Abstract
Background
Hepatocellular carcinoma (HCC) is one of the most common causes of cancer worldwide. Although many studies have focused on oncogene characteristics, the genomic landscape of Chinese HCC patients has not been fully clarified.
Methods
A total of 165 HCC patients, including 146 males and 19 females, were enrolled. The median age was 55 years (range, 27–78 years). Corresponding clinical and pathological information was collected for further analysis. A total of 168 tumor tissues from these patients were selected for next-generation sequencing (NGS)-based 450 panel gene sequencing. Genomic alterations including single nucleotide variations (SNV), short and long insertions and deletions (InDels), copy number variations, and gene rearrangements were analyzed. Tumor mutational burden (TMB) was measured by an algorithm developed in-house. The top quartile of HCC was classified as TMB high.
Results
A total of 1,004 genomic alterations were detected from 258 genes in 168 HCC tissues. TMB values were identified in 160 HCC specimens, with a median TMB of 5.4 Muts/Mb (range, 0–28.4 Muts/Mb) and a 75% TMB of 7.7 Muts/Mb. The most commonly mutated genes were TP53, TERT, CTNNB1, AXIN1, RB1, TSC2, CCND1, ARID1A, and FGF19. SNV was the most common mutation type and C:G>T:A and guanine transformation were the most common SNVs. Compared to wild-type patients, the proportion of Edmondson grade III–IV and microvascular invasion was significantly higher in TP53 mutated patients (P<0.05). The proportion of tumors invading the hepatic capsule was significantly higher in TERT mutated patients (P<0.05). The proportion of Edmondson grade I-II, alpha fetoprotein (AFP) <25 µmg/L, and those without a history of hepatitis B was significantly higher in CTNNB1 mutated patients (P<0.05). CTNNB1 mutations were associated with TMB high in HCC patients (P<0.05). Based on correlation analysis, the mutation of TP53 was independently correlated with microvascular invasion (P=0.002, OR =3.096) and Edmondson grade III–IV (P=0.008, OR =2.613). The mutation of TERT was independently correlated with tumor invasion of the liver capsule (P=0.001, OR =3.030), and the mutation of CTNNB1 was independently correlated with AFP (<25 µmg/L) (P=0.009, OR =3.414).
Conclusions
The most frequently mutated genes of HCC patients in China were TP53, TERT, and CTNNB1, which mainly lead to the occurrence and development of HCC by regulating the P53 pathway, Wnt pathway, and telomere repair pathway. There were more patients with microvascular invasion and Edmondson III–IV grade in TP53 mutated patients and more patients with hepatic capsule invasion in TERT mutated patients, while in CTNNB1 mutated patients, there were more patients with Edmondson I–II grade, AFP <25 µmg/L, and a non-hepatitis B background. Also, the TMB values were significantly higher in CTNNB1 mutated patients than in wild type patients.
Keywords: Hepatocellular carcinoma (HCC), gene mutation, next-generation sequencing (NGS), pathological features
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the fourth most common cause of cancer-related death worldwide (1). The occurrence and development of HCC is mostly related to hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, excessive alcohol intake, obesity, and rare genetic diseases (2). However, the key driving factors for HCC development are still unclear. Although considerable progress had been achieved in the clinical diagnosis and treatment of HCC, patients still have a poor prognosis (3). Further understanding of the pathogenesis mechanism of HCC could provide effective help for treating patients. Next-generation sequencing (NGS) technology is a revolutionary change in traditional sequencing methods because it can detect multiple genetic variations with high sensitivity (4). Although many studies have focused on oncogene characteristics, the genome alteration map of HCC patients has not been fully clarified. In this study we aimed to investigate the characteristics of HCC gene mutations and its relationship with clinical related factors by analyzing the results of gene mutation detection by NGS in 165 patients (168 HCC tissue specimens). We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/hbsn.2019.09.17).
Methods
Patient enrollment and sample collection
A total of 165 HCC patients who received genetic testing at the Affiliated Hospital of Qingdao University from January 2017 to December 2018 were enrolled. There were 146 males and 19 females with a median age of 55 years (range, 27–78 years). Among them, 140 cases were HBsAg positive, 3 cases were HCVAb positive, and 1 case was positive for both HBsAg and HCVAb. The remaining 21 cases were negative for hepatitis virus markers. The diagnosis of HCC in all specimens, including 158 resected specimens and 10 liver biopsy specimens, were pathologically confirmed. The patient information, serological test results, and pathological characteristics of tumors were collected from the Affiliated Hospital of Qingdao University. All patients signed a written informed consent. The study was conducted in strict compliance with the Helsinki Declaration and was approved by the Ethics Committee (ethics approval number: QDFYKYLLL-20161212).
Identification of genomic alterations, TMB, and MSI
The genomic profile was produced using the NGS-based YuanSuTM 450 gene panel (OrigiMed, the College of American Pathologists (CAP) accredited and Clinical Laboratory Improvement Amendments (CLIA) certified laboratory, Shanghai, China) (5). The genes were captured and sequenced with a mean coverage of 900× by using Illumina NextSeq 500. The sequence results were further analyzed for genomic alterations, including single nucleotide variants (SNV), short and long insertions/deletions (InDels), copy number variations (CNV), and structural variants of gene rearrangement/fusion and long InDels. Tumor mutational burden (TMB) and microsatellite instability (MSI) were measured by an algorithm developed in-house.
Statistical analysis
Statistical analyses were performed using SPSS. Chi-square test was used for qualitative data and t-test was used for quantitative data. P<0.05 was considered to be statistically significant.
Results
Characterization of genomic alterations in Chinese HCC patients
In this study, 2 isolated tumors with different genome characteristics were detected in 3 patients. Based on the experience of Duan et al., these 2 tumors from 1 patient were regarded as an independent lesion in this study (6). A total of 1004 genomic alterations from 258 genes were detected in 168 HCC tumors from 165 patients. The average alteration frequency was 5.8 alterations per patient. Out of 168 HCC specimens, TMB values were calculated in 160 HCC specimens, and the median TMB was 5.4 Muts/Mb (range, 0–28.4 Muts/Mb) and the 75% TMB was 7.7 Muts/Mb. MSI was not found in this group of patients.
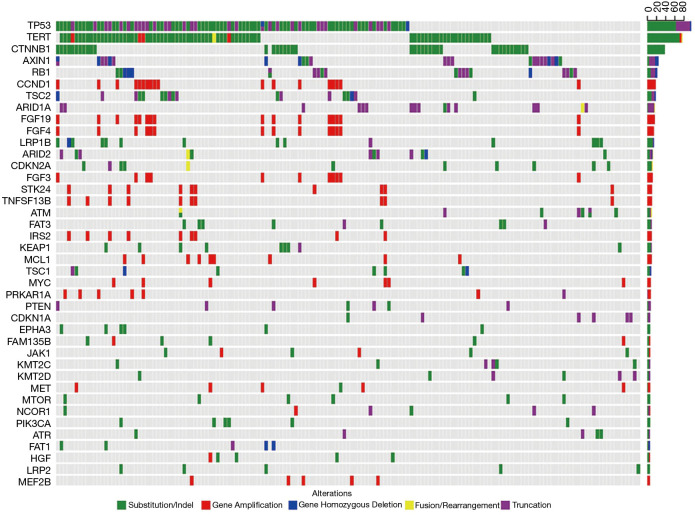
The most commonly mutated genes of Chinese HCC patients were TP53 (56.5%), TERT (45.2%), CTNNB1 (22.6%), AXIN1 (13.7%), RB1 (11.9%), TSC2 (10.7%), CCND1 (10.7%), ARID1A (9.5%), and FGF19 (9.5%). The co-mutation of TP53 and TERT was detected in 54 (32.1%) patients (Figure 1). Among the 1,004 genomic alterations from 258 genes, 68.2% were SNV (685/1,004), 26.0% were CNV (261/1,004), 3.5% were short or long segment insertion deletions (35/1,004), and 2.3% were gene rearrangement/fusions (23/1,004). Among 668 SNV mutations, 580 were base pair substitutions, of which C:G>T:A conversion (23.1%, 134/580) was the most frequent mutation, followed by G:C>T:A conversion (18.6%, 108/580) and G:C>A:T conversion (16.4%, 95/580).
Figure 1.
The mutational landscape of 168 HCC specimens. The X-axis represents each case sample and the Y-axis represents each mutated gene. The bar graph on the right shows the mutation frequency of each mutated gene in 168 samples. Green represents substitution/Indel mutations, red represents gene amplification mutations, blue represents gene homozygous deletion mutations, yellow represents fusion/rearrangement mutations, and purple represents truncation mutations. HCC, hepatocellular carcinoma.
The mutations of TP53 and CTNNB1 were the most frequent SNVs. The most frequent mutation type of TP53, CTNNB1 and TERT were G:C>T:A (38.9%, 28/97), C:G>T:A (20.9%, 9/43) and C:G>T:A (89.6%, 69/77), respectively. Gene variation types of FGF signaling pathway related genes and CCND1, IRS2, MYC, STK24, TNFSF13B were all CNV.
The abnormal carcinogenic pathways in Chinese HCC patients
According to the classification and statistical analysis of carcinogenic pathways, the most common affected pathway was the P53 pathway. Out of 168 samples, 113 (67.3%) had P53 pathway related gene mutations, followed by 80 (47.6%) in the Wnt pathway, 76 (45.2%) in telomere repair related genes, 45 (26.8%) in the PI3K/mTOR pathway, 43 (25.6%) in chromatin remodeling related pathways, and 31 (18.5%) in cell cycle related pathway.
The correlation between TMB-H and mutated genes in Chinese HCC patients
TMB values were calculated in 160 HCC specimens, and we found the median TMB was 5.4 Muts/Mb (range, 0–28.4 Muts/Mb) and the 75% TMB was 7.7 Muts/Mb. TMB values higher than 7.7 Muts/Mb were considered to be TMB high (TMB-H), while TMB values lower than 7.7 Muts/Mb were considered to be TMB low (TMB-L). Based on statistical analysis, the mutation frequency of CTNNB1 (35.7% vs. 18.6%, P=0.024) was significantly higher in the TMB-H group than that in the TMB-L group, while the mutation frequencies of TP53 (45.2% vs. 61.9%, P=0.061) and TERT (42.9% vs. 47.5%, P=0.608) were slightly lower in the TMB-H group, but the difference was not significant.
Germline mutations in Chinese HCC patients
Among the 165 Chinese HCC patients, 10 were detected as germline variation positive, and 1 was identified with germline variations in 2 genes. The germline variation genes were PMS2 (twice), SDHB (twice), NBN, BARD1, BLM, BRCA2, NF1, FANCD2, and RAD51D.
The correlation between mutated genes and clinicopathological features
Out of 168 specimens, 51 (28.7%) were TNM stage III–IV and 74 (37.8%) were BCLC stage II–III. There were 91 cases with grade 4 hepatic fibrosis and 89 cases with grade III–IV Edmondson classification.
Compared to patients without a TP53 mutation, the proportion of patients with Edmondson grade III–IV and microvascular invasion was significantly higher in patients with a TP53 mutation (P<0.001 and P<0.001, respectively). The mutation of TERT was significantly correlated with the invasion of the hepatic capsule (P=0.001). Among the patients with co-mutations of TP53 and TERT, there was a significantly increased proportion of patients with Edmondson grade III–IV, hepatic capsule invasion, and microvascular invasion by tumors (P=0.018, P=0.001, and P=0.003, respectively). Among the patients with a CTNNB1 mutation, the proportion of Edmondson grade I–II, AFP<25 µmg/L, and those without a background of hepatitis B were higher than those without a CTNNB1 mutation (P=0.020, P=0.003, and P=0.016, respectively) (Table 1).
Table 1. Analysis of the correlation between mutated genes and clinical characteristics.
| Characteristic | TP53 | TERT | TP53 + TERT | CTNNB1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | Mutant | P value | WT | Mutant | P value | WT | Mutant | P value | WT | Mutant | P value | ||||
| Gender (male/female) | 65/8 | 84/11 | 0.900 | 84/8 | 65/11 | 0.239 | 102/12 | 47/7 | 0.641 | 114/16 | 35/3 | 0.450 | |||
| Age (<55/≥55) | 30/43 | 47/48 | 0.280 | 41/51 | 36/40 | 0.717 | 52/62 | 25/29 | 0.934 | 61/69 | 16/22 | 0.600 | |||
| *Liver fibrosis (123/4) | 34/35 | 35/56 | 0.171 | 39/50 | 30/41 | 0.842 | 48/61 | 21/30 | 0.734 | 51/73 | 18/18 | 0.344 | |||
| *Differentiation (12/34) | 43/27 | 31/62 | 0.000 | 43/47 | 31/42 | 0.498 | 57/53 | 17/36 | 0.018 | 51/75 | 23/14 | 0.020 | |||
| TNM (12/34) | 52/21 | 65/30 | 0.694 | 66/26 | 51/25 | 0.516 | 82/32 | 35/19 | 0.349 | 90/40 | 27/11 | 0.830 | |||
| BCLC (01/23) | 43/30 | 51/44 | 0.499 | 56/36 | 38/38 | 0.158 | 8/46 | 26/28 | 0.161 | 72/58 | 22/16 | 0.784 | |||
| AFP (<25/≥25 ng/L) | 31/40 | 37/58 | 0.541 | 39/51 | 29/47 | 0.499 | 49/63 | 19/35 | 0.293 | 45/84 | 23/14 | 0.003 | |||
| Hepatitis B (without/with) | 11/62 | 13/82 | 0.799 | 12/80 | 12/64 | 0.613 | 15/99 | 9/45 | 0.544 | 14/116 | 10/28 | 0.016 | |||
| Tumor size (≤5/>5 cm) | 41/32 | 55/40 | 0.822 | 56/36 | 40/36 | 0.283 | 69/45 | 27/27 | 0.198 | 76/54 | 20/18 | 0.523 | |||
| Tumor number (single/multiple) | 48/25 | 57/38 | 0.445 | 58/34 | 47/29 | 0.873 | 71/43 | 34/20 | 0.932 | 82/48 | 23/15 | 0.775 | |||
| Hepatic capsule (without/with) | 38/31 | 42/49 | 0.264 | 55/34 | 25/46 | 0.001 | 64/45 | 16/35 | 0.001 | 60/64 | 20/16 | 0.449 | |||
| *Microcarcinoma suppository (without/with) | 43/23 | 29/60 | 0.000 | 44/41 | 28/42 | 0.144 | 57/47 | 15/36 | 0.003 | 58/62 | 14/21 | 0.384 | |||
*, a small amount of data is missing.
Logistic binary regression analysis showed that microvascular invasion (P=0.002, OR =3.096, 95% CI: 1.528–6.273) and Edmondson grade III-IV (P=0.008, OR =2.613, 95% CI: 1.289–5.297) were independently associated with the mutation of TP53. Tumor invasion of the liver capsule (P=0.001, OR =3.030, 95% CI: 1.557–5.899) was independently associated with the mutation of TERT. AFP (<25 µmg/L) (P=0.009, OR =3.414, 95% CI: 1.367–8.527) was independently associated with the mutation of CTNNB1.
Discussion
Based on NGS technology, we constructed a genomic profile of HCC patients in China. The most commonly mutated genes were TP53 (56.5%), TERT (45.2%), and CTNNB1 (22.6%), which were similar to those reported by Ahn et al., but different from the report of Totoki and Schulze, where TERT was the most common mutation (Table 2) (7-10). In this study, we analyzed the genome variation of HCC patients in The Cancer Genome Atlas (TCGA) database, and the results were similar to those of Japanese and European populations. We deduced that the etiology difference was a possible reason. There are studies that have reported that HCV infection and alcoholic cirrhosis (P=0.03) were the most common groups in TERT mutated HCC patients, but not in HBV infected patients (P<0.001) (11). In addition, studies also reported that the mutation frequency of TP53 was higher in HCC caused by HBV infection than those without HBV infection (41% vs. 16%, P<0.001), while the mutation of CTNNB1 was found more in patients without a HBV infection than those with an infection (15% vs. 44%, P<0.001) (12).
Table 2. Comparison of gene mutational frequencies in different ethnicities.
| Gene | Korean (7) (n=231) (%) | Japanese (8) (n=503) (%) | European (9) (n=243) (%) | TCGA (n=366) (%) | Chinese (n=168) (%) |
|---|---|---|---|---|---|
| TP53 | 32 | 31 | 24 | 33 | 57 |
| TERT | 23 | 55 | 60 | 44* | 45 |
| CTNNB1 | 23 | 31 | 35 | 27 | 23 |
| AXIN1 | 7 | 6 | 11 | 8 | 14 |
| RB1 | 8 | 4 | 4 | 11 | 12 |
| TSC2 | NA | 5 | 5 | 5 | 11 |
*, The TCGA database was sequenced by WES and TERT promoter mutations were not detected; 196 cases of Sanger sequencing results of TCGA database by David A. Wheeler et al. (10).
Tumors usually have its own unique mutation spectrum, and the substitution frequency of different base pairs is different. Studies have shown that C:G>T:A conversion is a common mutation type in many solid tumors (13). G:C>T:A conversion has been reported to be the main mutation in HCC (14). All of these reports have supported that G:C>T:A conversions are the most common mutations in HCC patients, with slightly varying proportions among different populations. According to the description by Li and Mao (15), G conversion (G>A, G>T, G>C) is common in HCC patients. In this study, the proportion of G conversion was 38.8% (225/580), which further confirmed that the most common mutation in Chinese HCC patients was G conversion. The C:G>A:T conversion seemed to occur more commonly in HBV-related HCC than in HCV-related HCC (16), even though there were limited C:G>A:T conversions in this study. Therefore, it was not possible to further explore relevant factors.
In this study, mutations in different carcinogenic pathways of HCC patients were obtained. However, due to the lack of sample information used for calculating the exact mutations in different carcinogenic pathways, we failed to compare mutated pathway information with the TCGA database. In Chinese HCC patients, the most common mutations were P53 pathway related genes, followed by the Wnt pathway, telomere repair pathway, PI3K/mTOR pathway, chromatin remodeling related pathways, and cell cycle related pathways. Mutations in these carcinogenic pathways are also commonly reported in many other studies on HCC among different populations (Table 3) (7-9). However, mutations in the P53 pathway, telomere repair, and Wnt pathway related genes are the most common in all studied populations, which indicates the common occurrence and development of HCC in all populations.
Table 3. Comparison of mutational frequencies of signal pathway-related genes in different ethnicities.
| Pathway | Korean (7) (n=231) (%) | Japanese (8) (n=503) (%) | European (9) (n=243) (%) | Chinese (n=168) (%) |
|---|---|---|---|---|
| P53 pathway | 37 | 72 | 49 | 67 |
| Telomere repair | NA | 68 | 60 | 45 |
| Wnt pathway | 37 | 66 | 54 | 48 |
| PI3K /mTOR pathway | NA | 45 | 51 | 27 |
| Cell cycle | 22 | NA | NA | 18 |
| Chromatin remodeling | 34 | 67 | 28 | 26 |
NA, TCGA database does not have mutation information.
TMB is defined as the number of somatic mutations, including point mutations and insertion deletions, within an average of 1 Mb in the sequenced cancer genome. It is becoming one of the important parameters used to predict the sensitivity of immunologic checkpoint inhibitors (17). For example, according to a report by Rizvi, the top quartile of the number of mutations, no less than 178 mutations, was considered as TMB high (18). Johnson found that high TMB (45.6 vs. 3.9 Muts/Mb in PD-L1 response/non-response patients, respectively), was associated with high sensitivity to PD-1 or PD-L1 immunotherapy strategies in non-small cell lung cancer and melanoma (19). Shrestha et al. reported that the overall prognosis of HCC patients with TMB-H (defined as a quantity of mutations one standard deviation above the average for the dataset) was significantly worse than that of patients with TMB-L (defined as a quantity of mutations one standard deviation below the average for the dataset) (20). TMB could be used as a biomarker to select immunotherapy for HCC patients. In this study, the mutation frequency of CTNNB1 in patients with TMB-H (≥7.7 Muts/Mb) was significantly increased. This is the first report of the correlation between CTNNB1 and TMB-H in HCC. However, its mechanism still needs further study. MSI, dysregulated by the mismatch repair (MMR) system, was first found in hereditary non-polyposis colorectal cancer, and has been involved in the pathogenesis of a variety of gastrointestinal cancers and other cancers (21). The detection rate of MSI-high (MSI-H) in colorectal cancer is about 15–20% (22). There are few reports of MSI-H status in HCC patients. Goumard et al. analyzed MSI in 164 HCC specimens, but did not find a specimen with an MSI-H phenotype (23). Here, we also did not find any MSI-H phenotypes in Chinese HCC patients. Fabrizio et al. identified the TMB and MSI in 6,004 paraffin specimens of colorectal cancer. The detection rate of MSI-H status was 5.0%, and the distribution range of TMB was 0–746.9 Muts/Mb, with a median of 4.5 Muts/Mb. There was a significant correlation between MSI-H status and TMB-H (>11.7 Muts/Mb) (P<0.0001) (24). In this study, the distribution of TMB in HCC ranged from 0.0 to 28.4 Muts/Mb, with a median of 5.4 Muts/Mb. Compared to colorectal cancer, HCC has the characteristics of centralized TMB distribution, small dispersion, and microsatellite stability.
As we know, TP53 is a tumor suppressor gene which can inhibit the occurrence and development of cancer. Calderaro et al. reported that TP53 mutations are associated with poor tumor differentiation (OR =6.41, P<0.001) and microvascular invasion (OR =2.03, P=0.02) (25). Consistent with previous studies, the mutation of TP53 in Chinese HCC patients was independently correlated with poor biological characteristics of tumors such as Edmondson grade III-IV and microvascular invasion. In this study, TERT was the second most commonly mutated gene. Except for the invasion of the liver capsule by tumors, there was no significant correlation between the mutation of TERT and age, sex, size of tumors, number of tumors, microvascular invasion, differentiation tumor grade, or TNM stage. This result is consistent with the report of Lee et al. (26). This suggests that TERT gene mutations are not easily expressed in the biological characteristics of tumors, which may be related to the fact that the mutation of TERT is an early step in the development of HCC (27). In addition, we found that TP53 was highly correlated with TERT, and that co-mutations of TP53 and TERT occurred in 32.1% (54/168) of patients. Moreover, the co-mutation of TP53 and TERT was more likely to occur in patients with Edmondson grade III–IV, invasion of the hepatic capsule, and microvascular invasion.
As a key gene in the Wnt/beta-catenin signaling pathway, CTNNB1 is one of the most common mutations in HCC in most populations. The mutation of CTNNB1 was reported to be significantly correlated with age in populations less than 60 years old (P=0.019) with moderately/poorly differentiated HCV-related HCC (P=0.015) (28). Cleary et al. also reported that CTNNB1 mutations were more common in HCV-related HCC than HBV-related HCC (37.5% vs. 62.5%, P=0.038) (29). Similarly, we found that the mutation frequency of CTNNB1 in HBV-infected patients was lower than that in non-HBV-infected patients (P=0.021). Also, the proportion of Edmondson grade I–II, AFP <25 µmg/L, and those without a background of hepatitis B was higher in the patients with a CTNNB1 mutation than those without a CTNNB1 mutation, indicating that the mutation of CTNNB1 was associated with better clinical characteristics. Additionally, in HCC cases with CTNNB1 mutations compared to those without mutations, Cleary et al. observed a lower recurrence rate (12.5% vs. 49.3%) and a prolonged disease-free survival (29), which is similar to the results of this study and also supports the inference that CTNNB1 mutations are related to a better prognosis.
The limited number of samples coming only from a single center is a shortcoming of this study. For future studies we will collect multi-center samples and try to describe the genomic alteration map of Chinese HCC patients.
In conclusion, the most common mutations were TP53, TERT, and CTNNB1 in Chinese HCC patients. The mutation frequency of TERT in Chinese HCC patients was different from that in European, and Japanese patients. The main mutation forms were SNV being C:G>T:A conversion and guanine conversion. The P53 pathway, Wnt pathway, and telomere repair pathway were the most mutagenic carcinogenic pathways in the development of HCC. The proportion of microvascular invasion and Edmondson III-IV grade was higher in HCC patients with TP53 mutations. The proportion of tumors invading the hepatic envelope was higher in HCC patients with TERT mutations. The proportion of Edmondson I-II grade, AFP <25 µmg/L, and a non-hepatitis B background, as well as a higher TMB value, was found to be higher in HCC patients with CTNNB1 gene mutations.
Acknowledgments
Thanks to OrigiMed for the technical support with NGS.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All patients signed a written informed consent. The study was conducted in strict compliance with the Helsinki Declaration and was approved by the Ethics Committee (ethics approval number: QDFYKYLLL-20161212).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/hbsn.2019.09.17
Data Sharing Statement: Available at http://dx.doi.org/10.21037/hbsn.2019.09.17
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/hbsn.2019.09.17). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. 10.1016/S0140-6736(11)61347-0 [DOI] [PubMed] [Google Scholar]
- 3.Graf D, Vallbohmer D, Knoefel WT, et al. Multimodal treatment of hepatocellular carcinoma. Eur J Intern Med 2014;25:430-7. 10.1016/j.ejim.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 4.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 2010;11:685-96. 10.1038/nrg2841 [DOI] [PubMed] [Google Scholar]
- 5.Tang B, Yan X, Sheng X, et al. Safety and clinical activity with an anti-PD-1 antibody JS001 in advanced melanoma or urologic cancer patients. Journal of Hematology & Oncology 2019;12:7. 10.1186/s13045-018-0693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan M, Hao J, Cui S, et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res 2018;28:359-73. 10.1038/cr.2018.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn SM, Jang SJ, Shim JH, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014;60:1972-82. 10.1002/hep.27198 [DOI] [PubMed] [Google Scholar]
- 8.Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267-73. 10.1038/ng.3126 [DOI] [PubMed] [Google Scholar]
- 9.Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. 10.1038/ng.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network . Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-1341.e23. 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013;4:2218. 10.1038/ncomms3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amaddeo G, Cao Q, Ladeiro Y, et al. Integration of tumour and viral genomic characterizations in HBV-related hepatocellular carcinomas. Gut 2015;64:820-9. 10.1136/gutjnl-2013-306228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature 2007;446:153-8. 10.1038/nature05610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8. 10.1038/ng.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Mao M. Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett 2013;340:247-53. 10.1016/j.canlet.2012.09.027 [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet 2012;44:1117-21. 10.1038/ng.2391 [DOI] [PubMed] [Google Scholar]
- 17.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 2016;17:e542-e551. 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DB, Frampton GM, Rioth MJ, et al. Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 2016;4:959-67. 10.1158/2326-6066.CIR-16-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shrestha R, Prithviraj P, Anaka M, et al. Monitoring Immune Checkpoint Regulators as Predictive Biomarkers in Hepatocellular Carcinoma. Front Oncol 2018;8:269. 10.3389/fonc.2018.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci U S A 2003;100:776-81. 10.1073/pnas.0334858100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005;352:1851-60. 10.1056/NEJMoa043146 [DOI] [PubMed] [Google Scholar]
- 23.Goumard C, Desbois-Mouthon C, Wendum D, et al. Low Levels of Microsatellite Instability at Simple Repeated Sequences Commonly Occur in Human Hepatocellular Carcinoma. Cancer Genomics Proteomics 2017;14:329-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabrizio DA, George TJ, Jr, Dunne RF, et al. Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J Gastrointest Oncol 2018;9:610-7. 10.21037/jgo.2018.05.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderaro J, Couchy G, Imbeaud S, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol 2017;67:727-38. 10.1016/j.jhep.2017.05.014 [DOI] [PubMed] [Google Scholar]
- 26.Lee SE, Chang SH, Kim WY, et al. Frequent somatic TERT promoter mutations and CTNNB1 mutations in hepatocellular carcinoma. Oncotarget 2016;7:69267-75. 10.18632/oncotarget.12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nault JC, Calderaro J, Di Tommaso L, et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 2014;60:1983-92. 10.1002/hep.27372 [DOI] [PubMed] [Google Scholar]
- 28.Tornesello ML, Buonaguro L, Tatangelo F, et al. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics 2013;102:74-83. 10.1016/j.ygeno.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 29.Cleary SP, Jeck WR, Zhao X, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693-702. 10.1002/hep.26540 [DOI] [PMC free article] [PubMed] [Google Scholar]