Abstract
Site-specific recombinases (SSRs) are invaluable genome engineering tools that have enormously boosted our understanding of gene functions and cell lineage relationships in developmental biology, stem cell biology, regenerative medicine, and multiple diseases. However, the ever-increasing complexity of biomedical research requires the development of novel site-specific genetic recombination technologies that can manipulate genomic DNA with high efficiency and fine spatiotemporal control. Here, we review the latest innovative strategies of the commonly used Cre-loxP recombination system and its combinatorial strategies with other site-specific recombinase systems. We also highlight recent progress with a focus on the new generation of chemical- and light-inducible genetic systems and discuss the merits and limitations of each new and established system. Finally, we provide the future perspectives of combining various recombination systems or improving well-established site-specific genetic tools to achieve more efficient and precise spatiotemporal genetic manipulation.
Keywords: site-specific recombinase, site-specific recombination, Cre-loxP, Dre-rox, inducible, genome engineering, gene manipulation, lineage tracing, optogenetics, photoactivatable
Abbreviations: 4-OHT, 4-hydroxytamoxifen; AR, androgen receptor; BphP1, bacterial phytochrome; CIB1, a basic helix-loop-helix protein; CreC, the C-terminal domain of Cre; CreN, the N-terminal domain of Cre; CrexER, a switchable CreER system with the Cre-rox-ER-rox construct; CRY2, cryptochrome 2; DHT, dihydrotestosterone; Di-Cre, dimerizable Cre; Dox, doxycycline; dRap, light-cleavable rapamycin dimer; ER, estrogen receptor; FISC system, far-red light-induced split Cre-loxP system; GR, glucocorticoid receptor; HR, homologous recombination; Hsp, heat shock protein; iSuRe-Cre, Cre/CreERT2-inducible dual Reporter-Cre-expressing mouse allele; LBD, ligand binding domain; Li-rtTA, light activated rtTA; LightOn, the light-on system; nMag, negative magnet; PA-Cre, photoactivatable Cre recombinase; PIF, photochrome-interacting factor; PhyB, photoreceptor phytochrome B; pMag, positive magnet; PpsR2, the natural partner of BphP1; PR, progesterone receptor; Roxed-Cre, a sequential binary SSR system with the Cre-N-rox-stop-rox-Cre-C strategy; rtTA, reverse tet-controlled transactivator; sCreER, self-cleaved inducible CreER; SSR, site-specific recombinase; Tet-On system, tetracycline-inducible gene expression system; TRE, Tet responsive element; VVD, photoreceptor Vivid
Precise genome engineering is indispensable for biomedical research. Site-specific recombination enables conditional genetic modifications and serves as an efficient complement to gene manipulation from point mutation to large chromosomal rearrangement by homologous recombination (HR) in murine embryonic stem cells (1). Although the site-specific recombinases (SSRs) have revolutionized biomedical science, the application of SSRs was limited by several technical caveats. First, the efficiency of temporal- and spatial-controlled inducible recombination needs to be enhanced. Second, the expression of a recombination reporter cannot reliably indicate deletion of target gene. Third, conventional lineage tracing used for cell fate mapping relies entirely on cell type–specific known markers. Finally, novel strategies are required to induce gene expression and deletion regionally and promptly. All of these limitations could be circumvented with remarkable improvements and progress of SSRs technologies (2, 3, 4).
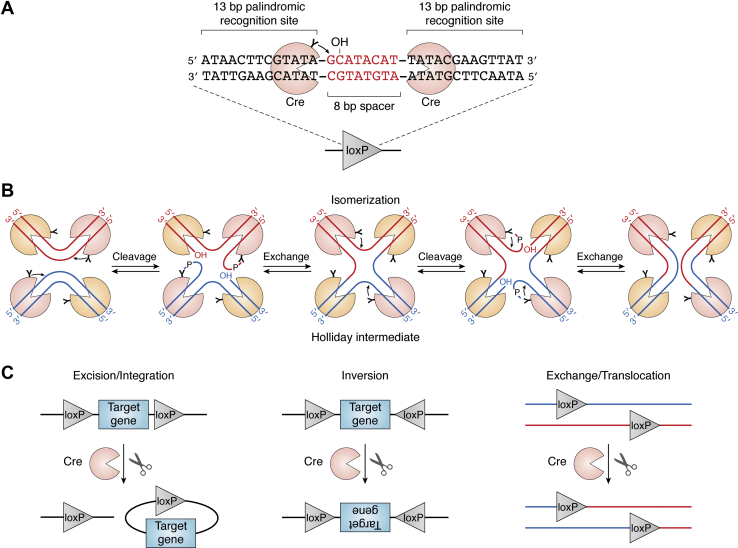
SSRs are invaluable genome engineering tools because of their exceptional ability to excise, integrate, inverse, and translocate genomic DNA in living organisms. The application of DNA site-specific recombination technologies in lineage tracing, gene activation and deletion, and cell lineage ablation have significantly advanced and refined our understanding of gene functions and cell behaviors in many biological processes (5). Cre (causes/cyclization recombination) recombinase is the most popular SSR and has been used extensively to manipulate many types of DNA such as linear, supercoiled, or circular in any cellular environment and a variety of organisms in vivo (6, 7, 8, 9). A site called loxP (locus of crossing over (x) in P1 bacteriophage) and a trans-acting function Cre are two components necessary for site-specific recombination (6). The loxP site is a 34 bp consensus sequence and consists of an 8 bp nonsymmetrical central region flanked by two 13 bp palindromic sequences. Cre recombinase is a 38 kD protein that catalyzes the recombination between two loxP recognition sites (Fig. 1A). Cre can recombine two loxP sites located on the same or different DNA strand.
Figure 1.
Schematic of the Cre-loxP recombination mechanism.A, the loxP site is a 34 bp consensus sequence and consists of a central 8 bp nonsymmetrical spacer flanked by two 13 bp palindromic recognition sites. Cre recombinase subunits bind each palindromic sequence and cleavage, exchange, and ligation DNA strand at the central spacer. The spacer provides the orientation of the loxP site. The nucleophilic tyrosine 324 in Cre recombinase attacks the phosphate forming a three phosphotyrosine bond and releasing a free 5′ OH. Cleavage position is indicated by arrow. B, a dimer of Cre subunits bind at the palindromic sequence of each loxP site. A tetrameric Cre structure arranges two loxP sites in an antiparallel fashion to stabilize a synaptic complex. Two opposite Cre subunits cleave and exchange one pair of strands. The released 5′ OH attacks the neighboring strand to form a Holliday junction intermediate. The second pair of Cre subunits is activated by an isomerization of the Holliday junction intermediate. The second pair of strands are cleaved and exchanged to complete the recombination and result in the recombinant products. C, different outcomes of a Cre-loxP recombination depending on the position and orientation of the two loxP sites. If the two loxP sites are in the same orientation, the recombination results in the excision or integration of the DNA segment (e.g., target gene) flanked by the two loxP sequences. If the orientation of loxP elements is in opposite, the result of the reaction is the inversion. Recombination between the two loxP sites located on different DNA strands results in the exchange or translocation.
The mechanism of Cre/loxP recombination is a multistep process. Cre recombinase catalyzes the DNA recombination reaction between loxP sites via a Holliday junction intermediate. A dimer of Cre recombinase subunits bind at the palindromic sequence of each loxP site. Two loxP sites are arranged in an antiparallel manner by a tetrameric Cre complex to form a “synapse”. The 8 bp nonsymmetrical central region provides the site of strand cleavage, exchange, and ligation. Two opposite active Cre recombinase subunits catalyze the cleavage and exchange of one pair of strands to produce a Holliday junction intermediate. The nucleophilic tyrosine 324 in Cre recombinase attacks the scissile phosphate forming a 3′ phosphototyrosine linkage and releasing a free 5′ OH. And strands exchange occurs when the free 5′ OH attacks the neighboring 3′ phosphotyrosine intermediate to form a Holliday junction intermediate. An isomerization of the Holliday junction induces the activation the second pair of Cre recombinase subunits that catalyze the same cleavage and exchange steps of the second pair of strands to result in the recombinant products (10, 11, 12) (Fig. 1B). Different outcomes of a Cre-loxP recombination depend on the position and orientation of the two loxP sites. If the loxP sequences have the same orientation, the recombination results in the excision of the DNA fragment flanked by the two loxP sequences. If the orientation of loxP elements is in opposite, the result of the reaction is the inversion of the DNA segment flanked by the two loxP sites. Recombination between the two loxP sites located on different DNA molecules produces strand exchange or translocation (10, 11) (Fig. 1C). The MGI (Mouse Genome Informatics, http://www.informatics.jax.org/home/recombinase) and IMPC (International Mouse Phenotype Consortium, https://www.mousephenotype.org) are the most comprehensive Cre database resources for academic community (13). To enhance the expression level of Cre recombinase in mammalian cells, the improved Cre was designed by reducing the high CpG content to constrain epigenetic silencing in mammals (14).
With the identification of novel recombinases, the diverse types of gene manipulation instruments are continuously increasing. Many other well-established recombinases, such as Flp (15), phiC31 (16), Dre (17), VCre, SCre (18), Vika (19), and Nigri (20), significantly enrich the genetic manipulation toolbox. In light of no cross-recombination among well-established SSRs, Cre and other analogous recombinases such as Flp, Dre, and Nigri have been successfully used in combination for some genetic strategies to achieve more precise and sophisticated experimental schemes that are required for addressing controversial or complicated biological issues (21, 22, 23, 24, 25, 26). The working principles and application guidelines of the new dual genetic approaches have been exhaustively described in other reviews (27, 28). Here, we review recent advances in more precise site-specific recombination systems that can control gene expression at a high spatiotemporal resolution, focusing on the new generation of chemical- and light-inducible recombination systems and discuss the merits and limitations of each new and established system that might be helpful to those considering the use of such strategies and technologies. The currently used and emerging recombination systems, which we review here, are summarized in Table 1.
Table 1.
An assessment of currently used and emerging recombination system
| Recombination systems | Requirements | Merits | Limitations | Improved production | Refs | |
|---|---|---|---|---|---|---|
| Chemical-inducible system | ||||||
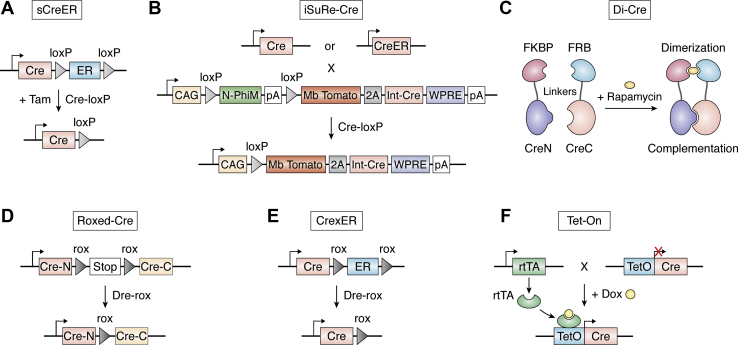
| sCreER | Tamoxifen induction | High efficiency; temporal control; reduced toxicity of tamoxifen | The efficiency of the initial CreER-loxP recombination-mediated switch of sCreER into Cre depends on the recombination efficiency of the loci where the sCreER knock-in. | Efficient and temporally controlled gene deletion for functional study. | (2) | |
| iSuRe-Cre | Cross w/other Cre/CreER lines; w/or w/o tamoxifen induction | High efficiency; temporal control; nontoxicity; no leakiness; reliably reports cells with gene deletion; compatible with the numerous existent loxP and Cre/CreERT2 alleles | Does not prevent the occurrence of gene-deletion false-negatives | Increasing the efficiency and reliability of Cre-dependent reporter and gene function analysis. | (3) | |
| Di-Cre | Rapamycin induction | Tight temporal control; rapid induction; low background Cre activity | Toxic during development; only can be used in adult animals | Tight temporal control of recombinase activity for conditional gene knock-out. | (47, 48) | |
| Roxed-Cre | Dual recombinases, Split-Cre and Dre | High-resolution recombination; ideal for sequential lineage tracing | A rox site remained in the coding sequence of Cre may decrease Cre activity; cannot be controlled temporally | The binary SSR system is ideal for sequential lineage tracing studies aimed at unraveling the relationships between cellular precursors and mature cell types. | (24) | |
| CrexER | Dual recombinases, Cre and Dre | Organ- or tissue-specific gene manipulation | Cannot be controlled temporally | The intersectional genetic system achieves both gene knockout and overexpression in vascular endothelial cells in an organ- or tissue-specific manner. | (25) | |
| Tet-On system | Genetically modified lines; Dox induction | Less toxic | Complexity of mice crossing; time-consuming | Temporal, spatial, and cell type-specific control of gene expression. | (49, 50, 51) | |
| Light-inducible system | ||||||
| System based on caged biomolecules | ||||||
| Photocaged 4-OHT or analogs | UV light illumination | Precise spatiotemporal control of gene expression | Cytotoxicity (DNA damage) by UV light; poor tissue penetration; limited recombination efficiency | Precise temporal- and location-specific control of CreER-mediated recombination in a light-dependent manner. | (61, 62, 63) | |
| Caged doxycycline/cyanodoxycycline | UV light or two-photon illumination | Precise spatial and temporal control of gene expression; the amount of UV light needed for induction is innocuous | Diffuse background fluorescence; low membrane permeability | High-resolution conditional transgene expression ranging from single cells to entire organisms. | (66) | |
| Near-IR uncaging strategy based on cyanine photochemistry | Near-IR light illumination | Cytocompatible; tissue penetration | Need to be validated in vivo | Spatial and temporal control of drug delivery. | (67) | |
| Light-cleavable dRap | UV light illumination | Simple to set up; temporal control of gene expression | Could not be readily placed under photochemical control | Protein dimerization induced by optically activated rapamycin dimer can be applied to control recombinase function. | (68) | |
| Photocaged Cre recombinase | Non-photodamaging UVA light illumination | Spatiotemporal control; background-free | Need to be validated in vivo | Tight spatiotemporal control of the activity of Cre recombinase and DNA recombination. | (70, 71) | |
| Genetically encoded system | ||||||
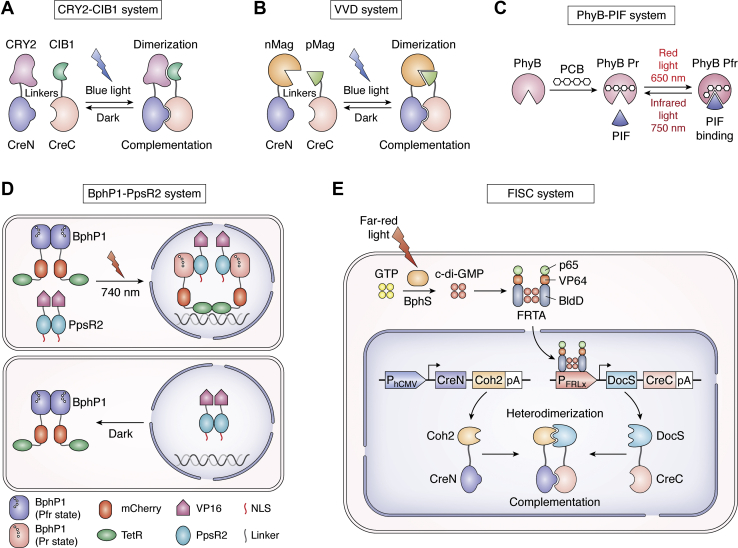
| CRY2-CIB1 system | PA-Cre 1.0 | Blue light illumination | Fast temporal; subcellular spatial resolution; reversible | Inefficient packaging; poor penetrative capacity | Fast temporal and spatial resolution without the need for exogenous cofactors. | (74) |
| PA-Cre 2.0 | Blue light illumination | High induced activity; low background; a single and brief light pulse; reduced light-mediated toxicity; reversible | Low recombination efficiency; need tuning nuclear import/export signals to reduce sensitivity to expression level differences to attain low background | Five-fold improved activity allowing precise spatial and temporal control of Cre recombinase ranging from single cells to whole organisms. | (92, 93) | |
| Li-rtTA | Dual induction of blue light and Dox | Reversible; spatiotemporal specific | Complexity of mice crossing; time-consuming | Genetical labeling and lineage tracing of multiple cell types in regional skin in a spatiotemporally specific manner. | (95) | |
| VVD system | LightOn | Blue light illumination | Simple and robust; spatiotemporal control; reversible | Poor penetrative capacity | A simple and robust system to quantitatively and spatiotemporally control gene expression and manipulate many biological processes in living cells and organisms. | (96) |
| Magnet | Blue light illumination | Low dimerization activity in the dark state; high spatiotemporal precision; reversible | Poor penetrative capacity | Spatially and temporally precise control over several signaling proteins in living mammalian cells with substantially enhanced dimerization efficiencies and accelerated switch-off kinetics. | (78) | |
| PA-Cre 3.0 | Blue light illumination | Reduced dark leak activity; improved efficiency; reversible | Poor penetrative capacity | Significantly addressed the issues of low recombination efficiency and dark leakiness. | (94) | |
| PhyB-PIF system | Red/far-red light illumination | Rapid stimulation and reversibility | Need for exogenous cofactor | Precisely and reversibly control gene expression and cell signaling. | (73, 100) | |
| BphP1-PpsR2 system | Near-IR light activation | Deep tissue penetration; low phototoxicity; tetracycline-independent | Minor interference with cellular metabolism | Targeting subcellular protein, inducing intracellular enzymatic activity, and activating gene expression with deep tissue penetration and low phototoxicity. | (101) | |
| FISC system | Far-red light excitation | High recombination efficiency; spatiotemporal precision; low background and photocytotoxicity; deep penetration capacity | Complexity; may require developing a vector with expanded packing capacity and small construct size to ensure efficient delivery in vivo | Precise control of genome engineering in target single cells or whole organisms in a spatiotemporal fashion with deep penetration, reduced toxicity, and invasiveness. | (102) | |
4-OHT, 4-hydroxytamoxifen; BphP1, bacterial phytochrome; CIB1, a basic helix-loop-helix protein; CrexER, a switchable CreER system with the Cre-rox-ER-rox construct; CRY2, cryptochrome 2; Di-Cre, dimerizable Cre; Dox, doxycycline; dRap, light-cleavable rapamycin dimer; FISC system, far-red light-induced split Cre-loxP system; iSuRe-Cre, a Cre/CreERT2-inducible dual Reporter-Cre-expressing mouse allele; Li-rtTA, light activated rtTA; LightOn, the light-on system; Near-IR, near-infrared; PA-Cre, photoactivatable Cre recombinase; PhyB, photoreceptor phytochrome B; PIF, photochrome-interacting factor; PpsR2, the natural partner of BphP1; Roxed-Cre, a sequential binary SSR system with the Cre-N-rox-stop-rox-Cre-C strategy; sCreER, self-cleaved inducible CreER with a Cre-loxP-ER-loxP construst; Tet-On system, tetracycline-inducible gene expression system; VVD, photoreceptor Vivid.
Chemical-inducible recombination systems
A conventional gene knockout, which eliminates a gene in the whole organism, may result in embryonic lethality in some cases, impeding the study of the gene’s function at later stages. This problem could be circumvented by using conditional gene targeting. By driving the expression of Cre recombinase under the control of a cell type- or tissue-specific promoter/enhancer, the conditional gene manipulation has been used for a tissue- or cell type-specific elimination of genes, fate mapping, and cell ablation. Special and temporal control of Cre activity is the key to successful conditional gene targeting. Given that the regulation of gene expression is dynamic over time, some Cre lines may target cells of interest at one time-point but in other undesired cells at a different stage. Some enhancers with constrained expression at embryonic stage may be reactivated in response to injury or stress in adult stage, confusing the results of lineage tracing. When tissue-specific gene knockout results in severe defects or embryonic lethality, spatial control of gene knockout alone may be insufficient to analyze gene function in postnatal period. Instead of constitutive/straight Cre lines, using an inducible Cre driven under a specific promoter could simultaneously achieve temporal and spatial control of DNA recombination in desired cell lineages (29, 30, 31).
Inducible system for Cre-mediated recombination
Some inducible systems for gene expression have been developed to control SSR activity temporally at the posttranslational level. In general, Cre or Flp recombinase is fused to a mutated ligand binding domain (LBD) of a nuclear receptor such as estrogen (ER), progesterone (PR), glucocorticoid (GR) or androgen (AR) receptors. These mutant LBDs can be activated by synthetic ligands but incapable of binding to physiological hormone. In the absence of synthetic agonists, the Cre fusion protein is sequestered in the cytoplasm by binding with heat shock protein (Hsp) and exhibits no recombinase activity in theory. Upon synthetic ligands administration, the confirmation of the receptor changes, leading to its release from Hsp. Activated Cre fusion protein translocate into the nucleus where Cre can execute recombination with loxP sites (1, 32). Tamoxifen and RU-486 are the most frequently used inducers for activating Cre-ER and Cre-PR, respectively (33, 34, 35). The Cre-ER and Cre-PR are the two most commonly used inducible systems of recombination. The inducers of the two systems anti-ER tamoxifen and anti-progestin RU486 (mifepristone) have side-effects on the health of pregnant mice and the development of embryos. Administration of high doses of tamoxifen can induce abortion in pregnant mice. Administration of tamoxifen to pregnant mice during early gestation can perturb embryonic development. RU486 had an all or none dose-dependent effect on fetoplacental development, resulting in either abortion or normal development of pregnancy. These facts should be taken into account when devising inducible SSR strategies (36, 37).
The first generation Cre-ER system, CreERTAM and Cre-ERT, was created by fusing of the Cre recombinase with a mutated LBD of the mouse and human ER (34, 38). Both the first generation Cre-ER systems required high tamoxifen levels to achieve sufficient induction. To avoid possible undesired tamoxifen-induced toxicity and abnormalities, the second generation Cre-ER system, Cre-ERT2, was constructed by the G400V/M543A/L544A triple mutation in the human ER LBD. The tamoxifen sensitivity of the Cre-ERT2 was ∼ 4-fold and ∼10-fold higher than that of Cre-ERT in cultured cells and engineered transgenic mice, respectively. Compared with Cre-ER, Cre-ERT2 is sensitive to low dosage of tamoxifen attaining reduced cytotoxicity and exhibits decreased background activity (leakiness) because it is not activated by natural mouse ER ligands (39). Cre-GRdex can be activated by synthetic ligands dexamethasone, triamcinolone acetonide, and RU38486 but not endogenous GR ligands corticosterone, cortisol, or aldosterone (40). Additionally, both dihydrotestosterone (DHT) and OH-flutamide are able to efficiently induce Cre-AR activity. DHT is a metabolite of testosterone. Both hormones are classified as ARs. DHT has similar but much stronger effects. Flutamide is in a class of medications called nonsteroidal antiandrogens. OH-flutamide is the active metabolite of flutamide. Both DHT and OH-flutamide are able to efficiently induce Cre-AR activity (41).
Induced activation of constitutive Cre for highly efficient gene manipulation
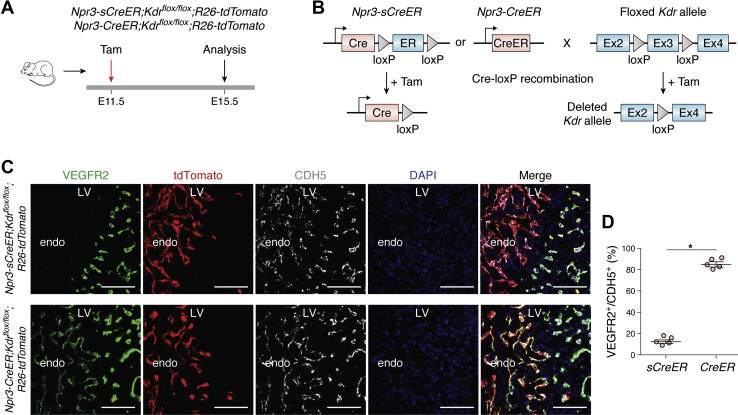
Despite that Cre activity can be controlled temporally and spatially by inducible recombination, inducible CreER often exhibits low efficiency in knockout of some genes. To enhance the efficiency of CreER, a self-cleaved inducible CreER (sCreER) with a Cre-loxP-ER-loxP system that switches inducible CreER into a constitutively active Cre by itself once tamoxifen induction has been generated (Fig. 2A). Compared with the conventional CreER, sCreER significantly improved the efficiency of genetic recombination of flox gene alleles that are inert for recombination. The advantage of sCreER over CreER is mainly reflected on genetic loci that are relatively inert or inaccessible for efficient recombination such as R26-Confetti and Kdrflox/flox alleles. Following one pulse of tamoxifen treatment, the efficiency of Kdr gene knockout with endocardial driver Npr3-sCreER is significantly higher than that with conventional Npr3-CreER (Fig. 3). In many cases, multiple times of tamoxifen administration have to be used for efficient gene deletion (42, 43), and long-term exposure to high-dose tamoxifen may be toxic to mice. sCreER alleviates the toxicity of tamoxifen, as only one dose is needed and allows efficient and temporally controlled gene deletion. The limitation of sCreER is the efficiency of sCreER switch into a constitutively active Cre mediated its initial CreER-loxP recombination. The efficiency of gene deletion may be affected when sCreER knock in some loci with low recombination efficiency. Even if the cells with recombination are of a limited amount, sCreER switch into Cre can be detected by reporter. A combination of temporal control and constitutive Cre activity makes sCreER suitable for lineage tracing and gene deletion simultaneously (2). Many studies have displayed that expression of a given Cre-loxP recombination reporter cannot reliably indicate deletion of target gene. It is unreliable to assume correlation between recombination of the reporter and the target allele because there is no genetic linkage between the reporter allele and other allele in the cell. To alleviate this problem, a Cre/CreERT2-inducible dual reporter-Cre-expressing mouse allele, iSuRe-Cre, has been generated. After Cre-loxP recombination, this new genetic tool co-expresses a fluorescent reporter and a constitutively active Cre, ensuring the efficiency gene deletion in reporter-expressing cells (Fig. 2B). In addition, the iSuRe-Cre is not leaky in the male germline and nontoxic. Fernández-Chacón et al. generated transgenic (Tg(iSuRe-Cre)) and ROSA26 targeted (Gt(ROSA)26Sor-iSuRe-Cre) mice and found that the Tg(iSuRe-Cre) transgenic allele is not leaky in the male germline. The Tg(iSuRe-Cre) transgenic allele is expressed and inducible in most tissues and is compatible with numerous existing Cre/CreERT2 and floxed mouse lines and significantly increases Cre activity in reporter-expressing cells. In theory, high Cre expression levels may result in cellular toxicity. However, the constitutive expression of Cre from the iSuRe-Cre is nontoxic, which is concluded after assessing potential toxicity related to high Cre expression. Gene-deletion false-negatives occur minimally with the iSuRe-Cre or other conventional Cre-reporters. Some reporter-negative cells may present deletion of other floxed genes that are easier to recombine (3).
Figure 2.
Chemical-inducible recombination systems.A, a self-cleaved inducible CreER (sCreER) with a Cre-loxP-ER-loxP construct that switches inducible CreER into a constitutively active Cre by itself once tamoxifen induction. B, the iSuRe-Cre is an inducible dual reporter-Cre, containing CAG promoter, N-PhiM and MbTomato reporter gene, and a constitutively active and permanently expressed Cre. After removal of the floxed N-PhiM cassette by Cre or CreER, iSuRe-Cre co-expresses MbTomato and a constitutively active Cre and significantly increases the efficiency and certainty of gene deletion in reporter-expressing cells. C, the DiCre in which Cre is split into two inactive moieties and fused with FKBP12 and FRB respectively. FKBP12 and FRB can be heterodimerized efficiently after rapamycin treatment, leading to the complementation of inactive fragments (CreN and CreC) and Cre activity restoration. D, the Roxed-Cre contains a Cre-N-rox-stop-rox-Cre-C construct. The reinstatement of Cre activity occurs after the removal of the rox-flanked STOP cassette by Dre-rox recombination. E, the CrexER with a Cre-rox-ER-rox construct executes Cre activity after the removal of ER by Dre-rox recombination. F, the Tet-On inducible system consists of two transgenes, a recombinase under the control of a TRE and a rtTA driven by a cell-specific promoter. After Dox administration, the rtTA is active and interacts with the TetO promoter. CreC, the C-terminal domain of Cre; CreN, the N-terminal domain of Cre; DiCre, dimerizable Cre; Dox, doxycycline; FKBP12, FK506-binding protein; FRB, binding domain of the FKBP12-rapamycin associated protein; rtTA, reverse tet-controlled transactivator; Tam, tamoxifen; TRE, Tet responsive element.
Figure 3.
Enhanced efficiency of gene knockout by sCreER, compared with conventional CreER.A, schematic figure showing experimental design. B, schematic diagram showing Kdr gene deletion by sCreER and CreER. C and D, immunostaining for tdTomato, VEGFR2, and CDH5 on heart sections from E15.5 Npr3-sCreER;Kdrflox/flox; R26-tdTomato and Npr3-CreER;Kdrflox/flox; R26-tdTomato mouse embryos. Quantification data showed the percentage of VEGFR2+ endocardial cells in CDH5+ endothelial cells in the inner myocardial wall. ∗p < 0.05; Data are mean ± SEM; n = 5. Scale bar, 50 μm. Each image is a representative of five individual mouse samples. endo, endocardium; Ex, exon; LV, left venticle; sCreER, self-cleaved inducible CreER; Tam, tamoxifen.
Dual promoter-mediated Cre activity for more precise genetic targeting
When using Cre-loxP recombination system for cell labeling and tracing, one need to vigilantly evaluate if the Cre-driving promoter also expresses in the undesired cell type. To more precisely target a specific cell population, a split-Cre approach has been designed to ensure expression of the Cre-driving promoter in the target cell type. In this strategy, two fragments of Cre recombinase coding sequence, the N terminus of Cre (Cre-N) and the C terminus of Cre (Cre-C), were driven by two independent promoters separately. Only when the two gene promoters are overlapped can the split-Cre be active in the cells of interest (44). An intein peptide that catalyzes self-excision and rejoins the flanking peptides was combined with split-Cre (Int-N and Int-C fragments) to increase the efficiency of Cre-N and Cre-C (45). This split-Cre approach has been utilized to trace adult neural stem cells (46). Jullien et al. (47) developed DiCre (dimerizable Cre) in which Cre is split into two inactive moieties and fused with FKBP12 (FK506-binding protein) and FRB (binding domain of the FKBP12-rapamycin associated protein), respectively. Following rapamycin treatment, FKBP12 and FRB can be heterodimerized efficiently, leading to the complementation of inactive fragments and the reconstitution of Cre activity (Fig. 2C). Given that rapamycin is toxic during embryonic and neonatal stage, it could be used cautiously as an inducer of DiCre in adults. The advantages of Di-Cre are tight temporal control, rapid induction, and low background Cre activity. The mechanism of action of the Di-Cre is complex. It involves a highly order protein–DNA complex between the two loxP sites, four enzymes interacting with each other, and a series of cleavage-religation steps (48). A sequential binary SSR system based on Split-Cre has been developed, with the Cre-N-rox-stop-rox-Cre-C (Roxed-Cre) strategy. In the Roxed-Cre system, the reinstatement of Cre activity occurs after the removal of the rox-flanked STOP cassette by Dre-rox recombination. In this system, the SSRs Dre and Cre are linked in a cascade that can be expressed from two individual tissue-specific promoters. The Roxed-Cre system has great potential for robust high-resolution recombination of conditional alleles and sequential lineage tracing studies on cell subpopulations. But a rox site remained in the coding sequence of Cre may decrease the activity of Cre recombinase (24) (Fig. 2D). Based on sequential intersectional recombination, a new switchable CreER system with the Cre-rox-ER-rox (CrexER) strategy was used for more precise and organ- or tissue-specific gene manipulation. In the CrexER system, Cre executes its activity after ER is removed by Dre-rox recombination (Fig. 2E). The fundamental principle of CrexER is the same as CreER, CrexER fusion protein is retained in the cytoplasm by binding with Hsp. Cre is released and translocated from the cytoplasm to nuclei to execute its activity after ER is removed by Dre-rox recombination. Taking advantage of the CrexER system, coronary or brain-specific vascular Cre lines were generated to achieve gene deletion or overexpression in coronary or brain vessels specifically and efficiently. Cre activity could be controlled temporally by using DreER and rox-stop-rox-Cre instead of Dre and CrexER. The limitation of CrexER is that Cre activity lacks temporal control because of constitutive Dre expression. This problem can be resolved using DreER and rox-stop-rox-Cre instead of Dre and CrexER (25).
Tet-regulated system for controlling Cre activity
An alternative approach of temporal control of gene expression is to regulate SSR activity at the transcriptional level. Transcriptional control approaches are based on the tetracycline (tet)-inducible binary system, which contains two transgenes, a recombinase under the control of a Tet responsive element and a tet-controlled transactivator (tTA) or “reverse” tet-controlled transactivator (rtTA) driven by a cell type- or tissue-specific promoter. Following tetracycline derivative doxycycline (Dox) induction, the tTA and rtTA are inactive and active, respectively. In the “Tet-off” or tTA system, transgene expression and the initiation of recombination are activated by removal of Dox. The prompt induction of gene deletion hinges on the rate of Dox clearance. Moreover, continuous long-term Dox administration may have side-effects on gene expression and organ function. Using a rtTA or “Tet-on” system (Fig. 2F) can minimize the disadvantages of a tTA or “Tet-off” system. Although Dox is less toxic than tamoxifen to embryos, the tet-on/off system requires the combination of three separate alleles, tissue-specific tTA/rtTA, Tet responsive element-Cre, and a floxed allele, thus increasing the complexity of mice crossing and time cost (49, 50, 51). The Dox inducible tet-on/off system has been successfully established and applied in various tissues and organs, such as heart (52), kidney (53, 54), neurons (55), intestinal epithelium (56), liver (51), and lung (57).
Light-inducible recombination systems
Light-inducible recombination systems based on caged biomolecules
During the past decades, chemical-inducible recombination systems have been extensively applied for temporal control of genome engineering in vivo. However, because chemical inducers diffuse freely and are hard to remove immediately, it is challenging to induce gene expression and deletion regionally and promptly. Some disadvantages of the chemical-inducible systems including leakiness, cytotoxicity, and potential off-target recombination constrain them to attain a high spatiotemporal gene manipulation (58, 59, 60). Based on UV, blue, or far-red light illumination, various optogenetics strategies were developed to enable genome engineering with high spatiotemporal precision.
Photoresponsive chemically caged inducers were the first optogenetic tools (61). Caging groups can be installed in a key position of chemical inducers or the catalytic site of Cre recombinase to render the biomolecules in an inactive state. The photocaged chemical inducers and caged Cre enzyme reactivation following the caging group is removed by UV light illumination. With covalent-modifying photocaged analog of 4-hydroxytamoxifen and the ligand-dependent recombinase Cre-ERT, gene expression can be permanently switched on or off by UV light activated recombination in cell culture systems (62). A caged tamoxifen analog 4-hydroxycyclofen was synthesized and applied to control CreER-mediated recombination upon light illumination in cell culture, organoid culture, and in adult mice (63). Inlay et al. designed photocaged tamoxifen or 4-hydroxytamoxifen (4-OHT) through covalent attachment to a photocleavable ortho-nitrobenzyl group. The free tamoxifen or 4-OHT release is attained after UV illumination, leading to Cre-mediated genetic recombination. In summary, the photocaged 4-OHT or analogs can be applied to achieve light-inducible precise spatiotemporal control of gene expression. However, there are some limitations of 4-OHT–based photocaging technology such as cytotoxicity (DNA damage) by UV light, poor tissue penetration, multiple irradiations, and limited recombination efficiency (64, 65). Based on Tet-on system, caged doxycycline derivatives were generated for precise spatiotemporal control of gene expression ranging from a single cell to organisms. As no signs of toxicity was detected, the amount of UV light needed for induction is innocuous. There was diffuse background fluorescence that could be discernible from the more intense signal. Because caged cyanodoxycycline has a low membrane permeability, it requires longer incubation times for full transgene expression after UV light illumination (66). Because the near-infrared (IR) light is cytocompatible and has significant tissue penetration, a near-IR uncaging strategy based on cyanine photochemistry has been designed to uncage small molecules. Gene expression can be controlled with near-IR light in CreERT/LoxP-reporter cell line derived from transgenic mice. Because near-IR light is cytocompatible and tissue penetrant, this uncaging strategy enable specific delivery of small molecules. Future applications of this near-IR uncaging strategy in complicated physiological settings need to validate in vivo (67). Brown et al. developed a new caged rapamycin, the light-cleavable rapamycin dimer (dRap) that induces FKBP12 and FRB heterodimerization through UV illumination. By fusing N terminus and C terminus of Cre to FKBP12 and FRB, respectively, Brown et al. achieved the photochemical control of Cre-catalyzed recombination by dRap. The light-cleavable rapamycin dimer dRaP is compatible with natural FKBP12 without the need for protein engineering of the FKBP domain. The synthesis of rapamycin dimer dRaP is simple in just two steps. The dRap can be applied to control biological processes that have responses to rapamycin but could not be readily placed under photochemical control (68).
A caging group was installed on the catalytic site of Cre to inhibit Cre activity and recombination. Exposure to nonphotodamaging UVA light removes the caging group and restores recombinase activity, thereby enabling tight spatiotemporal control of gene function is achieved in mammalian cell. To inhibit Cre activity, a light-responsive o-nitrobenzyl caging group was installed in the catalytic site of Cre. Upon UVA light triggering the removal of the caging group, Cre activity restores (69). Through the site-specific incorporation of photocaged tyrosine (ONBY) and photocaged lysine (PCK) into proteins in mammalian cells, Luo et al. developed two light-activated Cre recombinases. Photocaging provides highly stringent spatiotemporal and background-free control with high Cre recombinase activity. Additionally, this light-activated Cre recombinase significantly improved the efficiency of DNA recombination. Using photocaged Cre/loxP system to generate knock-in or knock-out organisms needs to be validated (70). Caging is a form of photoreversible chemical modification that has been used in the light-mediated activation of molecules (71). Applications of caging groups can be divided into two types: photocaged chemical inducers and caged Cre enzyme. Caging groups can be installed in a key position of chemical inducers or the catalytic site of Cre recombinase to render the biomolecules in an inactive state. The photocaged chemical inducers and caged Cre enzyme reactivation following the caging group is removed by UV light illumination. Application of photocaged biomolecules is limited by the difficulties associated with introducing these caged biomolecules into multicellular systems. The caged systems require the introduction of exogenous caged molecules and UV light illumination to trigger uncaging, yet UV can result in cytotoxicity by direct DNA damage. Also, the free diffusion of caged chemical inducers could lead to off-target effects, and limited recombination efficiency was detected following light illumination (62).
Genetically encoded light-inducible recombination systems
Various genetically encoded light-inducible Cre-loxP systems have been generated, making use of a variety of photoreceptors and split Cre recombinase. An array of optical dimerizer systems have been developed based on various light-sensing components: phytochromes (72, 73, 74), cryptochromes, light oxygen voltage (LOV domains) (75, 76, 77), Vivid (VVD) (78, 79), UVR8 (61, 80, 81), and EL222 (82) derived from prokaryotes, fungi, and plants. The cryptochrome 2 (CRY2–CIB1[basic helix-loop-helix protein]) and VVD (Magnet) systems are the most widely used optical dimerizers.
Generation and development of photoactivatable Cre recombinase
In 2010, Kennedy et al. developed genetically encoded blue light–inducible dimerization modules based on Arabidopsis thaliana CRY2 and CIB1 that require no exogenous cofactors. CRY2 and the N terminus of CIB1 were respectively fused to the N-terminal and C-terminal domain of Cre. Upon the illumination of blue light, the dimerization of CRY2 and N terminus of CIB1 leads to the reconstitution of split Cre recombinase activity. This is the first generation photoactivatable Cre recombinase called photoactivatable DNA recombinase, photoactivatable Cre recombinase (PA-Cre) 1.0. This system is fast temporal and subcellular spatial resolution without the need for exogenous chromophore. The CRY-CIB system could be improved by generating minimal domains for efficient packaging in viral vectors and constructing modules with modified dissociation kinetics and light sensitivities. Further optimization of the constructs of CRY-CIB modules may improve Cre recombinase–mediated DNA recombination efficiency (74) (Fig. 4A).
Figure 4.
Genetically encoded light-inducible recombination systems.A, CRY2 and the N terminus of CIB1 are fused to the CreN and CreC, respectively. Upon the illumination of blue light, the dimerization of CRY2 and CIBN leads to the reconstitution of split Cre recombinase activity. B, the photoreceptor Vivid (VVD) is designated as Magnets and comprises two photoswitches named pMag and nMag. The heterodimerization of pMag and nMag induced by the blue light illumination leads to the complementation of CreN and CreC and Cre activity reconstitution. C, the PhyB absorbs red and infrared light through the photoisomerization of a covalently bound PCB. The conformation of PhyB changes between the Pr (red-absorbing) and Pfr (far-red-absorbing) states catalyzed by red and infrared light. The PIF only associates with PhyB in Pfr state. The heterodimerization between PhyB and PIF is reversibly triggered by red (650 nm) and infrared (750 nm) light. D, a light-inducible transcription activation system based on BphP1-PpsR2 and TetRtetO. BphP1-mCherry and the C terminus of NLS-PpsR2 are fused to the TetR and VP16, respectively. Upon near-infrared light illumination, BphP1 converts into the Pr state and forms heterodimer with PpsR2. NLS facilitates the heterodimer translocates to the nucleus where BphP1 fusions interact with tetO DNA repeats via TetR. VP16 recruits the transcription initiation complex and triggers gene transcription. E, the FISC system is designed on the basis of the affinity of bacteriophytochrome Coh2 and DocS. In this system, DocS-CreC fusion protein is under the control of FRL-inducible promoter PFRL, CreN fused to Coh2 driven by a constitutive promoter PhCMV. Upon FRL exposure, the active photoreceptor BphS converts GTP into c-di-GMP which induces binding of FRTA (p65-VP64-BldD) to promoter PFRL to drive DocS-CreC expression. The interaction of Coh2 and DocS domains leads to the reunion of CreC and CreN and Cre activity reinstatement. c-di-GMP, cyclic diguanylate monophosphate; CreC, the C-terminal domain of Cre; CreN, the N-terminal domain of Cre; CRY2, cryptochrome 2; FISC system, far-red light-induced split Cre-loxP system; FRL, far-red light; FRTA, far-red light-dependent transactivator; GTP, guanylate triphosphate; NLS, nuclear localization signal; nMag, negative Magnet; PCB, chromophore phycocyanobilin; PhyB, photoreceptor phytochrome B; PIF, phytochrome interaction factor; pMag, positive Magnet; TetR, tetracycline repressor.
Integrating the customizable TALE DNA-binding domain with CRY2-CIB1, Konermann et al. (83) developed light-inducible transcriptional effectors to achieve optical control mammalian endogenous transcription and epigenetics states. By fusing CRY2 and CIB1 to the transactivation domain and the catalytically inactive form of Cas9, clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9–based photoactivatable systems have been established to control precise endogenous gene activation in mammalian cells (84, 85). In addition, the CRY2-CIB system has been used to optically regulate phosphoinositide levels (86, 87), cytoskeletal dynamics, and organelle distribution (88, 89). Combining Brainbow gene cassette encoding tricolor fluorescent proteins with PA-Cre, Boulina et al. (90) achieved live imaging of multicolor-labeled cells in Drosophila. Schindler et al. (91) demonstrated that PA-Cre recombinase can be activated by illumination from optical fibers or a two-photon microscope and permanently modify gene expression in the mouse brain.
Although CRY2-CIB optical dimerizer system has been extensively used as optogenetic tool, some limitations such as large size and dark background exist. Substitution of L348 F mutation in this system termed PA-Cre 1.5 with minimal background in dark, high induced activity, and sensitive response to a single brief light pulse. Using the long-lived L348F photocycle variant and truncated CRY2 and CIB constructs, Taslimi et al. generated an improved second-generation photoactivatable Cre DNA recombinase, PA-Cre 2.0. The PA-Cre 2.0 system exhibits minimal dark background and achieves 5-fold higher levels of activity than PA-Cre 1.0 (92). Meador et al. functionally characterized PA-Cre 2.0 in cultured cells and in vivo in rodent brain. The PA-Cre 2.0 system shows high induced activity, low background, and sensitive response to a single and shorter light pulse in vitro and in vivo. The low recombination efficiency of the PA-Cre based on the CRY2-CIB1 split Cre system is unsatisfactory. The low dark background of PA-Cre2.0 can be attained by tuning nuclear-cytosolic shuttling to reduce sensitivity to expression level differences. Combining two inducible systems, chemical-and-light dual regulated system has been designed and generated to achieve tight control of photoactivatable Cre recombinase gene switch (93).
By re-engineering the homodimer interface, Kawano et al. designed two distinct VVD variants, which is designated as Magnets. This blue light–dependent dimerization system comprises two photoswitches named positive Magnet (pMag) and negative Magnet (nMag). The synergistic activation of two photosensory units prevents dimerization in the dark state. The Magnets provide a powerful tool for spatiotemporal control of protein activities and related cellular functions (78). Based on the reconstitution of split Cre fragments induced by the blue light–dependent heterodimerization of the Magnet system, Kawano et al. (4) further succeeded in developing a genetically encoded photoactivatable Cre recombinase called PA-Cre to optogenetically control highly efficient induction of DNA recombination with high spatiotemporal precision in vivo (Fig. 4B). To reduce the unintentional Cre-loxP dark leak recombination, Morikawa et al. developed an improved version of Magnets-based PA-Cre called PA-Cre 3.0. A CAG promoter, Magnets, and 2A self-cleaving peptide have been used to optimize the expression of Magnets-based PA-Cre protein. Background recombination has been significantly reduced by codon modification in the dimerization domains. Compared with other photoreceptors-based PA-Cre systems, PA-Cre 3.0 have advantages with minimizing dark leakiness and high recombination efficiency. As new genetic tools, the PA-Cre 3.0 mouse lines and AAV-PA-Cre 3.0 could be applied to genetic study by spatiotemporal control of Cre-loxP recombination. The low penetrative capacity of blue light limits the application of the PA-Cre system in vivo (94).
Iteration of light-inducible recombination systems
By fusing the two functional domains of conventional rtTA to the light-sensitive proteins CRY2 and CIB1 separately, a novel light activated rtTA (Li-rtTA) system has been designed. Following the dual induction of blue light and Dox, the Li-rtTA system enables to activate and delete genes in a spatiotemporally specific fashion, e.g., in the regional skin of mice. There are several advantages of the Li-trTA system, such as tight spatiotemporal specific gene manipulation, many available existing genetic tools, and reversible activation of gene expression. The breeding scheme of Li-rtTA system requires to get both TetO-Cre and Li-rtTA alleles combined with the conditional gain or loss of function allele of interest. It is complex and time-consuming (95). Although the genetically encoded blue light–inducible CRY2-CIB1 split Cre system is noninvasive and requires no UV light illumination, its recombination efficiency still has much room to improve.
A blue light photoreceptor VVD derived from the filamentous fungus Neurospora crassa is one of the smallest photoreceptors and requires flavin adenine dinucleotide as its cofactor. Upon exposure to blue light, VVD undergoes switching from a monomer to a homodimer (79). Based on a genetically encoded light-switchable transactivator containing Gal4–VVD fusion protein, Wang et al. developed a simple and robust LightOn system that could be used to control gene expression spatiotemporally in mammalian cells and mice. The LightOn has low background expression, low toxicity, low interference with normal cellular function, and allows reversibility. Because only a single, genetically encoded photosensitive transactivator is required, the LightOn system is simple and easy to manipulate (96). Yao et al. generated light-inducible VVD-based Cre, Dre, and Flp recombinase system denoted as RecV. After inducing by one-photon or two-photon light illumination, RecV enables spatiotemporally precise optogenomic modifications of single cells or cell populations in mouse and zebrafish (97).
The inadequate transmittance of visible light through opaque tissues constrains the application of optogenetic approaches. The limitation could be broken through by using longer wavelength light. The far-red and near-infrared light spectrum within a spectral region (∼700–900 nm) can penetrate through deeper tissues or visceral organs (98, 99). The red/far-red light-switchable interaction of the photoreceptor phytochrome B (PhyB) and the photochrome-interacting factor (PIF) from A. thaliana was optimized to control gene expression at fine spatial and temporal resolutions in mammalian cells. Because using red/far-red light as an inducer, the PhyB-PIF system is rapidly reversible, precisely adjustable, nontoxic, space- and time-resolved, and has a low background expression. The PhyB-PIF system requires an exogenous phycocyanobilin chromophore or several bacterial enzymes producing chromophore. Because these exogenous proteins interfere with cellular metabolism, it limits the application of the PhyB-PIF system (73, 100) (Fig. 4C). A near-infrared light mediated optogenetic system based on the heterodimerization of bacterial phytochrome, BphP1, and its partner PpsR2 has been generated. This BphP-PpsR2 system enables activating gene expression in vivo and a signaling pathway in vitro. The BphP-PpsR2 system sensing near-infrared light is preferable for in vivo application because of its deep tissue penetration and low phototoxicity. This system is orthogonal to mammalian cells and minimally affects cellular metabolism (101) (Fig. 4D). More recently, based on the affinity of bacteriophytochrome Coh2 and DocS and split-Cre recombinase, a far-red light-induced split Cre-loxP system (FISC system) has been designed and enables efficient and precise control of DNA recombination in mammalian cells and in liver of mice. In the FISC system, the expression of the DocS-CreC fusion protein is under the control of far-red light–inducible promoter PFRL, CreN fused to Coh2 driven by a constitutive promoter PhCMV. Upon far-red light illumination, the interaction of Coh2 and DocS domains leads to the complementation of CreC and CreN and Cre activity reconstitution (Fig. 4E). In comparison to the UV and blue light–induced systems, the main advantages of the FISC system are low background and photocytotoxicity, deep penetration capacity, and high recombination efficiency with spatiotemporal precision. Owing to the packaging size limitation of AAV viral vectors, the FITC system was assigned in three separate AAV vectors for in vivo delivery. This obviously affects the future biomedical application of the FITC system. The FISC system still needs to be improved by developing a vector with expanded packing capacity and small construct size to ensure efficient delivery in vivo (102).
Comparison of the advantages and disadvantages of SSR and CRISPR-Cas9 system
SSR and CRISPR-Cas9 are the two ubiquitously used systems to achieve genome editing. SSRs are most suitable for insertion, deletion, or rearrangement of large fragments (e.g., hundreds or thousands of base pairs), rather than exquisitely targeted modifications such as point mutations. The off-site activity of SSRs is very low because they could tolerate minor variations of the site sequence. Under optimized conditions, the SSRs can complete site-specific recombination fast and efficiently in vivo and in vitro. Site-specific recombination requires only the SSRs and the two target sites. No other cofactors are required. In summary, the advantages of SSRs are specificity, simplicity, high efficiency, and fidelity. Nevertheless, there are some disadvantages such as toxicity, inefficiency, and off-target activity that can limit the applications of SSRs. Low levels of off-target recombination can be promoted by SSR such as Phi31 recombinase at a number of “pseudosites”. Low level but genome-wide off-target recombination could lead to manifold potential problems such as mutation, insertion, or deletion and failure of chromosome to segregate properly. SSRs expression in vivo can be toxic or even lethal. Persistence of recombination intermediates with strand break and DNA damage caused by covalent SSR-DNA linkages might also result in toxicity. Recombination reaction may be slow or incomplete because of inherent nature of the SSR or incompatibility with the condition of experimental system such as temperatures (103).
CRISPR/Cas (CRISPR-associated protein) is a novel and efficient genome editing tool. The widely used CRISPR-Cas9 system consists of two components: the Cas9 enzyme and a guide RNA (gRNA). The advantages of CRISPR-Cas9 system are simplicity and efficiency. This system is used for genome editing by introducing double-strand breaks (DSBs) which are repaired via error-prone nonhomologous end joining (NHEJ) or HR. HR precisely repairs the DSBs by making use of information from a homologous template strand and occurs only during the replicative phase of the cell cycle. Whereas NHEJ repairs the DSBs in a template-independent manner and thus results in introducing unspecified mutations at the target site, such as insertions, deletions, and substitutions. NHEJ is the predominant repair pathway because of it is function throughout the cell cycle and repairs the DSBs quickly to safeguard genomic integrity. The DNA ligase IV inhibitor SCR7 can increase the specificity of CRISPR/Cas9 genome editing by tackling NHEJ and favoring the more precise HR in vitro and in vivo (104, 105, 106).
Guo et al. found that NHEJ accurately repairs the DSBs in CRISPR/Cas9-mediated genome editing with precise deletion of a defined length. The complex HR and NHEJ require many host factors to complete the modification and may thus vary in efficiency depending on different factors, cell types, and host organisms. It is necessary to optimize the CRISPR-Cas9 system for desired genome editing. In addition, variants of Cas9 are still being generated to minimize off-target effect and enhance specificity. The CRISPR-Cas9 and the Cre-LoxP systems can be combined to generate conditional gene knockout and mutant knock-in (107, 108).
The CRISPR/Cas9 system has been enhanced by incorporating a catalytically dead Cas9 fused to transcriptional effector domains. The CRISPR interference (CRISPRi) system established by fusing dead Cas9 to a transcriptional repressor domain Kruppel associated box can efficiently and specifically repress transcription of endogenous genes. A key feature of CRISPRi is the low incidence of off-target effect. CRISPRi is inducible, reversible, and nontoxic and enables knockdown of noncoding RNAs. However, there are a few limitations of the CRISPRi. It can affect the adjacent genes, leading to unpredicted deleterious effects. Moreover, the dependence on the location of the nuclease-specific PAM (protospacer adjacent motif) sequences limits the number of genes targeted by CRISPRi (109).
Conclusions and perspectives
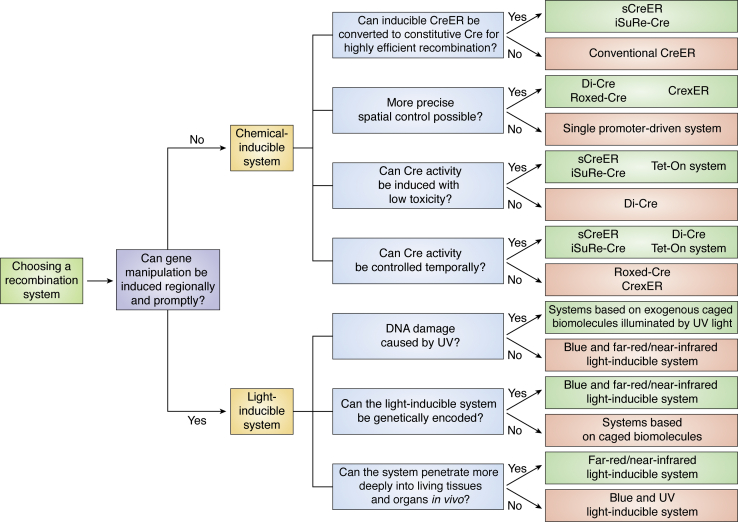
SSRs are instrumental genetic tools to precisely manipulate genomic DNA in multiple biomedical research disciplines. The application of SSRs has greatly facilitated our understanding of gene function and cell lineage relationship in the process of development, tissue homeostasis, regeneration, and diseases. The emerging new site-specific genetic recombination technologies enable efficient manipulation of genomic DNA with high efficiency and fine spatiotemporal control to meet the demand of the ever-increasing complexity of biomedical research. In this review, we have introduced conventional and recently developed site-specific recombination technologies, focusing on the chemical- and light-inducible recombination systems. Each recombination system has its own pros and cons. Thus, choosing an appropriate recombination system or combination strategy to address various biological questions needs to be considered. In Figure 5, we summarize some of the considerations that would help those who are trying to decide which strategy to use.
Figure 5.
Decision tree for choosing an appropriate site-specific recombination system or strategy. Flow chart summarizing site-specific recombination strategies discussed in the text and Table 1. CrexER, a switchable CreER system with the Cre-rox-ER-rox construct; Di-Cre, dimerizable Cre; iSuRe-Cre, a Cre/CreERT2-inducible dual Reporter-Cre-expressing mouse allele; Roxed-Cre, a sequential binary SSR system with the Cre-N-rox-stop-rox-Cre-C strategy; sCreER, self-cleaved inducible CreER with a Cre-loxP-ER-loxP construst; Tet-On system, tetracycline-inducible gene expression system.
Multiple recombinase–mediated genetic approaches facilitate simultaneously analyzing multiple cell lineages and their contribution to development and regeneration, reconstructing cell lineage trees in the developmental programs and unraveling cell lineage and cell fate determination, which is helpful for interrogation of stem cell function during regeneration and disease. The new dual genetic approaches combined Cre with other analogous recombinases such as Flp, Dre, and Nigri have been successfully utilized to design more complex and well-designed experiments for addressing controversial or complicated biological issues (21, 22, 23, 24, 25, 26). A combination of Dre and Cre could enhance the specificity of targeting cell subpopulations, avoid the ectopic expression of Cre recombinase by intersectional genetic approach, provide evidence for the long-standing controversial issues of stem cell differentiation without relying on cell type-specific known markers, and seamlessly record gene activity and cell proliferation during development, regeneration, and diseases (22, 110, 111, 112). In addition to more precise cell lineage tracing, dual recombination system can be used to implement gene knockout and functional analysis in specific cell lineages or subpopulations. The innovative site-specific recombination systems could also be combined with other emerging technologies such as single cell-RNA sequencing, DNA barcoding, and live imaging. Subtypes of new cell lineages and new markers identified by single cell-RNA sequencing provide valuable information for generating new Cre or Dre lines to specifically target the cell subpopulations. Over 70 Dre lines have been generated, which enlarge the pools of multiple recombinases mouse lines and provide a platform for the research of cell origin and fate plasticity via innovative strategies (113).
Further, the combinatorial strategy of chemical- and light-inducible recombination systems has been designed to spatiotemporally manipulate specific genes in vivo. Under the dual control of blue light illumination and doxycycline induction, the Li-rtTA system can be combined with many existing genetic tools to achieve diverse spatiotemporal-specific gene manipulation such as in the regional skin of a mouse. This region-specific light illumination system is also suitable for precise spatiotemporal control of gene expression and function analysis in specific brain regions and other organs (95). An optimal system should have the capacity for precise spatiotemporal control, low leakiness or background, reduced toxicity and invasiveness, and should be easy-to-use. The priorities of these requirements are more efficient and precise spatiotemporal control and low toxicity. Future iterations of site-specific recombination system approach will be widely used in multiply diverse fields of life science research that have great demand for high resolution spatiotemporal control of gene manipulation.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
The authors contributed equally to all aspects of the article.
Funding and additional information
This work was supported by the National Science Foundation of China (81872132 to X. T.; 31730112, 91849202, and 31625019 to B. Z.), National Key Research and Development Program of China (2018YFA0107900 and 2016YFC1300600 to X. T.; 2019YFA011040 and 2019YFA080200 to B. Z.), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000 and XDA16010507 to B. Z.), Ten Thousand Talent Program for Young Top-notch Talent to X. T., Royal Society-Newton Advanced Fellowship to B. Z.
Edited by Patrick Sung
Contributor Information
Xueying Tian, Email: xytian@jnu.edu.cn.
Bin Zhou, Email: zhoubin@sibs.ac.cn.
References
- 1.Nagy A. Cre recombinase: The universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 2.Tian X., He L., Liu K., Pu W., Zhao H., Li Y., Liu X., Tang M., Sun R., Fei J., Ji Y., Qiao Z., Lui K.O., Zhou B. Generation of a self-cleaved inducible Cre recombinase for efficient temporal genetic manipulation. EMBO J. 2020;39 doi: 10.15252/embj.2019102675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Chacon M., Casquero-Garcia V., Luo W., Francesca Lunella F., Ferreira Rocha S., Del Olmo-Cabrera S., Benedito R. iSuRe-Cre is a genetic tool to reliably induce and report Cre-dependent genetic modifications. Nat. Commun. 2019;10:2262. doi: 10.1038/s41467-019-10239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano F., Okazaki R., Yazawa M., Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 2016;12:1059–1064. doi: 10.1038/nchembio.2205. [DOI] [PubMed] [Google Scholar]
- 5.Lewandoski M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 6.Sternberg N.H.D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 7.Inoue S., Inoue M., Fujimura S., Nishinakamura R. A mouse line expressing Sall1-driven inducible Cre recombinase in the kidney mesenchyme. Genesis. 2010;48:207–212. doi: 10.1002/dvg.20603. [DOI] [PubMed] [Google Scholar]
- 8.Smedley D., Salimova E., Rosenthal N. Cre recombinase resources for conditional mouse mutagenesis. Methods. 2011;53:411–416. doi: 10.1016/j.ymeth.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Sauer B., Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U. S. A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tronche F., Casanova E., Turiault M., Sahly I., Kellendonk C. When reverse genetics meets physiology: The use of site-specific recombinases in mice. FEBS Lett. 2002;529:116–121. doi: 10.1016/s0014-5793(02)03266-0. [DOI] [PubMed] [Google Scholar]
- 11.García-Otín A.L., Guillou F. Mammalian genome targeting using site-specific recombinases. Front. Biosci. 2006;11:1108–1136. doi: 10.2741/1867. [DOI] [PubMed] [Google Scholar]
- 12.Meinke G., Bohm A., Hauber J., Pisabarro M.T., Buchholz F. Cre recombinase and other tyrosine recombinases. Chem. Rev. 2016;116:12785–12820. doi: 10.1021/acs.chemrev.6b00077. [DOI] [PubMed] [Google Scholar]
- 13.Murray S.A., Eppig J.T., Smedley D., Simpson E.M., Rosenthal N. Beyond knockouts: Cre resources for conditional mutagenesis. Mamm. Genome. 2012;23:587–599. doi: 10.1007/s00335-012-9430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimshek D.R., Kim J., Hubner M.R., Spergel D.J., Buchholz F., Casanova E., Stewart A.F., Seeburg P.H., Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 15.Andrews B.J., Proteau G.A., Beatty L.G., Sadowski P.D. The FLP recombinase of the 2 micron circle DNA of yeast: Interaction with its target sequences. Cell. 1985;40:795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 16.Kuhstoss S., Rao R.N. Analysis of the integration function of the streptomycete bacteriophage phi C31. J. Mol. Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 17.Sauer B., McDermott J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 2004;32:6086–6095. doi: 10.1093/nar/gkh941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki E., Nakayama M. VCre/VloxP and SCre/SloxP: New site-specific recombination systems for genome engineering. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karimova M., Abi-Ghanem J., Berger N., Surendranath V., Pisabarro M.T., Buchholz F. Vika/vox, a novel efficient and specific Cre/loxP-like site-specific recombination system. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karimova M., Splith V., Karpinski J., Pisabarro M.T., Buchholz F. Discovery of Nigri/nox and Panto/pox site-specific recombinase systems facilitates advanced genome engineering. Sci. Rep. 2016;6:30130. doi: 10.1038/srep30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He M., Tucciarone J., Lee S., Nigro M.J., Kim Y., Levine J.M., Kelly S.M., Krugikov I., Wu P., Chen Y., Gong L., Hou Y., Osten P., Rudy B., Huang Z.J. Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron. 2016;91:1228–1243. doi: 10.1016/j.neuron.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L., Li Y., Li Y., Pu W., Huang X., Tian X., Wang Y., Zhang H., Liu Q., Zhang L., Zhao H., Tang J., Ji H., Cai D., Han Z. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 2017;23:1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sajgo S., Ghinia M.G., Shi M., Liu P., Dong L., Parmhans N., Popescu O., Badea T.C. Dre - cre sequential recombination provides new tools for retinal ganglion cell labeling and manipulation in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermann M., Stillhard P., Wildner H., Seruggia D., Kapp V., Sánchez-Iranzo H., Mercader N., Montoliu L., Zeilhofer H.U., Pelczar P. Binary recombinase systems for high-resolution conditional mutagenesis. Nucleic Acids Res. 2014;42:3894–3907. doi: 10.1093/nar/gkt1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pu W., He L., Han X., Tian X., Li Y., Zhang H., Liu Q., Huang X., Zhang L., Wang Q.D., Yu Z., Yang X., Smart N., Zhou B. Genetic targeting of organ-specific blood vessels. Circ. Res. 2018;123:86–99. doi: 10.1161/CIRCRESAHA.118.312981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K., Yu W., Tang M., Tang J., Liu X., Liu Q., Li Y., He L., Zhang L., Evans S.M., Tian X., Lui K.O., Zhou B. A dual genetic tracing system identifies diverse and dynamic origins of cardiac valve mesenchyme. Development. 2018;145 doi: 10.1242/dev.167775. [DOI] [PubMed] [Google Scholar]
- 27.Zhao H., Zhou B. Dual genetic approaches for deciphering cell fate plasticity in vivo: More than double. Curr. Opin. Cell Biol. 2019;61:101–109. doi: 10.1016/j.ceb.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu K., Jin H., Zhou B. Genetic lineage tracing with multiple DNA recombinases: A user's guide for conducting more precise cell fate mapping studies. J. Biol. Chem. 2020;295:6413–6424. doi: 10.1074/jbc.REV120.011631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy A., Mar L., Watts G. Creation and use of a Cre recombinase transgenic database. In: Kuhn R., Wurst W., editors. Gene Knockout Protocols. 2nd Ed. Vol. 530. Humana Press, a part of Springer ScienceþBusiness Media, LLC; Totowa, NJ: 2009. pp. 365–378. [Google Scholar]
- 30.Hsu Y.-C. Theory and practice of lineage tracing. Stem Cells. 2015;33:3197–3204. doi: 10.1002/stem.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buch T., Heppner F.L., Tertilt C., Heinen T.J., Kremer M., Wunderlich F.T., Jung S., Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 32.Kretzschmar K., Watt F.M. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Metzger D., Ali S., Bornert J.M., Chambon P. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J. Biol. Chem. 1995;270:9535–9542. doi: 10.1074/jbc.270.16.9535. [DOI] [PubMed] [Google Scholar]
- 34.Feil R., Brocard J., Mascrez B., LeMeur M., Metzger D., Chambon P. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellendonk C., Tronche F., Casanova E., Anlag K., Opherk C., Schutz G. Inducible site-specific recombination in the brain. J. Mol. Biol. 1999;285:175–182. doi: 10.1006/jmbi.1998.2307. [DOI] [PubMed] [Google Scholar]
- 36.Ved N., Curran A., Ashcroft F.M., Sparrow D.B. Tamoxifen administration in pregnant mice can be deleterious to both mother and embryo. Lab. Anim. 2019;53:630–633. doi: 10.1177/0023677219856918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadek S., Bell S.C. The effects of the antihormones RU486 and tamoxifen on fetoplacental development and placental bed vascularization in the rat: A model for intrauterine fetal growth retardation. Br. J. Obstet. Gynaecol. 1996;103:630–641. doi: 10.1111/j.1471-0528.1996.tb09830.x. [DOI] [PubMed] [Google Scholar]
- 38.Littlewood T.D., Hancock D.C., Danielian P.S., Parker M.G., Evan G.I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Indra A.K., Warot X., Brocard J., Bornert J.M., Xiao J.H., Chambon P., Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brocard J., Feil R., Chambon P., Metzger D. A chimeric Cre recombinase inducible by synthetic, but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res. 1998;26:4086–4090. doi: 10.1093/nar/26.17.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaczmarczyk S.J. Induction of cre recombinase activity using modified androgen receptor ligand binding domains: A sensitive assay for ligand-receptor interactions. Nucleic Acids Res. 2003;31:86e–86. doi: 10.1093/nar/gng087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shankman L.S., Gomez D., Cherepanova O.A., Salmon M., Alencar G.F., Haskins R.M., Swiatlowska P., Newman A.A., Greene E.S., Straub A.C., Isakson B., Randolph G.J., Owens G.K. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Keymeulen A., Lee M.Y., Ousset M., Brohee S., Rorive S., Giraddi R.R., Wuidart A., Bouvencourt G., Dubois C., Salmon I., Sotiriou C., Phillips W.A., Blanpain C. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 44.Hirrlinger J., Requardt R.P., Winkler U., Wilhelm F., Schulze C., Hirrlinger P.G. Split-CreERT2: Temporal control of DNA recombination mediated by split-Cre protein fragment complementation. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P., Chen T., Sakurai K., Han B.X., He Z., Feng G., Wang F. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Sci. Rep. 2012;2:497. doi: 10.1038/srep00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beckervordersandforth R., Deshpande A., Schaffner I., Huttner H.B., Lepier A., Lie D.C., Gotz M. In vivo targeting of adult neural stem cells in the dentate gyrus by a split-cre approach. Stem Cell Rep. 2014;2:153–162. doi: 10.1016/j.stemcr.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jullien N., Sampieri F., Enjalbert A., Herman J.P. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jullien N., Goddard I., Selmi-Ruby S., Fina J.L., Cremer H., Herman J.P. Conditional transgenesis using dimerizable Cre (DiCre) PLoS One. 2007;2 doi: 10.1371/journal.pone.0001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Utomo A.R., Nikitin A.Y., Lee W.H. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat. Biotechnol. 1999;17:1091–1096. doi: 10.1038/15073. [DOI] [PubMed] [Google Scholar]
- 50.St-Onge L., Furth P.A., Gruss P. Temporal control of the Cre recombinase in transgenic mice by a tetracycline responsive promoter. Nucleic Acids Res. 1996;24:3875–3877. doi: 10.1093/nar/24.19.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schonig K., Schwenk F., Rajewsky K., Bujard H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/gnf134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu B., Zhou B., Wang Y., Cheng H.L., Hang C.T., Pu W.T., Chang C.P., Zhou B. Inducible cardiomyocyte-specific gene disruption directed by the rat Tnnt2 promoter in the mouse. Genesis. 2010;48:63–72. doi: 10.1002/dvg.20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traykova-Brauch M., Schonig K., Greiner O., Miloud T., Jauch A., Bode M., Felsher D.W., Glick A.B., Kwiatkowski D.J., Bujard H., Horst J., von Knebel Doeberitz M., Niggli F.K., Kriz W., Grone H.J. An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nat. Med. 2008;14:979–984. doi: 10.1038/nm.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan X., Small E.V., Igarashi P., Carroll T.J. Generation and characterization of KsprtTA and KsptTA transgenic mice. Genesis. 2013;51:430–435. doi: 10.1002/dvg.22381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindeberg J., Mattsson R., Ebendal T. Timing the doxycycline yields different patterns of genomic recombination in brain neurons with a new inducible Cre transgene. J. Neurosci. Res. 2002;68:248–253. doi: 10.1002/jnr.10213. [DOI] [PubMed] [Google Scholar]
- 56.Saam J.R., Gordon J.I. Inducible gene knockouts in the small intestinal and colonic epithelium. J. Biol. Chem. 1999;274:38071–38082. doi: 10.1074/jbc.274.53.38071. [DOI] [PubMed] [Google Scholar]
- 57.Perl A.K., Wert S.E., Nagy A., Lobe C.G., Whitsett J.A. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen D., Wu C.F., Shi B., Xu Y.M. Tamoxifen and toremifene cause impairment of learning and memory function in mice. Pharmacol. Biochem. Behav. 2002;71:269–276. doi: 10.1016/s0091-3057(01)00656-6. [DOI] [PubMed] [Google Scholar]
- 59.Roshangar L., Rad J.S., Afsordeh K. Maternal tamoxifen treatment alters oocyte differentiation in the neonatal mice: Inhibition of oocyte development and decreased folliculogenesis. J. Obstet. Gynaecol. Res. 2010;36:224–231. doi: 10.1111/j.1447-0756.2009.01129.x. [DOI] [PubMed] [Google Scholar]
- 60.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Müller K., Engesser R., Schulz S., Steinberg T., Tomakidi P., Weber C.C., Ulm R., Timmer J., Zurbriggen M.D., Weber W. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Link K.H., Shi Y., Koh J.T. Light activated recombination. J. Am. Chem. Soc. 2005;127:13088–13089. doi: 10.1021/ja0531226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu X., Agasti S.S., Vinegoni C., Waterman P., DePinho R.A., Weissleder R. Optochemogenetics (OCG) allows more precise control of genetic engineering in mice with CreER regulators. Bioconjug. Chem. 2012;23:1945–1951. doi: 10.1021/bc300319c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inlay M.A., Choe V., Bharathi S., Fernhoff N.B., Baker J.R., Jr., Weissman I.L., Choi S.K. Synthesis of a photocaged tamoxifen for light-dependent activation of Cre-ER recombinase-driven gene modification. Chem. Commun. (Camb.) 2013;49:4971–4973. doi: 10.1039/c3cc42179a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faal T., Wong P.T., Tang S., Coulter A., Chen Y., Tu C.H., Baker J.R., Choi S.K., Inlay M.A. 4-Hydroxytamoxifen probes for light-dependent spatiotemporal control of Cre-ER mediated reporter gene expression. Mol. Biosyst. 2015;11:783–790. doi: 10.1039/c4mb00581c. [DOI] [PubMed] [Google Scholar]
- 66.Cambridge S.B., Geissler D., Calegari F., Anastassiadis K., Hasan M.T., Stewart A.F., Huttner W.B., Hagen V., Bonhoeffer T. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat. Methods. 2009;6:527–531. doi: 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- 67.Gorka A.P., Nani R.R., Zhu J., Mackem S., Schnermann M.J. A near-IR uncaging strategy based on cyanine photochemistry. J. Am. Chem. Soc. 2014;136:14153–14159. doi: 10.1021/ja5065203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown K.A., Zou Y., Shirvanyants D., Zhang J., Samanta S., Mantravadi P.K., Dokholyan N.V., Deiters A. Light-cleavable rapamycin dimer as an optical trigger for protein dimerization. Chem. Commun. (Camb.) 2015;51:5702–5705. doi: 10.1039/c4cc09442e. [DOI] [PubMed] [Google Scholar]
- 69.Edwards W.F., Young D.D., Deiters A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem. Biol. 2009;4:441–445. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]
- 70.Luo J., Arbely E., Zhang J., Chou C., Uprety R., Chin J.W., Deiters A. Genetically encoded optical activation of DNA recombination in human cells. Chem. Commun. (Camb.) 2016;52:8529–8532. doi: 10.1039/c6cc03934k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cambridge S.B., Davis R.L., Minden J.S. Drosophila mitotic domain boundaries as cell fate boundaries. Science. 1997;277:825–828. doi: 10.1126/science.277.5327.825. [DOI] [PubMed] [Google Scholar]
- 72.Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. A light-switchable gene promoter system. Nat. Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 73.Levskaya A., Weiner O.D., Lim W.A., Voigt C.A. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yazawa M., Sadaghiani A.M., Hsueh B., Dolmetsch R.E. Induction of protein-protein interactions in live cells using light. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 76.Strickland D., Lin Y., Wagner E., Hope C.M., Zayner J., Antoniou C., Sosnick T.R., Weiss E.L., Glotzer M. TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guntas G., Hallett R.A., Zimmerman S.P., Williams T., Yumerefendi H., Bear J.E., Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. U. S. A. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawano F., Suzuki H., Furuya A., Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- 79.Zoltowski B.D., Schwerdtfeger C., Widom J., Loros J.J., Bilwes A.M., Dunlap J.C., Crane B.R. Conformational switching in the fungal light sensor vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen D., Gibson E.S., Kennedy M.J. A light-triggered protein secretion system. J. Cell Biol. 2013;201:631–640. doi: 10.1083/jcb.201210119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crefcoeur R.P., Yin R., Ulm R., Halazonetis T.D. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat. Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 82.Motta-Mena L.B., Reade A., Mallory M.J., Glantz S., Weiner O.D., Lynch K.W., Gardner K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Konermann S., Brigham M.D., Trevino A.E., Hsu P.D., Heidenreich M., Le C., Platt R.J., Scott D.A., Church G.M., Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Polstein L.R., Gersbach C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nihongaki Y., Yamamoto S., Kawano F., Suzuki H., Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 86.Idevall-Hagren O., Dickson E.J., Hille B., Toomre D.K., De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kakumoto T., Nakata T. Optogenetic control of PIP3: PIP3 is sufficient to induce the actin-based active part of growth cones and is regulated via endocytosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maiuri P., Rupprecht J.F., Wieser S., Ruprecht V., Benichou O., Carpi N., Coppey M., De Beco S., Gov N., Heisenberg C.P., Lage Crespo C., Lautenschlaeger F., Le Berre M., Lennon-Dumenil A.M., Raab M. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 89.Duan L., Che D., Zhang K., Ong Q., Guo S., Cui B. Optogenetic control of molecular motors and organelle distributions in cells. Chem. Biol. 2015;22:671–682. doi: 10.1016/j.chembiol.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boulina M., Samarajeewa H., Baker J.D., Kim M.D., Chiba A. Live imaging of multicolor-labeled cells in Drosophila. Development. 2013;140:1605–1613. doi: 10.1242/dev.088930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schindler S.E., McCall J.G., Yan P., Hyrc K.L., Li M., Tucker C.L., Lee J.M., Bruchas M.R., Diamond M.I. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci. Rep. 2015;5:13627. doi: 10.1038/srep13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taslimi A., Zoltowski B., Miranda J.G., Pathak G.P., Hughes R.M., Tucker C.L. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 2016;12:425–430. doi: 10.1038/nchembio.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meador K., Wysoczynski C.L., Norris A.J., Aoto J., Bruchas M.R., Tucker C.L. Achieving tight control of a photoactivatable Cre recombinase gene switch: New design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morikawa K., Furuhashi K., de Sena-Tomas C., Garcia-Garcia A.L., Bekdash R., Klein A.D., Gallerani N., Yamamoto H.E., Park S.E., Collins G.S., Kawano F., Sato M., Lin C.S., Targoff K.L., Au E. Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat. Commun. 2020;11:2141. doi: 10.1038/s41467-020-16030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li F., Lu Z., Wu W., Qian N., Wang F., Chen T. Optogenetic gene editing in regional skin. Cell Res. 2019;29:862–865. doi: 10.1038/s41422-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X., Chen X., Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 97.Yao S., Yuan P., Ouellette B., Zhou T., Mortrud M., Balaram P., Chatterjee S., Wang Y., Daigle T.L., Tasic B., Kuang X., Gong H., Luo Q., Zeng S., Curtright A. RecV recombinase system for in vivo targeted optogenomic modifications of single cells or cell populations. Nat. Methods. 2020;17:422–429. doi: 10.1038/s41592-020-0774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wan S., Parrish J.A., Anderson R.R., Madden M. Transmittance of nonionizing radiation in human tissues. Photochem. Photobiol. 1981;34:679–681. doi: 10.1111/j.1751-1097.1981.tb09063.x. [DOI] [PubMed] [Google Scholar]
- 99.Gomelsky M. Photoactivated cells link diagnosis and therapy. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan3936. [DOI] [PubMed] [Google Scholar]
- 100.Müller K., Engesser R., Metzger S., Schulz S., Kämpf M.M., Busacker M., Steinberg T., Tomakidi P., Ehrbar M., Nagy F., Timmer J., Zubriggen M.D., Weber W. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaberniuk A.A., Shemetov A.A., Verkhusha V.V. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat. Methods. 2016;13:591–597. doi: 10.1038/nmeth.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu J., Wang M., Yang X., Yi C., Jiang J., Yu Y., Ye H. A non-invasive far-red light-induced split-Cre recombinase system for controllable genome engineering in mice. Nat. Commun. 2020;11:3708. doi: 10.1038/s41467-020-17530-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olorunniji F.J., Rosser S.J., Stark W.M. Site-specific recombinases: Molecular machines for the genetic revolution. Biochem. J. 2016;473:673–684. doi: 10.1042/BJ20151112. [DOI] [PubMed] [Google Scholar]
- 104.Srivastava M., Nambiar M., Sharma S., Karki S.S., Goldsmith G., Hegde M., Kumar S., Pandey M., Singh R.K., Ray P., Natarajan R., Kelkar M., De A., Choudhary B., Raghavan S.C. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151:1474–1487. doi: 10.1016/j.cell.2012.11.054. [DOI] [PubMed] [Google Scholar]
- 105.Reid D.A., Conlin M.P., Yin Y., Chang H.H., Watanabe G., Lieber M.R., Ramsden D.A., Rothenberg E. Bridging of double-stranded breaks by the nonhomologous end-joining ligation complex is modulated by DNA end chemistry. Nucleic Acids Res. 2017;45:1872–1878. doi: 10.1093/nar/gkw1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reid D.A., Keegan S., Leo-Macias A., Watanabe G., Strande N.T., Chang H.H., Oksuz B.A., Fenyo D., Lieber M.R., Ramsden D.A., Rothenberg E. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E2575–E2584. doi: 10.1073/pnas.1420115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thakur V.S., Welford S.M. Generation of a conditional mutant knock-in under the control of the natural promoter using CRISPR-Cas9 and Cre-Lox systems. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Noiman T., Kahana C. A simple combined use of CRISPR-Cas9 and Cre-LoxP technologies for generating conditional gene knockouts in mammalian cells. CRISPR J. 2018;1:278–285. doi: 10.1089/crispr.2018.0010. [DOI] [PubMed] [Google Scholar]
- 109.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., Qi L.S., Kampmann M., Weissman J.S. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y., He L., Huang X., Bhaloo S.I., Zhao H., Zhang S., Pu W., Tian X., Li Y., Liu Q., Yu W., Zhang L., Liu X., Liu K., Tang J. Genetic lineage tracing of nonmyocyte population by dual recombinases. Circulation. 2018;138:793–805. doi: 10.1161/CIRCULATIONAHA.118.034250. [DOI] [PubMed] [Google Scholar]
- 111.Li Y., Lv Z., Zhang S., Wang Z., He L., Tang M., Pu W., Zhao H., Zhang Z., Shi Q., Cai D., Wu M., Hu G., Lui K.O., Feng J. Genetic fate mapping of transient cell fate reveals N-cadherin activity and function in tumor metastasis. Dev. Cell. 2020;54:593–607.e595. doi: 10.1016/j.devcel.2020.06.021. [DOI] [PubMed] [Google Scholar]
- 112.He L., Pu W., Liu X., Zhang Z., Han M., Li Y., Huang X., Han X., Li Y., Liu K., Shi M., Lai L., Sun R., Wang Q.D., Ji Y. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science. 2021;371 doi: 10.1126/science.abc4346. eabc4346. [DOI] [PubMed] [Google Scholar]
- 113.Han X., Zhang Z., He L., Zhu H., Li Y., Pu W., Han M., Zhao H., Liu K., Li Y., Huang X., Zhang M., Jin H., Lv Z., Tang J. A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.01.007. [DOI] [PubMed] [Google Scholar]