Abstract
The human and canine parasitic nematode Strongyloides stercoralis utilizes an XX/XO sex determination system, with parasitic females reproducing by mitotic parthenogenesis and free-living males and females reproducing sexually. However, the genes controlling S. stercoralis sex determination and male development are unknown. We observed precocious development of rhabditiform males in permissive hosts treated with corticosteroids, suggesting that steroid hormones can regulate male development. To examine differences in transcript abundance between free-living adult males and other developmental stages, we utilized RNA-Seq. We found two clusters of S. stercoralis-specific genes encoding predicted transmembrane proteins that are only expressed in free-living males. We additionally identified homologs of several genes important for sex determination in Caenorhabditis species, including mab-3, tra-1, fem-2, and sex-1, which may have similar functions. However, we identified three paralogs of gld-1; Ss-qki-1 transcripts were highly abundant in adult males, while Ss-qki-2 and Ss-qki-3 transcripts were highly abundant in adult females. We also identified paralogs of pumilio domain-containing proteins with sex-specific transcripts. Intriguingly, her-1 appears to have been lost in several parasite lineages, and we were unable to identify homologs of tra-2 outside of Caenorhabditis species. Together, our data suggest that different mechanisms control male development in S. stercoralis and Caenorhabditis species.
Subject terms: Parasitology, Parasite biology, Parasite development, Parasite evolution, Parasite genetics, Parasitic infection
Introduction
Strongyloides stercoralis is a skin-penetrating parasitic nematode that infects approximately 614 million people globally and causes the disease strongyloidiasis, which can persist for decades and is often asymptomatic1. However, S. stercoralis infection can result in chronic intestinal and/or respiratory issues, as well as death in cases of hyperinfection and dissemination, which can be triggered by either corticosteroid treatment or human T-lymphotropic virus type 1 (HTLV-1) co-infection2. S. stercoralis also infects dogs and non-human primates3, and the presence of genetically similar strains in dogs and humans indicate the potential for zoonotic transmission4. S. stercoralis is used as a model to study nematode parasitism, since it is one of the more tractable species for genetic manipulation due to the ability to insert transgenes in the free-living generation5.
The S. stercoralis life cycle is more complex than other obligate parasitic nematodes and can alternate between a parasitic and a single gonochoristic (with distinct male and female individuals) free-living generation3. The parasitic female, which lives in the crypts of the small intestine3, reproduces by mitotic parthenogenesis6. The post-parasitic generation, which can include both males and females7,8, develops either homogonically to infectious larvae or heterogonically to free-living adults3. Post-parasitic males invariably develop into free-living rhabditiform (short pharynx with two bulbs) larvae. By contrast, post-parasitic females can either develop into free-living rhabditiform larvae or dauer-like filariform (elongated and radially constricted pharynx) infectious third-stage larvae outside of the host, termed iL3, or inside the host as autoinfective larvae, termed aL33. Free-living adult males and females reproduce sexually9, producing post-free-living larvae that are all female and invariably develop into iL310. For iL3 to continue development, they must infect a permissive host—where they eventually mature into parasitic females3.
The switch controlling homogonic versus heterogonic development for S. stercoralis post-parasitic female larvae is triggered early in the first larval stage (L1)11. Factors controlling this switch include temperature12 and strain genetics13, and these signals are mediated, in part, by dafachronic acids14. By contrast, post-parasitic male development appears to be determined by the allocation of a single X chromosome to the egg12. Greater than 95% of post-parasitic larvae in the S. stercoralis strain used in this study develop via the heterogonic route when cultured at 22 °C.
Although most nematodes, including the model organism Caenorhabditis elegans, utilize an XX/XO mechanism for sex determination, other mechanisms have evolved, including Y chromosomes in some filarial species15 and chromosomal diminution in some Strongyloides species16,17. Furthermore, significant plasticity has been observed in nematode sex chromosomes, as new sex chromosomes have evolved in several lineages15. Sex determination in S. stercoralis is likely an XX/XO mechanism, similar to the closely-related S. ratti18. Both S. stercoralis and S. ratti have two pairs of autosomes, with free-living and parasitic females additionally possessing two X chromosomes (2n = 6) and males additionally possessing one X chromosome (2n = 5)6,18. However, in S. papillosus and S. vituli, chromosome I and X have fused, and males undergo a sex-specific diminution of the X-containing portion for just one of these fused chromosomes16,17. Since the more distantly related facultative parasite Parastrongyloides trichosuri has three pairs of chromosomes and also utilizes an XX/XO sex determination system, chromatin diminution in S. papillosus and S. vituli is likely a derived trait17. In contrast to C. elegans, where the X chromosome to autosome ratio is the initiating sex determination signal, environmental signals are sensed by the parasitic female and control the elimination of an X chromosome (in the case of S. stercoralis and S. ratti) or the portion of a chromosome (in the case of S. papillosus and S. vituli) and thus male development in Strongyloides species19.
The proportion of males and females in the post-parasitic generation is unequal in Strongyloides species, and the proportion of free-living males increases over the duration of an infection for both S. stercoralis and S. ratti12,20. Furthermore, immunosuppressing the host can decrease the proportion of larvae developing into free-living males in S. ratti20. The molecular mechanism behind this environmentally-influenced sex determination system is unknown. However, previous studies have hypothesized that S. ratti may use a modified form of mitotic parthenogenesis where only X chromosomes undergo a “mini meiosis,” resulting in XO oocytes and XXX polar bodies18. Chromosomal non-disjunction is also a possibility; however, this mechanism results in both XO male and XXX progeny in C. elegans21, and XXX progeny have not been observed in Strongyloides species (although they could be non-viable).
The mechanism by which post-free-living larvae invariably inherit the paternal X chromosome, in addition to a maternal X chromosome, is also unclear. One possibility is that spermatids receiving an X chromosome attract the bulk of the cytoplasm and organelles required for sperm function, similar to that observed in the trioecious nematode Auanema rhodensis22. In S. papillosus, mature sperm with a diminished X region are not formed16; however, nullo-X sperm have been observed in S. ratti23,24, and meiotic cells with either two or three chromosomes have been observed in the male testis of S. stercoralis6. In both S. ratti and S. stercoralis, free-living males contribute genetic material to the offspring, ruling out pseudogamy9,25.
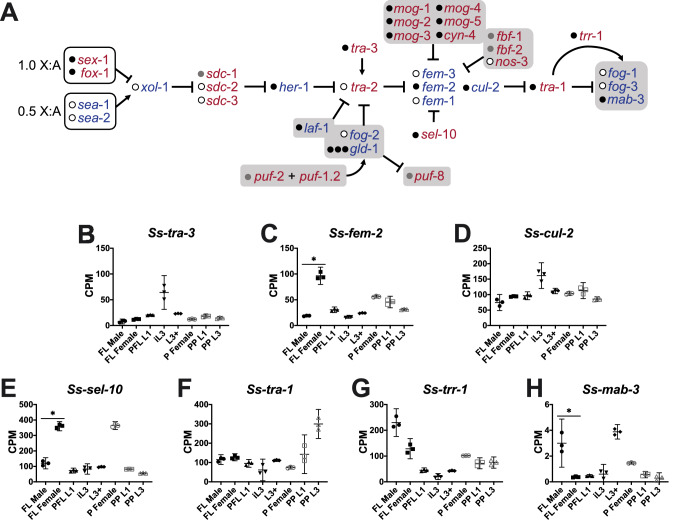
Caenorhabditis species have several genes that detect the ratio of autosomes to X chromosomes, genes that propagate this signal, and genes involved in tissue-specific responses for both somatic and germline sex determination (Fig. 1A). A few of these genes, including the transcription factors encoded by mab-3 and tra-1, appear to regulate sex determination in a broad range of species; however, there is some divergence in genes controlling sex determination at the periphery of the “core” pathway26 (Fig. 1A). As proposed by Wilkins (1995), the “bottom up” hypothesis posits that the most downstream genes in a sex determination pathway evolved before upstream elements and, therefore, should be more conserved between nematode species27.
Figure 1.
Components of the Caenorhabditis sex determination pathway are present in S. stercoralis, and key transcription factors likely have conserved function. (A) Genes that compose the “core” sex determination pathway in Caenorhabditis species, along with accessory genes that play a role in somatic (white boxes) and germline (gray boxes) sex determination, are diagrammed, with feminizing factors in red and masculinizing factors in blue. Homologs in S. stercoralis (black circle), homologs encoding a similar type of protein (gray circle), and genes for which no homolog was identified in the S. stercoralis genome (white circle), are indicated. Adapted from26. (B–H) Transcript abundances of the S. stercoralis homologs are represented using TMM-normalized counts per million (CPM) for the following developmental stages: free-living adult males (FL Male), free-living gravid adult females (FL Female), post-free-living first-stage larvae (PFL L1), developmentally arrested infectious third-stage larvae (iL3), L3 activated inside a permissive host (L3 +), parasitic gravid adult females (P Female), heterogonically-developing post-parasitic L1 (PP L1), and heterogonically-developing post-parasitic L3 enriched for females (PP L3). Graphs were constructed using GraphPad Prism v.9.0.0; bars indicate means (horizontal) and 95% confidence intervals (vertical) for each of the three biological replicates represented as individual data points. Asterisks (*) indicate a significant (fold change > 2.0; FDR < 0.05) difference between FL Male and FL Female.
The genes important for regulating the development of Strongyloides (clade IV28) free-living males and females, or their gametes, are not well-understood. Additionally, whether homologs of genes important for sex determination in Caenorhabditis species (clade V) have been conserved and have similar functions in Strongyloides species is also unknown. We therefore sought to identify S. stercoralis homologs of genes involved in sex determination and characterize the differences in gene expression in free-living adult males and females.
Materials and methods
S. stercoralis maintenance and experimental infections
We maintained S. stercoralis strain PV00129 in prednisolone-treated beagles, as previously described30, in accordance with protocols 702342, 801905, and 802593, approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC). We conducted experimental infections of Meriones unguiculatus (Mongolian gerbils) strain Crl:MON(Tum) (Charles River Laboratories, Wilmington, MA) by injecting 1,500 iL3 in phosphate buffered saline subcutaneously under the same IACUC protocols; we induced autoinfection by subcutaneously injecting 2 mg methylprednisolone acetate weekly starting with the day of infection31. We sacrificed M. unguiculatus after 16–18 days by CO2 asphyxiation in accordance with standards established by the American Veterinary Medical Association. We recovered the luminal contents of the small and large intestines by suspending the gut segments, split length-wise, in graduated cylinders with Dulbecco's Modified Eagle's Medium (DMEM), supplemented with gentamycin [100 μg/ml], for three hours at 37 °C, as previously described32. We performed all protocols and routine care of the animals in strict accordance with both the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the ARRIVE guidelines33.
Isolation of S. stercoralis free-living adult males for RNA-Seq
To first isolate S. stercoralis free-living adults, we incubated charcoal coprocultures34 (composed of fresh dog feces and bone charcoal) for 48 h at 21 °C and then used the Baermann technique, with water at 28–29 °C, to induce free-living adults to migrate from the cultures. We then removed a 30 ml aliquot of the Baermann effluent and allowed worms to sediment at 1 × g for 10 min. In order to separate the worms from most contaminating bacteria, we mixed ~ 1–2 ml of worms and water with an equal volume of 1% SeaKem low-melting temperature agarose at 30 °C (Lonza, Basel, Switzerland) and transferred this mixture to a petri dish. We then cooled the suspension on a glass slab at 4 °C until the agarose began to gel and then transferred it to room temperature for 10–15 min. We induced the mixture of free-living adult males, free-living adult females, and larvae to migrate from the agarose, which retained most contaminants, by adding 14 ml of BU Buffer35 and incubating at 28 °C for 30 min. We then manually isolated and transferred several hundred young free-living adult males to a 1.5 ml microfuge tube with a pipette and allowed them to settle at 1 × g for ~ 10 min. We then removed the supernatant and mixed the ~ 10 μl of adult males with 200 μl TRIzol reagent (Thermo Fisher Scientific, Waltham, MA) and snap froze the mixture in liquid nitrogen. This isolation of free-living adult male worms was performed in biological triplicate on different days.
RNA extraction, library preparation, and sequencing
We extracted total RNA from each replicate of free-living adult males using TRIzol and the manufacturer’s protocol, and we then quantified RNA concentration and determined the RNA integrity number (RIN) using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). All three samples had a RIN of 10.0. We then constructed libraries, each with unique indexes, using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA) using 500 ng of total RNA as starting material and a previously described protocol32. We determined the concentration of the three libraries using the Kapa SYBR Fast qPCR Kit for Library Quantification (Kapa Biosystems, Inc.) and pooled the three libraries as previously described. We then sequenced the three pooled libraries on an Illumina HiSeq 2000 with 100 base pair paired-end reads, and we then demultiplexed the reads using the unique indexes. Raw RNA-Seq reads for each of the three biological replicates of free-living adult males are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under BioProject ID: PRJNA689252.
RNA-Seq read processing and mapping
In addition to raw RNA-Seq reads from free-living adult males, we included raw RNA-Seq reads from S. stercoralis free-living adult females, post-free-living L1, iL3, L3 activated inside a permissive host (L3 +), parasitic adult females, post-parasitic L1 (primarily developing heterogonically), and post-parasitic L3 enriched for females (also developing heterogonically) in our analysis from previously described data32, available under BioProject ID: PRJEB3116. Since these libraries were constructed at the same time as those derived from free-living adult males and were sequenced using the same chemistry and instrument, but at an earlier date, batch effects should be minimized. To assess read quality, we used FastQC v.0.11.936. We then trimmed low-quality bases and index sequences with Trimmomatic v.0.3937 and the options ILLUMINACLIP:2:30:10:1 SLIDINGWINDOW:2:10 LEADING:10 TRAILING:10 MINLEN:40. We confirmed the removal of the low-quality bases and index sequences with FastQC. We subsequently removed contaminating rRNA sequences from the trimmed reads using bbduk.sh from bbtools v.38.8438 and a list of S. stercoralis 18S, 5.8S, 28S, and 5S rRNA sequences, including accession numbers M84229, AF279916, KU180693, DQ145710, EF653265, and sequences from the PV001 strain derived from previously de novo assembled RNA-Seq reads32. We mapped the processed reads to the S. stercoralis genome v2.0.439 using HISAT2 v.2.2.040 with the max intron length set to 50,000. We then converted the output SAM files to BAM files with SAMtools v.1.941.
Identification of S. stercoralis homologs
We identified S. stercoralis homologs of canonical C. elegans sex-determination genes by reciprocal Basic Local Alignment Search Tool (BLAST) searches. We first used C. elegans polypeptide sequences as queries for BLAST searches against the S. stercoralis genome39 using Geneious v.11.1.5 (Biomatters, Ltd., Auckland, New Zealand). We then corrected or confirmed the annotation for each S. stercoralis gene using RNA-Seq reads aligned to the genome and visualized using the Integrative Genome Viewer v.2.3.9442. We then predicted protein sequences of S. stercoralis homologs using the annotated transcripts in Geneious and subsequently used the predicted protein sequences as protein BLAST queries against the C. elegans database in NCBI to confirm gene identity. Only genes with top hits in reciprocal BLAST searches, where the second hit had a significantly higher e-value in both searches or was resolved by phylogenetic analysis (see below), were identified as one-to-one homologs. We manually adjusted any changes in the S. stercoralis genome annotations (Supplemental Data S1). All annotated S. stercoralis transcripts include the prefix Ss.
We used C. elegans and S. stercoralis predicted protein sequences as NCBI protein BLAST search queries to identify related predicted polypeptides in clade I, III, IV, and V nematodes28. We additionally performed BLAST searches of parasitic nematode genomes available on WormBase ParaSite43. We aligned the protein sequences with ClustalW and a BLOSUM matrix and then created neighbor-joining (N-J) phylogenetic trees with 100 iterations of bootstrapping in Geneious. Accession numbers or gene IDs for additional sequences are listed in the respective phylogenetic trees.
To determine whether predicted polypeptide sequences had a predicted signal peptide, we utilized SignalP v.5.0 server44. Similarly, to determine whether a predicted polypeptide had predicted transmembrane domains, we utilized TMHMM Server v.2.045.
Transcript abundance quantification and differential expression analysis
To quantify transcript abundance for each gene, we first converted the updated GFF3 annotation file to a GTF file using custom scripts and gffread v.0.11.746 and then quantified the paired-end fragment counts for the coding sequence (CDS) of each gene using featureCounts from SubReads v.2.0.147. We performed trimmed mean of M-values (TMM) normalization of counts per million (CPM) values for each gene in each biological replicate (Supplemental Data S2), using featureCounts summary data and EdgeR v.3.32.048,49. We performed differential gene expression analysis between developmental stages with glmQLFit and glmTreat functions in the EdgeR package using a significance threshold of: a minimum log2 fold change of one, a minimum mean log2 CPM of one in at least one developmental stage, a p-value adjusted threshold of < 0.05, and a p-value adjustment using Benjamini–Hochberg false-discovery-rate (FDR) (Supplemental Data S3).
Data visualization
Rhabditiform males recovered from experimentally infected Mongolian gerbils were wet-mounted without anesthetics using an Olympus BX60 compound microscope equipped with differential interference contrast (DIC) optics, a Spot RT Color digital camera, and Spot Advanced v5.1 image analysis software (Diagnostic Instruments, Inc., Sterling Heights, MI).
We used EdgeR to generate a multidimensional scaling (MDS) plot of all samples and a mean-difference (MD) plot of differential gene expression between free-living adult males and free-living adult females. We plotted the TMM-normalized CPM for each replicate, the mean TMM-normalized CPM, and error bars that represent 95% confidence intervals for the three replicates of each developmental stage in GraphPad Prism v.9.0.0 (GraphPad Software, Inc., San Diego, USA).
Results and discussion
S. stercoralis males are invariably rhabditiform
S. stercoralis, along with other Strongyloides species, has a unique life cycle with two types of adults: a parasitic form that is a parthenogenetic female, while the free-living forms are gonochoristic and reproduce sexually. Although Kreis (1932) detailed morphological descriptions of S. stercoralis parasitic males7, Faust (1933) described parasitic males in the ileum and rectum of the host8, and others have found rhabditiform males in the lungs50, it is now generally accepted that the parasitic generation is only female3. Interestingly, we have observed S. stercoralis L4/adult rhabditiform males, with characteristic enlarged pharyngeal bulbs and pair of copulatory spicules, in the large intestine of experimentally infected Mongolian gerbils that have been treated with corticosteroids and are undergoing hyperinfection (Supplemental Figure S1). These males are morphologically similar to those described by Kreis7.
We hypothesize that these rhabditiform males are precociously developing free-living males—an observation consistent with the male sex of the post-parasitic generation of S. stercoralis being determined by the inheritance of a single X chromosome, such that genetically male post-parasitic larvae invariably develop towards a rhabditiform free-living larva whether they are in the environment or in the host6. Furthermore, our findings suggest that corticosteroid treatment not only triggers precocious development of post-parasitic female larvae, but also post-parasitic male larvae. While the mechanism by which corticosteroid treatment triggers autoinfection is unknown, our findings suggest that corticosteroids may directly influence development of post-parasitic larvae. Whether precociously developing rhabditiform males up-regulate astacin-like metallopeptidases, which are up-regulated in iL339,51 and important for skin penetration52, or whether they are capable of penetrating the gut wall and migrating within the host also remains unknown.
The transcriptional profile of S. stercoralis free-living adult males is distinct from other developmental stages
Since little is known about the changes in gene expression that control development of post-parasitic larvae fated for free-living male or female development, we examined changes in transcript abundance between free-living adult males and free-living adult females using RNA-Seq. We isolated S. stercoralis free-living adult males in biological triplicate and used the three amassed cohorts to construct RNA-Seq libraries. We sequenced the libraries on an Illumina HiSeq 2000, generating a mean of 46.2 million paired-end reads (± 1.4 million, standard error of the mean) per sample.
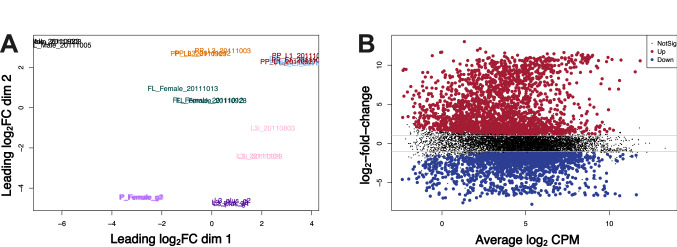
To identify differences in transcript abundance between S. stercoralis free-living adult males and other developmental stages32, we characterized each developmental stage in biological triplicate using RNA-Seq followed by TMM normalization and statistical testing using EdgeR. An MDS plot that arranged each sample in space based on similarities/differences in transcript abundance revealed that biological replicates grouped tightly together, while developmental stages were well separated (Fig. 2A). An MD plot comparing differences in transcript abundance between free-living adult males and free-living adult females revealed 2,215 transcripts that were significantly up-regulated in free-living adult males and 2,046 transcripts that were significantly up-regulated in free-living adult females (Fig. 2B). We also observed a subset of genes that were up-regulated to a greater extent in free-living adult males than genes up-regulated in free-living adult females (Fig. 2B). A similar observation has been made in males of several Caenorhabditis species53.
Figure 2.
S. stercoralis developmental stages have distinct transcript expression profiles. (A) S. stercoralis developmental stages examined by RNA-Seq are well-separated, and the three biological replicates group closely, by multidimensional scaling (MDS) analysis. (B) Numerous transcripts are significantly (log2 fold change > 1.0; FDR < 0.05) up-regulated (red) and down-regulated (blue) in free-living adult males, in comparison to free-living adult females, by mean-difference (MD) analysis. A subset of transcripts are significantly up-regulated in free-living adult males to a greater extent than transcripts are down-regulated. Plots were constructed using EdgeR v.3.32.0.
Transcripts encoding major sperm proteins and two clusters of predicted transmembrane polypeptides are among the most highly up-regulated transcripts in free-living adult males
To investigate which transcripts are most up-regulated in S. stercoralis free-living adult males in comparison to free-living adult females, we characterized the top 15 most up-regulated transcripts in free-living adult males (Table 1). Three of these transcripts are predicted to encode canonical 127 amino acid major sperm proteins (MSPs), which are conserved nematode-specific proteins that form stable dimers and assemble into actin-like filaments that are crucial for sperm motility54. In contrast to the C. elegans genome, which has 31 MSP-encoding genes55, we were only able to identify three canonical 127 amino acid MSP-encoding genes in the S. stercoralis genome.
Table 1.
Most up-regulated transcripts in S. stercoralis free-living adult males in comparison to free-living adult females.
| Gene name | *Log2 fold change | Predicted number of amino acids | Signal peptide likelihood | Predicted transmembrane domain(s) | Predicted homology and/or function |
|---|---|---|---|---|---|
| SSTP_0001266600 | 13.0 | 126 | 0.7582 | 102–124 aa |
C. elegans—no significant similarity S. ratti—similar proteins, but no 1-to-1 ortholog |
| SSTP_0000642100 | 12.1 | 208 | 0.9393 | – |
C. elegans—no significant similarity S. ratti—similar proteins, but no 1-to-1 ortholog |
| SSTP_0000017020 | 11.9 | 59 | 0.0005 | 18–40 aa |
C. elegans—no significant similarity S. ratti—no significant similarity |
| SSTP_0000031000 | 11.7 | 79 | 0.9996 | – |
C. elegans—no significant similarity S. ratti—hypothetical protein XP_024503864.1 (e-11) |
| SSTP_0001268100 | 11.6 | 182 | 0.9876 | 158–180 aa |
C. elegans—no significant similarity S. ratti—similar proteins, but no 1-to-1 ortholog |
| SSTP_0001268000 | 11.6 | 219 | 0.0041 | 125–147 aa |
C. elegans—no significant similarity S. ratti—similar proteins, but no 1-to-1 ortholog |
| SSTP_0000894400 | 11.6 | 223 | 0.9989 | 183–205 aa | emp24/gp25L/p24/GOLD family protein, which are involved in vesicular protein trafficking from the ER |
| SSTP_0000040300 | 11.6 | 90 | 0.0011 | – |
C. elegans—no significant similarity S. ratti—hypothetical protein XP_024505868.1 (e-23) |
| SSTP_0000384900 | 11.5 | 129 | 0.9783 | – |
C. elegans—no significant similarity S. ratti—no significant similarity |
| Ss_msp_3 | 11.4 | 127 | 0.0006 | – | major sperm protein (canonical) |
| SSTP_0001267400 | 11.4 | 190 | 0.9612 | 95–117 aa |
C. elegans—no significant similarity S. ratti—similar proteins, but no 1-to-1 ortholog |
| Ss_msp_1 | 11.4 | 127 | 0.0007 | – | major sperm protein (canonical) |
| SSTP_0001267900 | 11.4 | 189 | 0.9966 | 168–187 aa |
C. elegans—no significant similarity S. ratti—similar proteins, but no 1-to-1 ortholog |
| Ss_msp_2 | 11.3 | 127 | 0.0008 | – | major sperm protein (canonical) |
| SSTP_0000016700 | 11.3 | 110 | 0.0014 | 10–32 aa |
C. elegans—no significant similarity S. ratti—no significant similarity |
*For significance: FDR < 0.05, log2 fold change > 1.0, and log2 CPM > 1.0 in ≥ 1 developmental stage.
Intriguingly, we noted that several of the most up-regulated genes were clustered in the genome (Table 1). Upon further investigation, we identified two gene clusters, one on scaffold 5 and one on scaffold 9, where each gene in the cluster is highly expressed only in free-living adult males (Supplemental Figures S2 and S3). The cluster on scaffold 5 includes 12 genes, with the majority encoding predicted polypeptides that lack a signal peptide and are predicted Type I single-pass transmembrane proteins (N-term is extracellular and C-term is cytoplasmic) with the transmembrane domain located near the middle of the predicted polypeptide (Supplemental Table S1). None of these 12 predicted polypeptides have BLAST hits in S. ratti or C. elegans, nor do they have any clear phylogenetic relationships with each other. The cluster on scaffold 9 includes 14 genes and one pseudo-gene, with the majority encoding predicted polypeptides with both a predicted signal peptide and a predicted Type I single-pass transmembrane domain, with the transmembrane domain located near the C-terminus (Supplemental Table S2). Although we were able to identify seven homologs of these genes in S. ratti as well as one homolog in P. trichosuri, we were unable to identify any one-to-one homologs or clear phylogenetic relationships.
Our data suggest that the genes in these two clusters encode transmembrane proteins that are specific to S. stercoralis and are only present in free-living males. We hypothesize these transmembrane proteins may be important for sperm development or function, as they are up-regulated to a similar extent as transcripts encoding MSPs. Furthermore, these genes appear to be undergoing rapid sequence evolution, as they have little direct homology to each other or to genes outside of S. stercoralis. These genes may have evolved under similar selective pressures as the male secreted short (mss) genes that encode glycosylphosphatidylinositol (GPI)-anchored proteins found in spermatocytes of outcrossing Caenorhabditis species and are required for sperm competitiveness56.
Ss-srg-14 encodes a G protein-coupled receptor that is up-regulated in free-living adult males
In C. elegans, hermaphrodites produce ascaroside pheromones that attract males57, and pheromones have been described in P. trichosuri58. Since G protein-coupled receptors (GPCRs) play a role in sensing these pheromone cues59–61, we sought to determine whether any putative GPCR-encoding transcripts62 are up-regulated in S. stercoralis free-living males (Supplemental Figure S4). We found that Ss-srg-14 is highly expressed in males and 8.5-fold up-regulated in free-living adult males in comparison to free-living adult females (FDR < 0.05). We hypothesize that the GPCR encoded by Ss-srg-14 may be important for the detection of female pheromones. Future studies using CRISPR/Cas9 to knock out Ss-srg-14 may be informative in determining its biological function.
TRA-1 and MAB-3, which regulate sex determination in Caenorhabditis species, are present in S. stercoralis and may have similar functions
A variety of genetic techniques have been used to identify genes that regulate sex determination in C. elegans, C. briggsae, and other Caenorhabditis species26 (Fig. 1A); henceforth, Caenorhabditis genes do not have prefixes. These genes fall into several broad categories: genes that recognize the X chromosome to autosome ratio (e.g., sex-1 and fox-1) and integrate this signal (xol-1), genes that are part of the dosage compensation complex (e.g., sdc-1, sdc-2, and sdc-3), genes involved in a secreted ligand and membrane receptor pathway (e.g., her-1, tra-2, tra-3, fem-1, fem-2, and fem-3), the global regulator tra-1, and tissue-specific response genes (e.g., fog-1, fog-3, and mab-3)26. In order to identify S. stercoralis homologs of these genes, we utilized both BLAST searches and phylogenetic analyses. To determine whether our inability to find some S. stercoralis homologs of Caenorhabditis sex determination genes was due to an incomplete genome assembly, we performed BLAST searches of other clade III, IV, and V nematode species (Table 2). We then examined changes in transcript abundance of the S. stercoralis homologs throughout the life cycle (Fig. 1B-H; Supplemental Figure S5). Although differences in transcript abundance are not necessary for a gene to regulate sex determination, as Caenorhabditis species employ multiple post-transcriptional regulatory mechanisms in the sex determination pathway, differences in transcript abundance between free-living adult males and free-living adult females of S. stercoralis homologs help to elucidate transcripts that are sex-biased.
Table 2.
Homologs of genes involved in Caenorhabditis sex determination in representative clade III, IV, and V nematodes.
| xol-1 | sdc-1 | sdc-2 | sdc-3 | her-1 | tra-2 | tra-3 | fem-1 | fem-2 | fem-3 | tra-1 | mab-3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade V | ||||||||||||
| Caenorhabditis elegans | + | + | + | + | + | + | + | + | + | + | + | + |
| Caenorhabditis briggsae | + | + | + | + | + | + | + | + | + | + | + | + |
| Pristionchus pacificus | − | +/−* | − | − | + | − | + | + | + | − | + | + |
| Ancylostoma ceylancum | − | +/−* | + | − | + | − | + | + | + | + | + | + |
| Haemonchus contortus | − | +/−* | + | − | + | − | + | + | + | + | + | + |
| Clade IV | ||||||||||||
| Strongyloides stercoralis | − | +/−* | − | − | + | − | + | − | + | − | + | + |
| Strongyloides ratti | − | +/−* | − | − | − | − | + | − | + | − | + | + |
| Strongyloides papillosus | − | +/−* | − | − | + | − | + | − | + | − | + | + |
| Bursaphelenchus xylophilus | − | +/−* | − | − | − | − | + | − | + | − | + | + |
| Meloidogyne enterolobii | − | +/−* | - | − | − | − | + | − | + | − | + | + |
| Clade III | ||||||||||||
| Ascaris suum | − | +/−* | − | − | − | − | + | + | + | − | + | + |
| Brugia malayi | − | +/−* | − | − | + | − | + | + | + | − | + | + |
| Loa loa | − | +/−* | − | − | + | − | + | + | + | − | + | + |
| Toxocara canis | − | +/−* | − | − | + | − | + | + | + | − | + | + |
*Unclear whether these homologs are more similar to Caenorhabditis elegans sdc−1 or C04F5.9.
We identified S. stercoralis homologs of several “core” sex determination genes (Fig. 1A), including tra-1, which encodes a zinc-finger transcription factor related to Drosophila cubitus interruptus (Ci) that also plays a role in P. pacificus sex determination63,64, and mab-3, which encodes a doublesex (Dsx) -related transcription factor65. We found that the S. stercoralis homolog of the masculinizing mab-3 gene, Ss-mab-3, is 7.9-fold up-regulated in free-living adult males in comparison to free-living adult females (FDR < 0.05), which is consistent with mab-3 up-regulation in C. elegans males66. Furthermore, we identified homologs of genes encoding TRA-1 and MAB-3 in all clade III, IV, and V nematodes that we examined (Table 2). Due to the broad conservation of these genes across nematode species and their role in sex determination in other ecdysozoans, we hypothesize that tra-1 and mab-3 play a similar role in sex determination in S. stercoralis and other nematodes.
In Caenorhabditis species, TRA-1 activity is repressed in males by a FEM-1, FEM-2, FEM-3, and CUL-2 complex that ubiquitinates TRA-1 and targets it for degradation via the proteasome67. We identified S. stercoralis homologs of the genes fem-2, which encodes a PP2C phosphatase68, sel-10, which encodes an F-box protein that is a component of E3 ubiquitin ligases69, and cul-2, which encodes a cullin family protein that functions in ubiquitin ligase complexes67. S. stercoralis homologs of fem-2 and sel-10 are significantly up-regulated in free-living adult females in comparison to free-living adult males (5.1-fold and 3.0-fold up-regulated for Ss-fem-2 and Ss-sel-10, respectively; FDR < 0.05). Whether Ss-fem-2 and Ss-sel-10, which are members of gene families with functions outside of sex determination70, have a conserved role in S. stercoralis sex determination will require future functional studies.
We were only able to identify homologs of genes encoding FEM-3, which directly interacts with the C-terminal domain of TRA-2 in Caenorhabditis species71, in a few clade V nematodes (Table 2). As fem-3 appears to be rapidly evolving and has significant sequence divergence between Caenorhabditis species72, we may be unable to detect divergent homologs in clade III and IV nematodes by sequence homology. Interestingly, we were unable to identify homologs of genes encoding FEM-1, which is an E3 ubiquitin ligase subunit that recognizes substrates—including TRA-1—for targeted ubiquitination67, in clade IV parasites, but we were able to identify homologs in clade III and V nematodes (Table 2). Since Ss-tra-1 transcript abundance does not differ significantly between free-living adult males and females (Fig. 1F), we hypothesize that the S. stercoralis homolog of TRA-1 is still regulated post-translationally. However, the apparent loss of fem-1 in both S. stercoralis and other clade IV nematodes suggests that these parasites may have a modified mechanism of regulating TRA-1 activity.
The HER-1 ligand and TRA-2 receptor, which regulate sex determination in Caenorhabditis species, appear to have been lost in some parasitic nematode lineages
In C. elegans, the HER-1 ligand is expressed in males and represses the TRA-2A transmembrane receptor73, resulting in the repression of FEM-371. Additionally, TRA-3 can cleave the C-terminus of TRA-2A74, with the resulting fragment having feminizing activity via binding to FEM-371. We were unable to identify an S. stercoralis homolog of tra-2, which in C. elegans encodes a 1475 amino acid 12 transmembrane protein75 that is directly bound by the ligand encoded by her-173. Although we were able to identify conserved HER-1 homologs in S. stercoralis and several other clade III76, IV, and V nematodes (Supplemental Figure S6; Table 2), we were unable to identify TRA-2 homologs outside of the Caenorhabditis genus—despite the ready identification of the related Patched-1 and Patched-3 homologs75 in clade III, IV, and V nematodes (Supplemental Figure S7; Table 2). Intriguingly, Ss-her-1 was not expressed in any of the S. stercoralis developmental stages we examined, and the remnant of the S. ratti her-1 coding sequence, located between SRAE_2000507500 and SRAE_2000507600, has acquired several stop codons and deletions. In addition to S. ratti, we were unable to identify genes encoding HER-1 homologs in several disparate species (Table 2). By contrast, genes encoding TRA-3, a calpain protease that cleaves TRA-2A74, were broadly conserved among the parasitic nematode species we examined (Table 2).
Our observation that TRA-2 homologs are not readily identifiable by BLAST searches outside of the Caenorhabditis genus suggests that either TRA-2 has evolved so rapidly that it is not distinguishable by protein sequence homology in other species or that another protein functions as the receptor for HER-1 in other nematode species. We could not identify a TRA-2 homolog in either Diploscapter coronatus or D. pachys, but we were able to identify a TRA-2A homolog in C. parvicauda (14.5% identity to C. elegans TRA-2A), which is the most distantly-related Caenorhabditis species from C. elegans with a sequenced genome77. By contrast, the HER-1 sequence is conserved across the clade III, IV, and V nematode species in which it is present. Therefore, we speculate that TRA-2 may be a rapidly-evolving Patched-1/-3 paralog in Caenorhabditis species. Alternatively, homologs of the several hedgehog-like ligands in C. elegans78, of which HER-1 is a member, could function in sex determination in other nematode species. This is supported by the loss of her-1 in parasites such as S. ratti. Finally, it is also possible that HER-1 and TRA-2 have simply been co-opted into a role in sex determination in Caenorhabditis species.
Proteins that respond to the X chromosome to autosome ratio signal in Caenorhabditis species appear to be rapidly evolving
In Caenorhabditis species, XOL-1 is active only in males and integrates the X chromosome to autosome ratio signal, which is crucial for dosage compensation and down-regulation of gene expression from X chromosomes to male levels in XX worms79. Activated xol-1 represses the function of sdc-1, sdc-2, and sdc-3 (formerly dpy-29), which then permits her-1 expression80,81. We were unable to identify homologs of genes encoding either XOL-1, which is structurally related to GHMP small molecule kinases82, or SDC-3, which has zinc-finger motifs83, outside of Caenorhabditis species (Table 2). Similarly, we were only able to identify weak homologs of genes encoding SDC-2, which directly represses her-1 expression in C. elegans80, in a few clade V parasites (Table 2). Identifiable xol-1, sdc-2, and sdc-3 homologs also appear to be absent from the clade III parasitic nematode Brugia malayi15. We were unable to clearly identify an S. stercoralis homolog of sdc-1, which encodes a zinc-finger transcription factor84, because the top BLAST hit, SSTP_0000950300, encodes a protein that is equally related to SDC-1 and C04F5.9 in C. elegans and other Caenorhabditis species. Since xol-1 appears to be rapidly evolving and has significant sequence divergence between Caenorhabditis species82, and sdc-2 homologs appear to have similar sequence divergence, we may simply be unable to detect homologs of these genes in clade III and IV nematodes by sequence homology. Alternatively, a true loss of xol-1, sdc-2, and/or sdc-3 homologs in S. stercoralis may suggest a different mechanism of regulating dosage compensation of the X chromosome between XX and XO individuals. Whether S. stercoralis actually balances gene expression on the X chromosome between males and females is unknown, although sex-specific histone H3 modifications, which play a role in dosage compensation in other species, have been observed in the S. ratti male germline23.
Transcriptomic profiles of nuclear hormone receptors and cytochrome P450s are associated with sex
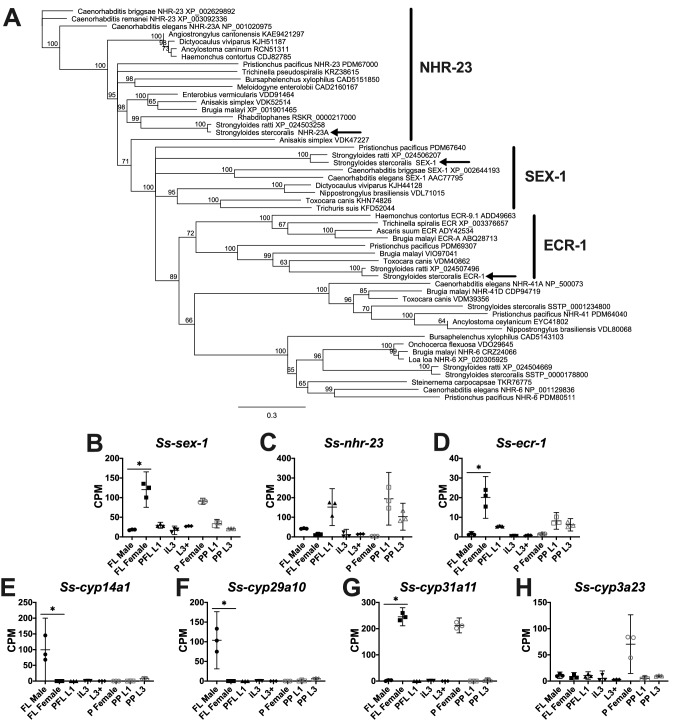
The C. elegans genome is predicted to encode 284 nuclear hormone receptors85, including SEX-1 and NHR-23, which function in sex determination and spermatogenesis, respectively, among other processes86,87. While both SEX-1 and NHR-23 are related to the ecdysone nuclear hormone receptor ECR-1, Caenorhabditis species do not have ECR-1 homologs, in contrast to most other nematode species88. Therefore, we sought to identify S. stercoralis homologs of sex-1, nhr-23, and ecr-1, and determine whether their transcripts were regulated in a sex-specific manner. Using reciprocal BLAST searches, we identified S. stercoralis homologs of these three putative nuclear hormone receptors (Fig. 3A). We found that both Ss-sex-1 and Ss-ecr-1 transcripts are significantly up-regulated in free-living adult females in comparison to free-living adult males (6.6-fold and 14.2-fold, respectively; FDR < 0.05), while transcripts for Ss-nhr-23 are up-regulated in developing larvae—consistent with its role in regulating molting in C. elegans89 (Fig. 3B-D).
Figure 3.
Transcripts encoding S. stercoralis nuclear hormone receptors and cytochrome P450s are differentially regulated between males and females. (A) Homologs of SEX-1, NHR-23, and ECR-1 in S. stercoralis (arrows) and other clade I, III, IV, and V nematodes are identified in a neighbor-joining phylogenetic tree with 100 iterations of bootstrapping constructed with Geneious v.11.1.5. Gene names and accession numbers are listed after the species names. (B-H) Transcript abundances of the S. stercoralis homologs encoding relevant nuclear hormone receptors and cytochrome P450s are represented using TMM-normalized counts per million (CPM) for the following developmental stages: free-living adult males (FL Male), free-living gravid adult females (FL Female), post-free-living first-stage larvae (PFL L1), developmentally arrested infectious third-stage larvae (iL3), L3 activated inside a permissive host (L3 +), parasitic gravid adult females (P Female), heterogonically-developing post-parasitic L1 (PP L1), and heterogonically-developing post-parasitic L3 enriched for females (PP L3). Graphs were constructed using GraphPad Prism v.9.0.0; bars indicate means (horizontal) and 95% confidence intervals (vertical) for each of the three biological replicates represented as individual data points. Asterisks (*) indicate a significant (fold change > 2.0; FDR < 0.05) difference between FL Male and FL Female.
While the native ligand for ECR-1 in parasitic nematodes is likely 20-hydroxyecdysone90, the endogenous ligands for SEX-1 and NHR-23 have not yet been described, but are hypothesized to be steroid hormones91. Therefore, we asked whether any of the 26 S. stercoralis cytochrome P450-encoding transcripts14 are regulated in a sex-specific manner and thus might produce a sex-specific hormone. We found that Ss-cyp14a1 and Ss-cyp29a10 transcripts were significantly up-regulated in free-living adult males in comparison to free-living adult females (565-fold and 832-fold, respectively; FDR < 0.05) (Fig. 3E-F). Consequently, we hypothesize these two genes may be responsible for the production of androgens. We also found that, of the developmental stages we examined, Ss-cyp31a11 transcripts were solely found in adult free-living and parasitic females and Ss-cyp3a23 transcripts were only found in parasitic females (Fig. 3G-H). We hypothesize that Ss-cyp-31a11 may encode the cytochrome P450 necessary for ecdysone biosynthesis in S. stercoralis, as its regulation through S. stercoralis development is consistent with that of Ss-ecr-1. Alternatively, these sex-specific cytochrome P450s may be involved in the biosynthesis of sex-specific pheromones or other signaling molecules.
Transcripts encoding putative mRNA-binding proteins are sex specific
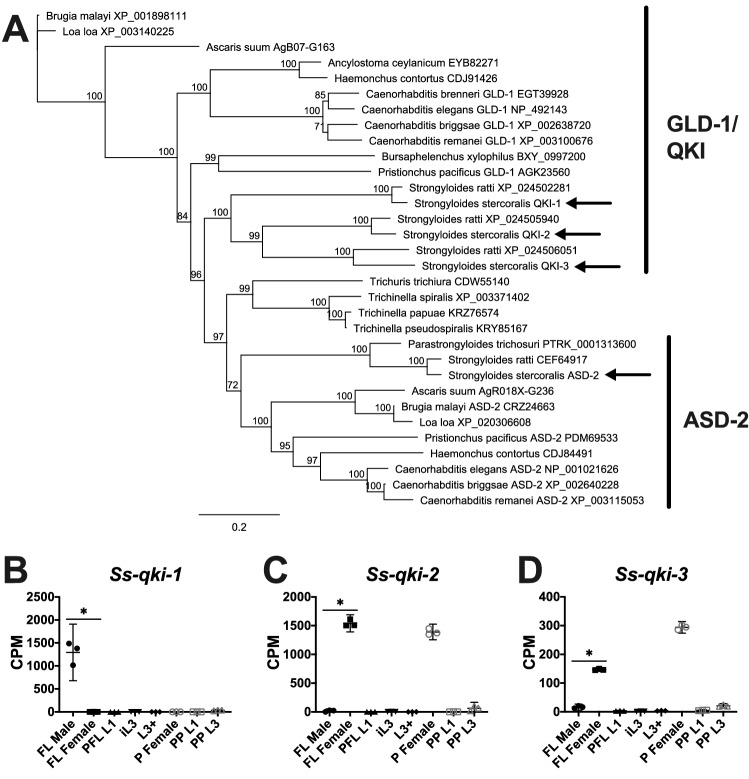
The STAR domain protein GLD-1 represses translation via mRNA 3′ UTR binding and regulates the transition of mitosis to meiosis and development of germ cells in Caenorhabditis species92, although specific molecules bound by C. elegans and C. briggsae GLD-1 homologs vary93. To identify a gld-1 homolog in S. stercoralis and differentiate it from the closely-related asd-2, we performed reciprocal BLAST searches and constructed a phylogenetic tree. We identified three gld-1 paralogs in the S. stercoralis genome (Fig. 4A), which we termed Ss-qki-1, Ss-qki-2, and Ss-qki-3. Strikingly, of the developmental stages we examined, Ss-qki-1 transcripts are only expressed in free-living adult males (~ 700-fold up-regulated in comparison to free-living adult females; FDR < 0.05), while Ss-qki-2 and Ss-qki-3 transcripts are specifically and highly expressed in both free-living and parasitic adult females (Fig. 4B-D). We hypothesize that an ancestral gld-1 homolog was duplicated twice, resulting in three paralogs in Strongyloides species, with Ss-qki-1 taking on adult male-specific functions and Ss-qki-2 and Ss-qki-3 taking on adult female-specific functions. Future studies to identify the molecular targets of the proteins these genes encode may aid in determining their functions.
Figure 4.
The S. stercoralis genome encodes three GLD-1 paralogs with transcripts that are sex specific. (A) Homologs of GLD-1 and ASD-2 in S. stercoralis (arrows) and other III, IV, and V nematodes are identified in a neighbor-joining phylogenetic tree with 100 iterations of bootstrapping constructed with Geneious v.11.1.5. Gene names and accession numbers are listed after the species names. (B-D) Transcript abundances of the three S. stercoralis paralogs are represented using TMM-normalized counts per million (CPM) for the following developmental stages: free-living adult males (FL Male), free-living gravid adult females (FL Female), post-free-living first-stage larvae (PFL L1), developmentally arrested infectious third-stage larvae (iL3), L3 activated inside a permissive host (L3 +), parasitic gravid adult females (P Female), heterogonically-developing post-parasitic L1 (PP L1), and heterogonically-developing post-parasitic L3 enriched for females (PP L3). Graphs were constructed using GraphPad Prism v.9.0.0; bars indicate means (horizontal) and 95% confidence intervals (vertical) for each of the three biological replicates represented as individual data points. Asterisks (*) indicate a significant (fold change > 2.0; FDR < 0.05) difference between FL Male and FL Female.
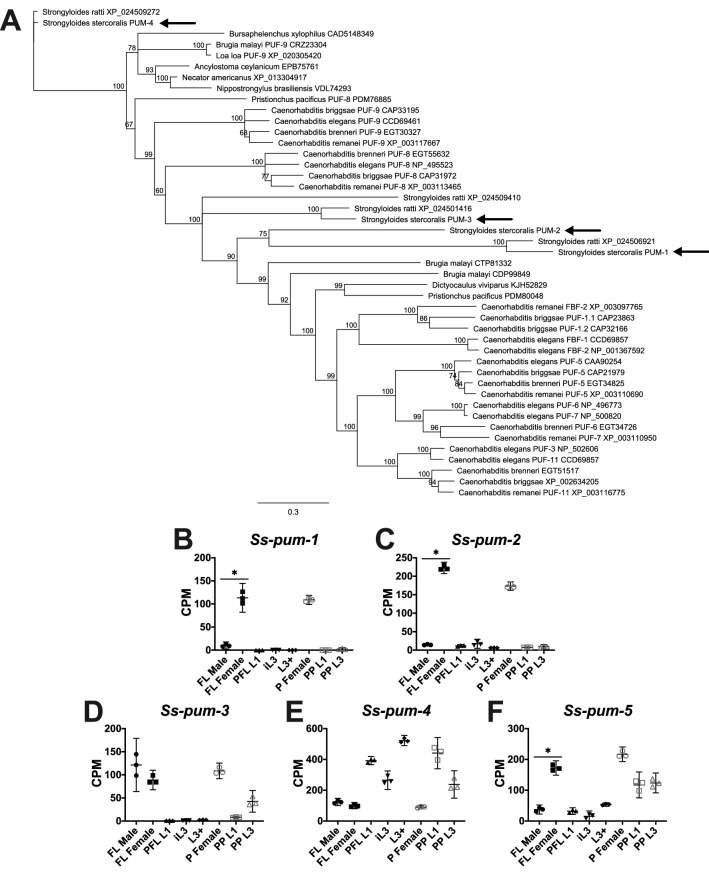
In Caenorhabditis species, the gld-1 mRNA is regulated by pumilio domain-containing mRNA-binding proteins94, which additionally regulate sex determination by binding to other mRNA targets26. We sought to identify S. stercoralis genes encoding pumilio family proteins by reciprocal BLAST searches and phylogenetic analyses. We found that the genes encoding pumilio family proteins appear to have been duplicated, not only in Caenorhabditis species95, but also in the common ancestor of Strongyloides species (Fig. 5A). The predicted polypeptides encoded by Ss-pum-1, Ss-pum-2, and Ss-pum-3 are more similar to each other than to other nematode pumilio family proteins, suggesting they are paralogs (Fig. 5A). Ss-pum-5 encodes a predicted polypeptide that is homologous to C. elegans PUF-12; these homologs are highly divergent from the other pumilio family proteins found in nematode species (data not shown). Ss-pum-1 and Ss-pum-2 transcripts are abundant in both free-living and parasitic adult females, but depleted in the other developmental stages we examined (Fig. 5B-C). Ss-pum-3, Ss-pum-4, and Ss-pum-5 transcripts have more complex patterns of developmental regulation (Fig. 5D-E), although Ss-pum-5 transcript abundance (Fig. 5F) is significantly lower in free-living adult males than free-living adult females (4.6-fold up-regulated in free-living adult females in comparison to free-living adult males; FDR < 0.05). We hypothesize that the proteins encoded by Ss-pum-1 and Ss-pum-2 play an important role in oogenesis or adult female development.
Figure 5.
Genes encoding pumilio family proteins appear to have been duplicated in Strongyloides species. (A) Homologs of pumilio family proteins in S. stercoralis (arrows) and other clade III, IV, and V nematodes are identified in a neighbor-joining phylogenetic tree with 100 iterations of bootstrapping constructed with Geneious v.11.1.5. The genes encoding Ss-PUM-1, Ss-PUM-2, and Ss-PUM-3 appear to have resulted from a duplication in an ancestor of Strongyloides species. Gene names and accession numbers are listed after the species names. (B-F) Transcript abundances of the S. stercoralis homologs are represented using TMM-normalized counts per million (CPM) for the following developmental stages: free-living adult males (FL Male), free-living gravid adult females (FL Female), post-free-living first-stage larvae (PFL L1), developmentally arrested infectious third-stage larvae (iL3), L3 activated inside a permissive host (L3 +), parasitic gravid adult females (P Female), heterogonically-developing post-parasitic L1 (PP L1), and heterogonically-developing post-parasitic L3 enriched for females (PP L3). Graphs were constructed using GraphPad Prism v.9.0.0; bars indicate means (horizontal) and 95% confidence intervals (vertical) for each of the three biological replicates represented as individual data points. Asterisks (*) indicate a significant (fold change > 2.0; FDR < 0.05) difference between FL Male and FL Female.
Conclusions
Our studies in S. stercoralis have revived the idea that post-parasitic males can precociously develop inside the host; whether these rhabditiform males are capable of tissue penetration warrants further study. Additionally, we have identified several S. stercoralis sex-specific transcripts that encode putative male-specific single-pass transmembrane domain proteins, cytochrome P450s, and a GPCR that may be involved in sex-specific functions. We have also identified S. stercoralis homologs of multiple genes that regulate sex determination in Caenorhabditis species as well as several specific differences, including three paralogs of gld-1 (Ss-qki-1, Ss-qki-2, and Ss-qki-3) and divergent homologs of pumilio domain-containing proteins (Ss-pum-1, Ss-pum-2, and Ss-pum-3). Several of these genes also have sex-specific transcripts. Although the molecular mechanisms controlling sex determination and male development in S. stercoralis remain far from resolved, our studies have identified several genes that warrant further study. Utilization of established techniques for transgenesis and targeted mutagenesis in S. stercoralis could help elucidate the biological functions of these genes in the parasite5.
More broadly, nematodes utilize a variety of mechanisms for sex determination. Although most nematodes use an XX/XO sex chromosome system, XX/XY systems also exist15, and the original signals that determine the combination of sex chromosomes in a developing embryo can be strictly genetic or environmentally-influenced19. Additionally, there are a few nematode species without autonomous X chromosomes; in S. papillosus and S. vituli, sex-specific chromatin diminution is necessary for male development19. In Caenorhabditis species, where sex is genetically determined, the genes responsible for sensing the X chromosome to autosome ratio, propagating this signal, and executing somatic and germline programs in the different sexes have been extensively studied26. Although core components are conserved between C. elegans and other Caenorhabditis species as well as P. pacificus, there are several differences26. These observations have led to the “bottom up” hypothesis, which posits that the most down-stream components are conserved and that up-stream components have evolved sequentially27. It has also been hypothesized that the genes from her-1 to tra-1 are derived from the hedgehog pathway and compose an indivisible “cassette”96.
Our data generally support the “bottom up” hypothesis, with the conservation of tra-1 and mab-3 across parasitic nematode species and less conservation (or at least more divergence in sequence) of more upstream members (e.g., xol-1, sdc-2, and sdc-3). However, the apparent loss of her-1 in several lineages, absence of fem-1 homologs in clade IV nematodes, and the potential absence of tra-2 outside of Caenorhabditis species, suggest that the “cassette” of genes from her-1 to tra-1 may not be indivisible. Even when genes are conserved across nematode species, it does not necessarily follow that they control sex determination in a particular species, since several genes in the Caenorhabditis sex determination pathway are pleiotropic and have functions outside sex determination. Future functional studies will be required to determine whether conserved genes indeed regulate sex determination in species distant from Caenorhabditis and whether other genes have been recruited to the sex determination pathway in these species. Together, these findings suggest that the mechanisms and genes controlling nematode sex determination may be more varied than previously appreciated.
Supplementary Information
Acknowledgements
This work was supported by the National Institutes of Health [Grant Numbers AI50688, AI22662, and AI105856 (JBL)]; the Pennsylvania State System of Higher Education (JDCS); and Millersville University of Pennsylvania (DGA, EJD, CRS, and JDCS). We also wish to thank Kristina B. Lewis and the anonymous reviewers for critical reading of the manuscript.
Author contributions
J.D.C.S., T.J.N., and J.B.L. cultured the parasite. J.D.C.S. isolated free-living adult males and constructed RNA-Seq libraries. D.G.A., E.J.D., C.R.S., and J.D.C.S analyzed the data. D.G.A. and J.D.C.S. constructed the figures. J.D.C.S. wrote the manuscript. The final version of the manuscript was approved by all authors.
Data availability
Raw RNA-Seq reads for free-living adult males are available in the NCBI SRA database under BioProject ID: PRJNA689252.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87478-3.
References
- 1.Buonfrate, D. et al. The global prevalence of Strongyloides stercoralis infection. Pathogens 9, (2020). [DOI] [PMC free article] [PubMed]
- 2.Nutman TB. Human infection with Strongyloides stercoralis and other related Strongyloides species. Parasitology. 2017;144:263–273. doi: 10.1017/S0031182016000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grove DI. Human strongyloidiasis. Adv. Parasitol. 1996;38:251–309. doi: 10.1016/S0065-308X(08)60036-6. [DOI] [PubMed] [Google Scholar]
- 4.Jaleta TG, et al. Different but overlapping populations of Strongyloides stercoralis in dogs and humans-Dogs as a possible source for zoonotic strongyloidiasis. PLoS Negl. Trop. Dis. 2017;11:e0005752. doi: 10.1371/journal.pntd.0005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaleta TG, Lok JB. Advances in the molecular and cellular biology of Strongyloides spp. Curr. Trop. Med. Rep. 2019;6:161–178. doi: 10.1007/s40475-019-00186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond MP, Robinson RD. Chromosome complement, gametogenesis, and development of Strongyloides stercoralis. J. Parasitol. 1994;80:689–695. doi: 10.2307/3283247. [DOI] [PubMed] [Google Scholar]
- 7.Kreis HA. Studies on the genus Strongyloides (Nematodes) Am. J. Epidemiol. 1932;16:450–491. doi: 10.1093/oxfordjournals.aje.a117869. [DOI] [Google Scholar]
- 8.Faust EC. Experimental studies on human and primate species of Strongyloides. II. The development of strongyloides in the experimental host. Am. J. Epidemiol. 1933;18:114–132. doi: 10.1093/oxfordjournals.aje.a117940. [DOI] [Google Scholar]
- 9.Shao H, Li X, Lok JB. Heritable genetic transformation of Strongyloides stercoralis by microinjection of plasmid DNA constructs into the male germline. Int. J. Parasitol. 2017;47:511–515. doi: 10.1016/j.ijpara.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada M, Matsuda S, Nakazawa M, Arizono N. Species-specific differences in heterogonic development of serially transferred free-living generations of Strongyloides planiceps and Strongyloides stercoralis. J. Parasitol. 1991;77:592–594. doi: 10.2307/3283165. [DOI] [PubMed] [Google Scholar]
- 11.Nolan TJ, et al. The amphidial neuron pair ALD controls the temperature-sensitive choice of alternative developmental pathways in the parasitic nematode Strongyloides stercoralis. Parasitology. 2004;129:753–759. doi: 10.1017/S0031182004006092. [DOI] [PubMed] [Google Scholar]
- 12.Shiwaku K, Chigusa Y, Kadosaka T, Kaneko K. Factors influencing development of free-living generations of Strongyloides stercoralis. Parasitology. 1988;97(Pt 1):129–138. doi: 10.1017/S0031182000066804. [DOI] [PubMed] [Google Scholar]
- 13.Faust EC, Kagy ES. Experimental studies on human and primate species of Strongyloides. Am. J. Trop. Med. Hyg. 1933;1–13:47–65. doi: 10.4269/ajtmh.1933.s1-13.47. [DOI] [Google Scholar]
- 14.Albarqi MMY, et al. Regulation of life cycle checkpoints and developmental activation of infective larvae in Strongyloides stercoralis by dafachronic acid. PLoS Pathog. 2016;12:e1005358. doi: 10.1371/journal.ppat.1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster JM, et al. Sex chromosome evolution in parasitic nematodes of humans. Nat. Commun. 2020;11:1964. doi: 10.1038/s41467-020-15654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemetschke L, Eberhardt AG, Hertzberg H, Streit A. Genetics, chromatin diminution, and sex chromosome evolution in the parasitic nematode genus Strongyloides. Curr. Biol. 2010;20:1687–1696. doi: 10.1016/j.cub.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Kulkarni A, Dyka A, Nemetschke L, Grant WN, Streit A. Parastrongyloides trichosuri suggests that XX/XO sex determination is ancestral in Strongyloididae (Nematoda) Parasitology. 2013;140:1822–1830. doi: 10.1017/S0031182013001315. [DOI] [PubMed] [Google Scholar]
- 18.Harvey SC, Viney ME. Sex determination in the parasitic nematode Strongyloides ratti. Genetics. 2001;158:1527–1533. doi: 10.1093/genetics/158.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streit A. How to become a parasite without sex chromosomes: a hypothesis for the evolution of Strongyloides spp. and related nematodes. Parasitology. 2014;141:1244–1254. doi: 10.1017/S003118201400064X. [DOI] [PubMed] [Google Scholar]
- 20.Harvey SC, Gemmill AW, Read AF, Viney ME. The control of morph development in the parasitic nematode Strongyloides ratti. Proc. Biol. Sci. 2000;267:2057–2063. doi: 10.1098/rspb.2000.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shakes DC, Neva BJ, Huynh H, Chaudhuri J, Pires-Dasilva A. Asymmetric spermatocyte division as a mechanism for controlling sex ratios. Nat. Commun. 2011;2:157. doi: 10.1038/ncomms1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni A, Holz A, Rödelsperger C, Harbecke D, Streit A. Differential chromatin amplification and chromosome complements in the germline of Strongyloididae (Nematoda) Chromosoma. 2016;125:125–136. doi: 10.1007/s00412-015-0532-y. [DOI] [PubMed] [Google Scholar]
- 24.Bolla RI, Roberts LS. Gametogenesis and chromosomal complement in Strongyloides ratti (Nematoda: Rhabdiasoidea) J. Parasitol. 1968;54:849–855. doi: 10.2307/3277110. [DOI] [PubMed] [Google Scholar]
- 25.Viney ME, Matthews BE, Walliker D. Mating in the nematode parasite Strongyloides ratti: proof of genetic exchange. Proc. Biol. Sci. 1993;254:213–219. doi: 10.1098/rspb.1993.0148. [DOI] [PubMed] [Google Scholar]
- 26.Haag ES, Fitch DHA, Delattre M. From ‘the worm’ to ‘the worms’ and back again: the evolutionary developmental biology of nematodes. Genetics. 2018;210:397–433. doi: 10.1534/genetics.118.300243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins AS. Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. BioEssays. 1995;17:71–77. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]
- 28.Blaxter M, Koutsovoulos G. The evolution of parasitism in Nematoda. Parasitology. 2015;142(Suppl 1):S26–39. doi: 10.1017/S0031182014000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoltzfus JD, Massey HC, Nolan TJ, Griffith SD, Lok JB. Strongyloides stercoralis age-1: a potential regulator of infective larval development in a parasitic nematode. PLoS ONE. 2012;7:e38587. doi: 10.1371/journal.pone.0038587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schad GA, Hellman ME, Muncey DW. Strongyloides stercoralis: hyperinfection in immunosuppressed dogs. Exp. Parasitol. 1984;57:287–296. doi: 10.1016/0014-4894(84)90103-6. [DOI] [PubMed] [Google Scholar]
- 31.Nolan TJ, Megyeri Z, Bhopale VM, Schad GA. Strongyloides stercoralis: the first rodent model for uncomplicated and hyperinfective strongyloidiasis, the Mongolian gerbil (Meriones unguiculatus) J. Infect. Dis. 1993;168:1479–1484. doi: 10.1093/infdis/168.6.1479. [DOI] [PubMed] [Google Scholar]
- 32.Stoltzfus JD, Minot S, Berriman M, Nolan TJ, Lok JB. RNAseq analysis of the parasitic nematode Strongyloides stercoralis reveals divergent regulation of canonical dauer pathways. PLoS Negl. Trop. Dis. 2012;6:e1854. doi: 10.1371/journal.pntd.0001854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lok, J. B. Strongyloides stercoralis: a model for translational research on parasitic nematode biology. WormBook 1–18 (2007). [DOI] [PMC free article] [PubMed]
- 35.Stiernagle, T. Maintenance of C. elegans. WormBook 1–11 (2006). [DOI] [PMC free article] [PubMed]
- 36.Andrews, S. FastQC: a quality control tool for high throughput sequence data. (2010).
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bushnell B, Rood J, Singer E. BBMerge - accurate paired shotgun read merging via overlap. PLoS ONE. 2017;12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt VL, et al. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat. Genet. 2016;48:299–307. doi: 10.1038/ng.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolt BJ, et al. Using wormbase parasite: an integrated platform for exploring helminth genomic data. Methods Mol. Biol. 2018;1757:471–491. doi: 10.1007/978-1-4939-7737-6_15. [DOI] [PubMed] [Google Scholar]
- 44.Almagro Armenteros JJ, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 45.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 46.Pertea, G. & Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Res 9, (2020). [DOI] [PMC free article] [PubMed]
- 47.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 48.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galliard H. Pathogenesis of Strongyloides. Helminthol. Abst. 1967;36:247–257. [Google Scholar]
- 51.Gomez Gallego S, et al. Identification of an astacin-like metallo-proteinase transcript from the infective larvae of Strongyloides stercoralis. Parasitol. Int. 2005;54:123–133. doi: 10.1016/j.parint.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 52.McKerrow JH, et al. Strongyloides stercoralis: identification of a protease that facilitates penetration of skin by the infective larvae. Exp. Parasitol. 1990;70:134–143. doi: 10.1016/0014-4894(90)90094-S. [DOI] [PubMed] [Google Scholar]
- 53.Thomas CG, et al. Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. 2012;22:2167–2172. doi: 10.1016/j.cub.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts TM, Stewart M. Role of major sperm protein (MSP) in the protrusion and retraction of Ascaris sperm. Int. Rev. Cell Mol. Biol. 2012;297:265–293. doi: 10.1016/B978-0-12-394308-8.00007-8. [DOI] [PubMed] [Google Scholar]
- 55.Kasimatis KR, Phillips PC. Rapid gene family evolution of a nematode sperm protein despite sequence hyper-conservation. G3 Bethesda. 2018;8:353–362. doi: 10.1534/g3.117.300281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin D, et al. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science. 2018;359:55–61. doi: 10.1126/science.aao0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasan J, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stasiuk SJ, Scott MJ, Grant WN. Developmental plasticity and the evolution of parasitism in an unusual nematode Parastrongyloides trichosuri. Evodevo. 2012;3:1. doi: 10.1186/2041-9139-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim K, et al. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGrath PT, et al. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park D, et al. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2012;109:9917–9922. doi: 10.1073/pnas.1202216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoltzfus JD, Bart SM, Lok JB. cGMP and NHR signaling co-regulate expression of insulin-like peptides and developmental activation of infective larvae in Strongyloides stercoralis. PLoS Pathog. 2014;10:e1004235. doi: 10.1371/journal.ppat.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-X. [DOI] [PubMed] [Google Scholar]
- 64.Pires-daSilva A, Sommer RJ. Conservation of the global sex determination gene tra-1 in distantly related nematodes. Genes Dev. 2004;18:1198–1208. doi: 10.1101/gad.293504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross JM, Kalis AK, Murphy MW, Zarkower D. The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev. Cell. 2005;8:881–892. doi: 10.1016/j.devcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 66.Kim B, Suo B, Emmons SW. Gene function prediction based on developmental transcriptomes of the two sexes in C. elegans. Cell Rep. 2016;17:917–928. doi: 10.1016/j.celrep.2016.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Starostina NG, et al. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell. 2007;13:127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pilgrim D, McGregor A, Jäckle P, Johnson T, Hansen DT. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol. Biol. Cell. 1995;6:1159–1171. doi: 10.1091/mbc.6.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hubbard EJ, Wu G, Kitajewski J, Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Refai O, Smit RB, Votra S, Pruyne D, Mains PE. Tissue-Specific Functions of fem-2/PP2c Phosphatase and fhod-1/formin During Caenorhabditis elegans Embryonic Morphogenesis. G3 Bethesda. 2018;8:2277–2290. doi: 10.1534/g3.118.200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mehra A, Gaudet J, Heck L, Kuwabara PE, Spence AM. Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev. 1999;13:1453–1463. doi: 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haag ES, Wang S, Kimble J. Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr. Biol. 2002;12:2035–2041. doi: 10.1016/S0960-9822(02)01333-7. [DOI] [PubMed] [Google Scholar]
- 73.Hamaoka BY, Dann CE, Geisbrecht BV, Leahy DJ. Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A. Proc. Natl. Acad. Sci. USA. 2004;101:11673–11678. doi: 10.1073/pnas.0402559101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokol SB, Kuwabara PE. Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 2000;14:901–906. [PMC free article] [PubMed] [Google Scholar]
- 75.Kuwabara PE, Okkema PG, Kimble J. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol. Biol. Cell. 1992;3:461–473. doi: 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Streit A, et al. Homologs of the Caenorhabditis elegans masculinizing gene her-1 in C. briggsae and the filarial parasite Brugia malayi. Genetics. 1999;152:1573–1584. doi: 10.1093/genetics/152.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stevens L, et al. Comparative genomics of 10 new Caenorhabditis species. Evol. Lett. 2019;3:217–236. doi: 10.1002/evl3.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bürglin, T. R. & Kuwabara, P. E. Homologs of the Hh signalling network in C. elegans. WormBook 1–14 (2006). [DOI] [PMC free article] [PubMed]
- 79.Rhind NR, Miller LM, Kopczynski JB, Meyer BJ. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell. 1995;80:71–82. doi: 10.1016/0092-8674(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 80.Chu DS, et al. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 2002;16:796–805. doi: 10.1101/gad.972702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dawes HE, et al. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- 82.Luz JG, et al. XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators. Genes Dev. 2003;17:977–990. doi: 10.1101/gad.1082303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–1031. doi: 10.1242/dev.124.5.1019. [DOI] [PubMed] [Google Scholar]
- 84.Nonet ML, Meyer BJ. Early aspects of Caenorhabditis elegans sex determination and dosage compensation are regulated by a zinc-finger protein. Nature. 1991;351:65–68. doi: 10.1038/351065a0. [DOI] [PubMed] [Google Scholar]
- 85.Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev. Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 86.Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- 87.Ragle, J. M. et al. The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis. Development 147, (2020). [DOI] [PMC free article] [PubMed]
- 88.Tzertzinis G, et al. Molecular evidence for a functional ecdysone signaling system in Brugia malayi. PLoS Negl. Trop. Dis. 2010;4:e625. doi: 10.1371/journal.pntd.0000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kouns NA, et al. NHR-23 dependent collagen and hedgehog-related genes required for molting. Biochem. Biophys. Res. Commun. 2011;413:515–520. doi: 10.1016/j.bbrc.2011.08.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mhashilkar AS, et al. Identification of ecdysone hormone receptor agonists as a therapeutic approach for treating filarial infections. PLoS Negl. Trop. Dis. 2016;10:e0004772. doi: 10.1371/journal.pntd.0004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Antebi, A. Nuclear receptor signal transduction in C. elegans. WormBook 1–49 (2015). [DOI] [PMC free article] [PubMed]
- 92.Lee M-H, Schedl TC. elegans star proteins, GLD-1 and ASD-2, regulate specific RNA targets to control development. Adv. Exp. Med. Biol. 2010;693:106–122. doi: 10.1007/978-1-4419-7005-3_8. [DOI] [PubMed] [Google Scholar]
- 93.Beadell AV, Haag ES. Evolutionary dynamics of GLD-1-mRNA complexes in Caenorhabditis nematodes. Genome Biol. Evol. 2014;7:314–335. doi: 10.1093/gbe/evu272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crittenden SL, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 95.Liu Q, Stumpf C, Thomas C, Wickens M, Haag ES. Context-dependent function of a conserved translational regulatory module. Development. 2012;139:1509–1521. doi: 10.1242/dev.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haag, E. S. The evolution of nematode sex determination: C. elegans as a reference point for comparative biology. WormBook 1–14 (2005). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-Seq reads for free-living adult males are available in the NCBI SRA database under BioProject ID: PRJNA689252.