Abstract
As people age, their tissues accumulate an increasing number of somatic mutations. While most of these mutations are of little or no functional consequence, a mutation may arise that confers a fitness advantage on a cell. When this process happens in the hematopoietic system, a substantial proportion of circulating blood cells may derive from a single mutated stem cell. This outgrowth, called “clonal hematopoiesis,” is highly prevalent in the elderly population. Here we discuss recent advances in our knowledge of clonal hematopoiesis, its relationship to malignancies, its link to non-malignant diseases of aging, and its potential impact on immune function. Clonal hematopoiesis provides a glimpse into the process of mutation and selection that likely occurs in all somatic tissues.
Introduction
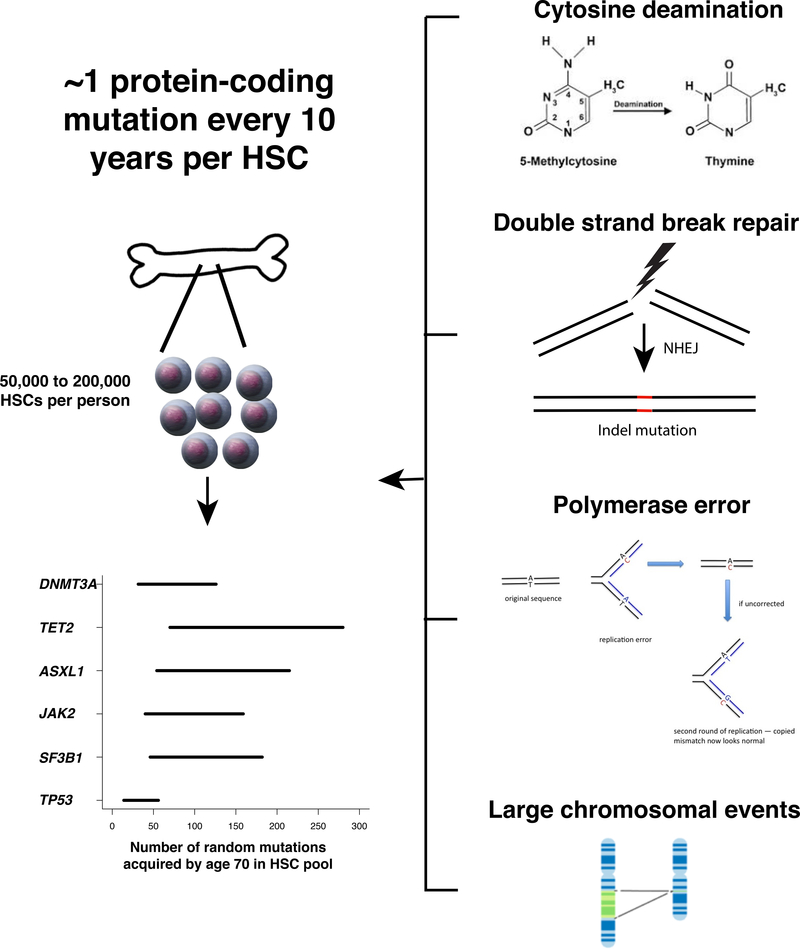
Aging is associated with a steady increase in the number of somatic mutations in virtually all tissues 1–5. These age-associated mutations fall into several classes. The most frequent class arises from the spontaneous deamination of 5-methylcytosine to thymine and primarily occurs at CpG dinucleotides, which are often in the methylated state 6. If a cell has not repaired this error before replication, one daughter cell will have a thymine:adenine pairing of DNA bases instead of the parental cytosine:guanine. This process occurs at a linear rate with respect to time and is therefore considered a signature of aging 7. A second class of mutation is small insertions and deletions (indels), which commonly arise from errors introduced during non-homologous end joining of DNA double-strand breaks and can result in frameshift mutations if the breaks occur in protein-coding portions of the genome 8. Evidence from model organisms suggests that double-strand breaks may become more common as cells age 9. A third mechanism of mutation is replication error by DNA polymerase, which also typically results in base substitutions or small indels. The error rate of eukaryotic DNA replication tends to be very low, except in the case where DNA mismatch repair function is compromised 10Sanders, 2018 #305}. The number of replication cycles that a cell has undergone typically increases with age; therefore the number of polymerase errors also cumulatively increases 11. The fourth type of age-associated mutation is large structural variation, such as insertions, deletions, loss-of-heterozygosity, or rearrangements spanning several kilobases or more, although these occur somewhat less commonly than base substitutions and small indels 12.
In combination, these mutational processes create a broad array of genetically distinct tissue stem cells. Through Darwinian selection, some of these stem cells gain a competitive advantage, a phenomenon that has been most extensively studied in the human hematopoietic system. It is estimated that humans have 50,000 to 200,000 hematopoietic stem cells (HSCs) 13. As each HSC acquires ~1 exonic mutation per decade of life 3, by the age of 70 an average person would be expected to harbor up to 1.4 million protein-coding variants, corresponding to an average of 70 mutations per gene, in at least one HSC (Fig. 1). If just one of these mutations is capable of imparting a fitness advantage to the cell in which it arose, expansions of mutated HSCs, termed “clones”, should be common in aging humans. Indeed, numerous studies have now shown that such outgrowths of mutated blood cells, termed “clonal hematopoiesis,” are highly prevalent in the elderly. An equivalent state is also pervasive in the epithelium of skin 14 and esophagus 15,16, suggesting that somatic mutation-driven clonal expansions may be a characteristic of aging in several tissues. Here, we review recent developments in our understanding of clonal hematopoiesis and its implications for human health.
Fig. 1.
A single HSC in a healthy person acquires approximately one protein-coding mutation per decade of life (00). Four mutational processes contribute to the bulk of these age-associated mutations (right). Assuming there are 50,000 to 200,000 HSCs in an average person (00), we estimate that by age 70, an average person will harbor 350,000 to 1.4 million protein coding mutations in his/her HSC pool. Shown in the bottom left is the expected range of random mutations in HSCs in the exons of DNMT3A, TET2, ASXL1, JAK2, SF3B1, and TP53 by age 70 per person. A subset of these mutations may lead to clonal expansions.
A brief history of clonal hematopoiesis
Clonal hematopoiesis is characterized by the over-representation of blood cells derived from a single clone. Blood cancers such as chronic myeloid leukemia were first demonstrated to be clonal from karyotypic analysis and are the prototypical example of clonal hematopoiesis 17. But in the 1990s, studies of non-random X-chromosome inactivation (XCI) in women led to the discovery that clonal hematopoiesis also occurs in individuals without cancer 18. One X-chromosome is randomly inactivated in each cell during female embryonic development and this epigenetic state is stably maintained across cell divisions. The demonstration of non-random XCI in a population of cells is therefore evidence of a clonal process. Importantly, non-random XCI in blood cells was observed to increase in frequency with age, although the underlying mechanism was unclear 19. A more complete catalog of the genetic framework of blood cancers allowed investigators to look for cancer-associated somatic mutations in these cases. In 2012, mutations in TET2 (a gene coding for an enzyme that oxidizes methylated DNA) were found in ~5% of elderly women with non-random XCI. This was the first demonstration that mutation-driven clonal hematopoiesis occurs in healthy persons 20.
A second line of evidence that supported the existence of pre-malignant clonal expansions in healthy individuals came from studies of patients who were in remission after being treated for acute myeloid leukemia (AML). Like most cancers, AML generally results from several driver mutations that occur sequentially in the same clone over time 3. If the first mutation to be acquired leads to clonal expansion without malignant transformation, it should be possible to identify a pre-malignant stage in which only the initiating lesion is present 21. This was first demonstrated in a case study of a patient with AML in which the driver mutation of the leukemia, an AML1/ETO translocation, could be detected in a fraction of phenotypically normal HSCs and mature hematopoietic cells in remission samples 22. In later studies, DNA sequencing of HSCs from AML patients in remission revealed that stem cells with only a single driver mutation were often present, suggesting that pre-malignant clonal hematopoiesis was a generalizable finding in AML 23–25. However, the extent of mutation-driven clonal hematopoiesis in the healthy population, its full genetic spectrum, and its natural history remained unknown.
Clonal hematopoiesis in the genome sequencing era
In 2014, three groups examined exome sequencing data from cases and controls within genetic association studies for diabetes, schizophrenia, and solid tumors that together comprised greater than 30,000 persons 26–28. Importantly, these individuals were unselected for hematological phenotypes and the source of DNA was peripheral blood cells, thus permitting the study of mutation-driven clonal hematopoiesis on a larger scale than was previously possible. These studies can be thought of as a natural experiment of saturation mutagenesis in humans, and can be summarized in a simple postulate—given a sufficiently large population, every possible mutation that can occur will occur in some HSCs. Those cells carrying mutations that are neutral or deleterious will not expand, and therefore will not be detectable from blood DNA. Those mutations that are detectable are the ones that cause clonal expansion, and these will point to biological pathways that increase the fitness of HSCs.
The surprising result of this experiment of nature was that clonal hematopoiesis largely results from mutations in a very restricted set of genes. Mutations in classical oncogenes and tumor suppressors, such as those involved in cellular growth signaling (JAK2, GNAS, GNB1, CBL) and the DNA damage response (TP53, PPM1D), were seen but were not the most common. Instead, nearly two-thirds of clonal hematopoiesis could be accounted for by loss-of-function mutations in just two enzymes involved in DNA methylation, DNMT3A and TET2. The third most commonly mutated gene was ASXL1, a chromatin regulator, while mutations in splicing factors (SF3B1, SRSF2, PRPF8, U2AF1) were also frequent. Why these mutations cause clonal expansion remains an intense area of investigation (see ‘Mechanisms of Clonal Expansion’ below).
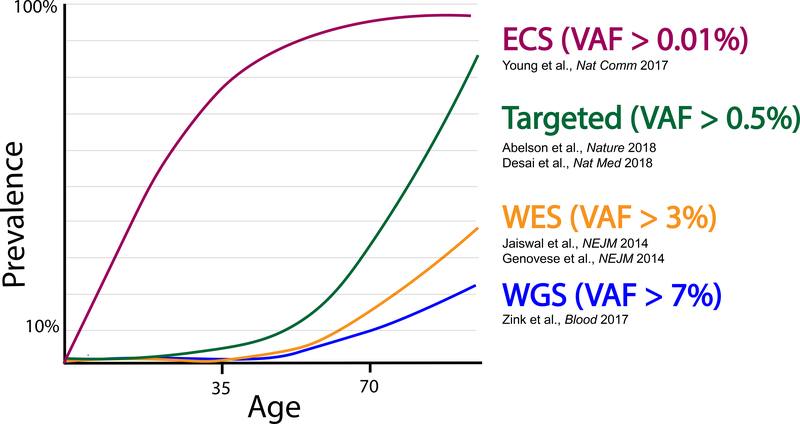
A second observation that emerged from these studies was that clonal hematopoiesis is an age-related phenomenon. Somatic clones were detectable in less than 1% of healthy individuals under age 40, but they increased in frequency with each decade of life. In contrast, 10–20% of individuals age 70 or older harbored a detectable clone. The size of these mutant clones was massive; in one study, a median of ~18% of blood cells carried the mutations 28. For comparison, previously described mutations in the blood of healthy persons such as BCR-ABL or BCL2 translocations were usually present in less than 0.01% of cells and were often transient 29,30. One important consideration is that the prevalence of clonal hematopoiesis is highly dependent on the sensitivity of the method used to detect it (Fig. 2). These initial studies used whole-exome sequencing, which is relatively insensitive to smaller clones. Subsequent studies in healthy persons using more sensitive approaches have found the prevalence of clonal hematopoiesis to be much higher, although the biological significance of the smaller clones is unknown 31–33.
Fig. 2.
The estimated prevalence of CHIP as a function of age varies according to the sequencing method used.
VAF, variant allele fraction. ECS, error corrected sequencing. WES, whole- exome sequencing. WGS, whole-genome sequencing.
Thus far our discussion of clonal hematopoiesis has focused on mutational changes that are limited to small stretches of DNA, such as base substitutions and small indels. But large structural variation, such as gains or losses of large segments of chromosomes, also increases with age and has clinically meaningful associations 34–36. While the accumulation of both small and large somatic variants are linked to aging, the underlying biology of the two is generally non-overlapping. For example, copy number gains or losses of DNMT3A, TET2, and ASXL1, are rarely found in surveys of somatic large structural variation 35. Instead, changes associated with chronic lymphocytic leukemia (CLL) and losses of sex chromosomes are the most common variants that accumulate in aging36. Mechanistic understanding of why these variants are positively selected during aging is lacking in the majority of cases. Further complicating the picture, clonal hematopoiesis has been observed in the absence of any known driver mutation 27,37. What causes apparent clonal expansion in these cases is unknown, but could be due to mutations in genes not previously queried in surveys of clonal hematopoiesis, mutations in the non-coding genome, or even genetic drift due to age-related constriction of the stem cell pool 37.
Clonal hematopoiesis of indeterminate potential (CHIP)
Clonal hematopoiesis generally refers to any clonal outgrowth of hematopoietic cells, regardless of cause or disease state. Thus, someone with a frank malignancy like acute myeloid leukemia would be considered to have clonal hematopoiesis. Stochastic processes such as constriction of the stem cell pool with aging may also lead to clonal hematopoiesis in the absence of a known driver mutation. The term “clonal hematopoiesis of indeterminate potential” (CHIP) was introduced to distinguish non-malignant clonal hematopoiesis that is clearly linked to cancer-associated mutations from other forms of clonal hematopoiesis 38. CHIP refers to the presence of a cancer-associated variant in the blood cells of a person without a frank malignancy or another recognized clonal entity, such as monoclonal B-lymphocytosis (MBL) or paroxysmal nocturnal hemoglobinuria 39. The term “indeterminate potential” is intended to invoke the medical uncertainty associated with such a state. By this definition, cancer-free persons with somatic mutations in genes such as TET2 or TP53 would be considered to have CHIP. This term would not apply to persons with copy number abnormalities associated with MBL/CLL or clonal expansion in the absence of a known driver mutation. To meet the definition of CHIP, the clones must also meet a certain size threshold. If sequenced deeply enough, a cancer-associated mutation may be detectable in most people over the age of 50 33, but only clones that reach a certain size are likely to be clinically meaningful. The threshold for CHIP was set at a variant allele fraction (VAF) of 2% (meaning 2% of the sequenced alleles contained the mutation, or roughly 4% of cells, assuming the mutation is heterozygous), but may be revised if it is demonstrated that there is prognostic significance for clones below this size.
Mechanisms of clonal expansion
The role of CHIP-associated genes in hematopoiesis has been extensively reviewed elsewhere 40,41. For some of these genes, clear mechanisms for clonal expansion have been found. HSCs from mice with mutations in both Tp53 42 and Ppm1d, a gene encoding a regulator of p53 and other members of the DNA damage response pathway, 43,44 are able to enter cell cycle despite the presence of DNA damage, leading to a stem cell advantage in the setting of cytotoxic drugs. Activating mutations in JAK2 allow constitutive signaling though growth factor receptors, thus leading to clonal expansion by enhanced proliferation of cells that carry this mutation 45.
Less clear is why mutations in TET2 or DNMT3A lead to clonal expansion. Especially perplexing is the fact that these two genes have ostensibly opposite biochemical functions. DNMT3A is one of the two enzymes responsible for de novo methylation of the fifth position in cytosine bases of DNA, a mark that is thought to influence gene expression 46,47. TET2 is one of three enzymes responsible for catalyzing the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine and further intermediates, which can eventually lead to demethylation 48,49. Mouse models carrying loss-of-function mutations in either of these genes clearly show an HSC competitive advantage in vivo as well as a propensity for leukemia when cooperating mutations are present 50–53. Stem cells from these mice are able to serially replate in colony forming assays over many generations, whereas wild-type stem cells quickly lose this capacity, suggestive of enhanced self-renewal capacity in the mutant stem cells 54,55. However, it is unknown why loss of cytosine methylation or hydroxymethylation, broadly or at specific genomic loci, leads to changes in self-renewal ability.
Mutations in genes encoding core members of the spliceosome are also common in CHIP. HSCs from mice mutant for these genes, Sf3b1 56, Srsf2 57, and U2af1 58, have a competitive disadvantage in vivo, unlike the clonal expansion observed in humans with these mutations. The reason for the disparity of phenotypes in humans and mice is unclear. Partly because of the inability to model clonal hematopoiesis in these mice, the exact mechanisms whereby these mutations lead to clonal expansion remains unknown.
Clonal hematopoiesis is also commonly found in aplastic anemia, a disorder caused by an autoimmune attack on bone marrow progenitor cells that results in severely depressed blood counts 59. In contrast to the mutations associated with CHIP, the mutations most commonly seen in aplastic anemia affect the genes PIGA, BCOR, and BCORL1. The PIGA gene is required for the synthesis of glycophosphotidylinositol (GPI) and its loss results in down-regulation of several cell surface proteins that are GPI anchored 60. Loss of some of these proteins may allow for immune escape, thus explaining selection for PIGA-mutated clones. The mechanism of selection for BCOR/BCORL1 mutations in this setting is unknown.
The risk of developing clonal hematopoiesis is largely related to the stochastic acquisition of somatic mutations in HSCs during aging. However, some epidemiological studies have implicated environmental and heritable components as well. For example, CHIP has been reported to be more common in older men 28 and smokers 27 and less common in Hispanics 28, although these associations are all relatively modest. Recent studies have implicated genetic predispositions 37,61 as well as the microbiome 62 in the etiology and progression of clonal hematopoiesis. More insights into the factors that influence clonal expansions are expected to emerge from genetic sequencing of large population cohorts with richly annotated clinical phenotypes.
CHIP and hematological malignancy
CHIP itself does not denote a malignancy, nor is it associated with clinically significant alterations in blood counts 28,63. But many of the most commonly seen mutations in CHIP are also recurrent drivers of AML 64, myelodysplastic syndrome (MDS) 65–67, myeloproliferative neoplasms 68, and certain lymphomas 69,70. Thus, one might predict that individuals with CHIP would develop hematological malignancies at a rate above background since they have the “first hit” needed for malignant transformation. Indeed, in population-based cohorts that underwent exome sequencing, the presence of CHIP was associated with ~10-fold increased relative risk of these malignancies over several years of follow-up 27,28. In one study, 4% of CHIP carriers developed a blood cancer over the subsequent 8 years, corresponding to ~0.5% of CHIP cases converting to malignancy per year 28. Notably, the risk of malignancy in the carriers of CHIP was associated with the size of the mutant clone, as those who went on to develop malignancy had substantially larger clone sizes than those who did not. Myeloid malignancies were most common, although some people with CHIP did develop lymphoid cancers 27,28.
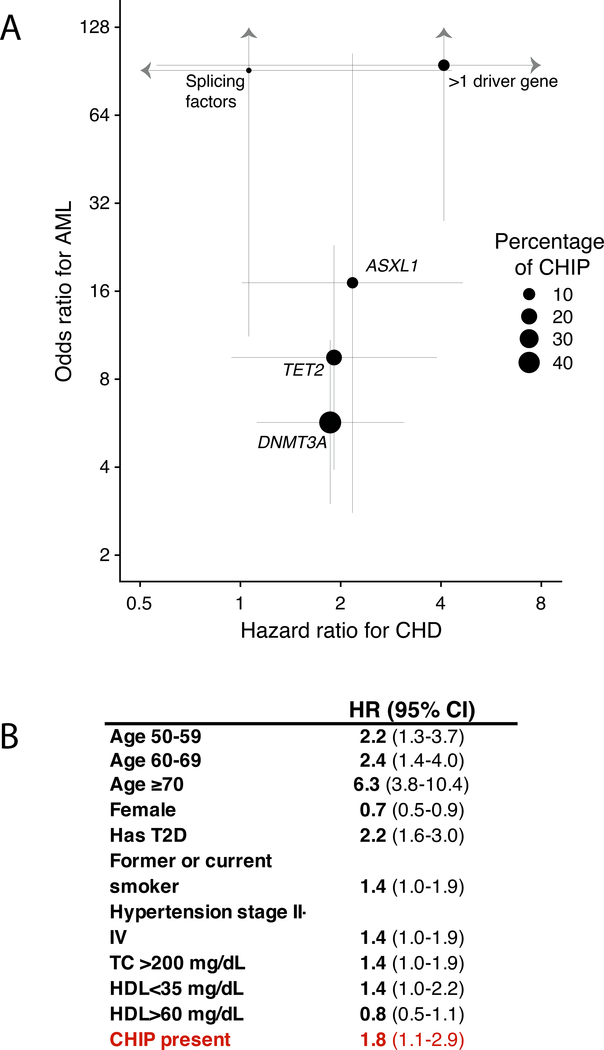
To refine risk estimates for developing AML associated with clonal hematopoiesis, two groups performed nested case-control studies within large population-based cohorts that had several years of follow-up 71,72. Both groups found that individuals with antecedent clonal hematopoiesis were at ~3 to 5-fold increased risk for developing AML in the subsequent years (Fig. 3A). The risk was lower than the ~10-fold increase seen in previous studies because clonal hematopoiesis was identified using methods that were more sensitive than exome sequencing, which resulted in the detection of more clones of smaller size, including clones below the size threshold for CHIP. Similar to previous studies, the risk of AML positively correlated with the size of the mutant clone. An intriguing finding from these studies was that mutations in TP53, JAK2, SF3B1, SRSF2 and U2AF1 were linked to a particularly high risk of developing AML; however, many of these individuals had multiple driver gene mutations, making the assessment of risk for singleton mutations challenging. Nonetheless, such studies could provide a rationale for population-wide screening for those at especially high risk for transformation, though it remains to be seen what interventions would be beneficial in these individuals.
Fig.3.
(A) Forest plots for odds ratio for developing acute myeloid leukemia (AML) and hazard ratio for developing coronary heart disease (CHD) in those with mutations in the genes listed, adapted from Abelson et al. Nature 2018 and Jaiswal et al., NEJM 2017. Only those mutations with a variant allele fraction greater than 2% (meeting the definition of CHIP) were included. Individuals with mutations in more than one driver gene were analyzed as a separate category. Lines represent the 95% confidence interval for odds or hazard ratios and the sizes of the dots are proportional to the number of mutations in each gene in the general population.
B) Hazard ratio (HR) and 95% confidence interval (CI) for developing CHD based on Framingham risk factors plus presence of CHIP mutations. Data taken from population-based cohorts unselected for CHD status (just give ref numbers).
A feared complication in patients who have been treated with cytotoxic drugs for solid cancers is therapy-related development of secondary AML and MDS. Several studies have found that patients with CHIP and solid tumors or lymphoma have an increased risk of these therapy-related myeloid neoplasms after treatment for the primary disease 73–75. It is hypothesized that pre-existing mutant HSC clones selectively expand under the pressure of cytotoxic therapy, and can cause cancer several years later with the acquisition of subsequent mutations 76. One study of a patient population treated for solid cancers found that CHIP mutations were more prevalent in this group than in populations not selected for cancer 77. Furthermore, the mutational spectrum in patients with non-hematologic cancers was altered: HSC mutations in DNA damage response genes such as TP53 and PPM1D were far more prevalent in this setting, likely due to strong selective pressure from exposure to cytotoxic therapies. Patients with CHIP also had increased mortality, most often due to progression of their primary malignancy. These studies indicate that CHIP in the setting of other cancers is likely to be especially pervasive and portends a poor prognosis.
CHIP and non-malignant disease
Most clonal expansion states are expected to increase the risk of neoplasia in the tissues in which they arise. But might there be consequences of the mutant clones apart from cancer? While it will be fascinating to determine these consequences in all tissues, some characteristics of the hematopoietic system make it particularly noteworthy as a potential cause of non-malignant disease. First, tissue architecture constrains the extent of clonal expansion within tissues such as gut or skin epithelium to patches that are rarely larger than a few square millimeters 15, but there is no such spatial restriction on HSCs, which freely admix throughout the bone marrow and body 78. Indeed, some individuals with clonal hematopoiesis have virtually all of their blood cells arising from a single mutated HSC 28. Second, alterations in hematopoietic cells have the potential to impact a wide range of disease states. In contrast to tissue-specific cells or epithelia, immune cells such as lymphocytes, granulocytes, and monocytes can migrate to and influence nearly every organ. These immune effector cells are derived from HSCs, so any mutations that occur in HSCs can also potentially alter the immune response or baseline inflammatory state.
These observations have prompted an examination of the effects of mutation-driven clonal hematopoiesis on human health and disease beyond blood cancer. Several studies have found that CHIP is associated with a 30–40% increased mortality risk 27,28,37. In an initial study, this risk could not be explained by cancer deaths but was instead related to increased cardiovascular mortality 28. Further analysis revealed that the risk of future ischemic stroke and coronary heart disease was more than doubled in carriers of CHIP (Fig. 3A). In this study of primarily middle-aged individuals, the risk of coronary heart disease and ischemic stroke associated with CHIP was as great or greater than that conferred by well-known risk factors for cardiovascular disease, such as circulating LDL-cholesterol levels, smoking, and blood pressure (Fig. 3B). Replication studies in additional cohorts confirmed and extended the early work. In studies of middle-aged and older individuals, the risk for coronary heart disease was nearly twice as high for individuals with CHIP compared to individuals without CHIP. The risk for early-onset myocardial infarction (MI), defined as heart attack before age 40 in men or age 50 in women, was four times higher in those with CHIP compared to those without CHIP. The relative risk for coronary heart disease in individuals bearing mutations in DNMT3A, TET2, or ASXL1 was roughly doubled compared to those without CHIP; in individuals bearing JAK2 mutations, the relative risk was ~12-fold higher compared to those without CHIP. In addition, individuals with larger mutant clones had the greatest risk of cardiovascular disease, mirroring the situation for malignancy risk 79. Just as CHIP is associated with an increased risk of cardiovascular disease, lower-risk subtypes of MDS are also associated with a doubling of the risk of dying from cardiovascular causes 80.
The link between CHIP and cardiovascular outcomes is not limited to atherosclerotic disease. Recent evidence suggests that individuals with post-MI related congestive heart failure who carry CHIP-associated DNMT3A or TET2 mutations have worse survival outcomes than individuals without CHIP 81. In a separate study (83), individuals with CHIP-associated JAK2 mutations were reported to have a ~12 times greater risk of developing venous thrombosis than those without CHIP, whereas individuals with mutations in other CHIP-associated genes had a doubling of the risk 82. Thus, CHIP is likely to be an indicator of poor prognosis for several distinct cardiovascular disorders.
While it is clear that CHIP-associated mutations play a causal role in the development of blood cancer, it is less clear, a priori, whether their role in non-malignant diseases is causal or merely correlative. Like several other well-described markers in blood cells, such as red cell distribution width 83, DNA methylation clocks 84, and loss of Y-chromosome 85, CHIP is associated with multiple adverse outcomes in epidemiological studies. One potential explanation is that all of these biomarkers are measures of some aspect of biological aging but are themselves not directly causal for health outcomes. What distinguishes CHIP from these other measures is the ability to manipulate model organisms experimentally to test causality.
In 2017, two research groups used mouse models to establish a causal role for CHIP in atherosclerosis. Both groups found that loss of Tet2 in bone marrow cells led to an increase in the size of atherosclerotic lesions in hyperlipidemic mice, an effect that could not be explained by quantitative changes in blood cell parameters of the mutant mice 79,86. Rather, the mutant bone marrow-derived macrophages upregulated many pro-inflammatory molecules, suggesting a potential mechanism for the increase in atherosclerosis. In support of this hypothesis, loss of Tet2 in myeloid cells was sufficient to confer enhanced atherosclerosis. In addition, blockade of the pro-inflammatory cytokine IL-1B reversed the accelerated atherosclerosis seen in Tet2 mutant mice 86.
Subsequent studies showed that loss of Tet2 or Dnmt3a led to worsening heart function in a mouse model of congestive heart failure, corroborating the human genetic association 87,88. Heart function in these studies was also improved with blockade of IL-1B, suggesting that this may be a common pathway for reversing the effects of CHIP in the heart. There is also evidence that Jak2 mutations enhance atherosclerosis in mouse models by altering macrophage function 89. Mutations in JAK2 have a well-described role in activating STAT transcription factors, which are central to immune response in several cell types involved in atherosclerosis. Furthermore, JAK2 mutations prime neutrophils to form neutrophil extracellular traps, leading to thrombosis, which may also contribute to poor cardiovascular outcomes 82. Together, these studies provide strong evidence that somatic mutation-driven clonal hematopoiesis has a causal role in cardiovascular disease.
To date, few studies have demonstrated an unequivocal link between CHIP and other diseases of aging. CHIP has been found to be associated with a 30% increase in the likelihood of having type 2 diabetes (T2D) 28. However, causality could not be established, and this could represent a case of reverse causation, as hyperglycemia might influence the development or expansion of clones by interfering with TET2 function 90. Other studies have found links between CHIP and chronic obstructive pulmonary disease (COPD) 37,63. However, CHIP was also strongly linked to smoking in these studies, so the result could be confounded by this association.
Immune function and CHIP
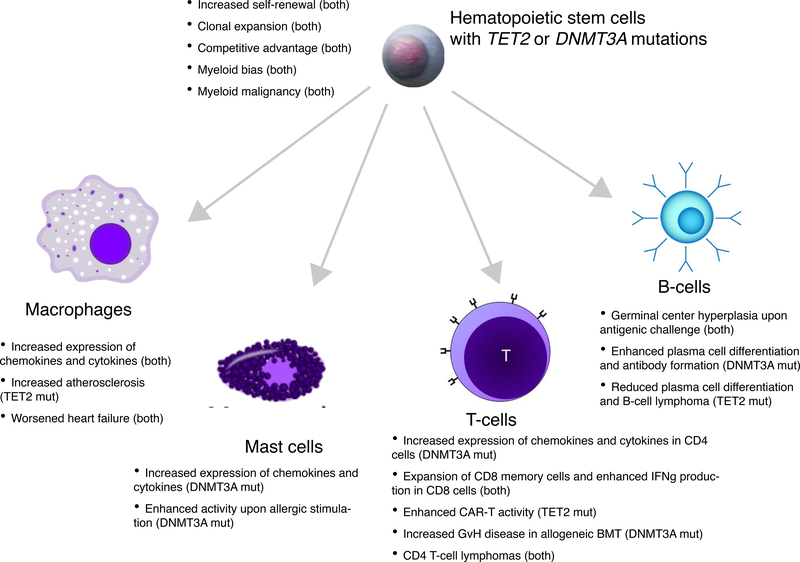
As many of the genes associated with CHIP are involved in transcriptional regulation, one might expect them to have broad effects on immune function. Several recent studies have examined the role of TET2 and DNMT3A in immunity (Fig. 4). One group found that mice deficient in Tet2 developed more severe inflammation in several tissues upon challenge with bacterial endotoxin, and this was partially explained by increased expression of the pro-inflammatory cytokine Il6 in dendritic cells and macrophages 91. Surprisingly, this effect on Il6 expression was independent of the catalytic function of Tet2, and was instead reported to be related to a direct interaction between Tet2 and histone deacetylases, resulting in transcriptional repression. Subsequent studies have confirmed the over-expression of Il6 and also found that Il1b, IL8 family chemokines, and other inflammatory mediators show increased expression in Tet2-deficient macrophages challenged with low-density lipoprotein or endotoxin 79,86,92. Humans with mutations in TET2 are also reported to have increased levels of circulating IL8 protein 79. The changes in expression are relatively modest for each gene (~2 to 3-fold increase) but appear to be biologically significant given the number of genes that are dysregulated and the breadth of phenotypes seen in Tet2 knockout mice. These molecules are thought to enhance inflammation locally within atherosclerotic plaques by increasing chemotaxis of leukocytes to arterial intima, which potentially explains the accelerated atherosclerosis seen with loss of Tet2 93.
Fig. 4.
Phenotypic changes in HSCs and immune cells with TET2 or DNMT3A mutations.
Less is known about the role of DNMT3A in innate immune function, but most studies to date have found evidence of enhanced inflammation when its function is perturbed. For example, one study found that mast cells from mice that lacked Dnmt3a produced higher levels of IL-6, TNF-alpha, and IL-13 in response to stimulation with IgE in vitro, and enhanced mast cell activity was also seen in a mouse model of allergy 94. Another study used CRISPR to mutate Dnmt3a in mouse RAW 264.7 macrophages, and observed increased expression of Cxcl1, Cxcl2, and Il6 in response to endotoxin 87. Mechanistically, very little is known about why these specific gene expression changes are seen with loss of Dnmt3a.
CHIP may be relevant in the adaptive immune response as well. Bone marrow transplant recipients with hematological malignancies who received donor marrow from carriers of CHIP had higher rates of graft-versus-host disease and reduced relapse rates 95. One possible explanation for this finding is that the donor cells harboring the mutations were capable of mounting stronger immune responses against both normal host tissues and the tumor. There is also a report of an exceptional response in a patient with CLL treated by infusion of chimeric antigen receptor (CAR) T-cells in which the CAR construct disrupted one copy of TET2, while the other copy had been previously mutated somatically 96. The authors speculate that the resulting TET2-deficient CAR-T clone was more effective at eliminating the tumor cells because of an expanded central memory CD8+ T-cell population, reduced T-cell exhaustion in response to stimulus, as well as enhanced cytokine production in T-cells. Several studies have found additional roles for TET2 and DNMT3A in normal T- and B-cell function (Figure 4).
In summary, growing evidence supports a role for the commonly mutated CHIP genes in immune function, and CHIP may underlie some part of the phenomenon termed “inflammaging”, the age-associated increase in systemic inflammation 97.
Future outlook
Clonal hematopoiesis provides a fascinating glimpse into the end result of decades of mutation and natural selection within a tissue. The potential health implications of CHIP are broad. It is associated with blood cancers, cardiovascular disease, and overall mortality. While our knowledge of this condition has increased exponentially over the last several years, this research has also highlighted fundamental biological questions and opened new pathways for translational discovery.
A major area of uncertainty is the range of disease states associated with CHIP. As CHIP is linked to enhanced inflammation, it is possible that links between CHIP and several diseases of aging will be found. Does CHIP influence the risk of Alzheimer’s disease, autoimmunity, liver disease, or others? As ever larger genetic cohorts with rich phenotypic information are assembled, these links may be systematically discovered.
While CHIP as a whole is clearly linked to cancer, cardiovascular disease, or death, the level of risk for any given individual is likely to be variable. The effect of specific gene variants on various outcomes may be quite different, as has been found in AML 71. Furthermore, the interaction of CHIP with other risk factors, such as having type 2 diabetes or elevated serum levels of C-reactive protein, may be synergistic for adverse outcomes. To have power to detect such associations, very large population-based cohorts are needed. Additional biomarkers such as plasma proteins, metabolites, and DNA methylation may prove useful for risk stratification as well. Studies of serial samples have shown that the trajectory of a clone can vary between people. Some people have clones that remain stagnant in size for many years, whereas other people have clones that show steady growth 28,33. Since clone size associates with the risk of leukemia and other adverse outcomes, it is imperative to understand this situation. It is likely that cell extrinsic factors, like the microenvironment, the microbiome, or diet, will play a role.
The mechanisms by which the CHIP-associated mutations cause clonal expansion and enhanced inflammation is also a central unanswered question. It is striking that the two most commonly mutated genes in CHIP, DNMT3A and TET2, are opposing enzymes in DNA methylation, yet both lead to convergent phenotypes in stem cell biology and immunity. It is also uncertain why some people have apparent clonal hematopoiesis in the absence of a known driver mutation. This may be due to undiscovered mutations, but could some of these clonal expansions result purely from selection on a heritable epigenetic state?
Most importantly, we need to find ways to reverse the pathogenic effects of CHIP. Blockade of downstream inflammatory molecules may be one way to treat CHIP-associated atherosclerosis. But ideally drugs will be found that can suppress the mutant clones directly, which could potentially mitigate the risk of both cancer and cardiovascular disease. The positive effects of CHIP on anti-tumor immunity may also one day be harnessed for therapy.
The process of mutation and clonal selection is likely to be universal across all organs and tissues. Understanding the causes and consequences of clonal hematopoiesis may provide a framework to understand this process, and aging, more broadly.
Acknowledgements
Funding:
S.J. is supported by the Burroughs Wellcome Fund, Evans Foundation, Ludwig Center for Cancer Stem Cell Research, and the Leducq Foundation. B.L.E. is an Investigator of the Howard Hughes Medical Institute and received funding from the NIH (P01CA066996, P50CA206963, and R01HL082945). B.L.E. has also received research funding from the biopharmaceutical company Celgene and from the DEERFIELD Foundation.
Footnotes
Competing interests: S.J. and B.L.E. have received consulting fees from GRAIL, a company developing methods for early detection of cancer. S.J. and B.L.E. are inventors on patent applications filed by Harvard University Medical School that are related to risk assessment of cardiovascular disease and therapeutic interventions in individuals with clonal hematopoiesis.
References
- 1.Blokzijl FJ et al. ,Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoang ML et al. , Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc Natl Acad Sci U S A 113, 9846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welch JS et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278, doi: 10.1016/j.cell.2012.06.023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martincorena I & Campbell PJ Somatic mutation in cancer and normal cells. Science 349, 1483–1489, doi: 10.1126/science.aab4082 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Yizhak K et al. , RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, doi: 10.1126/science.aaw0726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan BK & Miller JH Mutagenic deamination of cytosine residues in DNA. Nature 287, 560–561 (1980). [DOI] [PubMed] [Google Scholar]
- 7.Alexandrov LB et al. Signatures of mutational processes in human cancer. Nature 500, 415–421, doi: 10.1038/nature12477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodgers K & McVey M Error-Prone Repair of DNA Double-Strand Breaks. J Cell Physiol 231, 15–24, doi: 10.1002/jcp.25053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerman I, Seita J, Inlay MA, Weissman IL & Rossi DJ Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50, doi: 10.1016/j.stem.2014.04.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel TA DNA replication fidelity. J Biol Chem 279, 16895–16898, doi: 10.1074/jbc.R400006200 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Tomasetti C & Vogelstein B Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81, doi: 10.1126/science.1260825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurie CC et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nature genetics 44, 642–650, doi: 10.1038/ng.2271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Six H et al. Population dynamics of normal human blood inferred from somatic mutations. Nature, doi: 10.1038/s41586-018-0497-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martincorena I et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886, doi: 10.1126/science.aaa6806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917, doi: 10.1126/science.aau3879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama A et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317, doi: 10.1038/s41586-018-0811-x (2019). [DOI] [PubMed] [Google Scholar]
- 17.Rowley JD Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973). [DOI] [PubMed] [Google Scholar]
- 18.Fey MF et al. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood 83, 931–938 (1994). [PubMed] [Google Scholar]
- 19.Champion KM, Gilbert JG, Asimakopoulos FA, Hinshelwood S & Green AR Clonal haemopoiesis in normal elderly women: implications for the myeloproliferative disorders and myelodysplastic syndromes. British journal of haematology 97, 920–926 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Busque L et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature genetics 44, 1179–1181, doi: 10.1038/ng.2413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reya T, Morrison SJ, Clarke MF & Weissman IL Stem cells, cancer, and cancer stem cells. Nature 414, 105–111, doi: 10.1038/35102167 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto T, Weissman IL & Akashi K AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A 97, 7521–7526 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jan M et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Science translational medicine 4, 149ra118, doi: 10.1126/scitranslmed.3004315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlush LI et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333, doi: 10.1038/nature13038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC & Majeti R Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A 111, 2548–2553, doi: 10.1073/pnas.1324297111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie M et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20, 1472–1478, doi: 10.1038/nm.3733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genovese G et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371, 2477–2487, doi: 10.1056/NEJMoa1409405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371, 2488–2498, doi: 10.1056/NEJMoa1408617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biernaux C, Loos M, Sels A, Huez G & Stryckmans P Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood 86, 3118–3122 (1995). [PubMed] [Google Scholar]
- 30.Liu Y, Hernandez AM, Shibata D & Cortopassi GA BCL2 translocation frequency rises with age in humans. Proc Natl Acad Sci U S A 91, 8910–8914 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKerrell T et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep 10, 1239–1245, doi: 10.1016/j.celrep.2015.02.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Acuna-Hidalgo R et al. Ultra-sensitive Sequencing Identifies High Prevalence of Clonal Hematopoiesis-Associated Mutations throughout Adult Life. Am J Hum Genet 101, 50–64, doi: 10.1016/j.ajhg.2017.05.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young AL, Challen GA, Birmann BM & Druley TE Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 7, 12484, doi: 10.1038/ncomms12484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonnefond A et al. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nature genetics 45, 1040–1043, doi: 10.1038/ng.2700 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Jacobs KB et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nature genetics 44, 651–658, doi: 10.1038/ng.2270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loh PR et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 559, 350–355, doi: 10.1038/s41586-018-0321-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zink F et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752, doi: 10.1182/blood-2017-02-769869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silver AJ & Jaiswal S Clonal hematopoiesis: Pre-cancer PLUS. Adv Cancer Res 141, 85–128, doi: 10.1016/bs.acr.2018.12.003 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Steensma DP et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16, doi: 10.1182/blood-2015-03-631747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperling AS, Gibson CJ & Ebert BL The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer 17, 5–19, doi: 10.1038/nrc.2016.112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowman RL, Busque L & Levine RL Clonal Hematopoiesis and Evolution to Hematopoietic Malignancies. Cell Stem Cell 22, 157–170, doi: 10.1016/j.stem.2018.01.011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bondar T & Medzhitov R p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 6, 309–322, doi: 10.1016/j.stem.2010.03.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahn JD et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 132, 1095–1105, doi: 10.1182/blood-2018-05-850339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu JI et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 23, 700–713 e706, doi: 10.1016/j.stem.2018.10.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kralovics R et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352, 1779–1790, doi: 10.1056/NEJMoa051113 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Okano M, Xie S & Li E Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature genetics 19, 219–220, doi: 10.1038/890 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Schubeler D Function and information content of DNA methylation. Nature 517, 321–326, doi: 10.1038/nature14192 (2015). [DOI] [PubMed] [Google Scholar]
- 48.He YF et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307, doi: 10.1126/science.1210944 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito S et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303, doi: 10.1126/science.1210597 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ko M et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A 108, 14566–14571, doi: 10.1073/pnas.1112317108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Challen GA et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics 44, 23–31, doi: 10.1038/ng.1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guryanova OA et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med 22, 1488–1495, doi: 10.1038/nm.4210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cole CB et al. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J Clin Invest 127, 3657–3674, doi: 10.1172/JCI93041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moran-Crusio K et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell 20, 11–24, doi: 10.1016/j.ccr.2011.06.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celik H et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood 125, 619–628, doi: 10.1182/blood-2014-08-594564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obeng EA et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer cell 30, 404–417, doi: 10.1016/j.ccell.2016.08.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim E et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer cell 27, 617–630, doi: 10.1016/j.ccell.2015.04.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shirai CL et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer cell 27, 631–643, doi: 10.1016/j.ccell.2015.04.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshizato T et al. Somatic Mutations and Clonal Hematopoiesis in Aplastic Anemia. N Engl J Med 373, 35–47, doi: 10.1056/NEJMoa1414799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brodsky RA Paroxysmal nocturnal hemoglobinuria. Blood 124, 2804–2811, doi: 10.1182/blood-2014-02-522128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hinds DA et al. Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood 128, 1121–1128, doi: 10.1182/blood-2015-06-652941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meisel M et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557, 580–584, doi: 10.1038/s41586-018-0125-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buscarlet M et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130, 753–762, doi: 10.1182/blood-2017-04-777029 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Research N Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368, 2059–2074, doi: 10.1056/NEJMoa1301689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bejar R et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364, 2496–2506, doi: 10.1056/NEJMoa1013343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papaemmanuil E et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627; quiz 3699, doi: 10.1182/blood-2013-08-518886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindsley RC et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N Engl J Med 376, 536–547, doi: 10.1056/NEJMoa1611604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nangalia J et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 369, 2391–2405, doi: 10.1056/NEJMoa1312542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Couronne L, Bastard C & Bernard OA TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med 366, 95–96, doi: 10.1056/NEJMc1111708 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Reddy A et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171, 481–494 e415, doi: 10.1016/j.cell.2017.09.027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abelson S et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404, doi: 10.1038/s41586-018-0317-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desai P et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 24, 1015–1023, doi: 10.1038/s41591-018-0081-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibson CJ et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J Clin Oncol 35, 1598–1605, doi: 10.1200/JCO.2016.71.6712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gillis NK et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol 18, 112–121, doi: 10.1016/S1470-2045(16)30627-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takahashi K et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol 18, 100–111, doi: 10.1016/S1470-2045(16)30626-X (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wong TN et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 518, 552–555, doi: 10.1038/nature13968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coombs CC et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 21, 374–382 e374, doi: 10.1016/j.stem.2017.07.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright DE, Wagers AJ, Gulati AP, Johnson FL & Weissman IL Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933–1936, doi: 10.1126/science.1064081 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Jaiswal S et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med 377, 111–121, doi: 10.1056/NEJMoa1701719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brunner AM et al. Risk and timing of cardiovascular death among patients with myelodysplastic syndromes. Blood Adv 1, 2032–2040, doi: 10.1182/bloodadvances.2017010165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dorsheimer L et al. Association of Mutations Contributing to Clonal Hematopoiesis With Prognosis in Chronic Ischemic Heart Failure. JAMA Cardiol 4, 25–33, doi: 10.1001/jamacardio.2018.3965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolach O et al. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Science translational medicine 10, doi: 10.1126/scitranslmed.aan8292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patel KV, Ferrucci L, Ershler WB, Longo DL & Guralnik JM Red blood cell distribution width and the risk of death in middle-aged and older adults. Archives of internal medicine 169, 515–523, doi: 10.1001/archinternmed.2009.11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Horvath S & Raj K DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 19, 371–384, doi: 10.1038/s41576-018-0004-3 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Haitjema S et al. Loss of Y Chromosome in Blood Is Associated With Major Cardiovascular Events During Follow-Up in Men After Carotid Endarterectomy. Circ Cardiovasc Genet 10, e001544, doi: 10.1161/CIRCGENETICS.116.001544 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Fuster JJ et al. Clonal hematopoiesis associated with Tet2 deficiency accelerates atherosclerosis development in mice. Science, doi: 10.1126/science.aag1381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sano S et al. CRISPR-Mediated Gene Editing to Assess the Roles of Tet2 and Dnmt3a in Clonal Hematopoiesis and Cardiovascular Disease. Circ Res 123, 335–341, doi: 10.1161/CIRCRESAHA.118.313225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sano S et al. Tet2-Mediated Clonal Hematopoiesis Accelerates Heart Failure Through a Mechanism Involving the IL-1beta/NLRP3 Inflammasome. J Am Coll Cardiol 71, 875–886, doi: 10.1016/j.jacc.2017.12.037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang W et al. Macrophage Inflammation, Erythrophagocytosis, and Accelerated Atherosclerosis in Jak2 (V617F) Mice. Circ Res 123, e35–e47, doi: 10.1161/CIRCRESAHA.118.313283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu D et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature 559, 637–641, doi: 10.1038/s41586-018-0350-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Q et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525, 389–393, doi: 10.1038/nature15252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cull AH, Snetsinger B, Buckstein R, Wells RA & Rauh MJ Tet2 restrains inflammatory gene expression in macrophages. Exp Hematol 55, 56–70 e13, doi: 10.1016/j.exphem.2017.08.001 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Natarajan P, Jaiswal S & Kathiresan S Clonal Hematopoiesis: Somatic Mutations in Blood Cells and Atherosclerosis. Circ Genom Precis Med 11, e001926, doi: 10.1161/CIRCGEN.118.001926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leoni C et al. Dnmt3a restrains mast cell inflammatory responses. Proc Natl Acad Sci U S A 114, E1490–E1499, doi: 10.1073/pnas.1616420114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frick M et al. Role of Donor Clonal Hematopoiesis in Allogeneic Hematopoietic Stem-Cell Transplantation. J Clin Oncol, JCO2018792184, doi: 10.1200/JCO.2018.79.2184 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Fraietta JA et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312, doi: 10.1038/s41586-018-0178-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Franceschi C & Campisi J Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci 69 Suppl 1, S4–9, doi: 10.1093/gerona/glu057 (2014). [DOI] [PubMed] [Google Scholar]