Abstract
The current pandemic of COVID-19, with its climbing number of cases and deaths, has us searching for tools for rapid, reliable, and affordable methods of detection on one hand, and novel, improved therapeutic strategies on the other. The currently employed RT-PCR method, despite its all-encompassing utility, has its shortcomings. Newer diagnostic tools, based on the Clustered Regularly Interspaced Short Palindromic Repeats/Cas(CRISPR-Cas) system, with its better diagnostic accuracy measures, have come up to fill that void. These assay platforms are expected to slowly take up the place of COVID-19 diagnostics. Further, the current therapeutic options focus mainly on counteracting the viral proteins and components and their entry into host cells. The CRISPR-based system, especially through the RNA-guided Cas13 approach, can identify the genomic characteristics of SARS-CoV-2 and provide a novel inhibition strategy for coronaviruses. In this mini-review, we have discussed the available and upcoming CRISPR-based diagnostic assays and the potential of the CRISPR/Cas system as a therapeutic or prevention strategy in COVID-19. CRISPR-Cas system shows promise in both diagnostics as well as therapeutics and may as well change the face of molecular diagnosis and precision medicine.
Keywords: COVID-19, SHERLOCK, CRISPR-Cas, Cas13, PAC-MAN
Introduction
With the declaration of the Nobel Prize 2020 in Chemistry to Jennifer Doudna and Emmanuelle Charpentier, there has been a lot of curiosity around their discovered—new molecular scissors CRISPR/Cas system. The Clustered Regularly Interspaced Short Palindromic Repeats/Cas (CRISPR-Cas) system was first identified in bacteria, where it provides innate protection to the host cell against invading viruses. It continues to drive major advances in the field of molecular technology with newer genome editing tools [1, 2]. The currently available tools for modifying genomes in experimental systems can be divided into four classes—nucleases, base editors, transposases/recombinases and prime editors [3]. The CRISPR-Cas system includes both DNA- and RNA-targeting nucleases such as Cas9 and Cas13, respectively.
The CRISPR-Cas bacterial immune system is broadly divided into classes 1 and 2. The class 1 CRISPR system is composed of multiple Cas proteins, mediating pre-crRNA processing and binding, and interference by cleavage. Class 1 can further be divided into three types, namely types I, III, and IV. With recent computational advances, 33 novel variants or subtypes of CRISPR system have been identified in class 1 category [4]. Class 2, on the other hand, is considerably simple. It includes single, multidomain crRNA binding Cas proteins. This system, consists of both DNA- and RNA-targeting Cas proteins, is further subdivided into types II, V, and VI. Among these, RNA-targeting CRISPR-Cas13 is categorized under type VI, and it is the only known RNA-targeting-resistant mechanism in bacteria in addition to the type III CRISPR system in class 1 [5, 6] The year 2020 has be-fallen us with a global crisis of COVID-19 and till date there is a lack of a robust diagnostic method with high sensitivity as well as specificity; and a lack of efficient therapeutics that has shown a good recovery-rate. CRISP/Cas holds the potential to be both an excellent diagnostic tool as well as be the treatment/ vaccine that has been eluding scientists worldwide. We have compiled this mini-review with an aim to give a brief yet updated opinion on CRISP/Cas promise for COVID-19 pandemic. (Table 1 and Fig. 1).
Table 1.
Comparative analysis of COVID-19 diagnostic method
| Viral genome (NGS) | Viral RNA Test (RT-PCR) |
Viral RNA test (CRISPR) |
Serological antibody tests | Viral antigen test | |
|---|---|---|---|---|---|
| Accessibility | low | low | Medium | High | High |
| Accuracy | High | High | High | Medium | low |
| Affordability | Very low | low | Medium | High | High |
| Airway swab or sputum | Yes | Yes | Yes | No | No |
| Blood/Serum | High | Medium | Medium | High | High |
| Cross reactivity | Low | Low | Low | Medium | Medium |
| Genome sequencing | yes | No | No | No | No |
| Immunity status | No | No | No | Yes | No |
| Infection status | Yes | yes | yes | No | Yes |
| Point of care | Very low | low | Medium | High | High |
| Sample preparation | High | High | Medium | Low | Low |
| Test time | High | Medium | low | Very low | Very low |
| User friendly | low | low | Medium | High | High |
1. High or Yes; 2. Medium; 3. Low; 4. Very low or No
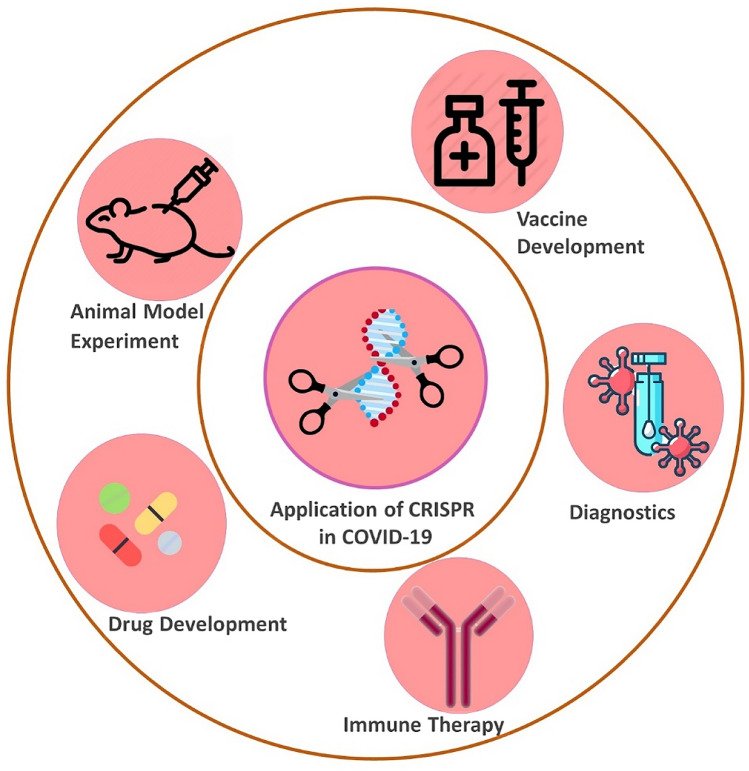
Fig. 1.
Application of CRISPR technology for COVID 19 research
In this mini-review, we have searched through the Pubmed database for articles on the applications of CRISPR/Cas in Coronavirus disease 2019 (COVID-19) with the keywords “CRISPR/Cas”, “CRISPR”, “COVID-19”, “SARS-CoV-2”, “diagnostics”, and “therapeutics” and briefly reviewed the existing CRISPR/Cas-based diagnostic tools in COVID-19 and further elaborated on its possible therapeutic applications.
As Diagnostic Tool for COVID 19
COVID-19 is caused by the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), which is a novel pathogenic member of the betacoronavirus genus, having 79% genetic similarity with the SARS-CoV, as discovered by metagenomic next-generation sequencing (mNGS) [7, 8]. The major complications associated with COVID-19 were pneumonia, respiratory failure, acute kidney injury [9], and sepsis or systemic inflammation [10]. Viricidal agents play important role in the prevention of SARS-CoV-2 transmission [11]. Currently, two molecular diagnostic approaches in detection of COVID-19 are- metagenomics and RT-PCR [12, 13]. However, several other molecular methods are being considered and under investigation, one of them being CRISPR/Cas, the widely used gene editing tool. The collateral cleavage activities of a unique group of Cas nucleases which form the basis of this tool was discovered not very long ago and has been tested for in vitro nucleicaciddetection since [14]. After the outbreak of the SARS-CoV-2 pandemic, rapid detection of the virus to hasten subsequent implementation of treatment strategies, to control the spread of the infection, and to detect asymptomatic infections and possible mutant strains of the virus, has become a matter of utmost importance. However, the mainstay of diagnosis at this time-point, RT-PCR, may not detect the virus as viral load fluctuates in different stages of infection, and further depends on pre-analytical factors is major diagnostic challenges include time-consuming, errors during sample collection, sample transport, shaking of blood samples, leading to hemolysis while pre-analytical inconsistencies have been demonstrated to reduce the stability of extracted viral RNA and PCR conditions [15–17]. Further, a rapid diagnostic tool is in demand, which can shorten the turnaround time and reduce the resources for COVID-19 testing.
CRISPR methods have come forward in this regard [18, 19]. CRISPR diagnostic tests can potentially reduce laboratory load and patient costs, as it can be performed using simple reagents and point-of-care devices such as paper-based lateral flow assays, such as the sensitivity enzymatic reporter unlocking (SHERLOCK)-based STOPCovid [20]. CRISPR-based diagnostic methods rely primarily on the principle that nucleic acids are effective biomarkers for diseases. It works by identifying specific sequences (Cas-SgRNA can cleave a target specifically) associated with the infective virus and cleaving it by virtue of Cas nucleases to generate as readable signal [21] The specificity of SgRNA targets mainly requires for the protospacer motif make it possible for the CRISPR system to distinguish between various strains of viruses [22]. This differentiation of viral strains is important specificity.
(Fig. 2). Recently, Broughton et al.showedits promising potential for in vitro diagnostics using Cas12 [23]. Hou et al. have exploited the polymerase-mediated DNA amplification by Recombinase Polymerase Amplification (RPA) as well as the CRISPR/Cas-mediated enzymatic signal amplification to develop a rapid, and highly sensitive, easy-to-use assay [14]. An Indian-based assay, the FELUDA test has come up as a faster and cheaper alternative with 96% sensitivity and 98% specificity [24]. (Table 2).
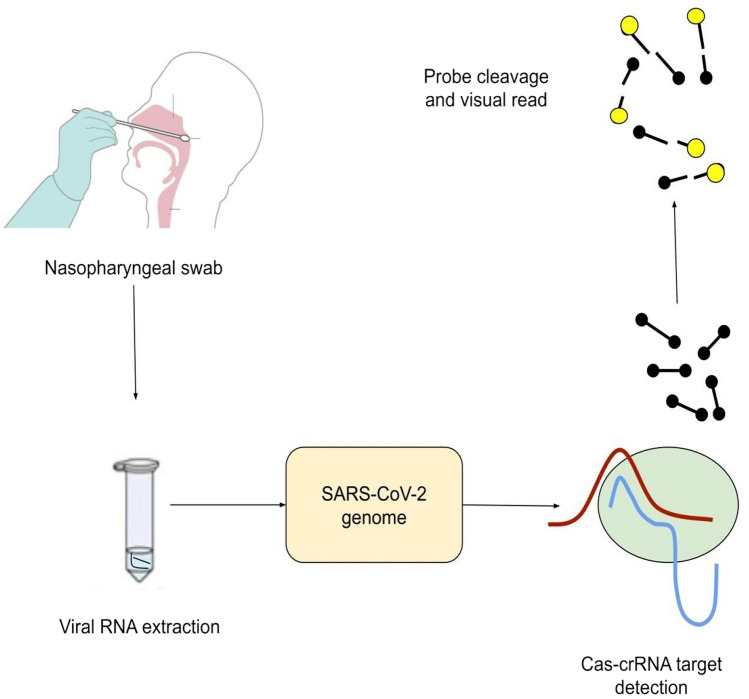
Fig. 2.
A simplified flow of detection of COVID-19 by CRISPR-Cas method
Table 2.
CRISPR technology used for diagnostic method in various countries
| Countries | Name of the method | Clinical diagnostic samples | Cas enzyme | Target region | Amplification method | Limit of detection (copy/μL) | Readout method | Sensitivity (%) | Specificity (%) | Total assay time (min) | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| China | CRISPR/Cas12a-NER | 31 | LbCas12a | E | RT-RAA | 10 | FL / NE | 100 | 100 | 45 | [43] |
| China | SENA | 295 | LbCas12a | N/O | RT-qPCR | < 2 | FL | 100 | 100 | 60 | [44] |
| China | CRISPR-COVID | 114 | LwCas13a | ORF1ab | RT-RPA | 0.3 | FL | 100 | 100 | 40 | [14] |
| Germany | – | 24 | LwCas13a | ORF1a/N/S | RT-RPA | – | Lf | 55.5 | 100 | 60 | [45] |
| Japan | CONAN | 31 | Cas3 and LbCas12a | N | RT-LAMP | < 100 | FL/Lf | 90 | 95 | 40 | [46] |
| Netherlands | DETECTR | 378 | LbCas12a | N | RT-LAMP | – | FL/Lf | 93 | 95.5 | 30 | [47] |
| Saudi Arabia | iSCAN | 24 | LbCas12a and AapCas12b | N/E | RT-LAMP | 0.2 | FL/Lf | 86 | 100 | 60 | [48] |
| Thailand | SHERLOCK | 154 | LwCas13a | S | RT-RPA | 2.1 | FL/Lf | 96 | 100 | 55–85 | [49] |
| United Kingdom | – | 3 | LwCas13a | N | RT-PCR/RT-RPA | 0.5 | FL | 100 | NA | 75 | [50] |
| USA | DETECTR | 82 | LbCas12a | N/E | RT-LAMP | 10 | FL/Lf | 95 | 100 | 30–40 | [51] |
| USA | STOPCovid | 17 | AapCas12b | N | RT-LAMP | 2 | FL/Lf | 91.7 | 100 | 40–70 | [20] |
| USA | ITP-CRISPR | 8 | LbCas12a | N/E | RT-LAMP | 10 | FL | 75 | 100 | 25 | [52] |
| USA | CRISPR-FDS | 35 | LbCas12a | ORF1ab/N | RT-PCR/RT-RPA | 2 | FL | 100 | 71.4 | 50 | [53, 54] |
| USA | SHINE | 50 | LwCas13a | ORF1a | RT-RPA | 10 | FL/Lf | 90 | 100 | 40 | [39, 55] |
| USA | CREST | 1808 | LwCas13a | N | RT-PCR | 10 | FL/Lf | 88.8 | 100 | 110 | [56, 57] |
Some of the advantages of the CRISPR/Cas-based technology over qRT–PCR include- single target specificity, reduced turnaround time, isothermal signal amplification, no requirement of a complex laboratory setup, and easily-accessible reporting with lateral flow strips [18]. Further, in comparison to mNGS and the PCR, the CRISPR-based method had a 100% specificity, while PCR could detect 90.4% (47/52) of the positive cases, with Ct’s ranging from 28.8 to 40.4 and a median Ct of 35.8, and requires more time than the former method. CRISPR was also highly sensitive for all 52 cases (100%), with fold change ranging from 5.0 to 66.3 and a median fold change of 22.8. Altogether, CRISPR-based assays have better diagnostic value without the need of thermal cyclers [14].
Advanced Approaches for CRISPR/Cas Mediated Diagnosis
The two CRISPR based methods namely, DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) and SHERLOCK methods have been proposed as diagnostic methods for SARS-CoV-2, and the protocols designed specifically for SARS-CoV-2 detection can be accomplished in an hour or less, and at relatively low cost [25, 26]. Recently, Zhang et al. reported specific SHERLOCK technology which is a CRISPR/Cas13 based nucleic acid detection technique for rapid detection of SARS-CoV-2 [26]. In this technique, S and ORF1ab protein genes of coronavirus genome have been targeted. Cas13 identifies and binds to previously determined target sequence which leads to random cleavage of surrounding ssRNA molecules. SHERLOCK technology used a quenched fluorescent ssRNA reporter. The presence of ssRNA coronavirus genome in samples activated Cas13, which resulted in the production of quantifiable signals. Its sensitivity has been improved through amplification of targeted DNA or RNA by RPA or reverse transcriptase-RPA (RT-RPA) prior to the start of reaction. Subsequently, amplified DNA is converted to RNA by combination of RPA and T7 transcription. Finally, detection is made by simultaneous incorporation of the ssRNA reporter (Biotin-RNA-FITC). Viral genome is detected at attomolar concentration using SHERLOCK technology. The test can be carried out starting with RNA isolated from patient samples, as used for qRT-PCR assays, and can be read out using a dipstick in less than an hour, without requiring various instrumentation. As a result, application of SHERLOCK as a diagnostic tool for SARS-CoV-2 detection is much faster than qRT-PCR and has high sensitivity of 93.1%. Consequently, SHERLOCK technology could slowly replace qRT-PCR technique considering the high demand for rapid diagnosis tests in current global pandemic state of COVID-19 [27]. The proposed SHERLOCK method generates a positive result for SARS-CoV-2 when the S and Orflab gene sequences are detected [28].
To overcome the difficulties faced in qRT–PCR-based assays, there is combined isothermal amplification with CRISPR–Cas12 DETECTR technology was developed for rapid (30–40 min) detection of SARS-CoV-2 in clinical samples. This assay performed simultaneous reverse transcription and isothermal amplification using loop-mediated amplification (RT–LAMP) [28] for RNA extracted from nasopharyngeal or oropharyngeal swabs and followed by Cas12 detection of predefined coronavirus sequences, after which cleaved reporter molecule confirms detection of the virus. The DETECTR assay developed gives comparable accuracy to qRT–PCR and used routine protocols and commercially available reagents. However, some key advantages of the approach over qRT–PCR include isothermal signal amplification avoiding the need for thermocycling, fast turnaround time, single nucleotide target specificity, integration with accessible and easy-to-use reporting formats such as lateral flow strips and no requirement for complex laboratory instrumentations. The time taken to develop and validate this SARS-CoV-2 DETECTR assay showed that this technology can be quickly mobilized to diagnose infections from emerging zoonotic viruses [21]. The DETECTR method has been focused to identify the presence of the N and E gene variants specific to SARS-CoV-2. A positive result is generated if both genes are detected, and the procedure has been optimized to exclude false positives resulting from related coronaviruses [19, 21].
Recent Advances in Therapeutics
The diagnostic applicability of CRISPR/Cas against SARS-CoV-2 in already under progress. Keeping up with the pace, researchers have adopted the CRISPR-Casmechanism as a therapeutic strategy to correct genetic abnormalities in human cells in hope of treating diseases or as an invaluable tool in chemical biology [29–32]. Since it confers protective immunity against invading pathogens such as bacteriophages, it has been co-opted to use as a potential antiviral therapy [33–36]. Due to its RNA targeting properties, the CRISPR-Cas13 has been successfully demonstrated as an antiviral strategy against single-strand RNA viruses such as lymphocytic choriomeningitis virus (LCMV), influenza A virus (IAV), and vesicular stomatitis virus (VSV) in human cells. Thus, the Cas13-assisted restriction of viral expression and readout (CARVER) had earlier been developed which demonstrated RNA-targeting capacity [37]. Recently, Abbott et al., using an RNA-guided RNA endonuclease Cas13d, have established that the CRISPR technology can be used to cleave SARS-CoV-2 genetic material in mammalian cells [38, 39]. Cas13d is much smaller than the other members of the Cas13 family and known for having strong target cleavage activity [40]. Further, by aligning the SARS-CoV-2 genome sequences from 47 patients with those of SARS-CoV and MERS, high levels of conservation in the RNA-dependent RNA polymerase (RdRP) gene and the Nucleocapsid (N) gene were observed. With the aid of a bioinformatic pipeline, a crRNA library was built with less off-target binding to the human genome, to target different coronaviruses in the RdRP and N protein coding regions. Indeed, in lung epithelial cell line A549, it has showcased the ability to degrade viral genetic material.
This new technology has been coined Prophylactic Antiviral CRISPR in huMAN cells (PAC-MAN). A key advantage is that it can be a potential pan-coronavirus inhibition strategy by targeting conserved regions across different coronaviruses using a single cocktail containing different crRNAs. Computationally, it has been shown that just three crRNAs are enough to target all betacoronaviruses implicated in SARS, MERS, and COVID-19 [41].
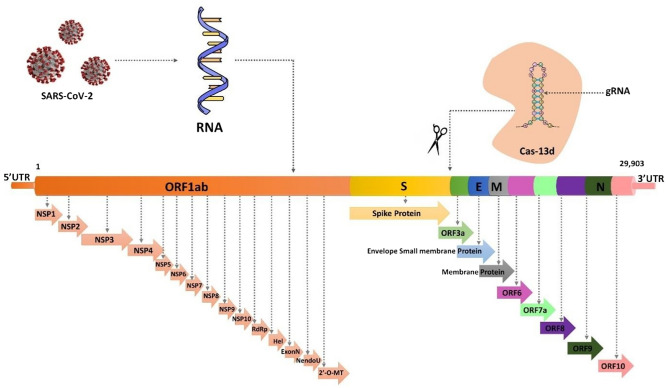
Lotfi et al. discussed the application of CRISPR/Cas13 in lieu of the potential therapeutic targets in the SARS-CoV-2 viral genome (Fig. 3). Several non-structural, accessory, and structural proteins have been evaluated which can be targeted by the CRISPR/Cas13 system with more sensitivity and specificity [42].
Fig. 3.
SARS-CoV-2 structure-based function annotation of genome. The variable coloured regions represent the various appropriate targeting sites of SARS-CoV-2
Limitations
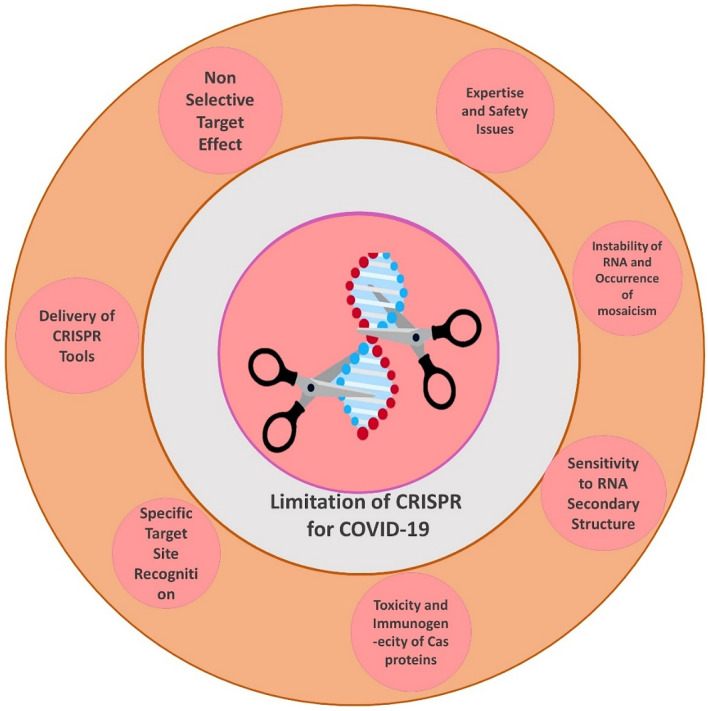
One short coming of this strategy is the potential crRNA target site mutations, which may decrease its potency as these viruses tend to alter their sequences in response to therapy. But, a simple alteration in the crRNA sequence may solve this road block and will further enable the designing of tools against other viruses. Viral vectors are the most common delivery method for CRISPR/Cas material but many time difficult to deliver the CRISPR/Cas material to mature cells, which are problem for therapeutic clinical applications. In this regard, a synergistic approach with already available clinically administered drugs may be considered. (Fig. 4).
Fig. 4.
Limitation of CRISPR for COVID-19 research
Conclusion
CRISPR-Cas system shows promise in both diagnostics as well as therapeutics and may as well change the face of molecular diagnosis and precision medicine.
Abbreviations
- CARVER
Cas13-assisted restriction of viral expression and readout
- Cas
CRISPR-associated complex
- COVID-19
Coronavirus disease 2019
- CRISPR
Clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- DETECTR
DNA Endonuclease-Targeted CRISPR Trans Reporter
- IAV
Influenza virus
- LCMV
Lymphocytic choriomeningitis virus
- MERS
Middle East Respiratory Syndrome
- PACMAN
Prophylactic Antiviral CRISPR in huMAN cells
- RPA
Recombinase Polymerase Amplification
- RT-LAMP
Reverse transcription and loop-mediated amplification
- RT-PCR
Reverse transcriptase- polymerase chain reaction
- SARS
Severe Acute Respiratory Syndrome
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SHERLOCK
Specific High Sensitivity Enzymatic Reporter UnLOCKing
- STOPCovid
SHERLOCK Testing in One Pot
- VSV
Vesicular stomatitis virus
Funding
None.
Declarations
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2(7):17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 3.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren L-L, Wang Y-M, Wu Z-Q, Xiang Z-C, Guo L, Xu T, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133(9):1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khokhar M, Purohit P, Roy D, Tomo S, Gadwal A, Modi A, et al. Acute kidney injury in COVID 19–an update on pathophysiology and management modalities. Arch Physiol Biochem. 2020;15:1–14. doi: 10.1080/13813455.2020.1856141. [DOI] [PubMed] [Google Scholar]
- 10.Tomo S, Kumar KP, Roy D, Sankanagoudar S, Purohit P, Yadav D, et al. Complement activation and coagulopathy-an ominous duo in COVID19. Expert Rev Hematol. 2021;14(2):155–173. doi: 10.1080/17474086.2021.1875813. [DOI] [PubMed] [Google Scholar]
- 11.Khokhar M, Roy D, Purohit P, Goyal M, Setia P. Viricidal treatments for prevention of coronavirus infection. Pathog Glob Health. 2020;2:1–11. doi: 10.1080/20477724.2020.1807177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29(1):23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 13.Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. 2019;24(14):319–338. doi: 10.1146/annurev-pathmechdis-012418-012751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou T, Zeng W, Yang M, Chen W, Ren L, Ai J, et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16(8):e1008705. doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payne D, Newton D, Evans P, Osman H, Baretto R. Preanalytical issues affecting the diagnosis of COVID-19. J Clin Pathol. 2021;74(4):207–208. doi: 10.1136/jclinpath-2020-206751. [DOI] [PubMed] [Google Scholar]
- 16.Malentacchi F, Pazzagli M, Simi L, Orlando C, Wyrich R, Günther K, et al. SPIDIA-RNA: Second external quality assessment for the pre-analytical phase of blood samples used for RNA based analyses. PLoS ONE. 2014;9(11):e112293. doi: 10.1371/journal.pone.0112293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng W, Newbigging AM, Le C, Pang B, Peng H, Cao Y, et al. Molecular diagnosis of COVID-19: challenges and research needs. Anal Chem. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 18.Huang C-H, Lee K-C, Doudna JA. Applications of CRISPR-Cas enzymes in cancer therapeutics and detection. Trends Cancer. 2018;4(7):499–512. doi: 10.1016/j.trecan.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jolany Vangah S, Katalani C, Booneh HA, Hajizade A, Sijercic A, Ahmadian G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol Proced Online. 2020;22:22. doi: 10.1186/s12575-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang MW, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics [Internet]. Infectious Diseases (except HIV/AIDS); 2020 May [cited 2020 Dec 8]. Available from: http://medrxiv.org/lookup/doi/10.1101/2020.05.04.20091231
- 21.Uppada V, Gokara M, Rasineni GK. Diagnosis and therapy with CRISPR advanced CRISPR based tools for point of care diagnostics and early therapies. Gene. 2018;20(656):22–29. doi: 10.1016/j.gene.2018.02.066. [DOI] [PubMed] [Google Scholar]
- 22.Karvelis T, Gasiunas G, Young J, Bigelyte G, Silanskas A, Cigan M, et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16(1):253. doi: 10.1186/s13059-015-0818-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azhar Mohd, Phutela R, Ansari AH, Sinha D, Sharma N, Kumar M, et al. Rapid, field-deployable nucleobase detection and identification using FnCas9 [Internet]. Molecular Biology; 2020 [cited 2020 Dec 8]. 10.1101/2020.04.07.028167
- 25.Barrangou R. CRISPR-Cas systems and RNA-guided interference: CRISPR-Cas systems and RNA-guided interference. WIREs RNA. 2013;4(3):267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR diagnostics. Synthego. 2020; 8. https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf. Accessed 12 April 2021.
- 27.Dara M, Talebzadeh M. CRISPR/Cas as a potential diagnosis technique for COVID-19. Avicenna J Med Biotechnol. 2020;12(3):201–202. [PMC free article] [PubMed] [Google Scholar]
- 28.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 31.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol. 2015;96(8):2252–2261. doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- 34.Ophinni Y, Inoue M, Kotaki T, Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci Rep. 2018;8(1):7784. doi: 10.1038/s41598-018-26190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roehm PC, Shekarabi M, Wollebo HS, Bellizzi A, He L, Salkind J, et al. Inhibition of HSV-1 replication by gene editing strategy. Sci Rep. 2016;6(1):23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Wang W, Cui YC, Pan Q, Zhu W, Gendron P, et al. HIV-1 Employs multiple mechanisms to resist Cas9/single guide RNA targeting the viral primer binding site. J Virol. 2018;92(20):01135–18. doi: 10.1128/JVI.01135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(5):826–837.e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metsky HC, Freije CA, Kosoko-Thoroddsen T-SF, Sabeti PC, Myhrvold C. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design [Internet]. Genomics; 2020 [cited 2020 Dec 8]. Available from: 10.1101/2020.02.26.967026
- 40.Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nalawansha DA, Samarasinghe KTG. Double-barreled CRISPR technology as a novel treatment strategy for COVID-19. ACS Pharmacol Transl Sci. 2020;3(5):790–800. doi: 10.1021/acsptsci.0c00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lotfi M, Rezaei N. CRISPR/Cas13: a potential therapeutic option of COVID-19. Biomed Pharmacother. 2020;131:110738. doi: 10.1016/j.biopha.2020.110738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Zhong M, Liu Y, Ma P, Dang L, Meng Q, et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci Bull. 2020;65(17):1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W, Yu L, Wen D, Wei D, Sun Y, Zhao H, et al. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine. 2020;61:103036. doi: 10.1016/j.ebiom.2020.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schermer B, Fabretti F, Damagnez M, Di Cristanziano V, Heger E, Arjune S, et al. Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS ONE. 2020;15(11):e0238612. doi: 10.1371/journal.pone.0238612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimi K, Takeshita K, Yamayoshi S, Shibumura S, Yamauchi Y, Yamamoto M, et al. Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3 [Internet]. Infectious Diseases (except HIV/AIDS); 2020 [cited 2021 Feb 4]. Available from: 10.1101/2020.06.02.20119875
- 47.Brandsma E, Verhagen HJMP, van de Laar TJW, Claas ECJ, Cornelissen M, van den Akker E. Rapid, sensitive and specific SARS coronavirus-2 detection: a multi-center comparison between standard qRT-PCR and CRISPR based DETECTR [Internet]. Infectious Diseases (except HIV/AIDS); 2020 [cited 2021 Feb 4]. Available from: 10.1101/2020.07.27.20147249 [DOI] [PMC free article] [PubMed]
- 48.Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 50.Crone MA, Priestman M, Ciechonska M, Jensen K, Sharp DJ, Anand A, et al. A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat Commun. 2020;11(1):4464. doi: 10.1038/s41467-020-18130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramachandran A, Huyke DA, Sharma E, Sahoo MK, Huang C, Banaei N, et al. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117(47):29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Z, Tian D, Liu Y, Lin Z, Lyon CJ, Lai W, et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niu A, McDougal A, Ning B, Safa F, Luk A, Mushatt DM, et al. COVID-19 in allogeneic stem cell transplant: high false-negative probability and role of CRISPR and convalescent plasma. Bone Marrow Transplant. 2020;55(12):2354–2356. doi: 10.1038/s41409-020-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arizti-Sanz J, Freije CA, Stanton AC, Petros BA, Boehm CK, Siddiqui S, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rauch JN, Valois E, Solley SC, Braig F, Lach RS, Audouard M, et al. A Scalable, Easy-to-Deploy, Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material [Internet]. Molecular Biology; 2020 [cited 2021 Feb 4]. Available from: 10.1101/2020.04.20.052159 [DOI] [PMC free article] [PubMed]
- 57.Rauch JN, Valois E, Ponce-Rojas JC, Aralis Z, Lach RS, Zappa F, et al. CRISPR-based and RT-qPCR surveillance of SARS-CoV-2 in asymptomatic individuals uncovers a shift in viral prevalence among a university population [Internet]. Infectious Diseases (except HIV/AIDS); 2020 [cited 2021 Feb 4]. Available from: 10.1101/2020.08.06.20169771