Abstract
Introduction:
Smoking remains a worldwide epidemic, and despite an increase in public acceptance of the harms of tobacco use, it remains the leading cause of preventable death. It is estimated that up to 70% of all smokers express a desire to quit, but only 3–5% of them are successful.
Areas covered:
The goal of this review was to evaluate the current status of smoking cessation treatments and the feasibility of implementing personalized-medicine approaches to these pharmacotherapies. We evaluated the genetics associated with higher levels of nicotine addiction and follow with an analysis of the genetic variants that affect the nicotine metabolic ratio (NMR) and the FDA approved treatments for smoking cessation. We also highlighted the gaps in the process of translating current laboratory understanding into clinical practice, and the benefits of personalized treatment approaches for a successful smoking cessation strategy.
Expert opinion:
Evidence supports the use of tailored therapies to ensure that the most efficient treatments are utilized in an individual’s smoking cessation efforts. An understanding of the genetic effects on the efficacy of individualized smoking cessation pharmacotherapies is key to smoking cessation, ideally utilizing a polygenetic risk score that considers all genetic variation.
Keywords: nicotine, metabolism, cytochrome P450, UDP glucuronosyltransferase, flavin monooxygenase, nicotine metabolism ratio, bupropion, varenicline, nicotine replacement therapy, tobacco cessation, pharmacogenetics
1. Current tobacco use
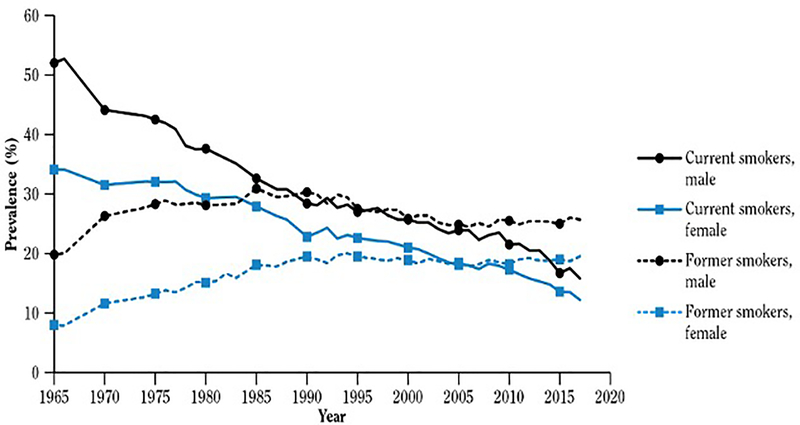
Tobacco remains the number one cause of preventable death in adults worldwide. Approximately 34 million American adults currently smoke cigarettes, accounting for around 14% of the US population. [1] Cigarette smoking rates have been decreasing over the last few decades, attributed to diverse policies such as increased tobacco taxes, tobacco-free indoor air regulations, awareness of second-hand smoking, educational programs on the health consequences of tobacco use, and accessible options for behavioral smoking cessation treatment options (Figure 1). [1] Reflecting the shifting public perceptions regarding the safety of smoking, more than half of all smokers make a serious attempt to quit once a year. However, only 3–5% maintain abstinence after one year. [2] Only one-third of those who attempt to quit have used a pharmacotherapeutic aid, [3] even though the quit rate among those utilizing pharmacotherapy is 2–3-fold higher as compared to subjects taking placebo. [4–6] This low success rate encompasses many factors including a wide variety of behavioral and genetic components. However, the presence of genetic variants may affect an individual’s response to cessation treatments, highlighting the need for an approach that is more reliant on personalized medicine.
Figure 1.
Trends in the prevalence of cigarette smoking among adults 18 years and older. National Health Interview Survey (NHIS) 1965–2017 US. Adapted from [1].
Source: NHIS, National Center for Health Statistics, public use data, 1965–2017.
Note: From 1965 to 2017, data were reported for the following years: 1965, 1966, 1970, 1974, 1976–1980, 1983, 1985, 1987, 1988, 1990–1995, and 1997–2017.
Below we review three main areas: nicotine dependence, nicotine metabolism, and personalized approaches to pharmacotherapy options aimed at improving smoking cessation success.
2. Nicotine dependence
2.1. Nicotine pharmacodynamics
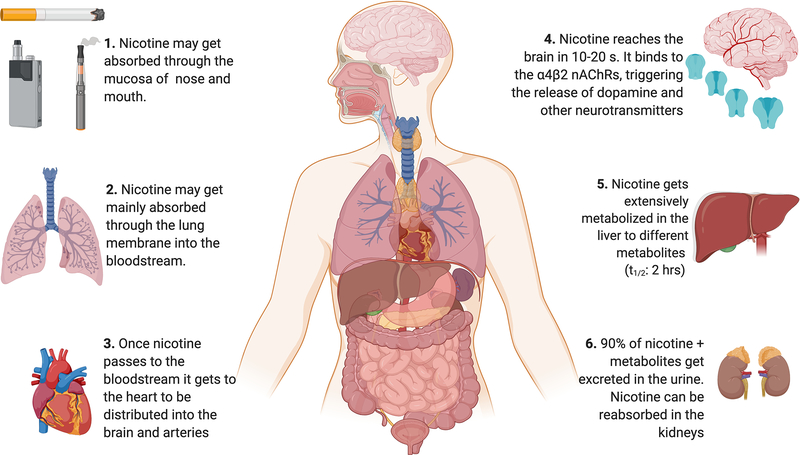
Nicotine (1-methyl-2-[3-pyridyl] pyrrolidine) is a tertiary amine alkaloid present in the leaves of the tobacco plant Nicotiana tabacum and is the ultimate cause of smoking addiction. [7] In smoked tobacco, nicotine volatilizes in the droplets of tar as free nicotine, whereas in smokeless tobacco itis solubilized in the moisture of the tobacco product. [8] Nicotine is easily absorbed through the nasal/oral mucosa or the skin, with the most effective absorption through pulmonary routes. [9] Non-ionized nicotine can cross the blood-brain barrier, and in alkaline solutions nicotine is easily absorbed through membranes. [10] The average intake of nicotine per cigarette is 1 mg, 3.5 mg per 2.5 g for snuff and 4.5 mg per 7.9 g of chewing tobacco for 30 min. [11,12]
Nicotine binds to the heterogenous nicotinic acetylcholine receptors (nAChRs) in the brain, ganglia, and neuromuscular junctions.[13] The (S)-nicotine isomer binds with high specificity to the nAChRs in the brain while the (R)-nicotine isomer binds only weakly to the same receptors. [14] (S)-nicotine shows a very high affinity (Ki = 1.1 nM) for the α4β2-subunit-containing nAChRs (Figure 2). [15] The nAChRs are a class of ligand-gated ion channels, and when nicotine is bound allosteric changes occur, causing modifications to the adjacent ion channels. (7) Bound nicotine opens the channel, allowing the entry of cations such as sodium and calcium. This stimulates the release of dopamine, resulting in the nicotine rewarding effect. [16,17] Besides dopamine, activation of these receptors also triggers the release of norepinephrine, acetylcholine, serotonin, γ-aminobutyric acid (GABA), glutamate, and endorphins. [18,19] The release of these neurotransmitters triggers a list of behaviors that include, but are not limited to, enhanced performance, mood modulation, reversal of withdrawal symptoms, and nicotine self-administration. [13,20] Additionally, sustained use of nicotine increases the number of nAChRs and also desensitizes the receptor towards the agonist, a phenomenon known as neuroadaptation [21,22] which is believed to play a role in nicotine withdrawal symptoms. [23]
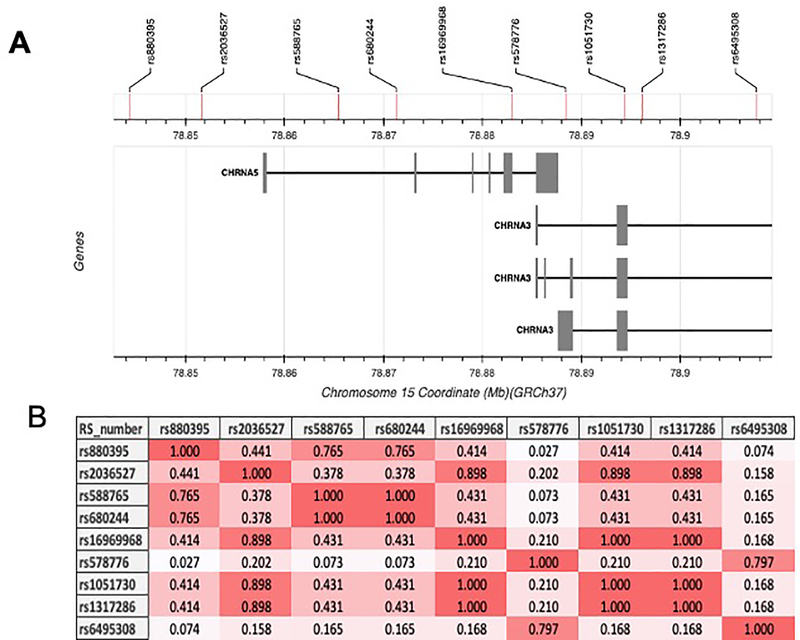
Figure 2.
Genetic variants in chromosome 15 associated with nicotine dependence. (A) Architecture of the genetic variants in chromosome 15q25. (B) Linkage disequilibrium between the genetic variation within the 15q25.1 locus associated with nicotine dependence in Northern Europeans from Utah (CEU) population. The color intensity indicates LD levels among variants. Using the LDlink tool from the National Institutes of Health.
2.2. Genetics of nicotine dependence
The level to which a person can become addicted to nicotine varies widely among individuals and populations based on their genetic disposition. A large proportion of variation in nicotine addiction can be explained by the genetic locus on chromosome 15q25.1, with risk alleles in this locus increasing nicotine dependence by 30–40%. [24,25] This locus includes the CHRNA5, CHRNB4, and CHRNA3 genes that code for the nAChRs, each of which contain variants known to modify nicotine dependence. [26]
When a smoker first decides to quit, they may experience heightened responses of withdrawal during early abstinence. [27] These feelings of less satiety in early abstinence are due to higher expression levels of nAChRs in smokers, resulting from the chronic exposure to nicotine. [28] Normal metabolizers have more expression of the nACHRs, so whenever they are craving (neural cues) nicotine their cravings are more than those in slow metabolizers. This results in higher amount of smoked cigarettes and as a consequence higher amounts of nicotine in the body. [29] In addition, normal metabolizers may benefit from adjunctive behavioral smoking cessation treatments (e.g., cue exposure therapy). [27] Differences in the amount and availability of nAChRs in the brain between slow and fast metabolizers have been linked to differences in rewarding effects. [30]Nicotine dependence has been evaluated among people of European descent, with the strongest associations in CHRNA3 and CHRNA5. [31–34] Multiple variants have been reported to be linked to these two genes (Figure 3). The variant rs16969968, located in CHRNA5, is a missense variant that alters conductance of alpha 5-containing nicotinic receptors in vivo. [35] This variant has been previously associated with delayed smoking cessation and earlier lung cancer diagnosis. [36–38] Genetic variation in chromosome 15 has been explored in different racial groups (European, Asian and African Americans), but only the rs16969968 variant conserves its association with heavy smoking across these populations. [39]
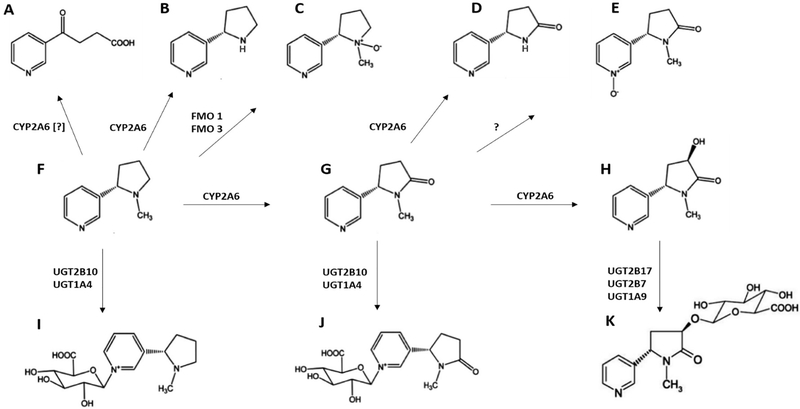
Figure 3.
Schematic of hepatic nicotine metabolism. (A) 4-hydroxy-4-(3-pyridyl)-butanoic acid, (B) nornicotine, (C) nicotine-N’-oxide, (D) norcotinine (E) cotinine-N-oxide, (F) nicotine, (G) cotinine, (H) trans-3’-hydroxycotinine, (I) nicotine-N-glucuronide, (J) cotinine-N-glucuronide, and (K) 3HC-O-glucuronide.
rs16969968 is a ‘defining variant’ in this locus since a number of additional variants that are tightly linked with it have been associated with nicotine dependence within populations with European ancestry. For example, the genetic variant rs1051730 located in the CHRNA3 gene is highly linked to rs16969968 [linkage disequilibrium (LD) r2=0.987] and is associated with smoking quantity and short-term smoking cessation among Europeans. [24,26,37] For example, when analyzing the (A) allelic variant among smokers of Europeans and Asian descent, it was strongly associated with heavy smoking (OR=1.33); however, this variant is only moderately linked with rs16969968 in African Americans (LDr2 =0.4), and thereby shows a lower effect (OR=1.15) when compared to that observed for the rs16969968 (G) allelic variant (OR=1.62) in this population. [39] In Europeans, the variant rs1317286 is also highly linked with rs16969968 (LD r2=0.975) and is highly associated with cigarettes per day (CPD). [40] Also linked is the rs2036527 variant (LD r2 =0.917), which is associated with heavy smoking and CPD in African Americans. [41]
A second defining variant, which is independent from rs16969968 (LD r2=0.226), is rs578776. This variant is located in the untranslated region of the CHRNA3 gene and is also strongly linked with nicotine dependence in smokers of European descent. [37] Although this variant has been tested in Asian and African American populations, it has not been associated with heavy smoking among these specific populations. [39] Linked to this variant (LD r2=0.8) is the rs6495308 variant located in the CHRNA3 gene, which is associated with CPD among Europeans and Asian descendent populations, but not in African Americans. [39,40] The rs680244 variant is moderately linked with rs578776 (LD r2=0.363) in European populations and has also been associated with decreased mRNA levels of CHRNA5 in vivo. [42]
A third defining genetic variant located within the CHRNA5 gene, but independent from both the rs16969968 variant (r2=0.431) and the rs578776 variant (r2=0.109) is rs588765 (also known as rs880395). It is associated with differences in CHRNA5 expression and smoking rates among European and Asian populations. [43] When tested in African American populations, the rs588765 variant only showed a trend towards significance. [39]
In addition to the genetic locus 15q25 locus, there is strong evidence that neurexins (NRXN; presynaptic cell adhesion proteins), specifically NRXN 1 and 3, play a role in nicotine dependence and polysubstance addiction, respectively. [44–46] Neurexins are highly polymorphic genes and different polymorphisms have been associated with different smoking behaviors. For example, for NRXN1, the rs10865246 variant has been associated with higher nicotine addiction when compared to subjects with wt NRXN1. [47] Similarly, the NRXN1 rs985919 and rs1882296 variants were associated with tobacco consumption in a Mexican mestizo population. Interestingly, the rs221473 and rs221497 variants in NRXN3 were associated with lower risk of smoking among Spanish Caucasians. [48]
Identification of the genetic factors which influence an individual’s level of nicotine addiction provides an opportunity for smoking cessation treatments that are personalized or tailored to best suit their level of addiction. In addition to the genetic variants that govern the physical dependence to nicotine, there are a number of behavioral traits linked to an individual’s predisposition towards tobacco use and nicotine addiction. These include such factors as anxiety, [49] sensation-seeking, [50] and impulsivity. [51] Although not discussed in-depth in this review, they are important components of the biological effects of nicotine, and co-medications to treat these behaviors have the potential to negatively interact have with any prescribed smoking cessation pharmacotherapy, leading to unwanted drug-drug interactions (DDI).
3. Smoking cessation treatment options
The current accepted best practices for increasing smoking cessation rates is a pharmacotherapy treatment along with behavioral counseling. [52,53] In 2019, the U.S. Food and Drug Administration (FDA) approved five nicotine replacement therapies (NRTs) and two non-nicotine oral medications indicated to help smokers stop using tobacco products. Additionally, there are groups of smokers for which behavioral support only, and not concurrent pharmacotherapies, are recommended. These groups include pregnant women, smokeless tobacco users, light smokers (<10 cigarettes per day), and adolescents. [53,54] A combination of pharmacotherapy and behavioral or psychological support therapies is now considered to be the standard of care. [48]
3.1. Nicotine replacement therapy
Nicotine Replacement Therapy (NRT) is an alternative nicotine source meant to reduce nicotine withdrawal symptoms at a rate at least 2 times greater than a placebo group. [4] There are several versions of NRT and they can be classified as long-acting (nicotine transdermal patch) and short-acting (nicotine gum, nicotine lozenge, nicotine inhaler, and nicotine nasal spray). The first NRT on the market was nicotine gum, initially introduced in the 1970s in the United Kingdom. It received FDA approval as an over-the-counter medication in the U.S. in 1996 [55] and has been the most widely used form of NRT since its introduction. However, its efficacy can be affected by physiological factors such as impaired absorption due to an acidic diet or other gastrointestinal problems. [56] The main adverse effects reported for NRT are specific for each NRT product, ranging from skin irritation, irritation in the gums, and (rarely) chest pain and palpitations. [57]
4.2. Bupropion
Bupropion is an atypical antidepressant that weakly inhibits dopamine/norepinephrine re-uptake and has properties as a nicotinic acetylcholine receptor antagonist. [58] Bupropion has been commercialized as a sustained-release formula (Zyban®) since 1997, helping to lessen the severity of cravings and nicotine withdrawal symptoms. [59] In clinical trials, it has been shown to double the abstinence rate compared to placebo or the nicotine patch groups. [5] It prevents relapses and increases quitting rates by 25–30%. [5,60] Some of the adverse effects associated with bupropion are insomnia, dry mouth, and (rarely) seizures and hypersensitivity reactions. [61]
4.3. Varenicline
Varenicline (Chantix®) is a high-affinity partial agonist of the α4β2 nAChR subtype, resulting in reduced dopamine release and producing relief from withdrawal and craving symptoms. [62] Varenicline has been approved and commercialized since 2006 [63] and has shown efficacy by increasing smoking cessation rates 3-fold vs placebo and 2-fold vs bupropion- or NRT-treated groups. [6] Some of the most commonly reported adverse effects for varenicline are nausea, insomnia, abnormal dreams, headache, suicidal behaviors and (rarely) skin rash. [64–67] As mentioned above, varenicline binds with highly specificity to α4β2 receptors, but it also binds to the serotonin 5-HT3 receptor, and this binding mechanism of action is hypothesized to be involved in reducing nicotine withdrawal symptoms. [68]
4. Pharmacogenetic factors influencing smoking cessation pharmacotherapy
In the clinical setting, smoking cessation pharmacotherapies and dosages are chosen based on levels of nicotine addiction as assessed by tools such as the Fagerström Test for Nicotine Dependence (FTND), a validated, standardized smoking assessment questionnaire, and the nicotine metabolic ratio (NMR), an in vivo biomarker for the rate of nicotine metabolism (discussed in more detail below in section 7). The overall success of pharmacotherapy treatments in smoking cessation efforts are between 7–30% post-treatment. [4,69] There are many factors that may affect the efficacy of these treatments and an individual’s response to smoking cessation pharmacotherapy, including heritable variation in both nicotine receptor and metabolism genes. [70] Genetic variation has been associated with differences in smoking cessation treatment efficacy and adverse effects throughout an individual’s smoking cessation efforts, [71] with studies suggesting that genetic factors account for approximately 50% of the variability in smoking cessation success. [72,73]
4.1. NRT
The genetic factors affecting NRT efficacy are similar to those affecting levels of nicotine addiction. When analyzing genetic variation within populations and their response to smoking cessation pharmacotherapy, there is an association between CHRNA5 genetic variation and cessation success (or lack thereof) in patients treated with NRT. [74,75] In Caucasians classified as low-, medium- and high-risk haplotypes (combining the rs16969968, rs680244,rs588765,and rs1051730 variants within the CHRNA5-CHRNA3-CHRNB4 loci), the “high risk” haplotype was not only associated with high nicotine dependence but also with increased risk of NRT pharmacotherapy failure. [75,76] The rate of nicotine metabolism (measured by the NMR; reviewed in section 6) has been shown to be a good biomarker for NRT efficacy as compared to other treatments among slow metabolizers. [77–79] Genetic variation in metabolizing enzymes can also play an important role in NRT efficacy and will be discussed in more detail in section 5.0 of this review.
4.2. Bupropion
Bupropion exhibits high variability in treatment response, with 30–45% of users successfully achieving long-term abstinence. [5,80,81] Given bupropion’s mechanism of action within the brain, individuals with genotypes that predispose for higher dopamine availability have better responses to bupropion. For example, genetic variation in the promoter region (−141C Ins) of the dopamine D2 receptor (DRD2) translates to higher transcriptional efficiency, and subjects with the variant alleles how an association with higher quit rates when using bupropion vs NRT. [82–84] In addition, the dopamine transporter gene SLC6A3/DAT1 presents a variable number of tandem repeats (VNTRs), with the presence of 9 vs 10 VTNRs associated with an increased ability to stop smoking using either NRT or bupropion. [83,85] Also, the GG haplotype (rs737865 and rs165599) in the enzyme catechol-O-methyltransferase (COMT), which is involved in the metabolic inactivation of dopamine, has been associated with a favorable outcome when using bupropion for smoking cessation in a Caucasian population. [86]
In humans, bupropion is metabolized to an active metabolite, hydroxy-bupropion by CYP2B6.[87] The CYP2B6*6 variant has been shown to predict decreased CYP2B6 function against bupropion among Alaskan Native and American Indian populations, [88] and it has been shown that individuals homozygous for the CYP2B6*6 allele exhibit lower apparent renal clearance of hydroxy-bupropion than subjects homozygous for the CYP2B6*1 (wildtype) isoform. [89,90] The CYP2B6*6 and *18 alleles were associated with 33% less hydroxy-bupropion in plasma. [91] It has been reported that carriers of the CYP2B6*6 allele have significantly higher abstinence rates than CYP2B6 (*1/*1) subjects when treated with bupropion. [92] The CYP2B6 non-coding single nucleotide polymorphism (SNP; rs8109525; −5293 G>A) has been associated with CPD and continuous abstinence (52 weeks) when treated with bupropion. [68,93] Additionally, in a German population, carriers of the CYP2B6*4 (rs2279343; K262R) allele exhibited a significantly higher (1.66-fold) Cmax for bupropion as compared to CYP2B6 (*1/*1) subjects, [94] potentially leading to higher exposure of active drug and a predisposition to adverse effects consequently decreasing adherence to therapy. It was also reported that higher hydroxy-bupropion concentrations resulted in improved smoking cessation outcomes, suggesting that slow CYP2B6 metabolizers should receive a higher bupropion dose to achieve the desired outcome. [95] However, studies also showed that smokers with the defective CYP2B6 activity allele, CYP2B6*5 (rs3211371; 1459C>T), reported greater increases in cravings and higher relapse rates when treated with bupropion, [96] indicating some inconsistency between studies when examining the role of CYP2B6 and the effects of bupropion in tobacco cessation. While some studies have shown that polymorphisms in CYP2C19 affect bupropion pharmacokinetics, this difference was not observed with smoking cessation outcomes, [97] and additional studies did not find any significant correlations between CYP2C19 SNPs and levels of bupropion or its metabolites in plasma. [98]
Approximately 75%, 25% and 10% of the active bupropion metabolites hydroxy-bupropion, erythro-hydrobupropion, and threo-hydrobupropion, respectively, are excreted in the urine as their glucuronide conjugates. [91] It was recently reported that UGT2B7 glucuronidates (S,S)-, (S,R)- and (R,S)- hydrobupropion, whereas UGT1A9 catalyzes (R,R)-hydrobupropion glucuronidation formation. [99] While not yet studied, it is therefore possible that genetic variation in UGTs 2B7 and 1A9 may also play a role in response to buproprion.
4.3. Varenicline
Varenicline efficiency as a smoking cessation therapy is highly influenced by polymorphisms in the 15q25 locus. The rs7164594 SNP in the PSMA4 gene within the 15q25 locus was highly associated with sustained abstinence after varenicline treatment. [68] In addition, the same study reported that two SNPs in the CHRNB2 gene (rs3811450 and rs4262952) were associated with increased smoking abstinence when subjects were treated with varenicline. [68] CHRNA7 encodes the nAChR α7 subunit, and a variant in its promoter region (rs28531779) showed an association with nicotine addiction while its hybrid gene, [100] CHRFAM7A (result of a CHRNA7 partial duplication and a fusion with the FAM7A gene), showed an association with abstinence among Italian smokers treated with varenicline. [101] SNPs (rs11606194, rs3758987, and rs11607240) in the HTR3A and HTR3B genes that code for the serotonin receptor 5-HT3 and play a role in varenicline-induced nausea and are associated with early relapse. [68]
In humans, 81% of varenicline is excreted as parent drug, with the only urinary metabolite occurring via N-carbamoyl glucuronidation in a reaction catalyzed by UGT2B7. [102] The UGT2B7*2 allele (rs7439366) was associated with decreased clearance, explaining 9% of the inter-individual variability observed in varenicline clearance. [103] The transporter SLC22A2 (OCT2) is involved in the glomerular filtration of varenicline excretion into urine. [104,105] A variant in OCT2 (rs316006;1502–529 A>T) has been associated with a 36–50% increase in abstinence at the end of 6 months treatment with varenicline. [106] Subjects homozygous for another CHRNA4 variant (rs1044396;1629 C>T) were associated with a 30% lower success rate in terms of varenicline treatment in a Brazilian population. [107] In addition, SNP rs555018 in the CHRNA5 gene was associated with increased varenicline-induced toxicity including nausea. [68] This side effect may be explained by tolerance, those individuals that can tolerate higher nicotine concentrations do not experience nausea when treated with varenicline. [68] Consistent with this possibility, CHRND (codes for the delta subunit of the nAChR) and CHRNG (codes for the gamma subunit of the nAChRs) have also been associated with both nicotine dependence and nausea. [108]
The efficacy of tobacco cessation pharmacotherapies may therefore be greatly influenced by individual variation in a wide range of genes encoding phase I and phase II metabolic enzymes, transporters and nicotine receptor subunits. There is also the potential for the many genetic variants in these genes to greatly affect the success of any smoking cessation effort, be it a prescribed pharmacotherapy, an over-the-counter NRT, or behavioral counseling, and should better inform which cessation option, or options, have the potential to be most effective.
5. Nicotine metabolism
It is known that tobacco users titrate their tobacco intake to achieve the desired psychopharmacological effects of nicotine. Hence, faster rates of nicotine metabolism result in higher levels of tobacco consumption. [109,110] This is in part due to the relatively short half-life of nicotine, at approximately 2 hours in the average smoker. Nicotine undergoes extensive hepatic metabolism, involving both phase I and phase II enzymes. Phase I metabolism consists primarily of the C-oxidation of nicotine to cotinine by CYP2A6 and the N’-oxidization to nicotine N’-oxide (NOX) by FMO1, FMO2, and FMO3 (see Figure 4). [111–114] Nicotine is also converted to 4-hydroxy-4-(3-pyridyl)-butanoic acid (HPBA), possibly by CYP2A6, [115] and to nornicotine (NON) by CYP2B6 and CYP2A6. [116] Cotinine is further metabolized by hydroxylation to trans-3’-hydroxycotinine (3HC) by CYP2A6, [117] and N-oxidization to cotinine-N-oxide (COX) possibly by CYPs 2C19, 2A6, and 2B6 (Perez-Paramo YX, unpublished results). Cotinine is also converted to norcotinine (NOC) by CYP2A6. [118]
Figure 4.
Schematic of nicotine metabolism and disposition. Created with BioRender.com
The phase II metabolism of nicotine, cotinine and 3HC is via glucuronidation. Nicotine-N-glucuronide (nicotine-Gluc) and cotinine-N-glucuronide (cotinine-Gluc) are formed primarily by UGT2B10, with UGT1A4 playing a minor role. [119–121] One study suggested that UGTs 1A1 and 1A9 may play a role in nicotine and cotinine glucuronidation. [121,122] Unlike the N-glucuronidation that occurs for both nicotine and cotinine, 3HC undergoes O-glucuronidation to form trans-3’-hydroxycotinine-O-glucuronide (3HC-Gluc), which is mediated by multiple UGT enzymes including UGTs 1A9, 2B7, and 2B17. [123,124] Approximately 90% of nicotine and its metabolites are excreted in the urine. [125]
It has been reported that an individual’s exposure to tobacco can be assessed by measuring the total nicotine metabolites in their urine. [126] The distribution of the different nicotine metabolites in body fluids varies among race and ethnicity, mainly due to the genetic makeup of individuals, with an estimated 60% of nicotine metabolism believed to be heritable. [127] As mentioned above, CYP2A6 is the major enzyme involved in the metabolism of nicotine to cotinine, cotinine to 3HC, the conversion of nicotine to the minor metabolites nornicotine and norcotinine, and potentially the 2’-hydroxylation of nicotine to HPBA. These CYP2A6-derived metabolites account for 70–80% of nicotine metabolites in the urine of Caucasian smokers; however, in East Asian populations this percentage could be as low as 50%. [128–130] This disparity may be linked to the higher prevalence of low activity and non-functional variants in CYP2A6 in East Asian populations, which would drive nicotine metabolism into pathways not mediated by CYP2A6. For example, NOX and nicotine-Gluc levels are higher among East Asians populations, at almost double those observed for Caucasian and Latino populations. [131–133] In addition, African American smokers show higher levels of urinary cotinine, which has been associated with a higher prevalence (˜37%) of the non-functional splice variant (rs116294140) in the UGT2B10 gene in this population, resulting in defects in the cotinine and nicotine glucuronidation pathways. [126,130,134]
Nicotine metabolism is by far one of the most important variables in nicotine exposure among smokers. Understanding the enzymes involved in this process and the analysis of the effects of genetic variants in the genes that code for them on nicotine metabolism is important to better tailor personalized approaches to smoking cessation therapy.
6. NMR
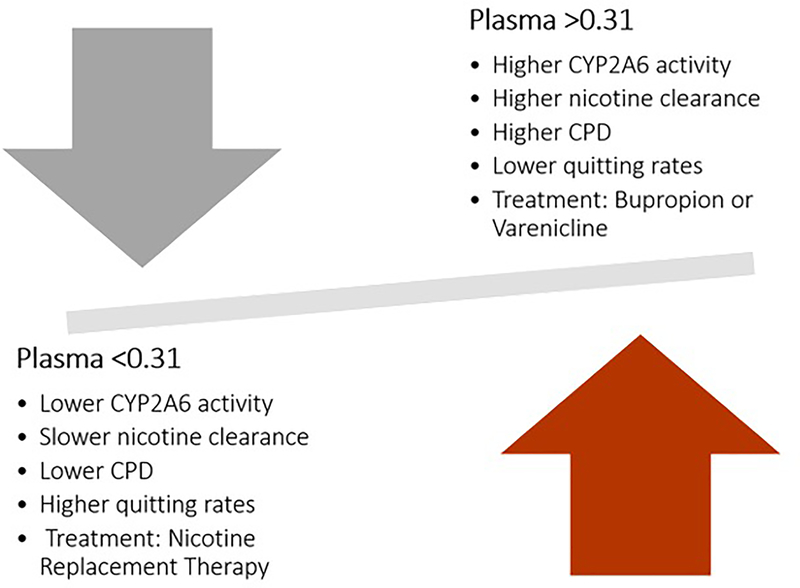
The NMR (3HC/COT) is a measure of nicotine clearance and a predictive biomarker of response to tobacco cessation pharmacotherapy. NMR was first described in 2003 as an in vivo biomarker for CYP2A6 activity and as a predictor of cigarette consumption. [135] The NMR is a stable measure regardless of time of day for smokers consuming at least 5 cigarettes per day, [136,137] and continues to be stable in ad-libitum smokers over a 44 week period. [138] Initially, the NMR was described in plasma; however, studies have tested its reproducibility in different body fluids such as saliva (r2= 0.95) and blood (r2=0.84) and found that urinary NMR (r2=0.76) is not quite as reliable a proxy as that observed in plasma. [139] In a clinical trial where smokers were randomized to treatment based on the NMR, the threshold of NMR in plasma defining a slow metabolizer was NMR < 0.31 while a fast metabolizer had an NMR > 0.31 (Figure 5). [140] It has been widely reported that nicotine metabolic rate is a good predictor of nicotine dependence and smoking cessation success. [136,141] For example, it was reported that when treating patients with the transdermal nicotine patch the percentage of abstinence was reduced by 30% in each increasing quartile of NMR, with patients in the highest NMR quartile reporting more severe cravings. [142] Similarly, NMR was shown to be a significant predictor of abstinence when treating patients with transdermal nicotine; faster metabolizers showed lower quit rates and higher levels of anxiety when compared to slow metabolizers, a result that has been repeated in multiple studies among Caucasians and African Americans. [79,143,144] Another study showed that among normal metabolizers, the smoking quit rates were higher when using varenicline as compared to the nicotine patch. [77,141,145] Similarly, fast, but not slow, metabolizers benefited from treatment with bupropion vs placebo. [78] Recently, a study showed that fast metabolizing women were 10 times less likely to stop using e-cigarettes than slow metabolizers. [146] In several studies, regardless of treatment option, slow metabolizers have been reported to have better quitting rates and fewer relapses, [147,148] and it was reported that normal metabolizers should be treated preferentially with a non-NRT pharmacotherapy option such as varenicline and bupropion. [149] However, other studies have reported that smokers with higher NMR (i.e., faster nicotine metabolizers) may be less likely to relapse after an attempt to quit smoking, [150] or that the use of NMR as a stratification tool does not impact the success of pharmacotherapy for smoking cessation. [145]
Figure 5.
The nicotine metabolic ratio (NMR) and its clinical interpretation.
It also has been suggested that the NMR is a better assessment of in vivo CYP2A6 activity than CYP2A6 genotyping alone, since the NMR incorporates not only genotype but also environmental factors affecting nicotine clearance such as CYP2A6 inducers or inhibitors, drug-drug interactions, as well as effects that other metabolizing enzymes may have on systemic levels of cotinine and 3HC. [136]
The NMR may potentially be affected by nicotine metabolism pathways other than CYP2A6. UGT2B10-associated increases in urinary cotinine have been associated with a lower NMR in African Americans as compared to Caucasians. [126] 3HC-Gluc formation is catalyzed by several UGTs including UGT2B17, which has a deletion variant whose prevalence varies widely different among different racial groups: ~30% in Caucasians, 25% in African American, and ˜80% among Asians and Native Hawaiians. [151] Variability in the prevalence of this deletion variant may potentially affect the NMR by interfering with 3HC metabolism, disproportionately increasing urinary 3HC levels (and the NMR ratio) in those smokers who carry the deletion. [152]
There are many non-genetic factors that may affect the NMR. For example, female smokers using estrogen therapies such as birth control or estrogen replacement therapy exhibit a higher NMR (19–29%) as a result of increased transcriptional activity for CYP2A6, induced via the estrogen receptor. [153–156] This results in pregnant women who are smokers exhibiting a higher NMR during pregnancy than one that is measured postpartum. [157] Another factor associated with the NMR is alcohol use, although the factors behind this effect are unknown. [153] Also, the NMR is negatively correlated with BMI, which could be due to the over-abundance of adipose tissue altering the activity of nicotine metabolic enzymes. [127,158,159] In addition, the NMR was 16% lower in users of mentholated cigarettes vs non-mentholated cigarettes. [153] It has been reported that the use of mentholated cigarettes inhibits CYP2A6 [160] and as a consequence reduces nicotine clearance in vivo by approximately 10%. [161] Nevertheless, up to 67% of NMR inter-individual variability can be explained by genetic variation in nicotine metabolizing genes, especially in CYP2A6. [127,152] While no correlation was observed between FTND and NMR, [135] smoking topography including puff volume has been correlated with NMR, with a puff volume increase of between 23 and 28% in each increasing NMR quartile. [162]
Multiple studies have already shown the NMR to be a useful tool in the clinical setting when a smoker must decide which pharmacotherapy aids to utilize when beginning their smoking cessation therapy (reviewed in ref. [74]). For example, Lerman et al., reported the successful use of NMR in a clinical trial examining the use of varenicline or NRT to maximize quitting rates while minimizing side effects. [77] Current best practices suggest that slow metabolizers utilize an NRT, while fast metabolizers will have the best success using bupropion and/or varenicline. These practices have thus far demonstrated promising results utilizing the NMR as a personalized approach, [141] indicating that the incorporation of precision medicine in deciding on smoking cessation therapy can maximize pharmacotherapy success while minimizing adverse effects among patients.
7. Pharmacogenetic factors influencing nicotine metabolism
7.1. Cytochrome P450 enzymes
CYP2A6 is a highly polymorphic enzyme with several loss-of-function alleles (e.g., *4, and *7), decreased function alleles, (e.g., *1A, *2, *9,and *12) and normal function alleles (e.g., *14 and *1). [163] The CYP2A6*1A allele (rs1137115) has been associated with lower mRNA expression, alternative splicing, and slower nicotine metabolism in European American smokers. [164,165] It has been shown that in Asian smokers, this variant reduces the NMR ratio by 10–20%, [166,167] and it has also been shown to be correlated with risk of early cigarette smoking in Mexican Mestizo smokers. [168] The missense variant CYP2A6*2 (L160H; rs1801272) located in exon 3, was associated with lower enzyme metabolism and fewer smoked cigarette pack-years. [109,169] The whole gene deletion CYP2A6*4 allele has been associated with reduced smoking and reduced NMR. [166,167,170] The CYP2A6*7 allele (I471T;rs5031016) is associated with decreased CYP2A6 activity, lower nicotine metabolism, fewer CPD, and reduced NMR. [166,170,171] The intron polymorphism (rs28399433) within the CYP2A6*9 allele has been associated with a >50% reduction in enzyme expression [172] and has been associated with higher nicotine addiction, tobacco dependence, and decreased nicotine metabolism in Spanish, [169] Mexican Mestizo, [168] and Japanese smokers. [173] CYP2A6*12, which codes for a hybrid allele between CYP2A6 and the CYP2A7 intron 2 sequence, results in approximately 60% reduced activity in vivo. [174] In addition, this allele has been associated with reduced 3HC levels in Mexican smokers. [175] The missense SNP(S29N; rs28399435) encoded by the CYP2A6*14 allele has a compensatory effect only when combined with the *1A variant in European Americans. [165] CYP2A6*17 (V365M; rs28399454), only present in African Americans, has been associated with decreased CYP2A6 enzyme expression and stability in vitro [176] with decreased nicotine and cotinine oxidation in vivo, accounting for up to 8% of the NMR variance in African Americans. [177,178] The missense polymorphisms encoded by the CYP2A6*23 (N203C; rs56256500) [179] and CYP2A6*35 (N438Y;rs143731390) [180] alleles have been associated with impaired CYP2A6 activity and lower NMR in smokers.
Several other non-coding polymorphisms identified in GWAS studies were shown to be associated with reduced CYP2A6 activity. Baurley et al., reported on several polymorphisms (rs12459249, rs56113850, rs4001926, rs7247098, and rs34226463) that were associated with reduced CYP2A6 transcription and reduced NMR in European, African, and Asian American smokers. [181] Patel et al., described two polymorphisms (rs35755165 and rs56113850) associated with low CYP2A6 activity in a Caucasian population, [182] while Loukola et al., demonstrated that several non-coding- polymorphisms (rs56113850, rs113288603, and rs12461964) were associated with decreased NMR in Finish smokers. [183] Chenoweth et al., reported several unique variants in CYP2A6 in African Americans that were not identified among Caucasians, with the rs12459249 variant demonstrating the strongest association with NMR variability in this population. [184] Recently, the largest GWAS study of a Caucasian population reported 14 putatively causal variants on chromosomes 19 and 4 that together explained ˜38% of NMR variation. [185]
CYP2B6 has been reported to be involved in the conversion of nicotine to cotinine but is 10-fold less active against nicotine than CYP2A6. [186–188] However, it has been reported that CYP2B6 is the main enzyme involved in NON formation, [116] and several polymorphisms in CYP2B6 have been reported to be associated with nicotine metabolism. The CYP2B6*5 (rs3211371) and CYP2B6*6 (rs3745274 and rs2279343) alleles have been associated with increased nicotine metabolism, [189] and the CYP2B6*6 allele was associated with faster nicotine and cotinine clearance in vivo. [190] While the CYP2B6*4 (rs2279343) allele showed similar activity as wild-type CYP2B6, the *5 and *9 (rs3745274) variants were correlated with decreased intrinsic clearance when compared to the wild-type isoform. [191] Other associations between CYP2B6 variants and smoking include the CYP2B6 intron rs7260329 SNP and decreased CPD, [93] a CYP2B6 promoter variant (rs8109525) and increased nicotine metabolism, [189] and an intron CYP2B6 variant, rs3786552, which was associated with nicotine metabolism and dependence. [189]
7.2. Flavin monooxygenase (FMO) enzymes
FMO involvement in the nicotine to NOX pathway was reported for the first time by Cashman et al., in 1992. [114] Later, Park et al., reported on the stereoselective production of S-(−)-nicotine-N’-oxide by FMO3. [112] The levels of NOX observed in the urine of smokers are attributed almost entirely to FMO3 since it is the major hepatically-expressed FMO involved in NOX formation. [112] However, a comprehensive study of all five FMO isoforms recently found that FMOs 1, 2 and 3 are all active in NOX formation. [111]
Polymorphisms in the FMO3 isoform have been widely associated with nicotine dependence and nicotine metabolism. The variant rs2266782 (E158K) has been associated with decreased NOX production in vitro (61%; [111]) and between 35–66% decreased NOX formation in vivo. [192] In addition, subjects with CYP2A6-defective alleles show an association between rs2266782 and the level of nicotine metabolism, but not with NMR, CPD or total nicotine metabolites. [193] This polymorphism has been associated with risk of hypertension [194] and sudden infant death syndrome (SIDS) due to an increased exposure to nicotine. [195]
Another FMO3 polymorphism associated with NOX production was rs2266780 (E308G). This polymorphism has been associated with aberrant splicing, resulting in decreased enzyme expression [196] and decreased NOX production in vitro (49%), [111] as well as with increased levels of nicotine dependence. [197] The FMO3 haplotype E158K/E308G (LD r2=0.98) has been analyzed in vitro, showing decreased enzyme stability. [198] This haplotype has been associated with the number of cigarettes consumed among nicotine dependent individuals. [199] A missense FMO3 variant, rs17366557 (V257M), has been associated with decreased NOX production in vitro but not in vivo, [196] while an intragenic variant in FMO3 (rs1736560) was also associated with nicotine dependence (evaluated using FTND/CPD). [200] Other missense FMO3 variants including rs12072582 (D132H), rs2066530 (V277A), and rs72549322 (N61S) were associated with decreased NOX production in vitro. [111]
Genetic variation in the FMO1 enzyme, which is expressed in the brain, has been widely associated with higher levels of nicotine dependence in Caucasians. [200] Polymorphisms (rs10912765 and rs4433435) in FMO1 have been associated with nicotine dependence (evaluated using FTND/CPD). [17] In addition, the I303V variant in FMO1 showed a 66% decreased activity against nicotine in vitro as compared to the wild-type FMO1 isoform. [111]
The major FMO2*1 allele in humans encodes for a truncated, non-functional enzyme with an stop codon at position Q472. [201] However, while the FMO2*2 allele that encodes a functional FMO2 isoform is not observed in Caucasians, its prevalence is 26% in African Americans and is preferentially expressed in the lung. The activity of the full length FMO2 enzyme against nicotine was reported for the first time by Perez-Paramo et al., in 2019. [111]
7.3. UDP-glucuronosyltransferase (UGT) enzymes
As described above, UGT2B10 is the major enzyme involved in cotinine-Gluc, and nicotine-Gluc formation. [120,121] UGT2B10 also exhibited activity in the N-glucuronidation of 3HC, a minor metabolite of nicotine not observed in the urine of smokers. [123] The splice variant defined by the UGT2B10 rs116294140 variant, which is most common in African Americans, has been widely associated with decreased levels nicotine-Gluc and cotinine-Gluc in the urine of smokers. [130,192] Moreover, it has been reported that carriers of this splice variant have significantly higher free cotinine levels. [134] Another important polymorphism in the UGT2B10 enzyme is the variant rs61750900 (D67Y), which is associated with significantly decreased glucuronidation activity among individuals homozygous for this variant. [121,130,202,203] Studies have suggested that this polymorphism may be associated with level of cigarette consumption. [203,204] This polymorphism was also associated with large decreases 3HC-N-glucuronide production in vitro. [123] The UGT2B10 genetic variant rs112561475 (N397D) was associated with enhanced glucuronidation activity against nicotine in Caucasians, [199] and several intergenic/intronic variants were associated with altered levels of urinary cotinine-Gluc (rs115765562 or rs34100980, rs141360540 orrs10028938, rs115219551or rs9997650, and rs294777), and nicotine-Gluc (rs116224959 or rs835315) in smokers. [205]
Chen et al., reported that UGT2B17 exhibited the highest 3HC-O-Gluc formation activity among all UGTs tested. [123] Consistent with this is the fact that Caucasian smokers homozygous for the UGT2B17 deletion exhibited a 42% decrease in urinary 3HC-Gluc levels. [206] A similar pattern was observed in African American smokers [152,192] but not in Mexican smokers. [175]
In addition to UGT2B10 and UGT2B17, several other UGTs play minor roles in nicotine metabolism. Studies in human liver microsomes suggested the involvement of UGT1A1 in nicotine-N-glucuronidation. [122] In an African American population, the intronic variants (rs6742078 and rs1018124)in the UGT1A1 gene were associated with altered cotinine-Gluc and nicotine-Gluc urinary levels in vivo. [207] In a separate study, the UGT1A1 intronic rs3771342 and the 3’-untranslated region rs10209214 variants were associated with decreased urinary levels of nicotine-Gluc and cotinine-Gluc, respectively. [207]
UGT1A4 has been shown to glucuronidate nicotine and cotinine at rates that are approximately 10-fold less than UGT2B10 in vitro. [119,208] Polymorphisms in the UGT1A4 gene have been associated with altered urinary levels of cotinine-Gluc and nicotine-Gluc in African Americans. [207] This includes the intronic variants rs3732220 and rs871514, which have been associated with increased levels of urinary cotinine-Gluc. [207] In addition, the intron rs13401281 UGT1A4 variant was associated with increased levels of nicotine-Gluc in African Americans. [207] Interestingly, in smokers taking olanzapine who were carriers of the UGT1A4 rs375836466 (L48V) variant, a 5.1-fold increase in olanzapine levels were observed as compared to non-smokers and non-carriers, indicating possible drug-drug interactions between nicotine or cotinine and olanzapine. [209]
UGT1A9 has been reported to metabolize 3HC to its O-glucuronide, but less efficiently than UGT2B7. [124] UGT1A9 was also suggested to catalyze nicotine-Gluc and cotinine-Gluc formation. [119] Recently, it was reported that an intronic UGT1A9 variant (rs12471326) was associated with high levels of urinary cotinine-Gluc in Mexican smokers. [175]
UGT2B7 exhibits low nicotine-Gluc formation activity and no activity against cotinine glucuronidation in vitro, [120] but has been shown to be active in catalyzing 3HC-Gluc formation. [124] The UGT2B7 intronic variants rs4535394 and rs57216626 have been associated with decreased 3HC-Gluc urinary levels in European and African American populations. [207] The intronic UGT2B7 variant rs4356975 was associated with increased urinary cotinine-Gluc levels in African Americans. [207]
7.4. OCT2 transporter
The transporter SLC22A2 (OCT2) is expressed in the renal proximal tubule cells and endothelial cells of the blood brain barrier, and has been reported to mediate the tubular secretion of nicotine. [210] The missense polymorphism rs316019 (S270A) is associated with increased nicotine and cotinine Cmax, and decreased nicotine clearance. [192] This polymorphism was also associated with a significant 6 month abstinence from smoking among 2233 participants who were treated with NRT. [106]
Summarized above are studies outlining the large number of genetic variants present in nicotine metabolizing enzymes and transporters that vary between populations and individuals and are linked to modifying the levels of nicotine exposure in smokers. The incorporation of this information in personalized approaches to smoking cessation treatment is essential to achieving maximum efficacy and decreasing adverse effects.
8. Conclusion
Smoking remains one of the biggest public health problems worldwide. Billions of dollars support smoking cessation programs in the United States, [1] yet a high percentage of relapse still occurs. A potential new tool to combat this crisis would be to utilize strategies employed by tailored or personalized treatment plans, which are based on a patient’s genetic profile. Such strategies are already in place in some smoking cessation programs, through the utilization of a patient’s NMR to guide which, if any, pharmacotherapies are used in optimizing and personalizing a patient’s smoking cessation treatment plan. One of the key steps in implementing this approach is performing a single patient stratification, instead of utilizing a reference population. The genotypic information from that patient could inform clinicians of which genetic variants might be playing the largest role in driving the patient’s nicotine addiction, and in turn which pharmacotherapies could be most effective in helping them reach their smoking cessation goals.
In this review, we highlight the current knowledge of the genes involved in nicotine, bupropion and varenicline metabolism and transport, receptor genes and the dopaminergic pathways. Detailed information on the genetic variants present in these genes are vital to implementing a tailored treatment strategy in clinical practice. One of the main paradigms is choosing the right biomarkers to provide adequate information to properly tailor the personalized treatment. Examining polymorphisms in nicotine metabolizing enzymes are essential in fully understanding the different enzymes at play in the metabolism of nicotine. Equally as important as the genetic variation that exists in the 15q25 locus, which has been demonstrated to strongly affect not only nicotine dependence, but also adherence to 3 different modes of pharmacotherapies. Finally, fully understanding the metabolism of the prescribed pharmacotherapies bupropion and varenicline are key in understanding the interplay of nicotine metabolism and smoking cessation pharmacotherapy. Therefore, examining a panel of key genetic variants in nicotine metabolism, nicotine dependence and pharmacotherapy metabolism is the optimal approach to developing a personalized treatment for smoking cessation.
Two of the key elements for the use of personalized treatment in the clinic is the cost effectiveness and implementation. With the recent advent of consumer driven genotyping services at a reasonable price, a future reality may include genotyping services provided to patients in the routine clinical practice. In addition, the NMR has been shown to be a consistent and reliable biomarker for nicotine metabolism rates in most populations. Currently, an NMR test costs approximately 25 USD and is available as a clinical laboratory test. [141] The current challenge in improving the utility of NMR in the clinical setting is the education and acceptance of this tool among clinical providers. [141,211]
Biomarkers commonly used to assess nicotine addiction and smoking topography are single phenotypic measures of a great many genetic variants, each influencing their own section of the nicotine metabolism pathway. In order for these biomarkers to be effectively utilized by clinicians to assist smokers in their cessation goals, a greater understanding of the molecular basis of addiction and metabolism is essential. Personalized treatments based on biomarker and genetic information must accurately reflect a smoker’s underlying biology in order to aid that smoker in their motivation to quit.
As reviewed in this study, nicotine metabolism exhibits great variability between individuals. The main variance in nicotine metabolism is due to genetic variants in the genes involved in its metabolism. While CYP2A6 catalysis to cotinine is the most important pathway in nicotine metabolism, there are other important enzymes including the FMOs, UGTs and other CYPs involved in nicotine metabolism. This review highlights the need to genotype individuals for all possible nicotine-related pathways for a better understanding on how biomarkers (NMR specifically) are reflecting an individual’s complex biology.
9. Expert opinion
Smoking remains one of the biggest public health problems worldwide. Billions of dollars are spent to support smoking cessation programs in the United States, yet a high percentage of relapse still occurs. This low success rate encompasses many factors including a wide variety of behavioral and genetic components. The efficacy of tobacco cessation pharmacotherapies (nicotine replacement therapy-NRT, varenicline and bupropion) is greatly influenced by individual variation in a wide range of genes encoding both phase I and phase II metabolizing enzymes and nicotine acetylcholine receptor subunits (nAChRs), highlighting the need for an approach that is more reliant on personalized medicine. The complex genetic matrix of variants in these genes has the potential to greatly affect the success of any smoking cessation effort, be it a prescribed pharmacotherapy, an over-the-counter nicotine replacement therapy (NRT; e.g., patches, gums, nasal sprays, etc.), or behavioral counseling, and should better inform which cessation option, or options, have the potential to be the most effective. The rate of nicotine metabolism varies between individuals and ethnicities, with large differences observed in urinary nicotine metabolite profiles between smokers. It has been reported that this factor plays an important role in smoking disparities and differences in smoking rates between people of different ethnicities.
The main variance in nicotine metabolism is due to genetic variants in the genes involved in its metabolism. While CYP2A6 catalysis of nicotine to cotinine is the primary pathway in nicotine metabolism, there are other important enzymes including the FMOs, UGTs and other CYPs involved in nicotine metabolism. Multiple studies have already shown that the nicotine metabolic ratio (NMR) is a useful tool in the clinical setting when decisions are made regarding which pharmacotherapy aids to utilize when beginning a smoking cessation therapy. [74] Current best practices suggest that slow CYP2A6 metabolizers (with a low NMR) utilize an NRT, while fast metabolizers (with a high NMR) will have the best success using bupropion and/or varenicline. [77,141,145,149] However, the NMR cutoff to stratify a population in slow vs. fast metabolizers needs to be explored among different ethnic populations. The current challenge in improving the utility of the NMR in the clinical setting is the education and acceptance of this tool among clinical providers. Also important is the need to genotype individuals for all possible nicotine-related pathways for a better understanding on how biomarkers (NMR specifically) are reflecting on an individual’s complex biology.
The need for prospective randomized trials to test the utility of the use of genetic marker and dosing options is imperative in order to provide solid evidence that the use of personalized therapy improves smoking cessation rates. In addition, bioinformatic tools that help clinicians to interpret and translate basic genetic findings into a more readily usable clinical interpretation would be of great help when implementing pharmacogenetics in the clinical setting. The goal of this review was to describe the current status of smoking cessation treatments and the feasibility of implementing personalized-medicine approaches to tobacco cessation pharmacotherapies, and to highlight the benefits of personalized treatment approaches for a successful smoking cessation strategy.
Article highlights.
Smoking is the leading preventable cause of death; although 70% of smokers desire to quit only 3–5% are successful.
Genetic variation in nicotine acetylcholine receptors results in differences in addiction to nicotine between individuals.
The metabolism of nicotine varies greatly between individuals due to genetic variation in these pathways, leading to unique nicotine clearance rates, and as a consequence results in varying levels of nicotine exposure between individuals.
Smoking cessation pharmacotherapies (nicotine replacement therapy, varenicline and bupropion) are metabolized differently among individuals, suggesting that personalized approaches to maximize smoking cessation outcomes are warranted.
More research is needed to achieve personalized smoking cessation approaches to improve smoking cessation outcomes. A polygenic risk score that incorporates all sources of variation in nicotine metabolism and target pathways is necessary to increase smoking cessation success.
Acknowledgements
The authors thank Gang Chen, PhD and Christy J.W. Watson, M.S. of Washington State University for their careful reading and advice to prepare this manuscript.
Funding
This paper was funded in part by a Health Sciences and Services Authority of Spokane, WA grant to WSU College of Pharmacy and Pharmaceutical Sciences (grant WSU002292). This paper was also funded in part by the National Institutes of Health, National Institutes of Environmental Health Sciences (grant R01ES025460 to P. Lazarus). This paper was also funded in part by the Fulbright-Garcia Robles Program and a CONACyT dissertation grant to Y.X. Perez-Paramo.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Articles of special interest have been highlighted as either of interest (*) or of considerable interest (**) to readers.
- [1].Department of Health U, Services H, for Disease Control C, et al. Smoking Cessation: A Report of the Surgeon General. 2020. [PubMed]
- [2].Hughes JR, Keely J, Naud S. Shape of the relapse curve and long-term abstinence among untreated smokers. Addiction. 2004;99:29–38. [DOI] [PubMed] [Google Scholar]
- [3].Steinberg MB, Akincigil A, Delnevo CD, et al. Gender and Age Disparities for Smoking-Cessation Treatment. Am. J. Prev. Med. 2006;30:405–412. [DOI] [PubMed] [Google Scholar]
- [4].Silagy C, Lancaster T, Stead LF, Mant DFG, Silagy. Nicotine replacement therapy for smoking cessation. Cochrane Database Syst. Rev. 2004;3:1–123. [DOI] [PubMed] [Google Scholar]
- [5].Jorenby DE, Leischow SJ, Nides MA, et al. A controlled trial of sustained-release bupropion, a nicotine patch, or both for smoking cessation. N. Engl. J. Med. 1999;340:685–691. [DOI] [PubMed] [Google Scholar]
- [6].Cahill K, Stevens S, Perera R, et al. Pharmacological interventions for smoking cessation: An overview and network meta-analysis. Cochrane Database Syst. Rev. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Posselt WR “Chemische Untersuchung des Tabaks und Darstellung eines eigenthümlich wirksamen Prinzips dieser Pflanze” [Chemical investigation of tobacco and preparation of a characteristically active constituent of this plant]. Mag. für Pharm. 1828;6:138–161. [Google Scholar]
- [8].Benowitz NL. Neurobiology of Nicotine Addiction: Implications for Smoking Cessation Treatment. Am. J. Med. 2008;121:S3–S10. [DOI] [PubMed] [Google Scholar]
- [9].Benowitz NL, Porchet H, Sheiner L, et al. Nicotine absorption and cardiovascular effects with smokeless tobacco use: Comparison with cigarettes and nicotine gum. Clin. Pharmacol. Ther. 1988;44:23–28. [DOI] [PubMed] [Google Scholar]
- [10].Armitage AKTD. Absorption of Nicotine Cigarette and Cigar smoke though the Oral Mucosa. Nature. 1970;226:1231–1232. [DOI] [PubMed] [Google Scholar]
- [11].Benowitz NL, III PJ. Daily intake of nicotine during cigarette smoking. Clin. Pharmacol. Ther. 1984;35:499–504. [DOI] [PubMed] [Google Scholar]
- [12].Benowitz NL. Nicotine and smokeless tobacco. CA. Cancer J. Clin. 1988;38:244–247. [DOI] [PubMed] [Google Scholar]
- [13].Dani JA, Heinemann S. Molecular and cellular aspects of nicotine abuse. Neuron. 1996;16:905–908. [DOI] [PubMed] [Google Scholar]
- [14].Benowitz NL. Pharmacology of Nicotine: Addiction and Therapeutics. Annu. Rev. Pharmacol. Toxicol. 1996;36:597–613. [DOI] [PubMed] [Google Scholar]
- [15].Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol. Sci. 2006;27:482–491. [DOI] [PubMed] [Google Scholar]
- [16].Dajas-Bailador F, Wonnacott S. Nicotinic acetylcholine receptors and the regulation of neuronal signalling. Trends Pharmacol. Sci. 2004;25:317–324. [DOI] [PubMed] [Google Scholar]
- [17].Dani JA, De Biasi M. Cellular mechanisms of nicotine addiction. Pharmacol. Biochem. Behav. 2001;70:439–446. [DOI] [PubMed] [Google Scholar]
- [18].Mansvelder HD, McGehee DS. Long-term potentiation of excitatory inputs to brain reward areas by nicotine. Neuron. 2000;27:349–357. [DOI] [PubMed] [Google Scholar]
- [19].Di Chiara G Role of dopamine in the behavioural actions of nicotine related to addiction. Eur. J. Pharmacol. 2000;393:295–314. [DOI] [PubMed] [Google Scholar]
- [20].Benowitz NL. Nicotine addiction. N. Engl. J. Med. 2010;362:2295.* This paper outlines the pharmacology of nicotine addiction.
- [21].Paradiso K, Brehm P. Long-term desensitization of nicotinic acetylcholine receptors is regulated via protein kinase A-mediated phosphorylation. J. Neurosci. 1998;18:9227–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang H, Sun X. Desensitized nicotinic receptors in brain. Brain Res. Rev. 2005;48:420–437. [DOI] [PubMed] [Google Scholar]
- [23].Brody AL, Mandelkern MA, London ED, et al. Cigarette smoking saturates brain α4β2 nicotinic acetylcholine receptors. Arch. Gen. Psychiatry. 2006;63:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fowler CD, Lu Q, Johnson PM, et al. Habenular α5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Thorgeirsson Thorgeir E.1*, Geller Frank1*, Sulem Patrick1*, Rafnar Thorunn1*, Wiste Anna1,2, Magnusson Kristinn P.1, Manolescu Andrei1, Thorleifsson Gudmar1, Stefansson Hreinn1, Ingason Andres1, Stacey Simon N.1, Bergthorsson Jon T.1, Thorlaciu AK Steinunn & KS. A Variant Associated with Nicotine Dependence, Lung Cancer and Peripheral Arterial Disease. Nature. 2008;452:638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Falcone M, Cao W, Bernardo L, et al. Brain Responses to Smoking Cues Differ Based on Nicotine Metabolism Rate. Biol. Psychiatry. 2015;1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dubroff JG, Doot RK, Falcone M, et al. Decreased nicotinic receptor availability in smokers with slow rates of nicotine metabolism. J. Nucl. Med. 2015;56:1724–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tang DW, Hello B, Mroziewicz M, et al. Genetic variation in CYP2A6 predicts neural reactivity to smoking cues as measured using fMRI. Neuroimage. 2012;60:2136–2143. [DOI] [PubMed] [Google Scholar]
- [30].Sofuoglu M, Herman AI, Nadim H, et al. Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology. 2012;37:1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Munafò MR, Johnstone EC, Walther D, et al. CHRNA3 rs1051730 genotype and short-term smoking cessation. Nicotine Tob. Res. 2011;13:982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Munafò MR, Timofeeva MN, Morris RW, et al. Association Between Genetic Variants on Chromosome 15q25 Locus and Objective Measures of Tobacco Exposure. Rev. | JNCI. 2012;104:740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bierut LJ. Nicotine Dependence and Genetic Variation in the Nicotinic Receptors. Drug Alcohol Depend. 2009;1:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sarginson JE, Killen JD, Lazzeroni LC, et al. Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2011;156:275–284. [DOI] [PubMed] [Google Scholar]
- [35].Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine receptor (AChR) α5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (α4β2) 2α5 AChR function. Mol. Pharmacol. 2011;79:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Furberg H, Kim Y, Dackor J, et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Scott F. Saccone, Anthony L. Hinrichs, Saccone Nancy L., Gary A. Chase, Karel Konvicka, Pamela A.F. Madden, Naomi Breslau, Eric O. Johnson, Dorothy Hatsukami, Ovide Pomerleau, Gary E. Swan, Alison M. Goate, Joni Rutter, 10 2 and Laura Jean Bierut, Saccone SF, Hinrichs AL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen LS, Hung RJ, Baker T, et al. CHRNA5 Risk variant predicts delayed smoking cessation and earlier lung cancer diagnosis-a meta-analysis. J. Natl. Cancer Inst. 2015;107:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen L-S, Saccone NL, Culverhouse RC, et al. Smoking and Genetic Risk Variation across Populations of European, Asian, and African-American Ancestry-A Meta-analysis of Chromosome 15q25 NIH Public Access Author Manuscript. Genet Epidemiol. 2012;36:340–351.** This paper summarizes the genetic variation that exists in chromosome 15 and its association with smoking across different populations.
- [40].Berrettini W, Yuan X, Tozzi F, et al. α−5/α−3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol. Psychiatry. 2008;13:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].David SP, Hamidovic A, Chen GK, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl. Psychiatry. 2012;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang JC, Cruchaga C, Saccone NL, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum. Mol. Genet. 2009;18:3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: A meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bierut LJ, Madden PAF, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Liu QR, Drgon T, Walther D, et al. Pooled association genome scanning: Validation and use to identify addiction vulnerability loci in two samples. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11864–11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Furberg H, Kim Y, Dackor J, et al. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pérez-Rubio G, Pérez-Rodríguez ME, Fernández-López JC, et al. SNPs in NRXN1 and CHRNA5 are associated to smoking and regulation of GABAergic and glutamatergic pathways. Pharmacogenomics. 2016;17:1145–1158. [DOI] [PubMed] [Google Scholar]
- [48].Docampo E, Ribasés M, Gratacòs M, et al. Association of Neurexin 3 polymorphisms with smoking behavior. Genes, Brain Behav. 2012;11:704–711. [DOI] [PubMed] [Google Scholar]
- [49].Grant BF, Hasin DS, Chou SP, et al. Nicotine dependence and psychiatric disorders in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch. Gen. Psychiatry. 2004;61:1107–1115. [DOI] [PubMed] [Google Scholar]
- [50].Zuckerman M Sensation seeking: A comparative approach to a human trait. Behav. Brain Sci. 1984;7:413–434. [Google Scholar]
- [51].Mitchell SH. Measures of impulsivity in cigarette smokers and non-smokers. Psychopharmacology (Berl). 1999;146:455–464. [DOI] [PubMed] [Google Scholar]
- [52].West R, Mcneill A, Raw M. Smoking cessation update Smoking cessation guidelines for health professionals: an update. Thorax. 2000;55:987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fiore MC, Jaén CR, Baker TB et al. A Clinical Practice Guideline for Treating Tobacco Use and Dependence: 2008 Update. A U.S. Public Health Service Report. Am. J. Prev. Med. 2008;35:158–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ashley Scherman JET and CM. Smoking cessation in pregnancy: a continuing challenge in the United States. Ther. Adv. Drug Saf. 2018;9:457–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].US Food and Drug Administration. Drug Approval Package: Nicorette (Nicotine Polacrilex Gum) NDA# 18–612/S25 & 20–066/S7 [Internet]. 1998. [cited 2020 Apr 21]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/98/18-612s025_Nicorette.cfm.
- [56].Cole PV Plasma nicotine levels after cigarette smoking and chewing nicotine gum. Br. Med. J. 1976;1:1043–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hartmann-Boyce J, Chepkin SC, Ye W, et al. Nicotine replacement therapy versus control for smoking cessation. Cochrane Database Syst. Rev. John Wiley and Sons Ltd; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cooper BR, Wang CM, Cox RF, et al. Evidence that the acute behavioral and electrophysiological effects of bupropion (Wellbutrin®) are mediated by a noradrenergic mechanism. Neuropsychopharmacology. 1994;11:133–141. [DOI] [PubMed] [Google Scholar]
- [59].US Food and Drug Administration. Drug Approval Package: Zyban (Bupropion Hydrochloride) NDA# 020711s002/s004 [Internet]. 1997. [cited 2020 Apr 21]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/99/020711_s002s004_ZybanTOC.cfm.
- [60].Hurt RD, Sachs DPL, Glover ED, et al. A Comparison of sustained-release bupropion and placebo for smoking cessation. N. Engl. J. Med. 1997;337:1995–1202. [DOI] [PubMed] [Google Scholar]
- [61].Aubin H-J. Tolerability and Safety of Sustained-Release Bupropion in the Management of Smoking Cessation. Drugs. 2002;62:45–52. [DOI] [PubMed] [Google Scholar]
- [62].Coe JW, Brooks PR, Vetelino MG, et al. Varenicline: An r42 Nicotinic Receptor Partial Agonist for Smoking Cessation. J Med Chem. 2005;48:3474–3477. [DOI] [PubMed] [Google Scholar]
- [63].US Food and Drug Administration. Drug Approval Package: Chantix (Varenicline) NDA #021928 [Internet]. 2006. [cited 2020 Apr 21]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2006/021928_s000_chantixtoc.cfm.
- [64].Jorenby DE, Hays JT, Rigotti NA, et al. Efficacy of varenicline, an α4β2 nicotinic acetylcholine receptor partial agonist, vs placebo or sustained-release bupropion for smoking cessation: A randomized controlled trial. J. Am. Med. Assoc. 2006;296:56–63. [DOI] [PubMed] [Google Scholar]
- [65].Oncken C, Gonzales D, Nides M, et al. Efficacy and safety of the novel selective nicotinic acetylcholine receptor partial agonist, varenicline, for smoking cessation. Arch. Intern. Med. 2006;166:1571–1577. [DOI] [PubMed] [Google Scholar]
- [66].Gonzales D, Rennard SI, Nides M, et al. Varenicline, an α4β2 nicotinic acetylcholine receptor partial agonist, vs sustained-release bupropion and placebo for smoking cessation: A randomized controlled trial. J. Am. Med. Assoc. 2006;296:47–55. [DOI] [PubMed] [Google Scholar]
- [67].Gunnell D, Irvine D, Wise L, et al. Varenicline and suicidal behaviour: A cohort study based on data from the General Practice Research Database. BMJ. 2009;339:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].King DP, Paciga S, Pickering E, et al. Smoking cessation pharmacogenetics: Analysis of varenicline and bupropion in placebo-controlled clinical trials. Neuropsychopharmacology. 2012;37:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bauld L, Bell K, McCullough L, et al. The effectiveness of NHS smoking cessation services: A systematic review. J. Public Health (Bangkok). 2010;32:71–82. [DOI] [PubMed] [Google Scholar]
- [70].Quaak M, Van Schayck CP, Knaapen AM, et al. Genetic variation as a predictor of smoking cessation success. A promising preventive and intervention tool for chronic respiratory diseases? Eur. Respir. J. 2009;33:468–480. [DOI] [PubMed] [Google Scholar]
- [71].Bough KJ, Lerman C, Rose JE, et al. Biomarkers for smoking cessation. Clin. Pharmacol. Ther. 2013;93:526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Quaak M, Van Schooten FJ, Van Schayck CP. Pharmacogenetics of smoking: How far to the clinic? Pharmacogenomics. 2014;15:723–726. [DOI] [PubMed] [Google Scholar]
- [73].Hong Xian 1, Jeffrey F Scherrer, Pamela A F Madden, Michael J Lyons, Ming Tsuang, William R True SAE. The Heritability of Failed Smoking Cessation and Nicotine Withdrawal in Twins Who Smoked and Attempted to Quit - PubMed. Nicotine Tob. Res. 2003;5:245–254. [PubMed] [Google Scholar]
- [74].Schuit E, Panagiotou OA, Munafò MR, et al. Pharmacotherapy for smoking cessation: Effects by subgroup defined by genetically informed biomarkers. Cochrane Database Syst. Rev. 2017;9:1–321. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [75].Chen L-S, Baker TB, Piper ME, et al. Interplay of Genetic Risk Factors (CHRNA5-CHRNA3-CHRNB4) and Cessation Treatments in Smoking Cessation Success. Am J Psychiatry. 2012;169:735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bergen AW, Javitz HS, Krasnow R, et al. Nicotinic Acetylcholine Receptor Variation and Response to Smoking Cessation Therapies. Pharmacogenet Genomics. 2013;23:94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lerman C, Schnoll RA, Hawk LW, et al. Use of the nicotine metabolite ratio as a genetically informed biomarker of response to nicotine patch or varenicline for smoking cessation: A randomised, double-blind placebo-controlled trial. Lancet Respir Med. 2015;3:131–138.** This paper proposes for the first time the use of the nicotine metabolic ratio as a stratification tool for smokers and smoking cessation.
- [78].Patterson F, Schnoll RA, Wileyto EP, et al. Toward personalized therapy for smoking cessation: A randomized placebo-controlled trial of bupropion. Clin. Pharmacol. Ther. 2008;84:320–325. [DOI] [PubMed] [Google Scholar]
- [79].Schnoll RA, Patterson F, Wileyto EP, et al. Nicotine metabolic rate predicts successful smoking cessation with transdermal nicotine: A validation study. Pharmacol. Biochem. Behav. 2009;92:6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hurt RD, Sachs DPL, Glover ED, et al. A Comparison of Sustained-Release Bupropion and Placebo for Smoking Cessation. N. Engl. J. Med. 1997;337:1195–1202. [DOI] [PubMed] [Google Scholar]
- [81].Dale LC, Glover ED, Sachs DPL, et al. Bupropion for smoking cessation: Predictors of successful outcome. Chest. 2001;119:1357–1364. [DOI] [PubMed] [Google Scholar]
- [82].Lerman C, Jepson C, Wileyto EP, et al. Role of Functional Genetic Variation in the Dopamine D2 Receptor (DRD2) in Response to Bupropion and Nicotine Replacement Therapy for Tobacco Dependence: Results of Two Randomized Clinical Trials. Neuropsychopharmacology. 2006;31:231–242. [DOI] [PubMed] [Google Scholar]
- [83].Lerman C, Wileyto EP, Audrain J, et al. Effects of dopamine transporter and receptor polymorphisms on smoking cessation in a bupropion clinical trial. Heal. Psychol. 2003;22:541–548. [DOI] [PubMed] [Google Scholar]
- [84].Swan GE, Jack LM, Valdes AM, et al. Joint effect of dopaminergic genes on likelihood of smoking following treatment with bupropion SR. Heal. Psychol. 2007;26:361–368. [DOI] [PubMed] [Google Scholar]
- [85].O’Gara C, Stapleton J, Sutherland G, et al. Dopamine transporter polymorphisms are associated with short-term response to smoking cessation treatment. Pharmacogenet. Genomics. 2007;17:61–67. [DOI] [PubMed] [Google Scholar]
- [86].Berrettini WH, Wileyto EP, Epstein L, et al. Catechol-O-Methyltransferase (COMT) Gene Variants Predict Response to Bupropion Therapy for Tobacco Dependence. Biol. Psychiatry. 2007;61:111–118. [DOI] [PubMed] [Google Scholar]
- [87].Faucette SR, Hawke RL, Lecluyse EL, et al. Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. DRUG Metab. Dispos. Drug Metab Dispos. 2000;28:1222–1230. [PubMed] [Google Scholar]
- [88].Claw KG, Beans JA, Lee S-B, et al. Pharmacogenomics of nicotine metabolism: novel CYP2A6 and CYP2B6 genetic variation patterns in Alaska Native and American Indian populations. Nicotine Tob. Res. 2019;22:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kharasch ED, Crafford A. Common Polymorphisms of CYP2B6 Influence Stereoselective Bupropion Disposition. Clin. Pharmacol. Ther. 2019;105:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Hesse LM, He P, Krishnaswamy S, et al. Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics. 2004;14:225–238. [DOI] [PubMed] [Google Scholar]
- [91].Benowitz NL, Zhu AZXX, Tyndale RF, et al. Influence of CYP2B6 genetic variants on plasma and urine concentrations of bupropion and metabolites at steady state. Pharmacogenet. Genomics. 2013;23:135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lee AM, Jepson C, Hoffmann E, et al. CYP2B6 Genotype Alters Abstinence Rates in a Bupropion Smoking Cessation Trial. Biol. Psychiatry. 2007;62:635–641. [DOI] [PubMed] [Google Scholar]
- [93].Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Kirchheiner J, Klein C, Meineke I, et al. Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics. 2003;13:619–626. [DOI] [PubMed] [Google Scholar]
- [95].Zhu AZX, Cox LS, Nollen N, et al. CYP2B6 and bupropion s smoking-cessation pharmacology: The role of hydroxybupropion. Clin. Pharmacol. Ther. 2012;92:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lerman C, Shields PG, Wileyto EP, et al. Pharmacogenetic investigation of smoking cessation treatment. Pharmacogenetics. 2002;12:627–634. [DOI] [PubMed] [Google Scholar]
- [97].Zhu AZX, Zhou Q, Cox LS, et al. Gene variants in CYP2C19 are associated with altered in vivo bupropion pharmacokinetics but not bupropion-assisted smoking cessation outcomes. Drug Metab. Dispos. 2014;42:1971–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Connarn JN, Flowers S, Kelly M, et al. Pharmacokinetics and Pharmacogenomics of Bupropion in Three Different Formulations with Different Release Kinetics in Healthy Human Volunteers. AAPS J. 2017;19:1513–1522. [DOI] [PubMed] [Google Scholar]
- [99].Gufford BT, Lu JBL, Metzger IF, et al. Stereoselective glucuronidation of bupropion metabolites in vitro and in vivo. Drug Metab. Dispos. 2016;44:544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gault J, Robinson M, Berger R, et al. Genomic organization and partial duplication of the human α7 neuronal nicotinic acetylcholine receptor gene (CHRNA7). Genomics. 1998;52:173–185. [DOI] [PubMed] [Google Scholar]
- [101].Cameli C, Bacchelli E, De Paola M, et al. Genetic variation in CHRNA7 and CHRFAM7A is associated with nicotine dependence and response to varenicline treatment. Eur. J. Hum. Genet. 2018;26:1824–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Scott Obach R, Reed-Hagen AE, Krueger SS, et al. Metabolism and disposition of varenicline, a selective α4β2 acetylcholine receptor partial agonist, in vivo and in vitro. Drug Metab. Dispos. 2006;34:121–130. [DOI] [PubMed] [Google Scholar]
- [103].Glatard A, Guidi M, Dobrinas M, et al. Influence of body weight and UGT2B7 polymorphism on varenicline exposure in a cohort of smokers from the general population. Eur. J. Clin. Pharmacol. 2019;75:939–949. [DOI] [PubMed] [Google Scholar]
- [104].Feng B, Obach R, Burstein A, et al. Effect of Human Renal Cationic Transporter Inhibition on the Pharmacokinetics of Varenicline, a New Therapy for Smoking Cessation: An In Vitro–In Vivo Study. Clin. Pharmacol. Ther. 2008;83:567–576. [DOI] [PubMed] [Google Scholar]
- [105].Faessel HM, Obach RS, Rollema H, et al. A review of the clinical pharmacokinetics and pharmacodynamics of Varenicline for smoking cessation. Clin. Pharmacokinet. Springer; September 30, 2010. p. 799–816. [DOI] [PubMed] [Google Scholar]
- [106].Bergen AW, Javitz HS, Krasnow R, et al. Organic Cation transporter variation and response to smoking Cessation therapies. Nicotine Tob. Res. 2014;16:1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Rocha Santos J, Tomaz PRX, Issa JS, et al. CHRNA4 rs1044396 is associated with smoking cessation in varenicline therapy. Front. Genet. 2015;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Saccone NL, Saccone SF, Hinrichs AL, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009;150:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Pianezza Michael L., Sellers RFT Edward M.. Nicotine metabolism defect reduces smoking. Nature. 1998;393:750. [DOI] [PubMed] [Google Scholar]
- [110].Benowitz NL. Nicotine addiction. Prim. Care - Clin. Off. Pract. 1999;26:611–631. [DOI] [PubMed] [Google Scholar]
- [111].Perez-Paramo YX, Chen G, Ashmore JH, et al. Nicotine-N’-oxidation by flavin monooxygenase enzymes. Cancer Epidemiol. Biomarkers Prev. 2019;28:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Park SB, Jacob P, Benowitz NL, et al. Stereoselective metabolism of (S)-(−)-nicotine in humans: Formation of trans-(S)-(−)-nicotine N-1’-oxide. Chem. Res. Toxicol. 1993;6:880–888. [DOI] [PubMed] [Google Scholar]
- [113].Nakajima M, Yoshihiko T, Noriaki K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab. Dispos. 1996;24:1212–1217. [PubMed] [Google Scholar]
- [114].Cashman JR, Park SB, Yang ZC, et al. Metabolism of nicotine by human liver microsomes: stereoselective formation of trans-nicotine N’-oxide. Chem. Res. Toxicol. 1992;5:639–646. [DOI] [PubMed] [Google Scholar]
- [115].Hecht SS, Hochalter BJ, Villalta PW, et al. 2’-Hydroxylation of nicotine by cytochrome P450 2A6 and human liver microsomes: Formation of a lung carcinogen precursor. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12493–12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Yamanaka H, Nakajima M, Fukami T, et al. CYP2A6 and CYP2B6 are involved in nornicotine formation from nicotine in humans: Interindividual differences in these contributions. Drug Metab. Dispos. 2005;33:1811–1818. [DOI] [PubMed] [Google Scholar]
- [117].Miki Nakajima, Toshinori Yamamoto, Ken-Ichi Nunoya, Tsuyoshi Yokoi, Kazuo Nagashima, Kazuaki Inoue, Yoshihiko Funae, Noriaki Shimada TK and YK. Characterization of CYP2A6 Involved in 3’-Hydroxylation of Cotinine in human Liver microsomes. Pharmacol. Exp. Ther. 1996;277:1010–1015. [PubMed] [Google Scholar]
- [118].Murphy SE, Johnson LM, Pullo DA. Characterization of multiple products of cytochrome P450 2A6-catalyzed cotinine metabolism. Chem. Res. Toxicol. 1999;12:639–645. [DOI] [PubMed] [Google Scholar]
- [119].Kuehl GE, Murphy SE. N-glucuronidation of nicotine and cotinine by human liver microsomes and heterologously expressed UDP-glucuronosyltransferases. Drug Metab. Dispos. 2003;31:1361–1368. [DOI] [PubMed] [Google Scholar]
- [120].Kaivosaari S, Toivonen P ivi, Hesse LM, et al. Nicotine Glucuronidation and the Human UDP-Glucuronosyltransferase UGT2B10. Mol. Pharmacol. 2007;72:761–768. [DOI] [PubMed] [Google Scholar]
- [121].Chen G, Blevins-Primeau AS, Dellinger RW, et al. Glucuronidation of Nicotine and Cotinine by UGT2B10 : Loss of Function by the UGT2B10 Codon 67 ( Asp > Tyr ) Polymorphism Function by the UGT2B10 Codon 67 ( Asp > Tyr ) Polymorphism. Cancer Res. 2007;67:9024–9029.* This paper is an example of a study describing a loss of function allele in an enzyme, UGT2B10, that plays an important role in the metabolism and detoxification of nicotine and cotinine.
- [122].Nakajima M, Tanaka E, Kwon JT, et al. Characterization of nicotine and cotinine N-glucuronidations in human liver microsomes. Drug Metab. Dispos. 2002;30:1484–1490. [DOI] [PubMed] [Google Scholar]
- [123].Chen G, Giambrone NE, Lazarus P. Glucuronidation of trans-3’-hydroxycotinine by UGT2B17 and UGT2B10. Pharmacogenet. Genomics. 2012;22:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yamanaka H, Nakajima M, Katoh M, et al. Trans-3′-hydroxycotinine O- and N-glucuronidations in human liver microsomes. Drug Metab. Dispos. 2005;33:23–30. [DOI] [PubMed] [Google Scholar]
- [125].Benowitz NL, Jacob P. Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin. Pharmacol. Ther. 1994;56:483–493. [DOI] [PubMed] [Google Scholar]
- [126].Kandel DB, Hu M-C, Schaffran C, et al. Urine Nicotine Metabolites and Smoking Behavior in a Multiracial/Multiethnic National Sample of Young Adults. Am J Epidemiol. 2007;165:901–910. [DOI] [PubMed] [Google Scholar]
- [127].Swan GE, Lessov-Schlaggar CN, Bergen AW, et al. Genetic and Environmental Influences on the Ratio of 3 ‘ Hydroxycotinine To Cotinine in Plasma and Urine. Pharmacogenet Genomics. 2009;19:388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Nakajima M, Kuroiwa Y, Yokoi T. Interindividual differences in nicotine metabolism and genetic polymorphisms of human CYP2A6. Drug Metab. Rev. 2002;34:865–877. [DOI] [PubMed] [Google Scholar]
- [129].Rangiah K, Hwang W-T, Mesaros C, et al. Nicotine exposure and metabolizer phenotypes from ana lysis of urinary nicotine and its 15 metabolites by LC–MS. Bioanalysis. 2011;3:745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Murphy SE, Park SSL, Thompson EF, et al. Nicotine N-glucuronidation relative to N-oxidation and C-oxidation and UGT2B10 genotype in five ethnic/racial groups. Carcinogenesis. 2014;35:2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Nakajima M, Yokoi T. Interindividual variability in nicotine metabolism: C-oxidation and glucuronidation. Drug Metab. Pharmacokinet. 2005;20:227–235. [DOI] [PubMed] [Google Scholar]
- [132].Yamanaka H, Nakajima M, Nishimura K, et al. Metabolic profile of nicotine in subjects whose CYP2A6 gene is deleted. Eur. J. Pharm. Sci. 2004;22:419–425. [DOI] [PubMed] [Google Scholar]
- [133].Murphy SE. Nicotine Metabolism and Smoking: Ethnic Differences in the Role of P450 2A6. Chem. Res. Toxicol. 2017;30:410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Murphy SE, Sipe CJ, Choi K, et al. Low cotinine glucuronidation results in higher serum and saliva cotinine in African American compared to White smokers. Cancer Epidemiol Biomarkers Prev. 2017;26:1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Benowitz NL, Pomerleau OF, Pomerleau CS, et al. Nicotine metabolite ratio as a predictor of cigarette consumption. Nicotine Tob. Res. 2003;5:621–624. [DOI] [PubMed] [Google Scholar]
- [136].Dempsey D, Tutka P, Iii PJ, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin. Pharmacol. Ther. 2004;450:64–72. [DOI] [PubMed] [Google Scholar]
- [137].Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J. Anal. Toxicol. 30:386–389. [DOI] [PubMed] [Google Scholar]
- [138].St Helen G, Jacob P, Benowitz NL. Stability of the nicotine metabolite ratio in smokers of progressively reduced nicotine content cigarettes. Nicotine Tob. Res. 2013;15:1939–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].St.Helen G, Novalen M, Heitjan DF, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol. Biomarkers Prev. 2012;21:1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Ashare RL, Thompson M, Leone F, et al. Differences in the rate of nicotine metabolism among smokers with and without HIV. AIDS. 2019;33:1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Wells QS, Freiberg MS, Greevy RA, et al. Nicotine metabolism-informed care for smoking cessation: A pilot precision RCT. Nicotine Tob. Res. 2018;20:1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Lerman C, Tyndale R, Patterson F, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clin. Pharmacol. Ther. 2006;79:600–608. [DOI] [PubMed] [Google Scholar]
- [143].Kaufmann A, Hitsman B, Goelz PM, et al. Addictive Behaviors Rate of Nicotine Metabolism and Smoking Cessation Outcomes in a Community-based Sample of Treatment-Seeking Smokers. Addict. Behav. 2015;51:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ho MK, Mwenifumbo JC, Al Koudsi N, et al. Association of nicotine metabolite ratio and CYP2A6 genotype with smoking cessation treatment in African-American light smokers. Clin. Pharmacol. Ther. 2009;85:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Shahab L, Bauld L, McNeill A, et al. Does the nicotine metabolite ratio moderate smoking cessation treatment outcomes in real-world settings? A prospective study. Addiction. 2019;114:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Terril L Verplaetse, MacKenzie R Peltier, Walter Roberts, Kelly E Moore, Brian P Pittman, SAM. Associations Between Nicotine Metabolite Ratio and Gender With Transitions in Cigarette Smoking Status and E-Cigarette Use: Findings Across Waves 1 and 2 of the Population Assessment of Tobacco and Health (PATH) Study. Nicotine Tob. Res. 2020;22:1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].West O, Hajek P, Mcrobbie H, et al. Systematic review of the relationship between the 3-hydroxycotinine/ cotinine ratio and cigarette dependence. Psychopharmacology (Berl). 2011;218:313–322. [DOI] [PubMed] [Google Scholar]
- [148].Johnstone E, Benowitz N, Cargill A, et al. Determinants of the rate of nicotine metabolism and effects on smoking behavior. Clin. Pharmacol. Ther. 2006;80:319–330. [DOI] [PubMed] [Google Scholar]
- [149].Shahab L, Mortimer E, Bauld L, et al. Characterising the nicotine metabolite ratio and its association with treatment choice: A cross sectional analysis of Stop Smoking Services in England. Sci. Rep. 2017;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Fix B V, O’Connor RJ, Benowitz N, et al. Nicotine Metabolite Ratio (NMR) Prospectively Predicts Smoking Relapse: Longitudinal Findings From ITC Surveys in Five Countries. Nicotine Tob. Res. 2017;19:1040–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Xue Y, Sun D, Daly A, et al. Adaptive Evolution of UGT2B17 Copy-Number Variation. Am. J. Hum. Genet. 2008;83:337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Zhu AZX, Zhou Q, Cox LS, et al. Variation in Trans-3’-Hydroxycotinine Glucuronidation Does Not Alter the Nicotine Metabolite Ratio or Nicotine Intake. PLoS One. 2013;8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Chenoweth MJ, Novalen M, LWH Jr, et al. Known and Novel Sources of Variability in the Nicotine Metabolite Ratio in a Large Sample of Treatment-Seeking Smokers. Cancer Epidemiol Biomarkers Prev. 2014;23:1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Benowitz N, Lessovschlaggar C, Swan G, et al. Female sex and oral contraceptive use accelerate nicotine metabolism. Clin. Pharmacol. Ther. 2006;79:480–488. [DOI] [PubMed] [Google Scholar]
- [155].Higashi E, Fukami T, Itoh M, et al. Human CYP2A6 Is Induced by Estrogen via Estrogen Receptor. DRUG Metab. Dispos. Drug Metab Dispos. 2007;35:1935–1941. [DOI] [PubMed] [Google Scholar]
- [156].Mwenifumbo JC, Sellers EM, Tyndale RF. Nicotine metabolism and CYP2A6 activity in a population of black African descent: Impact of gender and light smoking. Drug Alcohol Depend. 2007;89:24–33. [DOI] [PubMed] [Google Scholar]
- [157].Bowker K, Lewis S, Coleman T, et al. Changes in the rate of nicotine metabolism across pregnancy: A longitudinal study. Addiction. 2015;110:1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Allenby Cheyenne E., Boylan Kelly A., Lerman Caryn and MF, Allenby CE, Boylan KA, et al. Precision Medicine for Tobacco Dependence: Development and Validation of the Nicotine Metabolite Ratio. J Neuroimmune Pharmacol. 2016;11:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Mooney ME, Li Z-Z, Murphy SE, et al. Stability of the Nicotine Metabolite Ratio in ad Libitum and Reducing Smokers. Cancer Epidemiol Biomarkers Prev. 2008;17:1396–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].MacDougall JM, Fandrick K, Zhang X, et al. Inhibition of human liver microsomal (S)-nicotine oxidation by (−)-menthol and analogues. Chem. Res. Toxicol. 2003;16:988–993. [DOI] [PubMed] [Google Scholar]
- [161].Benowitz NL, Herrera B, Jacob P. Mentholated cigarette smoking inhibits nicotine metabolism. J. Pharmacol. Exp. Ther. 2004;310:1208–1215. [DOI] [PubMed] [Google Scholar]
- [162].Strasser AA, Benowitz NL, Pinto AG, et al. Nicotine metabolite ratio predicts smoking topography and carcinogen biomarker level. Cancer Epidemiol. Biomarkers Prev. 2011;20:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Gaedigk et al. 2018, CPT 103:399; Gaedigk et al. 2019 C 105:29. Pharmacogene Variation Consortium (PharmVar) [Internet]. [cited 2020 Nov 18]. Available from: https://www.pharmvar.org/.
- [164].Bloom J, Hinrichs AL, Wang JC, et al. The Contribution of Common CYP2A6 Alleles to Variation in Nicotine Metabolism Among European Americans. Pharmacogenet. Genomics. 2012;21:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Bloom AJ, Harari O, Martinez M, et al. A compensatory effect upon splicing results in normal function of the CYP2A6*14 allele. Pharmacogenet. Genomics. 2013;23:107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Yuan J-MM, Nelson HH, Carmella SG, et al. CYP2A6 genetic polymorphisms and biomarkers of tobacco smoke constituents in relation to risk of lung cancer in the Singapore Chinese Health Study. Carcinogenesis. 2017;38:411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Yuan J-MM, Nelson HH, Butler LM, et al. Genetic determinants of cytochrome P450 2A6 activity and biomarkers of tobacco smoke exposure in relation to risk of lung cancer development in the Shanghai cohort study. Int. J. Cancer. 2016;138:2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Pérez-Rubio G, López-Flores LA, Ramírez-Venegas A, et al. Genetic polymorphisms in CYP2A6 are associated with a risk of cigarette smoking and predispose to smoking at younger ages. Gene. 2017;628:205–210. [DOI] [PubMed] [Google Scholar]
- [169].Verde Z, Santiago C, González-Moro JMR, et al. “Smoking Genes”: A genetic association study. PLoS One. 2011;6:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Minematsu N, Nakamura H, Furuuchi M, et al. Limitation of cigarette consumption by CYP2A6*4, *7 and *9 polymorphisms. Eur. Respir. J. 2006;27:289–292. [DOI] [PubMed] [Google Scholar]
- [171].Xu C, Rao YS, Xu B, et al. An in vivo pilot study characterizing the new CYP2A6*7, *8, and *10 alleles. Biochem. Biophys. Res. Commun. 2002;290:318–324. [DOI] [PubMed] [Google Scholar]
- [172].Pitarque M, Von Richter O, Oke B, et al. Identification of a single nucleotide polymorphism in the TATA box of the CYP2A6 gene: Impairment of its promoter activity. Biochem. Biophys. Res. Commun. 2001;284:455–460. [DOI] [PubMed] [Google Scholar]
- [173].Yoshida R, Nakajima M, Nishimura K, et al. Effects of polymorphism in promoter region of human CYP2A6 gene (CYP2A6*9) on expression level of messenger ribonucleic acid and enzymatic activity in vivo and in vitro. Clin. Pharmacol. Ther. 2003;74:69–76. [DOI] [PubMed] [Google Scholar]
- [174].Oscarson M, McLellan RA, Asp V, et al. Characterization of a novel CYP2A7/CYP2A6 hybrid allele (CYP2A6*12) that causes reduced CYP2A6 activity. Hum. Mutat. 2002;20:275–283. [DOI] [PubMed] [Google Scholar]
- [175].Borrego-Soto G, Perez-Paramo YX, Chen G, et al. Genetic variants in CYP2A6 and UGT1A9 genes associated with urinary nicotine metabolites in young Mexican smokers. Pharmacogenomics J. 2020;20:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Piliguian M, Zhu AZX, Zhou Q, et al. Novel CYP2A6 variants identified in African Americans are associated with slow nicotine metabolism in vitro and in vivo. Pharmacogenet. Genomics. 2014;24:118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Chenoweth MJ, Ware JJ, Zhu AZX, et al. Genome-wide association study of a nicotine metabolism biomarker in African American smokers: impact of chromosome 19 genetic influences. Addiction. 2018;113:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Fukami T, Nakajima M, Yoshida R, et al. A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clin. Pharmacol. Ther. 2004;76:519–527. [DOI] [PubMed] [Google Scholar]
- [179].Ho MK, Mwenifumbo JC, Zhao B, et al. A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenet. Genomics. 2008;18:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Al Koudsi N, Ahluwalia JS, Lin S-K, et al. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. Pharmacogenomics J. 2009;9:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Baurley JW, Edlund CK, Pardamean CI, et al. Genome-wide association of the laboratory- based nicotine metabolite ratio in three ancestries. Nicotine Tob. Res. 2016;18:1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Yesha M. Patel, Sunghim L. Park, Younghun Han, Lynne R. Wilkens, Heike Bickebooller, Albert Rosenberger, Neil Caporaso, Maria Teresa Landi, Irene Br€uske, Angela Risch, Yongyue Wei, David C. Christiani, Paul Brennan, Richard Houlston, J and LLM, Patel YM, Park SL, et al. Novel Association of Genetic Markers Affecting CYP2A6 Activity and Lung Cancer Risk. Cancer Res. 2016;76:5768–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Loukola A, Buchwald J, Gupta R, et al. A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism. PLoS Genet. 2015;11:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Chenoweth MJ, Ware JJ, Zhu AZX, et al. Genome-wide association study of a nicotine metabolism biomarker in African American smokers: impact of chromosome 19 genetic influences. Addiction. 2018;113:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Buchwald J, Chenoweth MJ, Palviainen T, et al. Genome-wide association meta-analysis of nicotine metabolism and cigarette consumption measures in smokers of European descent. Mol. Psychiatry. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Al Koudsi N, Tyndale RF. Hepatic CYP2B6 is altered by genetic, physiologic, and environmental factors but plays little role in nicotine metabolism. Xenobiotica. 2010;40:381–392. [DOI] [PubMed] [Google Scholar]
- [187].Dicke KE, Skrlin SM, Murphy SE. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-butanone metabolism by cytochrome P450 2B6. Drug Metab. Dispos. 2005;33:1760–1764. [DOI] [PubMed] [Google Scholar]
- [188].Yamazaki H, Inoue K, Hashimoto M, et al. Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch. Toxicol. 1999;73:65–70. [DOI] [PubMed] [Google Scholar]
- [189].Bloom AJ, Martinez M, Chen LS, et al. CYP2B6 non-coding variation associated with smoking cessation is also associated with differences in allelic expression, splicing, and nicotine metabolism independent of common amino-acid changes. PLoS One. 2013;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Ring HZ, Valdes AM, Nishita DM, et al. Gene-gene interactions between CYP2B6 and CYP2A6 in nicotine metabolism. Pharmacogenet. Genomics. 2007;17:1007–1015. [DOI] [PubMed] [Google Scholar]
- [191].Bloom AJ, Wang PF, Kharasch ED. Nicotine oxidation by genetic variants of CYP2B6 and in human brain microsomes. Pharmacol. Res. Perspect. 2019;7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Taraneh Taghavi, Gideon St. Helen, Neal L. Benowitz RFT. Effect of UGT2B10, UGT2B17, FMO3, and OCT2 Genetic Variation on Nicotine and Cotinine Pharmacokinetics and Smoking in African Americans. Pharmacogenet Genomics. 2017;20:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Chenoweth MJ, Zhu AZX, Sanderson L, et al. Variation in P450 oxidoreductase ( POR ) A503V and flavin-containing monooxygenase ( FMO ) −3 E158K is associated with minor alterations in nicotine metabolism, but does not alter cigarette consumption. Pharmacogenet. Genomics. 2013;24:172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Bushueva O, Solodilova M, Churnosov M, et al. The Flavin-Containing Monooxygenase 3 Gene and Essential Hypertension: The Joint Effect of Polymorphism E158K and Cigarette Smoking on Disease Susceptibility. Int. J. Hypertens. 2014;2014:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Poetsch M, Czerwinski M, Wingenfeld L, et al. A common FMO3 polymorphism may amplify the effect of nicotine exposure in sudden infant death syndrome (SIDS). Int. J. Legal Med. 2010;124:301–306. [DOI] [PubMed] [Google Scholar]
- [196].Teitelbaum AM, Murphy SE, Akk G, et al. Nicotine dependence is associated with functional variation in FMO3, an enzyme that metabolizes nicotine in the brain. Pharmacogenomics J. 2017;1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Saccone SF, Saccone NL, Swan GE, et al. Systematic biological prioritization after a genome-wide association study: An application to nicotine dependence. Bioinformatics. 2008;24:1805–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Xu M, Bhatt DK, Yeung CK, et al. Genetic and Non-genetic Factors Associated with Protein Abundance of Flavin-containing Monooxygenase 3 in Human Liver. J. Pharmacol. Exp. Ther. 2017;363:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Bloom AJ, Murphy SE, Martinez M, et al. Effects upon in-vivo nicotine metabolism reveal functional variation in FMO3 associated with cigarette consumption. Pharmacogenet. Genomics. 2013;23:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Hinrichs AL, Murphy SE, Wang JC, et al. Common polymorphisms in FMO1 are associated with nicotine dependence. Pharmacogenet. Genomics. 2011;21:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Dolphin CT, Beckett DJ, Janmohamed A, et al. The flavin-containing monooxygenase 2 gene (FMO2) of humans, but not of other primates, encodes a truncated, nonfunctional protein. J. Biol. Chem. 1998;273:30599–30607. [DOI] [PubMed] [Google Scholar]
- [202].Bloom AJ, von Weymarn LB, Martinez M, et al. The contribution of common UGT2B10 and CYP2A6 alleles to variation in nicotine glucuronidation among European Americans. Pharmacogenet. Genomics. 2013;23:706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Berg JZ, Von Weymarn LB, Thompson EA, et al. UGT2B10 genotype influences nicotine glucuronidation, oxidation, and consumption. Cancer Epidemiol. Biomarkers Prev. 2010;19:1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Berg JZ, Mason J, Boettcher AJ, et al. Nicotine metabolism in African Americans and European Americans: Variation in glucuronidation by ethnicity and UGT2B10 haplotype. J. Pharmacol. Exp. Ther. 2010;332:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Patel YM, Stram DO, Wilkens LR, et al. The contribution of common genetic variation to nicotine and cotinine glucuronidation in multiple ethnic/racial populations. Cancer Epidemiol. Biomarkers Prev. 2015;24:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Chen G, Giambrone NE Jr, Dluzen DF, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: Importance of UGT2B10 and UGT2B17 knock-out polymorphisms. Cancer Res. 2010;70:7543–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Wassenaar CA, Conti DV., Das S, et al. UGT1A and UGT2B genetic variation alters nicotine and nitrosamine glucuronidation in European and African American smokers. Cancer Epidemiol. Biomarkers Prev. 2015;24:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Kaivosaari S, Finel M, Koskinen M. N-glucuronidation of drugs and other xenobiotics by human and animal UDP-glucuronosyltransferases. Xenobiotica. 2011;41:652–669. [DOI] [PubMed] [Google Scholar]
- [209].Ghotbi R, Mannheimer B, Aklillu E, et al. Carriers of the UGT1A4 142T>G gene variant are predisposed to reduced olanzapine exposure - An impact similar to male gender or smoking in schizophrenic patients. Eur. J. Clin. Pharmacol. 2010;66:465–474. [DOI] [PubMed] [Google Scholar]
- [210].Urakami Y, Okuda M, Masuda S, et al. Functional characteristics and membrane localization of rat multispecific organic cation transporters, OCT1 and OCT2, mediating tubular secretion of cationic drugs. J. Pharmacol. Exp. Ther. 1998;287:800–805. [PubMed] [Google Scholar]
- [211].Kuehn BM. Pilot programs seek to integrate genomic data into practice. JAMA - J. Am. Med. Assoc. 2017. August 1;318:410–412. [DOI] [PubMed] [Google Scholar]