Abstract
Background:
Centiloid was introduced to harmonise β-Amyloid (Aβ) PET quantification across different tracers, scanners and analysis techniques. Unfortunately, Centiloid still suffers from some quantification disparities in longitudinal analysis when normalising data from different tracers or scanners. In this work, we aim to reduce this variability using a different analysis technique applied to the existing calibration data.
Method:
All PET images from the Centiloid calibration dataset, along with 3762 PET images from the AIBL study were analysed using the recommended SPM pipeline. The PET images were SUVR normalised using the whole cerebellum. All SUVR normalised PiB images from the calibration dataset were decomposed using non-negative matrix factorisation (NMF). The NMF coefficients related to the first component were strongly correlated with global SUVR and were subsequently used as a surrogate for Aβ retention. For each tracer of the calibration dataset, the components of the NMF were computed in a way such that the coefficients of the first component would match those of the corresponding PiB. Given the strong correlations between the SUVR and the NMF coefficients on the calibration dataset, all PET images from AIBL were subsequently decomposed using the computed NMF, and their coefficients transformed into Centiloids.
Results:
Using the AIBL data, the correlation between the standard Centiloid and the novel NMF-based Centiloid was high in each tracer. The NMF-based Centiloids showed a reduction of outliers, and improved longitudinal consistency. Furthermore, it removed the effects of switching tracers from the longitudinal variance of the Centiloid measure, when assessed using a linear mixed effects model.
Conclusion:
We here propose a novel image driven method to perform the Centiloid quantification. The methods is highly correlated with standard Centiloids while improving the longitudinal reliability when switching tracers. Implementation of this method across multiple studies may lend to more robust and comparable data for future research.
Keywords: Centiloid, Aβ Imaging
Introduction
Recent advances in PET imaging mean that β-Amyloid (Aβ), one of the hallmarks of Alzheimer’s disease (AD), can now be imaged in-vivo using a variety of PET tracers. There are currently five Aβ PET tracers which are commonly used in research settings or in the clinic: 11 C-PiB (PiB), 18F-NAV4694 (NAV), 18 F-Florbetaben (FBB), 18F-Flutemetamol (FLT) and 18 F-Florbetapir (FBP). Because of their pharmacokinetics differences, each of these tracers have a prescribed acquisition protocol, a recommended reference region to generate the Standardised Uptake Value Ratios (SUVR), and therefore a tracer-specific cut-off value used to establish Aβ positivity. Each research group also tend to use their own image processing pipeline for the SUVR normalisation of the images. As a result, these differences in pharmacokinetics, reference region, cut-off values and processing pipeline can lead to large discrepancies between different studies.
The international Centiloid Project working group (Klunk et al., 2015) was setup to harmonise the quantification of Aβ PET images with a standardised processing pipeline, and a set of equations used to transform SUVR into Centiloid for each available tracer. The Centiloid scale is anchored on a set of young healthy controls whose mean Centiloid value is set to 0, and a set of mild AD patients whose mean centiloid value is set to 100. This initial calibration was performed on an independent set of 79 subjects and was used to define the equation transforming SUVRPiB into CLPiB. Then for each 18F-tracer, pairs of PiB and 18F-tracer were acquired on a population of young healthy controls (YHC), older healthy controls, MCI and AD to cover a wide range of Centiloid values. SUVR18F-tracer were transformed into SUVRPiB using linear regression, and the resulting SUVRPiB transformed into CL. The combination of both transforms was used to define the equation transforming SUVR18F-Tracer into CL18F-Tracer (Rowe et al., 2016, 2017; Navitsky et al., 2016; Battle et al., 2018).
While the Centiloid Project provides a good framework to harmonise processing methods and quantification, it is hindered by its multi-site design. In order to accelerate the standardisation of all tracers, different sites were responsible for the acquisition of the pairs of head-to-head PET images. This resulted in a mix of scanners and reconstruction methods being used for each tracer. For instance, 5 different scanners were used for the acquisition of the original PiB dataset, 3 for FLT, 3 for FBP, 2 for FBB and only NAV used a single scanner, with little to no overlap in the model of scanner used across tracers. While PET imaging is quantitative, it suffers from bias when different scanner models and reconstruction method/parameters are used (Joshi et al., 2009; Bourgeat et al., 2014; Aide et al., 2017). As a result, the fundamental assumption that the PiB scans acquired in each of these calibration datasets are comparable and can reliably be transformed into Centiloid may not be valid. This was observed in the study by Su et al. (2019), where pairs of PiB/FBP were transformed into CL and compared. While they were in good agreement (R 2 = 0.91), the slope of the regression indicated that the CL from FBP were 11% higher than those of their matching PiB. Similar findings were reported by Cho et al. (2019), where matching pairs of FLT/FBB were transformed into CL and compared. There was again a very good agreement (R 2 = 0.97), but the slope indicated that the CL from FBB was 21% higher than those of their matching FLT. We have also observed such discrepancies in longitudinal studies involving multiple tracers (Bourgeat et al., 2019). This is a very important factor in international multicentre disease-specific therapeutic trials, where these discrepancies might preclude finding significant differences between treatment groups.
In this study, we quantified these discrepancies using the longitudinal data in the Australian Imaging, Biomarkers and Lifestyle study (AIBL), and propose a novel calibration method using the existing Centiloid calibration data, based on non-negative matrix factorisation.
Methods
Datasets
All the PET and MR images from the Centiloid calibration datasets (Klunk et al., 2015; Rowe et al., 2016, 2017; Navitsky et al., 2016; Battle et al., 2018) (289 PiB, 55 NAV, 35 FBB, 74 FLT, 46 FBP) were downloaded from the GAAIN website (http://www.gaain.org/centiloid-project). From the AIBL study (Ellis et al., 2009), 3762 PET images from 1543 subjects having undergone 2 or more PET scans with corresponding MRI were selected. The distribution of PET scans included 1503 PiB, 850 NAV, 244 FBB, 535 FLT and 630 FBP. All the images were spatially normalised to the SPM template using the prescribed SPM pipeline as described in the original Centiloid paper (Klunk et al., 2015), and SUVR normalised using the Centiloid Whole Cerebellum mask. Neocortical SUVR was computed using the Centiloid neocortical mask and was transformed into Centiloids using the existing calibration equations (Rowe et al., 2016 2017; Navitsky et al., 2016; Battle et al., 2018). A Centiloid value of 20 was used to separate Aβ negative from Aβ positive subjects (Amadoru et al., 2020).
Image decomposition
In this work, we aimed to develop a novel image-based Centiloid quantification method based on the image decomposition into a set of components. Given a matrix X composed of N vectors, the decomposition seeks to obtain 2 matrices W and H whose product can approximate X. We will here refer to H as the set of component images, and W the set of weights used to reconstruct X given H. Each image x i can then be approximated as
Where wk is the kth scalar weight, and hk is the kth component image. Image decomposition can be seen as a projection of the image in a different space, where the axes are defined by H, and the coordinates in that space defined by W. The idea of this research is to find a space where the coordinates of each tracer’s projections are comparable.
Decomposition of Aβ images into a set of components was first proposed by Fripp et al. (2008) using principal component analysis (PCA). They showed that 2 components were enough to represent 80% of the information contained in the population, and that the first component closely followed the expected pattern of deposition from histology and was strongly correlated with SUVR (R 2 = 0.83). It was later used to build an adaptive atlas for PET registration in PiB (Fripp et al., 2008) and FLT (Lilja et al., 2019). More recently, a voxel based logistic growth model was used to build a model of Aβ burden and non-specific binding in FBP images (Whittington et al., 2018). This model was subsequently shown to increase the effect size between clinical groups (Whittington and Gunn, 2019). Tanaka et al. (2020) later showed similar results using PCA decomposition, where the first component could be used to separate specific Aβ burden from non-specific uptake. This led to an improved quantification, with a stronger association with cognitive measures and an increased effect size between high and low Aβ.
Non-negative matrix factorisation
In this work, we treated the variability introduced by using different scanners in the Centiloid calibration set as an extra nuisance, such as the non-specific binding target targeted the work of Tanaka et al. (2020), and use the decomposition to measure the Aβ burden. To this end, the model is built using all the PiB images from the calibration dataset, which were acquired on 13 different scanners. Since these datasets were built to include the whole range of Aβ load, they are ideally suited to build a model of tracer retention. By including different scanners in the model, the first component of the model will need to be a common denominator to all scanners and is therefore more likely to be robust to the variations introduced by these scanner changes. While PCA has the advantage of maximising the compression of information, with components being orthogonal and representing the maximum variance in X, the decomposition can have negative weights W and components H. As a result, some components can be over-expressed, and be compensated by other smaller negative components. While this is useful to compress the information, it can be detrimental to the interpretability of the model (Tanaka et al., 2020). In this work, we use instead the NMF, which has similar use to the PCA, with the extra constraint that all the components and their associated coefficients need to be positive (Lee and Seung, 1999). As a result, each component can be seen as a separate tissue retention. This property was recently exploited for the partitioning of PiB images (Sotiras et al., 2018).
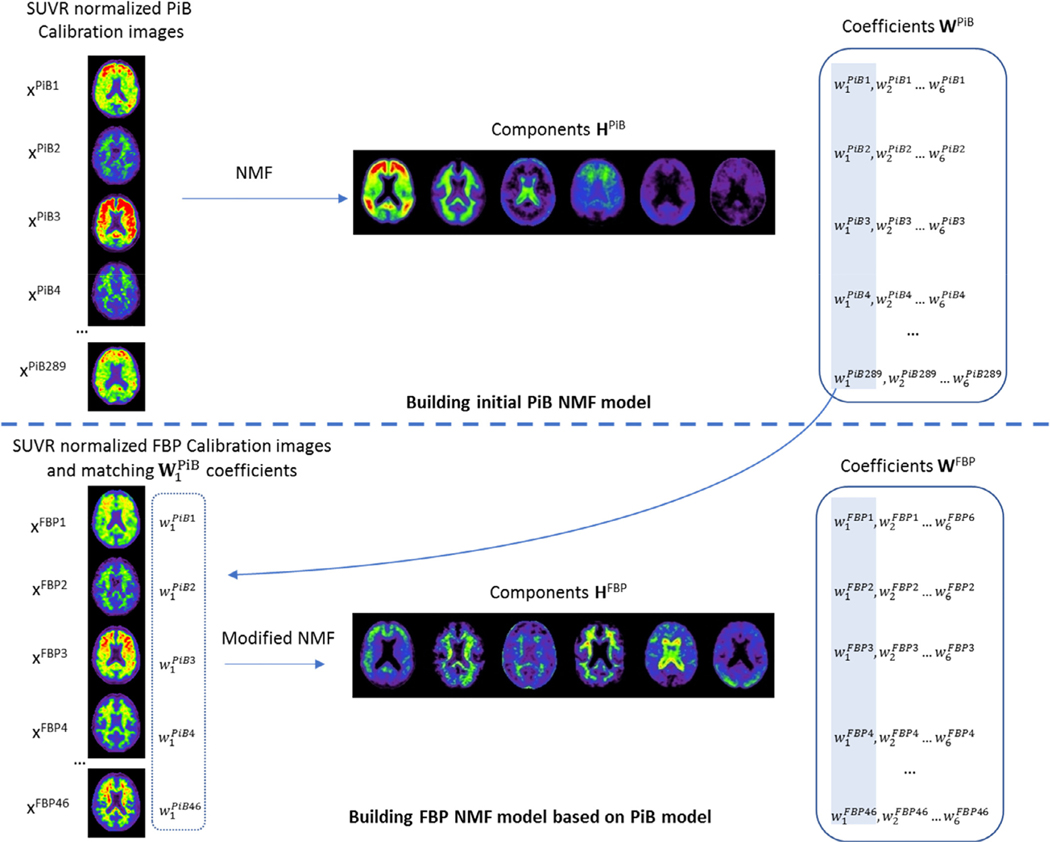
Unlike PCA, NMF is solved as an optimisation problem as no unique solution exists. This can be advantageous, as the decomposition can also be computed to find the optimal set of components H given a set of coefficients W. Here, we exploit this property to enforce that the coefficients of the first component of PiB match those of their corresponding 18F-tracers. In other words, for each calibration set where we have N PiB/18F-tracers pairs, we can compute the coefficients w1 of the first component h1 of the PiB NMF decomposition, and then compute the NMF decomposition of the matching 18F-tracer images while fixing its coefficients w1 so that they match with those computed from PiB. This way, the decomposition is computed with an extra constraint that w1 should match across each pair of tracers. The workflow used to compute the NMF model for PiB and the 18F-tracers is illustrated in Fig. 1. Since w1 of all tracers are intrinsically matched to those of PiB, a single transform can be used to convert all the coefficients w1 into Centiloids.
Fig. 1.
Steps involved to construct the PiB NMF model, and subsequent 18F models. We only show the construction of the FBP model using 6 components for simplicity, but the same steps are used for the other 18F tracers.
In this work, we used the scipy implementation of NMF. For each tracer, a range of different number of components was evaluated. For PiB, a random initialisation was used. For the other tracers, the NMF implementation was modified so that we could set and fix the coefficients of the first component based on the PiB model when computing the NMF decomposition of the 18F-tracers. In practice, this was implemented by randomly initialising W and H, setting w1 Tracer to w1 PiB, and then computing the update on all components of W and H at each iteration, but applying the update on all but the first component of W. All the images were SUVR normalised using the Centiloid Whole Cerebellum mask and skull-stripped before running the decomposition. The NMF coefficients were finally transformed into Centiloids using the anchoring to YHC and AD from the PiB calibration set:
| (1) |
Model built using a different dataset
To confirm that the proposed approach is more robust to the changes of scanner in the calibration set, a comparable PiB model was built using images from a single scanner from AIBL. All the images were SUVR matched to the ones in the calibration set. This model was used to compute the coefficients of the PiB images in the calibration set, which were then used to compute the decomposition of the 18F-tracers.
Number of components
As the NMF is solved as an optimisation problem, the number of components selected will determine the final results. Therefore, the number of components needs to be set apriori. In this work, we test a range of components from 2 to 7. The main metric is the correlation between the resulting CL of each 18F tracer and that of their matching 11C-PiB, as improving the matching between the tracers is the main aim. We also evaluate the correlation between the Centiloid derived from the NMF, referred to as CLNMF and that of the standard method CLStd.
Validation
While there is no ground truth to validate the new NMF-based Centiloid, a number of surrogate measures can be extracted based on the hypothesis that β-Amyloid accumulation is a relatively slow process, and the accumulation curves should be relatively smooth. Using consecutive imaging timepoints where there is no change of tracer or scanner, we can estimate the normal or expected range of rate of CL accumulation in both negative and positive subjects. These rates can be compared to the measured range of rates of accumulation on the entire cohort for the different methods.
Longitudinal consistency when switching tracer can also be used as a surrogate metric of generalisability of the Centiloid conversion. We here look at the average fitting error of a linear model, where we assume that Aβ accumulation is linear over a period < 10 years. The effect of the change of scanner/tracer on the slopes is also assessed using a linear mixed effects model for both standard and NMF-based Centiloid calculations.
To check the hypothesis that including multiple scanners in the initial PiB model can lessen the effect of having multiple scanners in the calibration dataset, a second model was built using PiB images from AIBL from a single scanner (Philips Allegro). The 289 PiB images from AIBL were SUVR matched to those in the calibration set and used to build the initial PiB NMF model. Using this model, the NMF coefficients were then computed on all PiB images from the calibration set and used to compute the matching models for each 18F tracers.
The NMF models and python code used to build and apply these models are available at https://doi.org/10.25919/5f8400a0b6a1e.
Results
Calibration dataset
The NMF decomposition was run on the Calibration dataset using a number of components ranging from 2 to 7. For each NMF model, the coefficients of the first component in the YHC and mild AD of the PiB dataset were anchored to the Centiloid scale using Eq. (1). The correlation between the NMF Centiloid for 11C-PiB and NMF Centiloid of their matching 18F tracer was the highest when 6 components were used with an overall R2 of 0.977, closely followed by 2 components with a R2 of 0.975 (Full results are available in Supplementary Table 1). The correlation of the CLNMF with the standard Centiloid was however the highest when 2 components were used with an overall R2 of 0.975, followed by 6 components with a R2 of 0.905 (Supplementary Table 2). While a good correlation between each tracer pairs is the most important factor in this work, a good correlation with the standard method is also important. Therefore, both sets of results were used for subsequent analysis. We will refer to the Centiloid derived from the 2 components NMF decomposition as CLNMF2 and the 6 components one as CLNMF6.
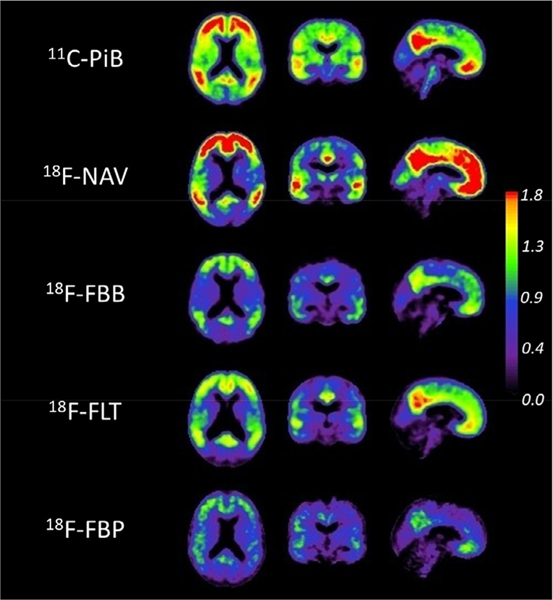
Fig. 2 illustrates the 6 components NMF decomposition, by showing the first component for each tracer. Since the NMF is driven by PiB, it results in forcing the decomposition of each tracer to be similar to that of PiB. This results in the first component of each tracer to mimic the appearance of PiB. It is most striking for FBP, which is typically characterised by lower specific binding which can be hard to visualise next to the non-specific WM binding, and only shows GM retention in the first component, with the non-specific binding being captured by other components of the model (Full model for 2 and 6 components decomposition can be seen in Supplementary Figs. 1 and 2). The correlation between the PiB CLNMF6 and the corresponding tracer CLNMF6 was comparable across each tracer with R2 = 0.99 in NAV, 0.98 in FBB, 0.98 in FLT and 0.97 in FBP, which was higher than the one obtained using the standard method with R2 = 0.99 in NAV, 0.95 in FBB, 0.96 in FLT and 0.89 in FBP. When using 2 components, the correlations were a bit lower with R2 = 0.99 in NAV, 0.97 in FBB, 0.98 in FLT and 0.96 in FBP, but still higher than using the Standard method.
Fig. 2.
First component of the 6 components NMF decomposition for each tracer in the calibration set.
The correlations with the Standard Centiloid for each tracer and both 2 and 6 components NMF are presented in Table 1. For both approaches, the correlation was the lowest for FBP. While the slopes of the transforms from CLStd to CLNMF were close to 1 for CLNMF2, they tended to be lower than 1 for CLNMF6, resulting in an underestimation of the Standard Centiloid.
Table 1.
Transforms from CLStd to CLNMF2 and CLNMF6 estimated on the Calibration dataset.
| 2 Components |
6 Components |
|||
|---|---|---|---|---|
| Tracer | Equation | R2 | Equation | R2 |
| PiB | CLNMF2 = 1.01 × CLStd − 0.85 | 0.99 | CLNMF6 = 0.94 × CLStd + 0.57 | 0.93 |
| NAV | CLNMF2 = 1.01 × CLStd − 2.19 | 0.99 | CLNMF6 = 0.84 × CLStd − 3.76 | 0.93 |
| FBB | CLNMF2 = 1.01 × CLStd − 0.58 | 0.97 | CLNMF6 = 0.95 × CLStd − 1.91 | 0.96 |
| FLT | CLNMF2 = 1.04 × CLStd + 0.11 | 0.98 | CLNMF6 = 0.86 × CLStd + 6.09 | 0.88 |
| FBP | CLNMF2 = 0.93 × CLStd + 0.30 | 0.90 | CLNMF6 = 0.84 × CLStd + 4.65 | 0.84 |
| Overall | CLNMF2 = 1.01 × CLStd − 0.72 | 0.98 | CLNMF6 = 0.92 × CLStd + 1.07 | 0.91 |
AIBL dataset
In AIBL, the correlation between the standard Centiloid CLStd and the novel CLNMF was high in each tracer, being the highest in NAV, and the lowest in FBP regardless of the number of components being used as shown in Table 2. Similarly to the results from the calibration dataset, there was no noticeable bias between CLStd and CLNMF2 with all the slopes being close to 1. With CLNMF6, there was however a difference in scale, with CLNMF being scaled down for each tracer compared to CLStd. This scaling, however, did not change the typical cut-off value, with 20 CLStd = 20.2 CLNMF6 using the overall equation in Table 2.
Table 2.
Transforms from CLStd to CLNMF2 and CLNMF6 estimated on the AIBL dataset.
| 2 Components |
6 Components |
|||
|---|---|---|---|---|
| Tracer | Equation | R2 | Equation | R2 |
| PiB | CLNMF2 = 1.03 × CLStd – 2.04 | 0.99 | CLNMF6 = 0.88 × CLStd + 0.65 | 0.93 |
| NAV | CLNMF2 = 1.01 × CLStd – 2.74 | 0.99 | CLNMF6 = 0.84 × CLStd + 1.76 | 0.97 |
| FBB | CLNMF2 = 0.95 × CLStd + 0.48 | 0.96 | CLNMF6 = 0.88 × CLStd + 0.12 | 0.96 |
| FLT | CLNMF2 = 0.98 × CLStd – 1.45 | 0.97 | CLNMF6 = 0.82 × CLStd + 4.16 | 0.91 |
| FBP | CLNMF2 = 0.93 × CLStd + 4.57 | 0.91 | CLNMF6 = 0.70 × CLStd + 11.74 | 0.87 |
| Overall | CLNMF2 = 0.99 × CLStd - 0.67 | 0.97 | CLNMF6 = 0.83 × CLStd + 3.60 | 0.93 |
Longitudinal consistency
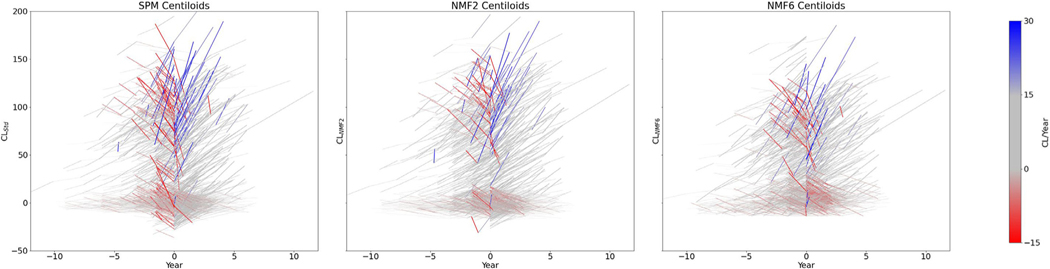
Fig. 3 illustrates the longitudinal consistency of both techniques. Since a change in scanner or a change in tracer could potentially introduce discontinuities in the longitudinal Centilloid measures for a given subject, the plots are centred on the first timepoint showing a change in scanner or tracer. Furthermore, each segment is colour-coded based on its slope to further highlight large positive changes in blue, and large negative in red. This visually emphasises these discontinuities, and better illustrates how the NMF-based Centiloid appears to be less susceptible to such changes. A qualitative observation of the graphs indicates that there are larger “jumps” using the Standard centiloids which appear to be reduced when using NMF.
Fig. 3.
Longitudinal plot of Centiloid value changes over time of all subjects in AIBL with longitudinal scans. Each line connects CL values from unique subjects. The plot is centred on the first change of scanner or tracer, to emphasise potential discontinuities. This is achieved by assigning year zero, for each subject, to their last imaging timepoint before a change of tracer of scanner occurred. Left plot shows CL values measured using the standard method, while the ones in the middle and on the right shows the CL values measured using the NMF2 and NMF6 method, respectively.
To quantitatively estimate the expected normal range of changes per year, the rate of CLStd change per year was computed between consecutive timepoints when there was no change in both scanner and tracer. Using the 95th percentile, we found that in subjects that were negative at baseline, 95% of changes were less than 6.56 CLStd/year and in the positive, it was less than 17.06 CLStd/year. Table 3 shows the percentage of changes that were above these thresholds for all methods when all timepoints were used.
Table 3.
Percentage of changes above the expected 95th percentile thresholds for each method.
| 95th percentile absolute change | CLStd | CLNMF2 | CLNMF6 | CLNMF6_Sc | |
|---|---|---|---|---|---|
| Negative | 6.56 CLStd/year | 8.90% | 6.36% | 6.43% | 9.24% |
| Positive | 17.06 CLStd/year | 9.73% | 6.53% | 5.33% | 7.60% |
In both negative and positive subjects, the percentage of “abnormal” changes using CLStd was almost double of what it was when there was no change of scanner or tracer. In contrast, there were much fewer “abnormal” changes when using either CLNMF2 or CLNMF6, with both of them being within 2% of the expected 5% proportion. It should however be noted that since CLNMF6 has a lower range than CLStd, it should be rescaled to the same scale as CLStd using the equation defined in Table 2 so that their results are comparable. Using this rescaled version (referred to as CLNMF6_Sc), the proportions of “abnormal” changes are comparable in negative subjects and only lower in positive subjects.
The fitting error was computed based on the hypothesis that Aβ accumulation is linear for each subject over the time-course of the study. The mean absolute difference between the actual and predicted Centiloid value when fitting a linear regression line to each subject’s Centiloid values against time, is used as a surrogate measure of longitudinal discrepancies. The mean absolute difference was 2.62CL with CLStd, and 2.23CL with CLNMF2 and 2.34CL with CLNMF6. If we use the rescaled version of CLNMF6, then the error increases to 2.63CL, a level comparable to that of the standard method.
Since the apparent difference in scale between CLStd and CLNMF6 can confound some of the error metric, we also used a linear mixed effect (LME) model to check the tracer and scanner interaction on the Centiloid variance over time in both methods. The LME model included a random slope and intercept for each subject, and the interaction between Scanner, Tracer, and number of days since baseline, as follows:
The step method from the lmertest R package was used to remove parameters that do not contribute to the outcome. Using this approach on the CLStd, all the parameters were found to contribute to the outcome, meaning that both Tracer and Scanner significantly contribute towards the variance seen in the longitudinal measures of Centiloid. Similar results were found when using CLNMF2. However, applying the same model on the CLNMF6 data, the Days*Scanner*Tracer interaction and Days*Tracer interactions did not significantly contribute to the model, meaning that the NMF method successfully removed the effects of using different Tracers in the longitudinal variance of the Centiloid measure. The Days*Scanner interaction, however, remained significant, meaning that it did not remove the effects of using different Scanners, which was still a significant contributor of the Centiloid variance over time.
Rate of change
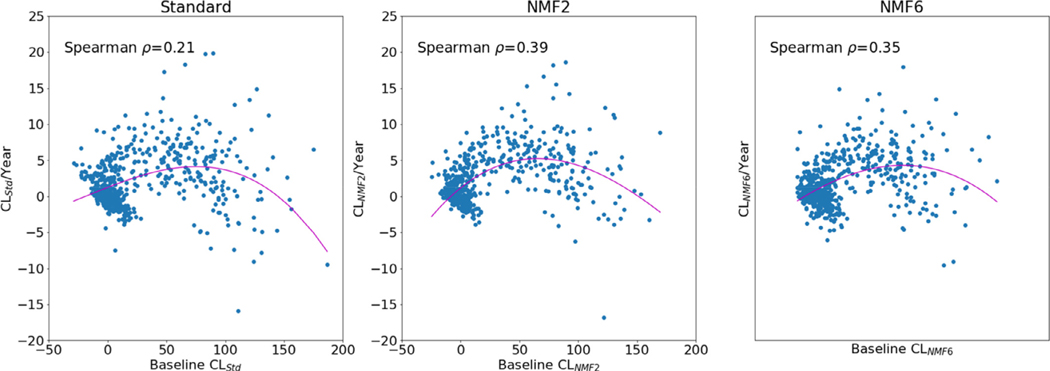
Lastly, we explored the relationship between the baseline Centiloid load and the rate of Centiloid change, assessed using robust linear regression on each subject with 3 or more timepoints. A third order non-linear regression was run on the rate of CL change against CL at baseline (Fig. 4). Spearman ρ was used to assess the correlation between the baseline Centiloid and the rate of change. There appear to be a stronger association using the NMF Centiloid, compared to the standard Centiloid, with ρ = 0.39 using CLNMF2 and ρ = 0.35 using CLNMF6, compared to ρ = 0.21 using CLStd. The mean rate of change in the subjects negative at baseline (defined as CLStd < 20) was smaller using NMF with 0.73 CLNMF2/y and 0.51 CLNMF6/y compared to the standard method where it was 0.90 CLStd/y. In the positive subjects (CLStd ≥ 20), it was higher using NMF with 4.82 CLNMF/y and 4.42 CLNMF6/y, compared to 3.94 CLStd/y.
Fig. 4.
Rate of change in CL/year against baseline CL value for CL measured using the standard method (left) and the NMF method using 2 components (middle) and 6 components (right).
Model built using single scanner PiB data
Using 289 PiB images from AIBL acquired on a single scanner, two NMF PiB models were built using 2 and 6 components respectively. Using these models, the PiB images from the Calibration set were decomposed and converted into Centiloids. The resulting coefficients from the PiB images were then used to build the corresponding NMF models using their matching 18 F images. These NMF models were then applied to the entire AIBL dataset. Using the 2 components model, there was a small increase in the number of jumps in the negative to 7.03% (compared to 6.36%), and in the positive to 6.93% (from 6.53%). The average fitting error also had a small increase to 2.32CL (compared to 2.23CL). Using the 6 components model, there was a very large increase in the number of jumps in the negative to 23.9% (compared to 6.43%), as well as in the positive with 10.80% (from 5.33%), and the average fitting error also had a large increase to 4.10CL (from 2.34CL). Visually, the longitudinal consistency was much worse using this single scanner model (results not shown). The step function applied to the matching LME failed to identify any term that did not contribute to either models, meaning that these models were not only more noisy, but also failed to remove the effect of using different tracers on the variance of the CL measures over time.
Discussion
We have developed a novel method to compute Centiloid values using the Non-negative Matrix Factorisation. We’ve shown that the NMF model was able to improve the correlation between the 11C-PiB and their matching 18F-tracers in the calibration dataset. Using a 6 components NMF showed the highest correlation between tracers, while using a 2 components NMF showed the highest correlation with the standard Centiloid values. The 2 component NMF also showed the lowest fitting error in longitudinal linear regression, and smallest proportion of “jumps” when applied to the AIBL dataset. This model also showed the strongest association between the baseline CL value and the rate of CL accumulation. The 6 components NMF model was also superior to the standard CL in terms of longitudinal consistency, and was the only model to remove the effect of using different tracers in a LME. However, this model suffers from a bias, with an approximately 17% under-quantification of the CLStd which made comparison more difficult.
It should be noted that this under-estimation of the CLStd was only observed when using the 6 components model, and was not present when using the 2 components model. The main difference between the 2 models is that the 6 components model has more degrees of freedom, and is able to better identify different patterns of Aβ retention. As illustrated in Fig. 1, we can see that not all cortical retention is captured by the first component, with some cortical retention visible in the 4th and the 5th components. A similar finding was observed in the work of Sotiras et al. (2018) where they identified different depositions profiles corresponding to different stages of Aβ deposition. For instance, they also identified the occipital cortices retention (which is visible in the 5th component of our PiB NMF6 model) as a late stage deposition. This could explain why the under-estimation of the CL values mostly happened in the high Centiloid values, as these are more likely to exhibit late stage patterns of accumulation. In contrast, the effect in the low CL values was not as strong, as illustrated by the rescaling of the 20CL cut-off which was barely changed. The Standard Centiloid mask does not attempt to separate different patterns of retention, but instead measures the global cortical retention. In that sense, it is more similar to the 2 component NMF model where all the cortical retention is captured by the first component. This also explains why CLNMF2 had a much stronger correlation with CLStd and no bias. In contrast, the 6 components NMF model only samples a subset of the cortical retention, which would underestimate the global cortical retention. It could be argued that the calibration performed using Eq. (1) should alleviate these differences, as it is meant to anchor all methods to the same scale. However, the dataset used to perform this calibration is composed of mild AD subjects, and these subjects do not give a good representation of the deposition patterns of Aβ seen in later stages of the disease. To alleviate these differences in the validation experiments, we had to apply a post-hoc rescaling. This greatly increased all the error metrics and made this approach less competitive. A larger set of calibration data, including subjects with high β-Amyloid load would likely benefit the 6 components NMF model and improve its performances. As it stands, however, this bias is likely to limit its usefulness as a replacement for the Standard method.
We also showed that including different scanners in the model helps to treat these as an additional nuisance term that is accounted for when building the model, especially with the 6 components model. This leads to a better calibration of the different tracers from the Calibration set, and in turns leads to more accurate longitudinal measures. When building a model with a single scanner, such results could not be achieved.
While the 2 components NMF failed to remove the effect of using different scanners or tracers in the LME experiment and did not give the best correlation between the different tracers in the calibration dataset, it provides the best compromise in terms of correlation with CLStd and improved longitudinal consistency and should therefore be the preferred quantification method.
Limitations
It should be noted that while the decomposition is by design driven by PiB, it means that the NMF components are only mathematically optimal for PiB, and a sub-optimal decomposition is built for each of the other 18F tracers. This can be beneficial in the case of tracers such as FBP or FLT which have a lot of non-specific binding, as the effect of this approach means that this binding is excluded from the fitting of the first component. However, if a tracer has specific binding affinity that does not correlate with the binding observed in PiB, it will not contribute to the computed Centiloid. Conversely, if the PiB model captures any non-specific binding, this will translate into a similar non-specific binding to be captured by the model built on the matching 18F tracers. Potentially, a single consensus NMF decomposition could be built using all the tracers together. The implementation of such decomposition is however non-trivial, due to each tracer being acquired on separate, non-overlapping, populations. The current implementation of the NMF decomposition is also non-optimal, as an ad-hoc approach is used to fix w1 during the optimisation. Such constraint could be directly added inside the cost function. While this would make the optimisation of the model more elegant, it may affect its convergence.
The improvements we have found were only demonstrated using the AIBL dataset. While the proposed NMF approach improves the robustness to scanner change in the calibration set, it does little to improve the robustness of scanner change in the test dataset. It is also possible that some of the improvement observed in AIBL are due to two of the scanners used in AIBL being also used in the calibration set of NAV and FBB. Future work validating this approach on external datasets is warranted to ensure the robustness of the method. Such validation is also warranted to check the longitudinal behaviours of Centiloid measures using FBP. It has been previously shown that FBP has poor longitudinal stability when the whole cerebellum is used as the reference region and that reference regions containing sub-cortical WM can help reduce the longitudinal variability (Landau et al., 2015; Chen et al., 2015). Using such region for the SUVR normalisation of FBP scans before the NMF decomposition might also help to improve the longitudinal Centiloid measures. However, since AIBL has only limited number of repeat FBP scans, we were not able to validate this approach which will be explored in future work using ADNI.
Another weakness of this method is that it does not explicitly model the differences in resolution, scatter correction or reconstruction method between the different scanners, which could also explain why the scanner effect remained in the AIBL dataset. Recent work on group NMF (Lee and Choi, 2009) integrate the group information when computing the NMF decomposition to extract features that are discriminant between different groups, while extracting features that capture the variability inside each group. Future work will focus on extending such method to extract components that are distinctive between different scanners.
Our validation framework relies on the assumption that Aβ accumulation is linear over a period < 10 years, when the accumulation is believed to follow trajectory close to a sigmoid (Villemagne et al., 2013). While we could have used a non-linear model to better account for this trajectory, it should be noted that half of the subjects had their last imaging session less than 3.2 years before their baseline, a timeframe where changes are expected to be linear. For those with longer imaging time-frame, more than half had their CL remain below 10, and had therefore very little changes over time.
Conclusions
Non-negative matrix factorisation can be used to compute Centiloid values on all Aβ tracers. By building a model on all the images from the calibration set, it can reduce the bias introduced by using different scanners. Such model can improve longitudinal consistency when multiple tracers are used such as in international multicentre disease-specific therapeutic trials.
Supplementary Material
Acknowledgements
This work was supported by the National Health and Medical Research Council [GA16788] and the National Institutes of Health [R01-AG058676-01A1].
Disclosures
Chris Rowe has received research grants from Piramal Imaging, GE Healthcare, Cerveau, Astra Zeneca, Biogen. Victor Villemagne is and has been a consultant or paid speaker at sponsored conference sessions for Eli Lilly, Piramal Imaging, GE Healthcare, Abbvie, Lundbeck, Shanghai Green Valley Pharmaceutical Co Ltd, and Hoffmann La Roche.
Footnotes
Data-code availability statement
The Centiloid calibration datasets can be downloaded at http://www.gaain.org/centiloid-project
The AIBL data can be downloaded through LONI after registration at http://adni.loni.usc.edu/category/aibl-study-data/
The python code used to build the NMF models as well as the NMF models can be downloaded from https://doi.org/10.25919/5f8400a0b6a1e
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.neuroimage.2020.117578.
References
- Aide N, Lasnon C, Veit-Haibach P, Sera T, Sattler B, Boellaard R, 2017. EANM/EARL harmonization strategies in PET quantification: from daily practice to multicentre oncological studies. Eur. J. Nucl. Med. Mol. Imaging 44 (Suppl 1), 17–31. doi: 10.1007/s00259-017-3740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amadoru S, Doré V, McLean CA, et al. , 2020. Comparison of amyloid PET measured in Centiloid units with neuropathological findings in Alzheimer’s disease. Alzheimer’s Res. Therapy 12 (1), 22. doi: 10.1186/s13195-020-00587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MR, Pillay LC, Lowe VJ, et al. , 2018. Centiloid scaling for quantification of brain amyloid with [18F]flutemetamol using multiple processing methods. EJNMMI Res. 8 (1), 107. doi: 10.1186/s13550-018-0456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeat P, Doré V, Williams S, et al. , 2019. Correcting for pet scanner changes in longitudinal studies. Alzheimer’s Dement. 15, P952. doi: 10.1016/j.jalz.2019.06.3094. [DOI] [Google Scholar]
- Bourgeat P, Williams R, Rowe C, et al. , 2014. Does enhanced reconstruction methodology change the quantification of amyloid PET with flumetamol. Alzheimer’s Dement.: J. Alzheimer’s Assoc. 10, P401–P402. [Google Scholar]
- Chen K, Roontiva A, Thiyyagura P, et al. , 2015. Improved power for characterizing longitudinal amyloid-β PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J. Nucl. Med. 56 (4), 560–566. doi: 10.2967/jnumed.114.149732. [DOI] [PubMed] [Google Scholar]
- Cho SH, Choe YS, Kim HJ, et al. , 2019. A new Centiloid method for 18F-florbetaben and 18F-flutemetamol PET without conversion to PiB. Eur. J. Nucl. Med. Mol. Imaging 13. doi: 10.1007/s00259-019-04596-x, Published online December. [DOI] [PubMed] [Google Scholar]
- Ellis KA, Bush AI, Darby D, et al. , 2009. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int. Psychogeriatr. 21 (4), 672–687. doi: 10.1017/S1041610209009405. [DOI] [PubMed] [Google Scholar]
- Fripp J, Bourgeat P, Acosta O, et al. , 2008a. Appearance modeling of 11C PiB PET images: characterizing amyloid deposition in Alzheimer’s disease, mild cognitive impairment and healthy aging. Neuroimage 43 (3), 430–439. doi: 10.1016/j.neuroimage.2008.07.053. [DOI] [PubMed] [Google Scholar]
- Fripp J, Bourgeat P, Raniga P, et al. , 2008b. MR-less high dimensional spatial normalization of 11C PiB PET images on a population of elderly, mild cognitive impaired and Alzheimer disease patients.. In: Proceedings of the 11th International Conference on Medical Image Computing and Computer-Assisted Intervention – Part I, MICCAI ‘08. Springer-Verlag, pp. 442–449. doi: 10.1007/978-3-540-85988-8_53. [DOI] [PubMed] [Google Scholar]
- Joshi A, Koeppe RA, Fessler JA, 2009. Reducing between scanner differences in multi-center PET studies. Neuroimage 46 (1), 154–159. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Koeppe RA, Price JC, et al. , 2015. The Centiloid project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 11 (1), 1–15. doi: 10.1016/j.jalz.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau SM, Fero A, Baker SL, et al. , 2015. Measurement of longitudinal β-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J. Nucl. Med. 56 (4), 567–574. doi: 10.2967/jnumed.114.148981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Choi S, 2009. Group nonnegative matrix factorization for EEG classification. Artif. Intell. Stat. 5, 320–327. http://proceedings.mlr.press/v5/lee09a.html. [Google Scholar]
- Lee DD, Seung HS., 1999. Learning the parts of objects by non-negative matrix factorization. Nature 401 (6755), 788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- Lilja J, Leuzy A, Chiotis K, Savitcheva I, Sörensen J, Nordberg A, 2019. Spatial normalization of 18F-flutemetamol PET images using an adaptive principal-component template. J. Nucl. Med. 60 (2), 285–291. doi: 10.2967/jnumed.118.207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navitsky M, Joshi AD, Devous MD, et al. , 2016. Conversion of amyloid quantitation with florbetapir SUVR to the centiloid scale. Alzheimer’s Dement.: J. Alzheimer’s Assoc. 12 (7), P25–P26. doi: 10.1016/j.jalz.2016.06.032. [DOI] [PubMed] [Google Scholar]
- Rowe CC, Doré V, Jones G, et al. , 2017. 18F-Florbetaben PET beta-amyloid binding expressed in Centiloids. Eur. J. Nucl. Med. Mol. Imaging 44 (12), 2053–2059. doi: 10.1007/s00259-017-3749-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe CC, Jones G, Doré V, et al. , 2016. Standardized expression of 18F-NAV4694 and 11C-PiB β-amyloid PET results with the Centiloid scale. J. Nucl. Med. 57 (8), 1233–1237. doi: 10.2967/jnumed.115.171595. [DOI] [PubMed] [Google Scholar]
- Sotiras A, Bilgel M, Erus G, et al. , 2018. Multivariate Pattern analysis on a longitudinal cohort of cognitively normal elderly reveals distinct stages of regional amyloid deposition. Alzheimer’s Dement.: J. Alzheimer’s Assoc. 14 (7), P26–P28. doi: 10.1016/j.jalz.2018.06.2085. [DOI] [Google Scholar]
- Su Y, Flores S, Wang G, et al. , 2019. Comparison of Pittsburgh compound B and florbetapir in cross-sectional and longitudinal studies. Alzheimers Dement. 11, 180–190. doi: 10.1016/j.dadm.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Stephenson MC, Nai Y-H, et al. , 2020. Improved quantification of amyloid burden and associated biomarker cut-off points: results from the first amyloid Singaporean cohort with overlapping cerebrovascular disease. Eur. J. Nucl. Med. Mol. Imaging 47 (2), 319–331. doi: 10.1007/s00259-019-04642-8. [DOI] [PubMed] [Google Scholar]
- Villemagne VL, Burnham S, Bourgeat P, et al. , 2013. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 12 (4), 357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- Whittington A, Gunn RN, 2019. Amyloid load: a more sensitive biomarker for amyloid imaging. J. Nucl. Med. 60 (4), 536–540. doi: 10.2967/jnumed.118.210518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington A, Sharp DJ, Gunn RN, 2018. Spatiotemporal distribution of β-amyloid in Alzheimer Disease is the result of heterogeneous regional carrying capacities. J. Nucl. Med. 59 (5), 822–827. doi: 10.2967/jnumed.117.194720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.