Abstract
PCR diagnosis has been considered as the gold standard for coronavirus disease 2019 (COVID-19) and other many diseases. However, there are many problems in using PCR, such as non-specific (i.e., false-positive) and false-negative amplifications, the limits of a target sample volume, deactivation of the enzymes used, complicated techniques, difficulty in designing probe sequences, and the expense. We, thus, need an alternative to PCR, for example an ultrasensitive antigen test. In the present review, we summarize the following three topics. (1) The problems of PCR are outlined. (2) The antigen tests are surveyed in the literature that was published in 2020, and their pros and cons are discussed for commercially available antigen tests. (3) Our own antigen test on the basis of an ultrasensitive enzyme-linked immunosorbent assay (ELISA) is introduced. Finally, we discuss the possibility that our antigen test by an ultrasensitive ELISA technique will become the gold standard for diagnosis of COVID-19 and other diseases.
Keywords: antigen test, COVID-19, real-time PCR, SARS-CoV-2, ultrasensitive ELISA
Significance
We have proposed an ultrasensitive antigen test for the diagnosis of COVID-19. Our detection system is based on an ultrasensitive ELISA, and its sensitivity is comparable to real-time PCR. In the present review, we surveyed the literature for the antigen tests for COVID-19 that was published in 2020. Then, we discussed the possibility that our ultrasensitive ELISA technique will become the gold standard for diagnosis of COVID-19 and other diseases.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic is still ongoing at the end of 2020. Genomic sequencing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes COVID-19, revealed high homology with SARS-related bat viruses belonging to the genus β-coronavirus [1]. This genus is composed of crown-like, enveloped, non-segmented, positive-sense single-stranded RNA (+ssRNA) viruses (Fig. 1) [2]. The 3' terminus of the genome encodes four major structural proteins, including a spike surface glycoprotein, small envelope protein, membrane protein, and nucleocapsid protein, as well as accessory proteins [3,4].
Figure 1 .
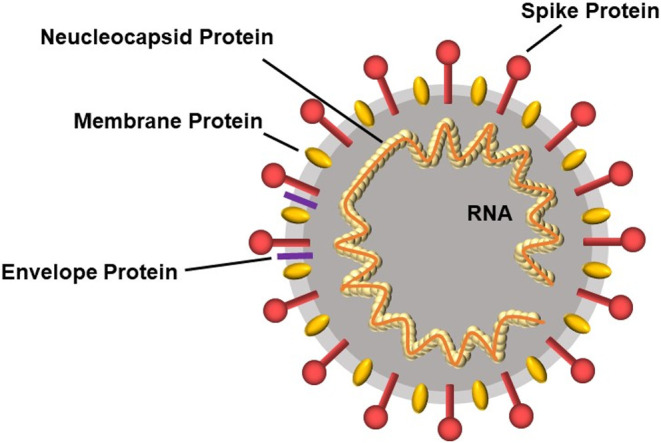
Scheme of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus belongs to the genus β-coronavirus, and it is a crown-like, enveloped, non-segmented, positive-sense single-stranded RNA (+ssRNA) virus. There are four major structural proteins, including a spike surface glycoprotein, small envelope protein, membrane protein, and nucleocapsid protein.
The nucleocapsid protein packages the RNA to form a helical nucleocapsid. The spike protein facilitates viral entry into host cells by using its receptor-binding domain region to bind to the host cell receptors through angiotensin-converting enzyme 2 (ACE2) as the main receptor [5]. Therefore, the receptor-binding domain of the spike protein is the major target for COVID-19 therapy. For example, an anti-spike protein antibody assay is used to screen serum containing high titers of SARS-CoV-2 neutralizing antibodies targeting the spike proteins, and this serum can be used to treat severe COVID-19 patients [6,7].
Since the appearance of COVID-19 in 2019 [8], many useful diagnostic methods have been proposed [9–11]. Many university researchers as well as many governmental organizations indicated that real-time reverse transcription-polymerase chain reaction (PCR) for RNA in SARS-CoV-2 was the “gold standard” for the definite diagnosis of COVID-19 [12–17]. However, there are many comments regarding the limitations of PCR in the diagnosis, and these comments sometimes recommended to combine chest computed tomography (CT) together with PCR [18–21]. Chest CT has been established to play an important role in detecting lung abnormalities, allowing for precise treatment.
However, because chest CT is not easy to use, the improvement of PCR, without the chest CT, has been conducted by various approaches in COVID-19 diagnosis [9–11,13,22]. These approaches include, for example, digital PCR and clustered regularly interspaced short palindromic repeats (CRISPR). We ourselves began to develop a new method without the amplification of nucleic acids to overcome the limitations of PCR [23,24]. This non-amplification method for the detection of nucleic acids will be described elsewhere; however, in the present review, the limitations of PCR are discussed in detail.
Following the discussion of PCR tests, we will describe antigen tests, in which the proteins contained in SARS-CoV-2 are detected for diagnosis of COVID-19. Antigen tests are rapid, low-priced, and easy-to-use in the diagnosis of COVID-19 [25]. However, the sensitivity of antigen tests is generally worse than the PCR tests, even though the specificity is high enough compared with PCR [26]. To overcome this disadvantage, various approaches, including our new system, have attempted to improve the sensitivity in antigen tests [27]. The results of this improvement are discussed in the present review. We hope that the improvement of antigen tests will promote their use. The use of antigen tests is clearly beneficial.
From the viewpoint of biophysics, the development of precise measurements of nucleic acids and proteins is so important for all the fields of biology and medicine. For example, after PCR was developed by Kary Banks Mullis [28], the progress in biology and medicine has been striking. In addition, PCR can amplify nucleic acids, resulting in detection of a small number of copies, whereas we have no methods to amplify proteins, failing to measure a trace amount of proteins. However, we have to take notice that proteins, not nucleic acids, function almost all cases in organisms. Thus, we have to develop precise, quantitative measurement methods for a trace amount of proteins to promote biology and medicine.
Limitations of PCR
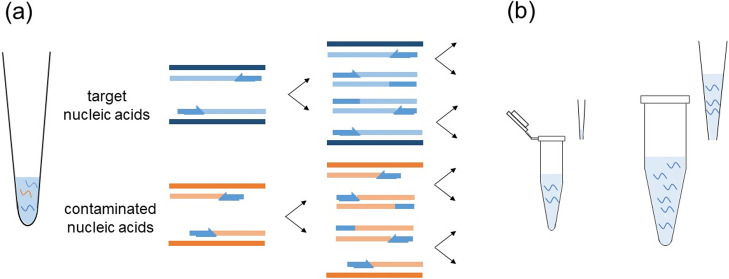
The PCR tests have the advantage in the limit of detection (LOD), e.g., a few copies/assay for various nucleic acids [29], including SARS-CoV-2 RNA [30]. However, the weakness of PCR is indicated in the following points [31]. (1) Non-specific (i.e., false-positive) and false-negative amplifications are often obtained. Designed primers may cross-react with non-specific nucleic acids of other viruses, bacteria, and contaminations in a laboratory, possibly raising the false-positive results. On the other hand, false-negative results are easily produced by unskilled technicians. The false-negative results may also be caused by mutations in the target gene. That is, the designed primers or probes may not recognize the targets. Designing primer and probe sequences is not easy (Fig. 2).
Figure 2 .
Some limitations of PCR. (a) Non-specific (i.e., false-positive) amplifications are easily obtained by the effects of other viruses, bacteria, and contaminations in a laboratory. (b) The handling volume for PCR is usually a μL order, possibly failing to capture the target nucleic acids (see the left panel). If it becomes a 100 μL order or more, like that of a microplate reader well, the chance of capturing the target nucleic acids becomes larger (see the right panel).
(2) The limit of a target sample volume may cause issues. If the target nucleic acids are included in a small amount of assay volume (e.g., microliter order), PCR can detect the target nucleic acids. However, it is typically difficult to sample the target nucleic acids in a microliter handling volume, whereas it is much easier to obtain them in a milliliter volume (Fig. 2). In addition, (3) PCR takes time; (4) PCR is expensive; (5) PCR techniques are complicated; and (6) the enzymes used are deactivated during a PCR procedure, and so forth. The weakness of PCR has thus motivated many researchers to develop antigen tests for the proteins in SARS-CoV-2.
Antigen Tests for COVID-19
The World Health Organization (WHO) stated in April, 2020 that it did not “recommend the use of antigen-detecting rapid diagnostic tests for patient care, although research into their performance and potential diagnostic utility is highly encouraged” [32]. In this WHO judgement, the main weakness of antigen tests is the low sensitivity, resulting in many false-negative responses. In addition, an important editorial published in Science in May, 2020 concluded that, although coronavirus antigen tests are quick and cheap, they are too often wrong [33]. So far, the antigen tests, for example for influenza viruses, have been reported to have poor sensitivity in comparison with PCR [34,35].
However, in 2020, the opinion was divided on the usefulness of antigen tests. There were many negative reports. Scohy et al. pointed out that a rapid immunochromatographic test for the detection of SARS-CoV-2 antigen provided the poor sensitivity (30.2%) compared with ‘gold standard’ PCR, suggesting that it should not be used alone as the frontline testing for COVID-19 diagnosis [36]. Mak et al. also indicated that a rapid immunochromatographic antigen test was 105-fold less detection-sensitive than PCR for SARS-CoV-2, and it detected only between 11.1% and 45.7% of PCR-positive samples obtained from COVID-19 patients [37]. They concluded that a rapid antigen test can serve only as an adjunct to the PCR test.
On the other hand, there are some papers insisting that rapid antigen tests should be used for the first-line diagnosis of COVID-19. Mertens and colleagues used an immunochromatographic assay for the rapid detection of SARS-CoV-2 antigen and found 57.6% sensitivity and 99.5% specificity, with an accuracy of 82.6% [38]. Even though the sensitivity appeared low, the specificity was high enough, and the detection time was only 15 min. Thus, they suggested that the antigen tests are complementary to the PCR. There was another report to recommend the use of antigen tests in the diagnosis of SARS-CoV-2 infection to control the spread of infection [26].
Various other reports regarding the performance of the commercially available rapid antigen tests have been published from Germany [39], India [40], Italy [41], Spain [42,43], Thailand [44], and USA [45]. All these results indicated that the sensitivity was decent (70.6%–100%) and the specificity was high (96%–100%). The results from a meta-analysis of published papers showed that the average sensitivity was 56.2% (95% CI: 29.5% to 79.8%), and the average specificity was 99.5% (95% CI: 98.1% to 99.9%) [25]. That is, the high specificity of antigen tests for COVID-19 was remarkable. From the viewpoint of the emergency use listing procedure (EUL) by the WHO [46], we could find the 2 antigen tests: Panbio COVID-19 Ag Rapid Test by Abbott Rapid Diagnostics and STANDARD Q COVID-19 Ag Test by SD BIOSENSOR [47,48]. Some papers reported that the sensitivity was not so high for these antigen tests even though they were a little better than the tests that were not listed on EUL [49–51].
As can be seen, the antigen tests generally had low sensitivity and high specificity. Regarding the problem of specificity, we noted two papers on a commercially available antigen test in Japan [52]. One is the paper reported by Hirotsu and colleagues, describing that, in comparison with the PCR results, the antigen test based on chemiluminescence enzyme immunoassay exhibited 55.2% sensitivity and 99.6% specificity, with a 91.4% overall agreement rate [53]. They concluded that this antigen test may be helpful for monitoring viral clearance in hospitalized patients, because the specificity was so high (i.e., 99.6%). In other words, the false-positive results seemed very rare. However, the other paper reported by Ogawa et al. excited attention that this antigen test may offer some false-positive results [54]. The Ministry of Health, Labour, and Welfare of Japan permits the definitive diagnosis of COVID-19 without PCR when the antigen test shows positive [55]. Therefore, Ogawa et al. warned that the use of antigen tests in Japan, which are prone to false positive, may create a situation in which false-positive patients who are not infected with SARS-CoV-2 are at risk of nosocomial infection [54]. That is, the use of antigen tests is dangerous for admission into a hospital.
Recently, new techniques have been used for antigen tests of SARS-CoV-2. A study using a fluorescence immunochromatographic antigen test for SARS-CoV-2 showed 93.9% sensitivity and 100% specificity for the patients who displayed the symptoms within a week [56]. The authors suggested that this antigen test will become an important tool in situations with limited access to molecular methods. Another study using a fluorescence immunochromatographic assay also provided good data regarding both the sensitivity and specificity [57]. This showed that the sensitivity was 75.6%, and the specificity was 100%. These results suggest that fluorescence immunochromatographic antigen tests are potentially useful for COVID-19 diagnosis in the early phases of infection. In addition, there is a rapid detection system achieved by an integration of nanozyme and enzymatic chemiluminescence immunoassay with a lateral flow strip [58]. This antigen test targeted the spike protein, even though almost all the tests used the nucleocapsid protein. This system provides a high-sensitive point-of-care testing (POCT) approach for SARS-CoV-2 antigen detection.
Principles of Ultrasensitive Enzyme-Linked Immunosorbent Assay (ELISA)
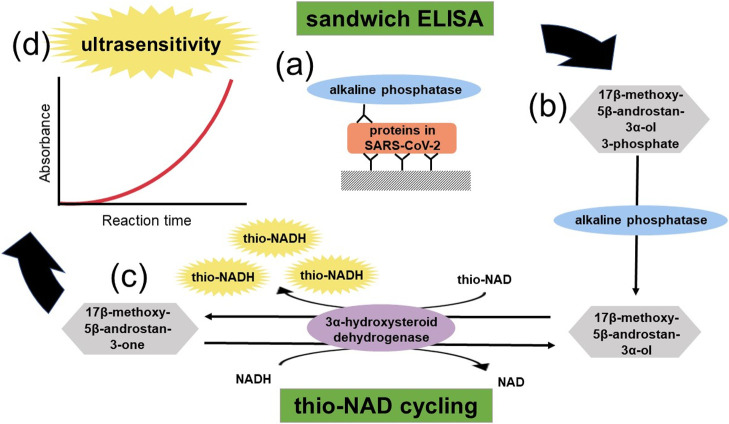
In the following two sections, we introduce our own antigen test for COVID-19. Antigen tests detect proteins. Regarding proteins, they cannot be amplified, whereas nucleic acids can be amplified in a PCR procedure [28]. Our idea, thus, amplifies the detectable signal for a trace amount of proteins (Fig. 3). The quantitative detection for a trace amount of proteins is achieved by sandwich ELISA with two different antibodies specific to the target protein. ELISA is an easy, rapid, specific, and highly sensitive detection method and, thus, has been widely used as a diagnostic tool in medicine and for quality-control checks in various industries [59].
Figure 3 .
Schematics of ultrasensitive enzyme-linked immunosorbent assay (ELISA) by combining sandwich ELISA with thio-NAD cycling. (a) Two antibodies used in ELISA specifically target a pathogenic protein. (b) The second antibody is labeled with alkaline phosphatase, which hydrolyzes a substrate containing phosphate. The hydrolyzed substrates are used in thio-NAD cycling. (c) Thio-NAD cycling employs 3α-hydroxysteroid dehydrogenase (3α-HSD) and its coenzymes (NADH and thio-NAD). (d) Thio-NADH accumulates in a triangular manner and can be measured at 405 nm.
The primary antibody is used for immobilization, and the secondary antibody is linked with an enzyme that converts a substrate to another form. For example, when alkaline phosphatase (ALP) is used as a labeled enzyme, it dephosphorylates a phosphorylated molecule as the applied substrate. Most commonly, this produces a color change in the substrate—that is, a detectable signal. This detectable signal changes linearly with time, and thus it is difficult to achieve high detection sensitivity.
Therefore, we combined a sandwich ELISA with another method (Fig. 3). This is an amplification technique of molecules (i.e., substrates) as a result of the continuous reaction of the enzyme function, and this is called an enzyme cycling method [60,61]. Generally, this technique uses two different enzyme reaction systems in which each enzyme independently and cooperatively acts on the same substrate in a different way. However, we decided to use only one enzyme to reduce the number of enzymes for an easy decision regarding the optimal conditions, as we use another one, ALP, for the ELISA reaction.
This cycling enzyme is 3α-hydroxysteroid dehydrogenase (3α-HSD) [62]. In this cycling reaction, 3α-HSD catalyzes a substrate cycling between 3α-hydroxysteroid and its corresponding 3-ketosteroid in the presence of an excess amount of NADH and thionicotinamide-adenine dinucleotide (thio-NAD), and both of them are cofactors of 3α-HSD [63]. That is, in each turn of the cycle, the 3α-hydroxysteroid is subsequently oxidized to a 3-ketosteroid via 3α-HSD using thio-NAD as a cofactor. In the reverse reaction, the 3-ketosteroid is reduced to 3α-hydroxysteroid by 3α-HSD using NADH as the cofactor.
Thio-NAD is reduced to thio-NADH, which can be measured directly by an increase in the absorbance at 400 nm (11900 M–1 cm–1), e.g., 405 nm with a commercially available microplate reader, without any interference from other cofactors, such as thio-NAD, NAD, and NADH, the absorbance maximums of which are all under 340 nm. This method is now referred to as thio-NAD cycling. These features make it possible to determine the amount of 3α-hydroxysteroids with high sensitivity by measuring the cumulative quantity of thio-NADH. However, this detectable signal also changes linearly with time.
Let us combine a sandwich ELISA and thio-NAD cycling (Fig. 3). We call this detection system ‘ultrasensitive ELISA’, because it can detect ALP at the order of 10–20 moles [23,64]. The thio-NADH signal intensity is expressed as
where a is the turnover ratio of ALP per minute, b is the cycling ratio of 3α-HSD per minute, and n is measurement time in minutes. In our experiments performed recently, the primary substrate applied to ultrasensitive ELISA was 17β-methoxy-5β-androstan-3α-ol 3-phosphate, and the signal generated at 405 nm was normalized to a signal at 660 nm [24]. This ultrasensitive approach required minimal equipment other than a relatively inexpensive microplate reader, thus, making it feasible for most laboratories.
A De Novo Antigen Test for COVID-19 Using Ultrasensitive ELISA
In 2020, we attempted to develop a de novo antigen test for COVID-19 diagnosis [27]. An increase in the ‘sensitivity’ of antigen tests is an important issue as described above. For this purpose, we attempted to increase the ‘detection sensitivity’ of our antigen test using an ultrasensitive ELISA that is hopefully comparable to that of PCR. Then, we noticed the spike proteins, not the nucleocapsid proteins, because the nucleocapsid proteins of SARS-CoV-2 also recognize those of SARS-CoV (Fig. 1). Thus, these two characteristics in our antigen test brought about striking results.
When the antibodies for S1 spike proteins were used, the LOD was 2.3 × 10–18 moles/assay (i.e., 1.8 pg/mL) [27]. This value was determined using the calibration curve for the target protein. Next, we consider how ultrasensitive our developed antigen test is. It was reported that there are about 25 S1 spike proteins on one virus [65]. Therefore, the detection of the S1 spike proteins by our antigen test means that the virus itself can be detected at about 10–20 moles/assay. In other words, the RNA of the virus can be detected at about 104 copies/assay.
This detection sensitivity closes in that of the PCR tests, as it is difficult to detect naso- and oro-pharyngeal specimens at <105 virus RNA copies/swab by a PCR test in a laboratory [66,67]. The potential cross reactions of our SARS-CoV-2 detection system were examined by using another coronavirus, SARS-CoV. The absorbance for SARS-CoV-2 was significantly higher than that of SARS-CoV, and thus our system distinguished SARS-CoV-2 from SARS-CoV [27]. In other words, our ultrasensitive detection method opened up the possibility of overcoming the drawbacks of PCR and establishing a new antigen test specific to COVID-19.
Various State of the Art Diagnostic Methods
In addition to the PCR and antigen tests, many unique approaches have emerged for COVID-19 [31]. One of these is isothermal amplification methods [68]. They do not require any thermal cycling, leading to easy operation and requiring less energy than PCR, which requires rapid heating and cooling steps. These include loop-mediated isothermal amplification (LAMP) [69], multiple displacement amplification (MDA) [70], recombinase polymerase amplification (RPA) [71], circle-to-circle amplification (C2CA), and rolling circle amplification (RCA) [72]. The sensitivity and specificity are properly 100% [71,73].
Another new method is a CRISPR assay. This assay detects DNA or RNA using nucleic acid pre-amplification combined with CRISPR-associated nuclease (Cas) enzymology for the specific recognition of sequences (see [74] for detailed method of CRISPR assays). The usefulness of CRISPR-Cas assays for COVID-19 diagnosis has been strongly indicated [75–77]. The versatility of Cas effectors, including Cas9, Cas12, Cas13, and Cas14, was confirmed to work for nucleotide sensing with high sensitivity at zeptomolar concentrations (10–21 M) and good selectivity (i.e., single nucleotide resolution) [77].
Let us return to the limits of PCR. PCR requires a long duration. To reduce this time, methods are needed to release and enrich SARS-CoV-2 RNA effectively [78,79]. For example, the use of selective electrokinetic concentration enabled to achieve one-step, liquid-phase nucleic acid purification that is simpler and faster than conventional solid-phase extraction [80]. This method performed PCR in a microfluidic chamber, likely offering a POC diagnosis in the future.
Specimens for COVID-19 Diagnosis
Although the diagnosis methods described above are different in their detection mechanisms, the quality of specimens collected from the patients of COVID-19 is very important in all diagnoses. Here, three types of specimens are discussed.
Naso- and oro-pharyngeal swabs
The PCR and antigen tests typically use naso- and oro-pharyngeal swabs for the examination of upper respiratory specimens to detect SARS-CoV-2. The nasopharyngeal specimens are thought to be more sensitive than the oropharyngeal ones [67,81]. Flocked nylon swabs are thought to be suitable, because they can collect a higher volume of specimens compared with cotton, polyester, or rayon swabs [82]. Naso- and oro-pharyngeal swabs should be used for early diagnosis [83].
Saliva
Saliva is now being used for the PCR diagnosis. Saliva collection is easier than swab sampling, because it can be done by the patient. However, the sensitivity compared with nasopharyngeal swabs was divided in every report. Whereas some studies reported that the use of saliva showed a similar sensitivity to nasopharyngeal swabs for COVID-19 diagnostic tests [84–86], others reported slightly lower sensitivity [87].
Sputum
Sputum collection is also easier than swab sampling. However, because not every patient produces sputum, in this case, we must use a clinical induction method [88]. A higher chance to detect SARS-CoV-2 by using sputum compared with naso- and oro-pharyngeal swabs [67,89] was reported, even in the clinically induced sputum [90].
Symptoms in COVID-19 and Expected Diagnosis
The WHO announced that COVID-19 affects different people in different ways. Most infected people will develop mild to moderate illness and recover without hospitalization [91]. According to the WHO, the most common symptoms are fever, dry cough, and tiredness. The less common symptoms include aches and pains, sore throat, diarrhea, conjunctivitis, headache, loss of taste or smell, and a rash on skin or discoloration of the fingers or toes.
The serious symptoms are difficulty breathing or shortness of breath, chest pain or pressure, and loss of speech or movement. Some reports showed that the symptoms varied from mild flu-like symptoms to very severe respiratory symptoms [92,93]. In any case, it is not so easy to diagnose which infectious disease the patient contracts. For this purpose, our diagnosis system based on an ultrasensitive ELISA using microplates is suitable, because an ELISA can be used for a multiplex, simultaneous, high throughput detection.
In particular, middle-aged and elderly people with chronic diseases should be diagnosed quickly and accurately. Children and youth have lower rates of COVID-19 infection compared to middle-aged and elderly people [94–96]. If the patients have chronic diseases, for example hypertension [97,98], diabetes [99,100], and chronic obstructive pulmonary disease [101], they are also vulnerable [102–104]. Thus, a multiplex, simultaneous diagnosis, like our system, becomes a key tool for COVID-19.
Versatility of Our Antigen Test Based on Ultrasensitive ELISA
The antigen test based on an ultrasensitive ELISA method that we developed is extremely versatile if the antibodies against the target proteins are available. For tuberculosis, this ultrasensitive ELISA provided almost the same detection sensitivity as the culture test that is considered a definitive diagnosis (i.e., the gold standard). However, the culture tests take too much time for the bacteria (Mycobacterium tuberculosis) to increase. In our diagnosis, heat treatment induced the bacteria to secrete a very small amount of protein called MPT64 [105]. Therefore, the detection of MPT64 provides the diagnosis of tuberculosis in a few hours [106].
On the other hand, for the diagnosis of acquired immunodeficiency syndrome (AIDS), the detection of the human immunodeficiency virus (HIV)-1 p24 antigen is required in a fourth generation HIV test [107]. The LOD of HIV-1 p24 in our ultrasensitive ELISA was 0.0065 IU/assay (i.e., ca. 10–18 moles/assay) [108]. Because the HIV-1 p24 antigen is thought to be present in the virion in much greater numbers than viral RNA copies, the value of 10–18 moles of the p24/assay corresponds to ca. 103 copies of the HIV-1 RNA/assay. This data is comparable to that of the PCR.
Our ultrasensitive ELISA is useful not only for the diagnosis of infectious diseases but also for various tests for lifestyle-related diseases, for example diabetes [64,109]. Adiponectin is a protein, hormone, and adipokine that is involved in regulating the glucose levels and fatty acid breakdown, and it is decreased in the plasma of patients with visceral obesity and type 2 diabetes [110]. Our system enabled examination of the progression of chronic kidney disease derived from type 2 diabetes by a non-invasive test measuring a trace amount of adiponectin in urine [111,112].
Conclusions
At present, we would like to state that a gold standard for COVID-19 diagnosis has not yet been established. In other words, the PCR methods are not the gold standard [113,114]. Researchers confirmed that PCR has limitations in the sensitivity [58]. In the present review, we characterized PCR tests and antigen tests and introduced our new, ultrasensitive ELISA method as an antigen test for COVID-19 diagnosis with the aim to be an alternative of PCR. By making an increased effort with an increase in the sensitivity and specificity of our new ultrasensitive ELISA-based antigen test, we believe that our diagnosis system can become a gold standard for diagnosis.
In the end of 2020, the world has run into some new variants of SARS-CoV-2 [115]. Analysis of genome sequences for SARS-CoV-2 obtained in a rapid increase of COVID-19 in the United Kingdom clarified that a large portion of cases belonged to a new single phylogenic cluster [116]. The nomenclature of one variant was determined as VOC-202012/01 [117]. This variant was significantly more transmissible than previously circulating variants. For example, this variant was estimated to increase the productive number by 0.4 and to increase transmissibility of up to 70% [116]. This variant is especially characterized by multiple spike protein mutations (deletion 69-60, deletion 144, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H) [115,117]. That is, when we use the antigen tests targeting the spike proteins in SARS-CoV-2, we must note these mutations. The European Centre for Disease Prevention and Control explained one possibility for the emergence of this variant due to the prolonged SARS-CoV-2 infection in a single patient, potentially with reduced immunocompetence. Such prolonged infection may lead to accumulation of immune escape mutations at an elevated rate [116].
Therefore, because the mutation regions in the virus appear mainly in the spike protein, we do not know whether the antibodies produced against the original spike protein can recognize the new variant spike proteins. On the other hand, the mutations rarely occur in the nucleocapsid proteins. Thus, we should target both spike proteins (i.e., proteins outside the envelop) and nucleocapsid proteins (i.e., proteins inside the envelop) to detect variants.
Although we focused on antigen tests for COVID-19 in the present review, the fact that proteins can be measured with ultra-high sensitivity indicates that the proteins contained in a single cell are likely measured directly [118]. Furthermore, we believe that we have to offer a versatile and easy-to-use method to the public. When a great measurement method is developed but it is not easy to use, we do not think it makes sufficient contribution to science. The cost for ultrasensitive ELISA is estimated to be 10 USD at the highest per test. Because that of PCR is now about 20 USD per test, the cost of ultrasensitive ELISA is more inexpensive. In this way, our ultrasensitive ELISA can be widely applied from clinical applications to basic biology. Finally, regarding antibody tests for COVID-19, there are several good review articles [119–121].
Acknowledgments
We appreciate the support from the both Editorial Boards of SEIBUTSU BUTSURI and Biophysics and Physicobiology.
This work was partly supported by the Waseda University grants for Specific Research Projects (grant numbers 2017A-015 and 2019C-123) and the A-STEP Program from JST (grant number AS3015096U).
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author Contributions
Conceptualization and funding acquisition, E.I.; original data collection, Y.K., S.Y., and M.N.; methodology, T.Y., S.W., and E.I.; interpretation and administration, Y.K., T.Y., S.W., and E.I.; writing the original draft, Y.K. and E.I.; editing the draft, Y.K., S.Y., M.N., T.Y., and S.W. All authors have read and agreed to the published version of the manuscript.
References
- [1].Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). DOI: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gupta, P., Goyal, K., Kanta, P., Ghosh, A. & Singh, M. P.. Novel 2019-coronavirus on new year’s Eve. Indian J. Med. Microbiol. 37, 459–477 (2019). DOI: 10.4103/ijmm.IJMM_20_54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wu, A., Peng, Y., Huang, B., Ding, X., Wang, X., Niu, P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 (2020). DOI: 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhang Lab, Genome-wide structure and function modeling of SARS-CoV-2. Available online: https://zhanglab.ccmb.med.umich.edu/COVID-19/ (accessed on January 25, 2021).
- [5].Matsuyama, S., Nao, N., Shirato, K., Kawase, M., Saito, S., Takayama, I., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 117, 7001–7003 (2020). DOI: 10.1073/pnas.2002589117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bloch, E. M., Shoham, S., Casadevall, A., Sachais, B. S., Shaz, B., Winters, J. L., et al. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 130, 2757–2765 (2020). DOI: 10.1172/JCI138745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Okba, N. M. A., Müller, M. A., Li, W., Wang, C., GeurtsvanKessel, C. H., Corman, V. M., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 26, 1478–1488 (2020). DOI: 10.3201/eid2607.200841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].World Health Organization, Timeline: WHO’s COVID-19 response. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline (accessed on January 25, 2021).
- [9].Dhamad, A. E. & Abdal Rhida, M. A.. COVID-19: molecular and serological detection methods. PeerJ 8, e10180 (2020). DOI: 10.7717/peerj.10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jayamohan, H., Lambert, C. J., Sant, H. J., Jafek, A., Patel, D., Feng, H., et al. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 413, 49–71 (2021). DOI: 10.1007/s00216-020-02958-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Prabhakar, P. K. & Lakhanpal, J.. Recent advances in the nucleic acid-based diagnostic tool for coronavirus. Mol. Biol. Rep. 47, 9033–9041 (2020). DOI: 10.1007/s11033-020-05889-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bordi, L., Sberna, G., Lalle, E., Piselli, P., Colavita, F., Nicastri, E., et al. Frequency and duration of SARS-CoV-2 shedding in oral fluid samples assessed by a modified commercial rapid molecular assay. Viruses 12, 1184 (2020). DOI: 10.3390/v12101184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goudouris, E. S. Laboratory diagnosis of COVID-19. J. Pediatr. (Rio J) 97, 7–12 (2021). DOI: 10.1016/j.jped.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Österdahl, M. F., Lee, K. A., Ni Lochlainn, M., Wilson, S., Douthwaite, S., Horsfall, R., et al. Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect Dis. 20, 783 (2020). DOI: 10.1186/s12879-020-05484-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhu, H., Zhang, H., Xu, Y., Laššáková, S., Korabečná, M. & Neužil, P.. PCR past, present and future. Biotechniques 69, 317–325 (2020). DOI: 10.2144/btn-2020-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ministry of Health, Labour and Welfare of Japan, COVID-19 tests. Available online: https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/0000121431_00182.html (accessed on January 25, 2021).
- [17].U.S. Department of Health & Human Services, COVID-19 Diagnostic Laboratory Testing (PCR Testing) Time Series. Available online: https://healthdata.gov/dataset/covid-19-diagnostic-laboratory-testing-pcr-testing-time-series (accessed on January 25, 2021).
- [18].Alsharif, W. & Qurashi, A.. Effectiveness of COVID-19 diagnosis and management tools: A review. Radiography (in press). DOI: 10.1016/j.radi.2020.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hossein, H., Ali, K. M., Hosseini, M., Sarveazad, A., Safari, S. & Yousefifard, M.. Value of chest computed tomography scan in diagnosis of COVID-19; A systematic review and meta-analysis. Clin. Transl. Imaging 8, 469–481 (2020). DOI: 10.1007/s40336-020-00387-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kovács, A., Palásti, P., Veréb, D., Bozsik, B., Palkó, A. & Kincses, Z. T.. The sensitivity and specificity of chest CT in the diagnosis of COVID-19. Eur. Radiol. (in press). DOI: 10.1007/s00330-020-07347-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ria, F., Fu, W., Chalian, H., Abadi, E., Segars, P. W., Fricks, R., et al. A comparison of COVID-19 and imaging radiation risk in clinical patient populations. J. Radiol. Prot. 40, 1336–1345 (2020). DOI: 10.1088/1361-6498/abbf3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu, M., Wang, D., Wang, H., Zhang, X., Liang, T., Dai, J., et al. COVID-19 diagnostic testing: Technology perspective. Clin. Transl. Med. 10, e158 (2020). DOI: 10.1002/ctm2.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Iha, K., Inada, M., Kawada, N., Nakaishi, K., Watabe, S., Tan, Y. H., et al. Ultrasensitive ELISA developed for diagnosis. Diagnostics 9, 78 (2019). DOI: 10.3390/diagnostics9030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ito, E., Iha, K., Yoshimura, T., Nakaishi, K. & Watabe, S.. Early diagnosis with ultrasensitive ELISA. Adv. Clin. Chem. (in press). DOI: 10.1016/bs.acc.2020.06.002 [DOI] [PubMed] [Google Scholar]
- [25].Dinnes, J., Deeks, J. J., Adriano, A., Berhane, S., Davenport, C., Dittrich, S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 8, CD013705 (2020). DOI: 10.1002/14651858.CD013705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Candel, F. J., Barreiro, P., San Román, J., Abanades, J. C., Barba, R., Barberán, J., et al. Recommendations for use of antigenic tests in the diagnosis of acute SARS-CoV-2 infection in the second pandemic wave: attitude in different clinical settings. Rev. Esp. Quimioter. 33, 466–484 (2020). DOI: 10.37201/req/120.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kyosei, Y., Namba, M., Yamura, S., Takeuchi, R., Aoki, N., Nakaishi, K., et al. Proposal of de novo antigen test for COVID-19: Ultrasensitive detection of spike proteins of SARS-CoV-2. Diagnostics 10, 594 (2020). DOI: 10.3390/diagnostics10080594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mullis, K. B. & Faloona, F. A.. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 155, 335–350 (1987). DOI: 10.1016/0076-6879(87)55023-6 [DOI] [PubMed] [Google Scholar]
- [29].Wagatsuma, A., Sadamoto, H., Kitahashi, T., Lukowiak, K., Urano, A. & Ito, E.. Determination of the exact copy numbers of particular mRNAs in a single cell by quantitative real-time RT-PCR. J. Exp. Biol. 208, 2389–2398 (2005). DOI: 10.1242/jeb.01625 [DOI] [PubMed] [Google Scholar]
- [30].Chu, D. K. W., Pan, Y., Cheng, S. M. S., Hui, K. P. Y., Krishnan, P., Liu, Y., et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 66, 549–555 (2020). DOI: 10.1093/clinchem/hvaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].D’Cruz, R. J., Currier, A. W. & Sampson, V. B.. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Front. Cell Dev. Biol. 8, 468 (2020). DOI: 10.3389/fcell.2020.00468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].World Health Organization, Advice on the use of point-of-care immunodiagnostic tests for COVID-19. Available online: https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19 (accessed on January 25, 2021).
- [33].Science, Coronavirus antigen tests: quick and cheap, but too often wrong? Available online: https://www.sciencemag.org/news/2020/05/coronavirus-antigen-tests-quick-and-cheap-too-often-wrong (accessed on January 25, 2021).
- [34].Drexler, J. F., Helmer, A., Kirberg, H., Reber, U., Panning, M., Müller, M., et al. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg. Infect. Dis. 15, 1662–1664 (2009). DOI: 10.3201/eid1510.091186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Magnard, C., Valette, M., Aymard, M. & Lina, B.. Comparison of two nested PCR, cell culture, and antigen detection for the diagnosis of upper respiratory tract infections due to influenza viruses. J. Med. Virol. 59, 215–220 (1999). DOI: 10.1002/(sici)1096-9071(199910)59:2<215::aid-jmv15>3.0.co;2-j [DOI] [PubMed] [Google Scholar]
- [36].Scohy, A., Anantharajah, A., Bodéus, M., Kabamba-Mukadi, B., Verroken, A. & Rodriguez-Villalobos, H.. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 129, 104455 (2020). DOI: 10.1016/j.jcv.2020.104455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mak, G. C., Cheng, P. K., Lau, S. S., Wong, K. K., Lau, C. S., Lam, E. T., et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 129, 104500 (2020). DOI: 10.1016/j.jcv.2020.104500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mertens, P., De Vos, N., Martiny, D., Jassoy, C., Mirazimi, A., Cuypers, L., et al. Development and potential usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a pandemic context. Front. Med. 7, 225 (2020). DOI: 10.3389/fmed.2020.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Krüttgen, A., Cornelissen, C. G., Dreher, M., Hornef, M. W., Imöhl, M. & Kleines, M.. Comparison of the SARS-CoV-2 rapid antigen test to the real star SARS-CoV-2 RT PCR kit. J. Virol. Methods 288, 114024 (2021). DOI: 10.1016/j.jviromet.2020.114024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gupta, A., Khurana, S., Das, R., Srigyan, D., Singh, A., Mittal, A., et al. Rapid chromatographic immunoassay-based evaluation of COVID-19: A cross-sectional, diagnostic test accuracy study & its implications for COVID-19 management in India. Indian J. Med. Res. (in press). DOI: 10.4103/ijmr.IJMR_3305_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cerutti, F., Burdino, E., Milia, M. G., Allice, T., Gregori, G., Bruzzone, B., et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 132, 104654 (2020). DOI: 10.1016/j.jcv.2020.104654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Albert, E., Torres, I., Bueno, F., Huntley, D., Molla, E., Fernández-Fuentes, M. A., et al. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centers. Clin. Microbiol. Infect. 27, 472.e7–472.e10 (2021). DOI: 10.1016/j.cmi.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Linares, M., Pérez-Tanoira, R., Carrero, A., Romanyk, J., Pérez-García, F., Gómez-Herruz, P., et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J. Clin. Virol. 133, 104659 (2020). DOI: 10.1016/j.jcv.2020.104659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chaimayo, C., Kaewnaphan, B., Tanlieng, N., Athipanyasilp, N., Sirijatuphat, R., Chayakulkeeree, M., et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 17, 177 (2020). DOI: 10.1186/s12985-020-01452-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Young, S., Taylor, S. N., Cammarata, C. L., Varnado, K. G., Roger-Dalbert, C., Montano, A., et al. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS Antigen point-of-care test. J. Clin. Microbiol. 59, e02338-20 (2020). DOI: 10.1128/JCM.02338-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].World Health Organization, Coronavirus disease (COVID-19) Pandemic—Emergency Use Listing Procedure (EUL) open for IVDs. Available online: https://extranet.who.int/pqweb/vitro-diagnostics/coronavirus-disease-covid-19-pandemic-%E2%80%94-emergency-use-listing-procedure-eul-open (accessed on January 25, 2021).
- [47].Abbott, PANBIOTM COVID-19 Ag RAPID TEST DEVICE. Available online: https://www.globalpointofcare.abbott/en/product-details/panbio-covid-19-ag-antigen-test.html (accessed on January 25, 2021).
- [48].SD BIOSENSOR, STANDARD Q COVID-19 Ag, Available online: http://sdbiosensor.com/xe/product/7672 (accessed on January 25, 2021).
- [49].Nicol, T., Lefeuvre, C., Serri, O., Pivert, A., Joubaud, F., Dubée, V., et al. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J. Clin. Virol. 129, 104511 (2020). DOI: 10.1016/j.jcv.2020.104511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schildgen, V., Demuth, S., Lüsebrink, J. & Schildgen, O.. Limits and opportunities of SARS-CoV-2 antigen rapid tests: An experienced-based perspective. Pathogens 10, 38 (2021). DOI: 10.3390/pathogens10010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Khairat, S. M., Guindy, N. E. L., Motaleb, M. S. E. A. & Soliman, N. S.. Evaluation of two rapid antigen tests for detection of SARS-CoV-2 virus. Int. J. Microbiol. Biotechnol. 5, 131–134 (2020). DOI: 10.11648/j.ijmb.20200503.18 [Google Scholar]
- [52].Ministry of Health, Labour and Welfare of Japan, Approval of In Vitro Diagnostics for the novel coronavirus infection. Available online: https://www.pmda.go.jp/files/000235116.pdf (accessed on January 25, 2021).
- [53].Hirotsu, Y., Maejima, M., Shibusawa, M., Nagakubo, Y., Hosaka, K., Amemiya, K., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. 99, 397–402 (2020). DOI: 10.1016/j.ijid.2020.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ogawa, T., Fukumori, T., Nishihara, Y., Sekine, T., Okuda, N., Nishimura, T., et al. Another false-positive problem for a SARS-CoV-2 antigen test in Japan. J. Clin. Virol. 131, 104612 (2020). DOI: 10.1016/j.jcv.2020.104612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ministry of Health, Labour and Welfare of Japan, Clinical Management of Patients with COVID-19. Available online: https://www.mhlw.go.jp/content/000646531.pdf (accessed on January 25, 2021).
- [56].Porte, L., Legarraga, P., Vollrath, V., Aguilera, X., Munita, J. M., Araos, R., et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 99, 328–333 (2020). DOI: 10.1016/j.ijid.2020.05.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Diao, B., Wen, K., Zhang, J., Chen, J., Han, C., Chen, Y., et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 27, 289.e1–289.e4 (2021). DOI: 10.1016/j.cmi.2020.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu, D., Ju, C., Han, C., Shi, R., Chen, X., Duan, D., et al. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 173, 112817 (2020). DOI: 10.1016/j.bios.2020.112817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 72, 4–15 (2015). DOI: 10.1016/j.peptides.2015.04.012 [DOI] [PubMed] [Google Scholar]
- [60].Lowry, O. H., Passonneau, J. V., Schulz, D. W. & Rock, M. K.. The measurement of pyridine nucleotides by enzymatic cycling. J. Biol. Chem. 236, 2746–2755 (1961). DOI: 10.1016/S0021-9258(19)61729-1 [PubMed] [Google Scholar]
- [61].Kato, T., Berger, S. J., Carter, J. A. & Lowry, O. H.. An enzymatic cycling method for nicotinamide-adenine dinucleotide with malic and alcohol dehydrogenases. Anal. Biochem. 53, 86–97 (1973). DOI: 10.1016/0003-2697(73)90409-0 [DOI] [PubMed] [Google Scholar]
- [62].Iwai, A., Yoshimura, T., Wada, K., Watabe, S., Sakamoto, Y., Ito, E., et al. Spectrophotometric method for the assay of steroid 5α-reductase activity of rat liver and prostate microsomes. Anal. Sci. 29, 455–459 (2013). DOI: 10.2116/analsci.29.455 [DOI] [PubMed] [Google Scholar]
- [63].Skålhegg, B. A. 3α-hydroxysteroid dehydrogenase from Pseudomonas testosteroni: Kinetic properties with NAD and its thionicotinamide analogue. Eur. J. Biochem. 50, 603–609 (1975). DOI: 10.1111/j.1432-1033.1975.tb09901.x [DOI] [PubMed] [Google Scholar]
- [64].Watabe, S., Kodama, H., Kaneda, M., Morikawa, M., Nakaishi, K., Yoshimura, T., et al. Ultrasensitive enzyme-linked immunosorbent assay (ELISA) of proteins by combination with the thio-NAD cycling method. Biophysics 10, 49–54 (2014). DOI: 10.2142/biophysics.10.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ke, Z., Oton, J., Qu, K., Cortese, M., Zila, V., McKeane, L., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). DOI: 10.1038/s41586-020-2665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wölfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Müller, M. A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). DOI: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- [67].Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844 (2020). DOI: 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zanoli, L. M. & Spoto, G.. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 3, 18–43 (2013). DOI: 10.3390/bios3010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Augustine, R., Hasan, A., Das, S., Ahmed, R., Mori, Y., Notomi, T., et al. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology 9, 182 (2020). DOI: 10.3390/biology9080182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li, S., Jiang, W., Huang, J., Liu, Y., Ren, L., Zhuang, L., et al. Highly sensitive and specific diagnosis of COVID-19 by reverse transcription multiple cross displacement amplification-labelled nanoparticles biosensor. Eur. Respir. J. 56, 2002060 (2020). DOI: 10.1183/13993003.02060-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Behrmann, O., Bachmann, I., Spiegel, M., Schramm, M., El Wahed, A. B., Dobler, G., et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (Exo-IQ). Clin. Chem. 66, 1047–1054 (2020). DOI: 10.1093/clinchem/hvaa116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tian, B., Gao, F., Fock, J., Dufva, M. & Hansen, M. F.. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 165, 112356 (2020). DOI: 10.1016/j.bios.2020.112356 [DOI] [PubMed] [Google Scholar]
- [73].Yan, C., Cui, J., Huang, L., Du, B., Chen, L., Xue, G., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 26, 773–779 (2020). DOI: 10.1016/j.cmi.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ji, T., Liu, Z., Wang, G., Guo, X., Akbar Khan, S., Lai, C., et al. Detection of COVID-19: a review of the current literature and future perspectives. Biosens. Bioelectron. 166, 112455 (2020). DOI: 10.1016/j.bios.2020.112455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Javalkote, V. S., Kancharla, N., Bhadra, B., Shukla, M., Soni, B., Sapre, A., et al. CRISPR-based assays for rapid detection of SARS-CoV-2. Methods (in press). DOI: 10.1016/j.ymeth.2020.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kumar, P., Malik, Y. S., Ganesh, B., Rahangdale, S., Saurabh, S., Natesan, S., et al. CRISPR-Cas system: An approach with potentials for COVID-19 diagnosis and therapeutics. Front. Cell. Infect. Microbiol. 10, 576875 (2020). DOI: 10.3389/fcimb.2020.576875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].van Dongen, J. E., Berendsen, J. T. W., Steenbergen, R. D. M., Wolthuis, R. M. F., Eijkel, J. C. T. & Segerink, L. I.. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 166, 112445 (2020). DOI: 10.1016/j.bios.2020.112445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nyan, D. C., Ulitzky, L. E., Cehan, N., Williamson, P., Winkelman, V., Rios, M., et al. Rapid detection of hepatitis B virus in blood plasma by a specific and sensitive loop-mediated isothermal amplification assay. Clin. Infect. Dis. 59, 16–23 (2014). DOI: 10.1093/cid/ciu210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].He, H., Li, R., Chen, Y., Pan, P., Tong, W., Dong, X., et al. Integrated DNA and RNA extraction using magnetic beads from viral pathogens causing acute respiratory infections. Sci. Rep. 7, 45199 (2017). DOI: 10.1038/srep45199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ouyang, W. & Han, J.. One-step nucleic acid purification and noise-resistant polymerase chain reaction by electrokinetic concentration for ultralow-abundance nucleic acid detection. Angew. Chem. Int. Ed. Engl. 59, 10981–10988 (2020). DOI: 10.1002/anie.201915788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zou, L., Ruan, F., Huang, M., Liang, L., Huang, H., Hong, Z., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 382, 1177–1179 (2020). DOI: 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Daley, P., Castriciano, S., Chernesky, M. & Smieja, M.. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J. Clin. Microbiol. 44, 2265–2267 (2006). DOI: 10.1128/JCM.02055-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Sethuraman, N., Jeremiah, S. S. & Ryo, A.. Interpreting diagnostic tests for SARS-CoV-2. JAMA 323, 2249–2251 (2020). DOI: 10.1001/jama.2020.8259 [DOI] [PubMed] [Google Scholar]
- [84].Alizargar, J., Sh, M. E., Aghamohammadi, M. & Hatefi, S.. S aliva samples as an alternative for novel coronavirus (COVID-19) diagnosis. J. Formos. Med. Assoc. 119, 1234–1235 (2020). DOI: 10.1016/j.jfma.2020.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Azzi, L., Maurino, V., Baj, A., Dani, M., d’Aiuto, A., Fasano, M., et al. Diagnostic salivary tests for SARS-CoV-2. J. Dent. Res. 100, 115–123 (2021). DOI: 10.1177/0022034520969670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].To, K. K., Tsang, O. T., Yip, C. C., Chan, K. H., Wu, T. C., Chan, J. M., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 71, 841–843 (2020). DOI: 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Griesemer, S. B., Van Slyke, G., Ehrbar, D., Strle, K., Yildirim, T., Centurioni, D. A., et al. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. medRxiv (2020). DOI: 10.1101/2020.06.16.20133041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Weiszhar, Z. & Horvath, I.. Induced sputum analysis: step by step. Breathe 9, 300–306 (2013). DOI: 10.1183/20734735.042912 [Google Scholar]
- [89].Pan, Y., Zhang, D., Yang, P., Poon, L. L. M. & Wang, Q.. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 20, 411–412 (2020). DOI: 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Han, H., Luo, Q., Mo, F., Long, L. & Zheng, W.. SARS-CoV-2 RNA more readily detected in induced sputum than in throat swabs of convalescent COVID-19 patients. Lancet Infect. Dis. 20, 655–656 (2020). DOI: 10.1016/S1473-3099(20)30174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].World Health Organization, Coronavirus. Available online: https://www.who.int/health-topics/coronavirus#tab=tab_3 (accessed on January 25, 2021).
- [92].Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). DOI: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sohrabi, C., Alsafi, Z., O’Neill, N., Khan, M., Kerwan, A., Al-Jabir, A., et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 76, 71–76 (2020). DOI: 10.1016/j.ijsu.2020.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chan, J. F., Yuan, S., Kok, K. H., To, K. K., Chu, H., Yang, J., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 395, 514–523 (2020). DOI: 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Davies, N. G., Klepac, P., Liu, Y., Prem, K., Jit, M. & CMMID COVID-19 working group, et al. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 26, 1205–1211 (2020). DOI: 10.1038/s41591-020-0962-9 [DOI] [PubMed] [Google Scholar]
- [96].Tian, S., Hu, N., Lou, J., Chen, K., Kang, X., Xiang, Z., et al. Characteristics of COVID-19 infection in Beijing. J. Infect. 80, 401–406 (2020). DOI: 10.1016/j.jinf.2020.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kreutz, R., Algharably, E. A. E., Azizi, M., Dobrowolski, P., Guzik, T., Januszewicz, A., et al. Hypertension, the renin-angiotensin system, and the risk of lower respiratory tract infections and lung injury: implications for COVID-19. Cardiovasc. Res. 116, 1688–1699 (2020). DOI: 10.1093/cvr/cvaa097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Schiffrin, E. L., Flack, J. M., Ito, S., Muntner, P. & Webb, R. C.. Hypertension and COVID-19. Am. J. Hypertens. 33, 373–374 (2020). DOI: 10.1093/ajh/hpaa057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Fang, L., Karakiulakis, G. & Roth, M.. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 8, e21 (2020). DOI: 10.1016/S2213-2600(20)30116-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hussain, A., Bhowmik, B. & do Vale Moreira, N. C.. COVID-19 and diabetes: Knowledge in progress. Diabetes Res. Clin. Pract. 162, 108142 (2020). DOI: 10.1016/j.diabres.2020.108142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lippi, G. & Henry, B. M.. Chronic obstructive pulmonary disease is associated with severe coronavirus disease 2019 (COVID-19). Respir. Med. 167, 105941 (2020). DOI: 10.1016/j.rmed.2020.105941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Emami, A., Javanmardi, F., Pirbonyeh, N. & Akbari, A.. Prevalence of underlying diseases in hospitalized patients with COVID-19: A systematic review and meta-analysis. Arch. Acad. Emerg. Med. 8, e35 (2020). DOI: 10.22037/aaem.v8i1.600 [PMC free article] [PubMed] [Google Scholar]
- [103].Javanmardi, F., Keshavarzi, A., Akbari, A., Emami, A. & Pirbonyeh, N.. Prevalence of underlying diseases in died cases of COVID-19: A systematic review and meta-analysis. PLoS One 15, e0241265 (2020). DOI: 10.1371/journal.pone.0241265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Muniyappa, R. & Gubbi, S.. COVID-19 pandemic, coronaviruses, and diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 318, E736–E741 (2020). DOI: 10.1152/ajpendo.00124.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nakaishi, K., Watabe, S., Kitagawa, T. & Ito, E.. Immunochromatographic detection of MPB64 secreted from active BCG by heating: toward same-day diagnosis of tuberculosis. Biotechniques 66, 240–242 (2019). DOI: 10.2144/btn-2019-0020 [DOI] [PubMed] [Google Scholar]
- [106].Wang, W. H., Takeuchi, R., Jain, S. H., Jiang, Y. H., Watanuki, S., Ohtaki, Y., et al. A novel, rapid (within hours) culture-free diagnostic method for detecting live Mycobacterium tuberculosis with high sensitivity. EBioMedicine 60, 103007 (2020). DOI: 10.1016/j.ebiom.2020.103007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Mitchell, E. O., Stewart, G., Bajzik, O., Ferret, M., Bentsen, C. & Shriver, M. K.. Performance comparison of the 4th generation Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA on the EVOLIS automated system versus Abbott ARCHITECT HIV Ag/Ab Combo, Ortho Anti-HIV 1+2 EIA on Vitros ECi and Siemens HIV-1/O/2 enhanced on Advia Centaur. J. Clin. Virol. 58 Suppl 1, e79–e84 (2013). DOI: 10.1016/j.jcv.2013.08.009 [DOI] [PubMed] [Google Scholar]
- [108].Nakatsuma, A., Kaneda, M., Kodama, H., Morikawa, M., Watabe, S., Nakaishi, K., et al. Detection of HIV-1 p24 at attomole level by ultrasensitive ELISA with thio-NAD cycling. PLoS One 10, e0131319 (2015). DOI: 10.1371/journal.pone.0131319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ito, E., Kaneda, M., Kodama, H., Morikawa, M., Tai, M., Aoki, K., et al. Immunoreactive insulin in diabetes mellitus patient sera detected by ultrasensitive ELISA with thio-NAD cycling. Biotechniques 59, 361–367 (2015). DOI: 10.2144/000114355 [DOI] [PubMed] [Google Scholar]
- [110].Matsuzawa, Y., Shimomura, I., Kihara, S. & Funahashi, T.. Importance of adipocytokines in obesity-related diseases. Horm. Res. 60 Suppl 3, 56–59 (2003). DOI: 10.1159/000074502 [DOI] [PubMed] [Google Scholar]
- [111].Morikawa, M., Naito, R., Mita, K., Watabe, S., Nakaishi, K., Yoshimura, T., et al. Subattomole detection of adiponectin in urine by ultrasensitive ELISA coupled with thio-NAD cycling. Biophys. Physicobiol. 12, 79–86 (2015). DOI: 10.2142/biophysico.12.0_79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Yamakado, S., Cho, H., Inada, M., Morikawa, M., Jiang, Y. H., Saito, K., et al. Urinary adiponectin as a new diagnostic index for chronic kidney disease due to diabetic nephropathy. BMJ Open Diabetes Res. Care 7, e000661 (2019). DOI: 10.1136/bmjdrc-2019-000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Barra, G. B, Santa Rita, T. H., Mesquita, P. G., Jácomo, R. F. & Nery, L. F. A.. Analytical sensitivity and specificity of two RT-qPCR protocols for SARS-CoV-2 detection performed in an automated workflow. Genes 11, 1183 (2020). DOI: 10.3390/genes11101183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Tu, Y.-P. & O’Leary, T. J.. Testing for severe acute respiratory syndrome-coronavirus 2: challenges in getting good specimens, choosing the right test, and interpreting the results. Crit. Care Med. 48, 1680–1689 (2020). DOI: 10.1097/CCM.0000000000004594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].World Health Organization, SARS-CoV-2 variant - United Kingdom of Great Britain and Northern Ireland. Disease outbreak news. Available online: https://www.who.int/csr/don/21-december-2020-sars-cov2-variant-united-kingdom/en/ (accessed on January 25, 2021)
- [116].European Centre for Disease Prevention and Control, Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf (accessed on January 25, 2021)
- [117].Public Health England, Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/947048/Technical_Briefing_VOC_SH_NJL2_SH2.pdf (accessed on January 25, 2021)
- [118].Watabe, S., Morikawa, M., Kaneda, M., Nakaishi, K., Nakatsuma, A., Ninomiya, M., et al. Ultrasensitive detection of proteins and sugars at single-cell level. Commun. Integr. Biol. 9, e1124201 (2016). DOI: 10.1080/19420889.2015.1124201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Augustine, R., Das, S., Hasan, A., Abhilash, S., Salam, S. A., Augustine, P., et al. Rapid antibody-based COVID-19 mass surveillance: relevance, challenges, and prospects in a pandemic and post-pandemic world. J. Clin. Med. 9, 3372 (2020). DOI: 10.3390/jcm9103372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Benzigar, M. R., Bhattacharjee, R., Baharfar, M. & Liu, G.. Current methods for diagnosis of human coronaviruses: pros and cons. Anal. Bioanal. Chem. (in press). DOI: 10.1007/s00216-020-03046-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].West, R., Kobokovich, A., Connell, N. & Gronvall, G. K.. COVID-19 antibody tests: A valuable public health tool with limited relevance to individuals. Trends Microbiol. 29, 214–223 (2021). DOI: 10.1016/j.tim.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]