science, Purdue University, West Lafayette, IN, USA.
Desh Ranjan, Department of Computer Science, Old Dominion University, Norfolk, VA, USA.
Harold Riethman, School of Medical Diagnostic & Translational Sciences, Old Dominion University, Norfolk, VA, USA.
REFERENCES
- [1].Armanios M, Alder JK, Parry EM, Karim B, Strong MA, and Greider CW, “Short telomeres are sufficient to cause the degenerative defects associated with aging,” The American Journal of Human Genetics, vol. 85, no. 6, pp. 823–832, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Meier A et al. , “Spreading of mammalian DNA‐damage response factors studied by ChIP-chip at damaged telomeres,” The EMBO journal, vol. 26, no. 11, pp. 2707–2718, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Davalos AR, Coppe J-P, Campisi J, and Desprez P-Y, “Senescent cells as a source of inflammatory factors for tumor progression,” Cancer and Metastasis Reviews, vol. 29, no. 2, pp. 273283, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jaskelioff M et al. , “Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice,” Nature, vol. 469, no. 7328, p. 102, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, and Baird DM, “Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans,” Human molecular genetics, vol. 15, no. 5, pp. 725–733, 2006. [DOI] [PubMed] [Google Scholar]
- [6].McCaffrey J et al. , “High-throughput single-molecule telomere characterization,” Genome research, vol. 27, no. 11, pp. 1904–1915, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Consortium IHGS, “Finishing the euchromatic sequence of the human genome,” Nature, vol. 431, no. 7011, p. 931, 2004. [DOI] [PubMed] [Google Scholar]
- [8].Zheng GX et al. , “Haplotyping germline and cancer genomes with high-throughput linked-read sequencing,” Nature biotechnology, vol. 34, no. 3, p. 303, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Weisenfeld NI, Kumar V, Shah P, Church DM, and Jaffe DB, “Direct determination of diploid genome sequences,” Genome research, vol. 27, no. 5, pp. 757–767, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Islam T, Ranjan D, Young E, Xiao M, Zubair M, and Riethman H, “REXTAL: Regional Extension of Assemblies Using Linked-Reads,” in International Symposium on Bioinformatics Research and Applications, 2018, pp. 63–78: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Islam T, Ranjan D, Zubair M, Young E, Xiao M, and Riethman H, “Analysis of Subtelomeric REXTAL Assemblies Using QUAST,” IEEE/ACM transactions on computational biology and bioinformatics, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jain M et al. , “Nanopore sequencing and assembly of a human genome with ultra-long reads,” Nature biotechnology, vol. 36, no. 4, p. 338, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, and McCombie WR, “Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome,” Genome research, vol. 25, no. 11, pp. 1750–1756, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ma ZS, Li L, Ye C, Peng M, and Zhang Y-P, “Hybrid assembly of ultra-long Nanopore reads augmented with 10x-Genomics contigs: Demonstrated with a human genome,” Genomics, 2018. [DOI] [PubMed] [Google Scholar]
- [15].Vollger MR et al. , “Long-read sequence and assembly of segmental duplications,” Nature methods, vol. 16, no. 1, p. 88, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Stong N et al. , “Subtelomeric CTCF and cohesin binding site organization using improved subtelomere assemblies and a novel annotation pipeline,” Genome research, vol. 24, no. 6, pp. 1039–1050, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Makarov VL, Hirose Y, and Langmore JP, “Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening,” Cell, vol. 88, no. 5, pp. 657–666, 1997. [DOI] [PubMed] [Google Scholar]