Vessoni et al. demonstrate telomere shortening leads to a unique DDR response in human pluripotent stem cells. Unlike terminally differentiated cells, telomere shortening induces formation of single-stranded DNA telomere overhangs in hPSCs, which activate ATR signaling and lead to mitotic catastrophe and p53-dependent cell death.
Abstract
It is well established that short telomeres activate an ATM-driven DNA damage response that leads to senescence in terminally differentiated cells. However, technical limitations have hampered our understanding of how telomere shortening is signaled in human stem cells. Here, we show that telomere attrition induces ssDNA accumulation (G-strand) at telomeres in human pluripotent stem cells (hPSCs), but not in their differentiated progeny. This led to a unique role for ATR in the response of hPSCs to telomere shortening that culminated in an extended S/G2 cell cycle phase and a longer period of mitosis, which was associated with aneuploidy and mitotic catastrophe. Loss of p53 increased resistance to death, at the expense of increased mitotic abnormalities in hPSCs. Taken together, our data reveal an unexpected dominant role of ATR in hPSCs, combined with unique cell cycle abnormalities and, ultimately, consequences distinct from those observed in their isogenic differentiated counterparts.
Introduction
Mammalian telomeres are composed of TTAGGG repeats bound by shelterin, a protein complex that protects chromosome ends and safeguards genome integrity (Palm and de Lange, 2008; Shay and Wright, 2019). Telomeres shorten due to both the “end-replication problem” and postreplicative end-processing (Shay and Wright, 2019). While adult stem cells express telomerase, a large ribonucleoprotein complex capable of lengthening telomeres, this expression is not sufficient to prevent telomere shortening throughout organismal life (Shay and Wright, 2019). The deleterious consequences of progressive telomere shortening are exemplified in patients harboring mutations in telomere-biology genes, whose symptoms range from skin and liver abnormalities to lung fibrosis, bone marrow failure, and high cancer rates (Armanios and Blackburn, 2012; Bertuch, 2016; Savage, 2018). Many of these symptoms are linked to stem cell dysfunction causing loss of homeostasis in highly proliferative tissues (Armanios et al., 2009; Hao et al., 2005; Lee et al., 1998).
In differentiated cells such as fibroblasts, telomere dysfunction triggers an ataxia telangiectasia mutated (ATM)–dependent DNA damage response (DDR) and the activation of the p53 and pRb pathways. These pathways converge to induce senescence, halting proliferation of genetically unstable cells (d’Adda di Fagagna et al., 2003; Guo et al., 2007; Shay, 2016). However, it remains unclear if telomere shortening in human stem cells activates similar molecular pathways or leads to similar phenotypes, hampering our ability to design better treatment strategies for telomere syndrome patients.
A significant hurdle has been a lack of suitable models to study signaling events activated by short telomeres in human stem cells. Here, we created human pluripotent stem cells (hPSCs) in which expression of TERT, the reverse transcriptase component of telomerase (Shay and Wright, 2019), can be conditionally regulated. With this system, we show that telomere attrition induces a unique DDR in hPSCs. Telomere shortening triggered accumulation of single-strand DNA (ssDNA; G-strand) at telomeres in hPSCs, leading to a dominant role of ataxia telangiectasia and Rad3-related (ATR), in contrast to the predominant role played by ATM in terminally differentiated cells with short telomeres (Herbig et al., 2004). In contrast with differentiated cells, telomere shortening led to an extended S/G2 phase and longer mitosis in hPSCs, which was associated with aneuploidy and mitotic catastrophe, triggering p53-dependent cell death. Our work reveals an unexpected dominant role of ATR in hPSCs with short telomeres, combined with unique cell cycle abnormalities and distinct consequences compared with their isogenic differentiated progeny.
Results and discussion
HPSCs and fibroblasts adopt different fates in response to telomere attrition
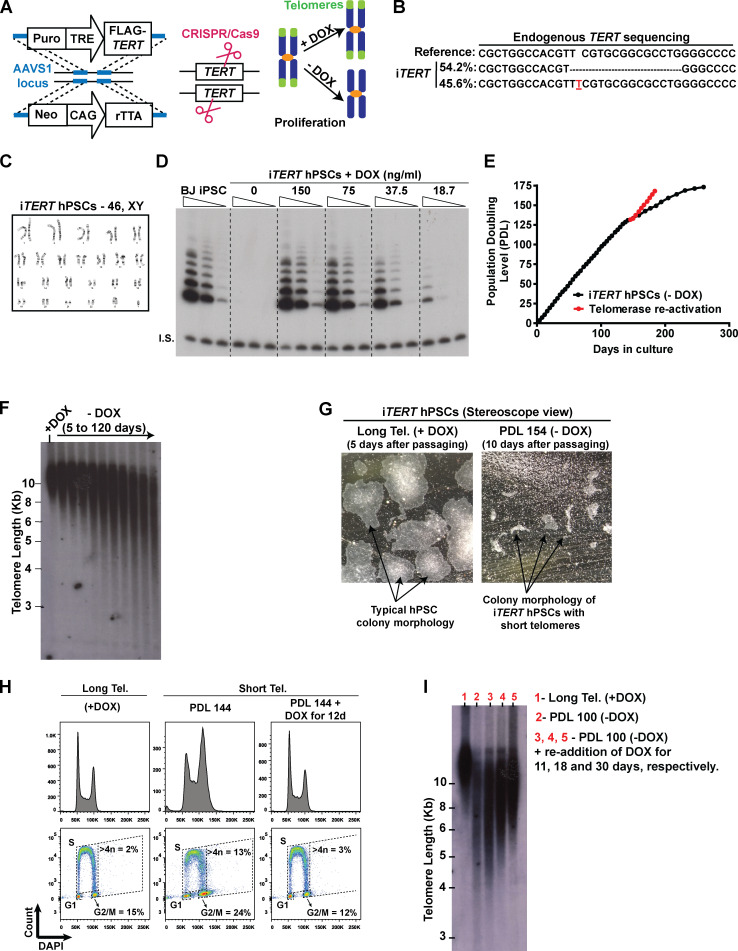
To study the effects of telomere shortening in human stem cells and their differentiated progeny, we engineered hPSCs in which telomerase activity could be conditionally regulated. We introduced a TRE-TIGHT-FLAG-TERT construct into the AAVS1 safe harbor locus followed by knockout (KO) of the endogenous TERT locus in hPSCs (Fig. S1, A and B). The resulting stem cell line (iTERT [inducible-TERT] hPSCs) retained a normal karyotype (Fig. S1 C) and activates telomerase in response to doxycycline (DOX; Fig. S1 D).
Figure S1.
Establishing a novel model to study telomere (Tel.) dysfunction in hPSCs. (A) Engineering iTERT hPSCs. First, TRE-Tight–regulated FLAG-TERT cDNA and CAG-driven rTTA sequences were inserted into the AAVS1 safe harbor locus of BJ-iPSCs using zinc fingers. After clonal selection, the endogenous TERT gene was knocked out using CRISPR/Cas9. Telomerase activity in the resulting cell (iTERT hPSC) is regulated by DOX. (B) NGS confirming the deletion of the TERT endogenous locus. (C) Karyotype analysis of iTERT hPSCs. (D) TRAP analysis of iTERT hPSCs treated with different concentrations of DOX. Unedited BJ-iPSCs were used as positive control. I.S., internal standard. (E) PDLs of iTERT hPSCs grown in the absence of DOX (black curve) or after telomerase reactivation (red curve). (F) Telomere length analysis by TRF in iTERT hPSCs kept with or without DOX for the indicated time. (G) Representative images of iTERT hPSCs colonies grown in the presence of DOX (long telomeres) or at PDL 154 after DOX withdrawal (short telomeres). (H) Readdition of DOX for 12 d fully rescued the cell cycle profile of iTERT hPSCs as detected by EdU and DAPI staining (flow cytometry). (I) Telomere length analysis by TRF in iTERT hPSCs grown continuously with DOX (long telomeres, lane 1), with short telomeres (PDL 100, lane 2), and after readdition of DOX for 11, 18, or 30 d (lanes 3, 4, and 5, respectively) to the culture media of the cells with short telomeres.
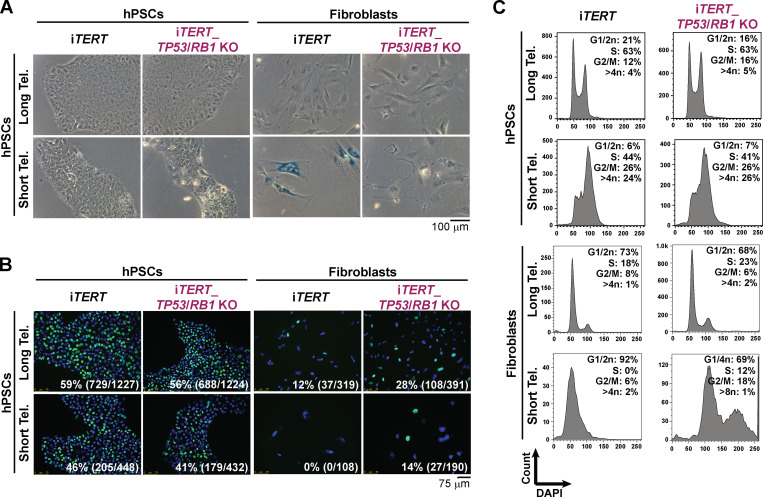
iTERT hPSCs grew normally in the absence of DOX (telomerase off) for ∼140 d (population doubling level [PDL] ∼130) without noticeable changes in proliferation (Fig. S1 E), despite progressive telomere shortening (Fig. S1 F). After PDL ∼140, iTERT hPSCs progressively slowed rates of population doubling, with impaired colony formation and cell detachment (Fig. S1, E and G). At PDL ∼159, iTERT hPSC culture was severely impaired (Fig. S1 E). Surprisingly, senescence was not detected at this stage, as iTERT hPSCs with short telomeres did not display β-galactosidase (β-gal) staining (Fig. 1 A) and continued to replicate DNA (Fig. 1 B). This is in strong contrast to the fate of isogenic fibroblasts differentiated from iTERT hPSCs, which showed strong β-gal staining and a complete block in DNA replication after growing in the absence of TERT (Fig. 1, A and B), in agreement with previous studies in human primary fibroblasts (d’Adda di Fagagna et al., 2003; Herbig et al., 2004; Jacobs and de Lange, 2004; Vaziri and Benchimol, 1996).
Figure 1.
hPSCs and fibroblasts adopt different fates in response to telomere (Tel.) attrition. (A) β-gal activity assay. (B and C) EdU incorporation (percentage and absolute number of EdU+ cells is indicated in each image; B) and cell cycle analysis (C) performed in iTERT and iTERT_TP53/RB1 KO hPSCs and their isogenic fibroblasts with long and short telomeres. PDL for cells with short telomeres: 149–159 (iTERT hPSCs); 205–215 (iTERT_TP53/RB1 KO hPSCs); 45–47 (iTERT fibroblasts); and 95–97 (iTERT_TP53/RB1 KO fibroblasts).
In addition, iTERT hPSCs with short telomeres displayed G2/M accumulation and aneuploidy (>4n DNA; Figs. 1 C and S1 H), in contrast to their isogenic fibroblasts, which showed a strong G1/S arrest (Fig. 1 C). Reactivation of telomerase in iTERT hPSCs with short telomeres reelongated telomeres and rescued proliferation and cell cycle profile (Fig. S1, E, H, and I), confirming that these phenotypes are due to telomere attrition. Combined, these results show that isogenic hPSCs and fibroblasts respond differently to telomere dysfunction. As other forms of DNA damage also fail to induce a robust G1/S arrest in hPSCs (Jaiswal et al., 2020; Momcilović et al., 2009), and the enforced arrest of the cell cycle in G1 in these cells leads to differentiation (Maimets et al., 2008; Neganova et al., 2009), it is tempting to speculate that hPSCs are physiologically unable to undergo senescence due to their unique cell cycle characteristics (Padgett and Santos, 2020).
p53 and pRb are not involved in checkpoint activation in hPSCs with short telomeres
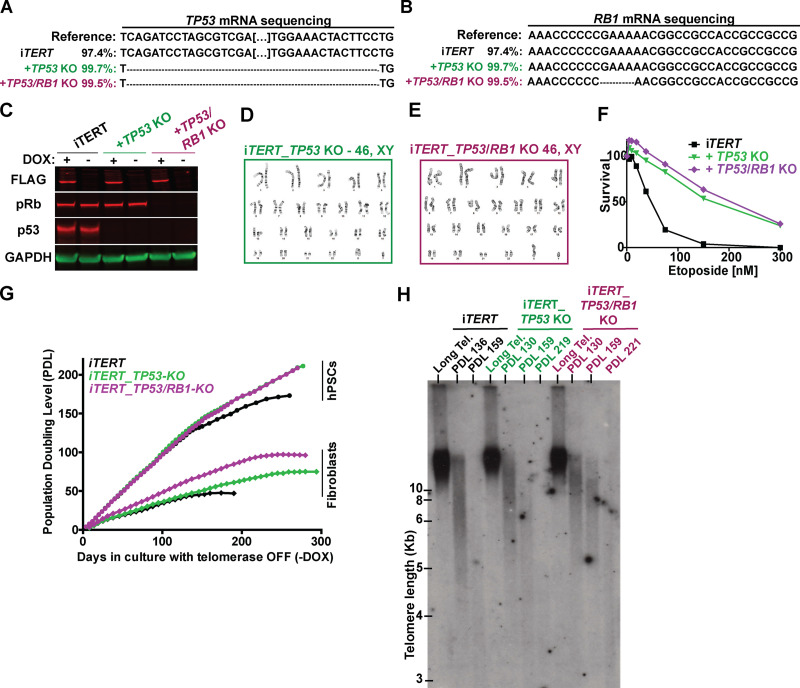
To gain molecular insights into the response of hPSCs to telomere shortening, we ablated TP53 or TP53 and Rb1, two major regulators of DDR in human cells (Hara et al., 1991; Shay et al., 1991; Smogorzewska and de Lange, 2002), in iTERT hPSCs (Fig. S2, A–C). The resulting cell lines (iTERT_TP53 KO and iTERT_TP53/Rb1 KO) have normal karyotypes and show increased resistance to etoposide treatment (Fig. S2, D–F).
Figure S2.
Generation of isogenic iTERT hPSCs knocked out for TP53 (iTERT_TP53 KO) or both TP53 and RB1 (iTERT_TP53/RB1 KO). (A) DNA sequencing analysis confirming ablation of TP53 in the indicated cell lines. (B) DNA sequencing data analysis confirming ablation of Rb1 in the indicated cell lines. (C) Western blotting analysis of FLAG-TERT, p53, and pRb in iTERT hPSCs and their TP53 KO and TP53/Rb1 KO derivatives. GAPDH is shown as loading control. (D and E) Karyotype analysis of iTERT_TP53 KO and iTERT_TP53/Rb1 KO hPSCs, respectively. (F) Survival analysis of iTERT hPSCs, TP53 KO, and TP53/Rb1 KO after treatment with different concentrations of etoposide. Analysis performed by Sapphire700/DRAQ5 staining. (G) PDLs of iTERT hPSCs and fibroblasts, and their TP53 and TP53/RB1 KO counterparts, grown in the absence of DOX. (H) TRF analysis in iTERT, iTERT_TP53 KO, and iTERT_TP53/RB1 KO hPSCs grown in the absence of DOX for the indicated PDLs. Tel., telomere.
TP53 ablation increased the proliferative capacity of iTERT hPSCs and fibroblasts growing without telomerase (Fig. S2 G). The combined ablation of TP53 and Rb1 allowed fibroblasts to bypass replicative senescence, as revealed by lack of β-gal staining and failure to induce G1/S arrest (Fig. 1, A–C; and Fig. S2 G), in agreement with previous literature (Davoli and de Lange, 2012; Girardi et al., 1965; Hayashi et al., 2015). Interestingly, ablation of Rb1 had no additional effect in p53-null hPSCs (Fig. S2, G and H), and the combined ablation of TP53 and Rb1 did not rescue G2/M accumulation induced by telomere shortening in these cells (Fig. 1 C), indicating that p53 and pRb do not control checkpoint activation in hPSCs with short telomeres, in strong contrast to their isogenic fibroblasts.
Telomere attrition prolongs S/G2 and mitosis in hPSCs, leading to p53-mediated death
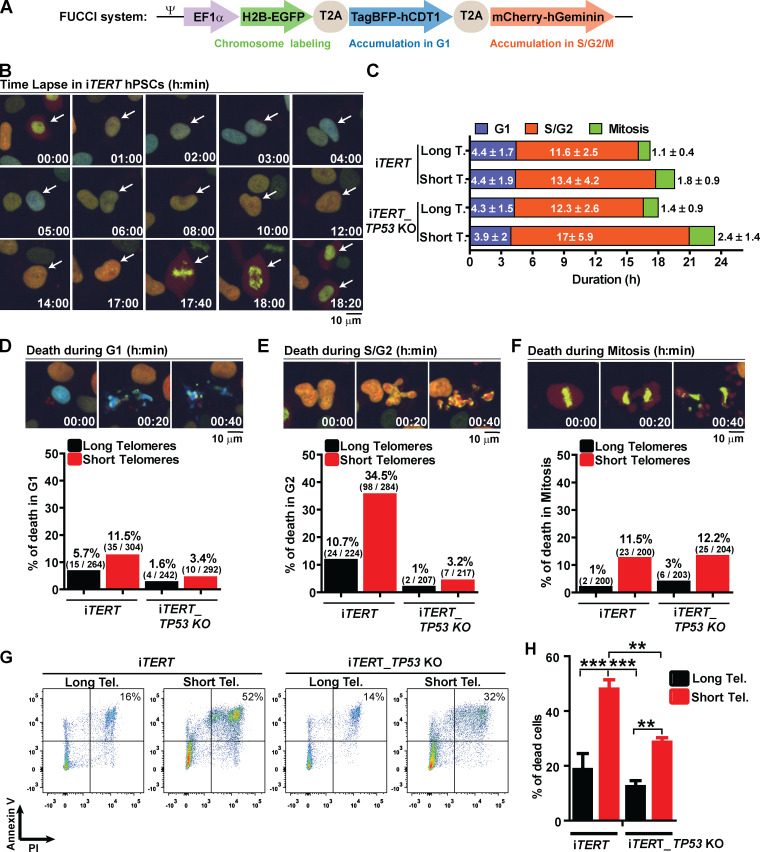
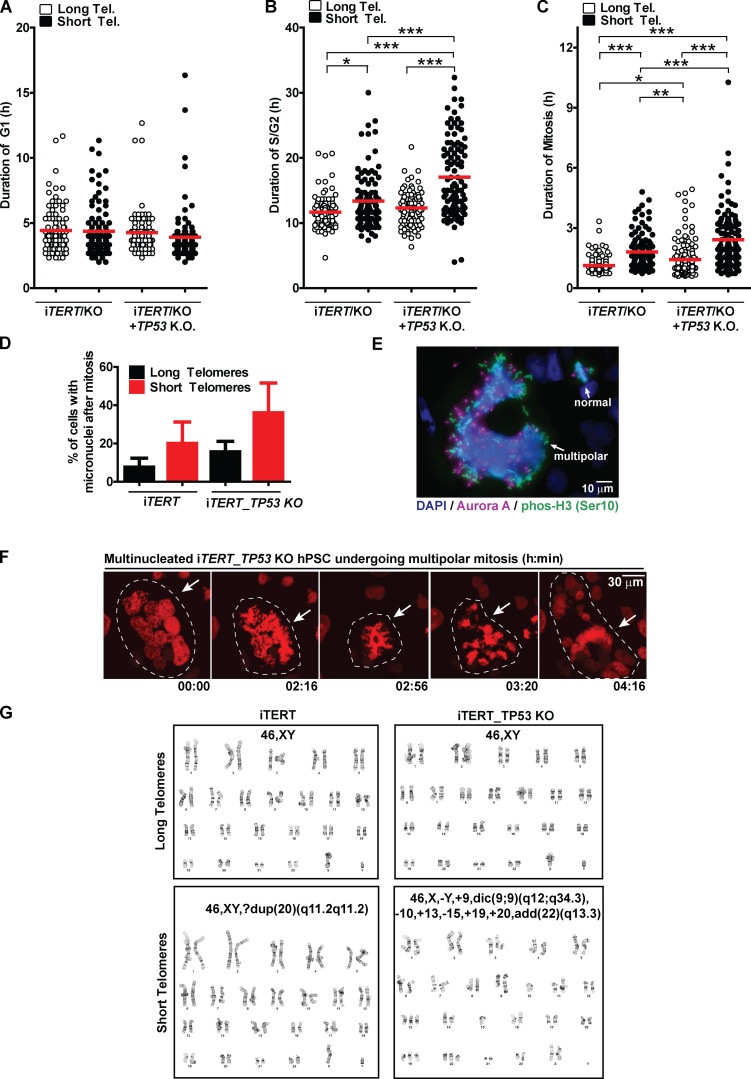
These results prompted us to better understand the fate of hPSCs with short telomeres. We used a Fluorescent Ubiquitination-based Cell Cycle Indicator (FUCCI) adapted from Sato et al. (2019) to track cell cycle dynamics and the fate of individual cells during live imaging (Fig. 2, A and B). Telomere erosion did not alter G1 duration in iTERT hPSCs (PDL ∼159; long telomere = 4.4 ± 1.7 h; short telomere = 4.4 ± 1.9 h; Figs. 2 C and S3 A). Instead, it significantly increased the average length of S/G2 (long telomere = 11.6 ± 2.5 h; short telomere = 13.4 ± 4.2 h; Figs. 2 C and S3 B) and mitosis (long telomere = 1.1 ± 0.4 h; short telomere = 1.8 ± 0.9 h; Figs. 2 C and S3 C) in iTERT hPSCs. Our FUCCI analysis also showed that longer S/G2 and mitosis were associated with an increase in hPSC death, specifically during these phases of the cell cycle (Fig. 2, D–F), which was confirmed by Annexin V and propidium iodide (PI) staining (Fig. 2, G and H).
Figure 2.
Telomere attrition prolongs S/G2 and mitosis duration in hPSCs, leading to p53-mediated cell death. (A) FUCCI system: Cells express EGFP-H2B (green fluorescence to track mitotic chromosomes), TagBFP (blue fluorescence in G1), and mCherry (red fluorescence in S/G2/M). (B) Representative time-lapse image of FUCCI live imaging. (C) Average duration (h) ± SEM of 100 events analyzed per phase (G1, S/G2, or M) per cell. (D–F) Illustrative images and percentage cell death during G1, S/G2, or mitosis, respectively. At least 200 events were analyzed per cell cycle phase per cell and scored as live (able to transit to the next cell cycle phase) or dead (died within that specific cell cycle phase). (G and H) Cell death analysis by Annexin V and PI staining. Average ± SD of three independent experiments is shown in H. Tel., telomere. Statistical analysis: one-way ANOVA followed by Bonferroni’s test. **, P < 0.01; ***, P < 0.001. PDLs for cells with short telomeres: 159 (iTERT hPSCs); 209 (iTERT_TP53 KO hPSCs).
Figure S3.
Cell cycle and mitotic abnormalities induced by telomere (Tel.) shortening in iTERT and iTERT_TP53 KO hPSCs. (A–C) Duration of G1, S/G2, and mitosis, respectively, analyzed during live imaging using FUCCI. Each dot represents an individual cell. Average ± SEM. (D) Percentage of cells displaying at least one micronucleus after mitosis is concluded. Graph shows average ± SD of three independent experiments. (E) Representative image of mitotic iTERT_TP53 KO hPSC with short telomeres with multiple centrosomes; a normal mitosis (two centrosomes) is also visible in the same image (top right). Mitotic chromosomes are positive for pHistone H3 (Ser10; green), centrosomes are positive for Aurora A (magenta), and DNA is stained with DAPI (blue). (F) Illustrative time-lapse image of a multinucleated iTERT_TP53 KO hPSC with short telomeres undergoing mitosis; DNA is visualized via expression of an H2B-mApple fusion protein. (G) Illustrative images of G-band karyotyping analysis in metaphase spreads of iTERT and iTERT_TP53 KO hPSCs, with long and short telomeres. Statistics: one-way ANOVA followed by Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P > 0.001. PDL for cells with short telomeres: 159 (iTERT hPSCs); 209 (iTERT_TP53 KO hPSCs).
To determine the consequences of the extended lifespan conferred by p53 loss in hPSCs with short telomeres, we tracked the behavior of these cells using FUCCI. p53 ablation significantly increased both S/G2 (long telomere = 12.3 ± 2.6 h; short telomere = 17 ± 5.9 h) and mitosis (long telomere = 1.4 ± 0.9 h; short telomere = 2.4 ±1.4 h) length in hPSCs with short telomeres (Fig. 2 C; and Fig. S3, B and C). Concomitantly, p53 ablation significantly protected hPSCs from death during S/G2 (long telomeres = 1%; short telomeres = 3.2%; Fig. 2 E), indicating that p53 eliminates hPSCs with short telomeres during S/G2. This increased protection against death due to p53 loss was confirmed by flow cytometry (Fig. 2, G and H). Our results establish that while the main role of p53 in fibroblasts with short telomeres is to induce an irreversible arrest at G1/S, in hPSCs p53 is rewired to regulate cell death during S/G2.
Loss of p53 exacerbates mitotic abnormalities induced by telomere attrition in hPSCs
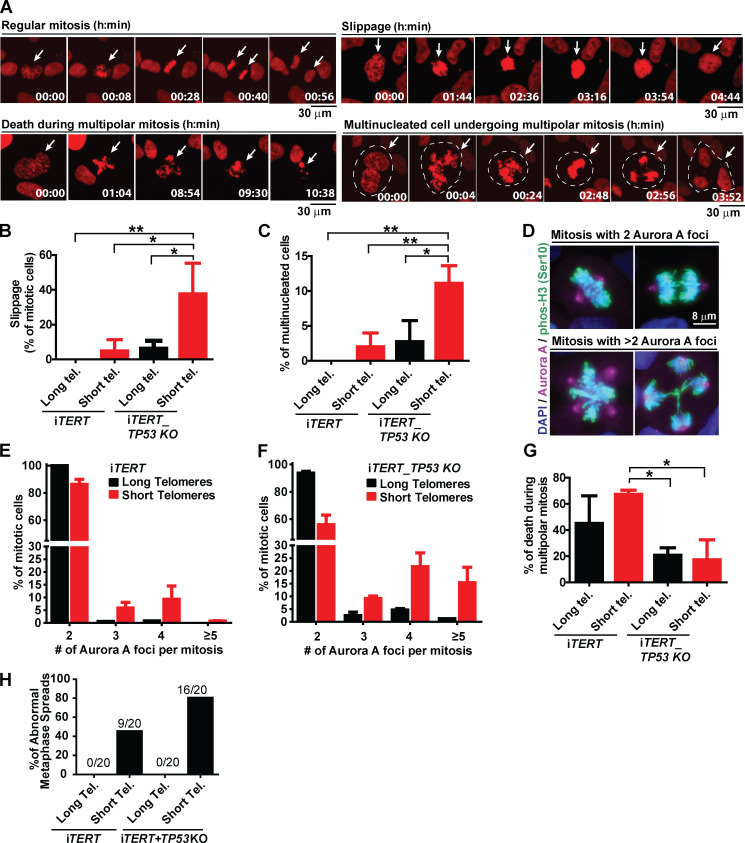
We next wanted to determine the consequences of telomere shortening specifically during mitosis. We performed live imaging using an H2B-mApple construct to track chromosomes during mitosis and observed the appearance of various mitotic abnormalities in iTERT hPSCs with short telomeres, including a significant increase in slippage (mitotic exit without proper chromosome segregation) and multinucleated cells with three to four centrosomes (Fig. 3, A–C). We also detected a trend of increased numbers of cells finishing mitosis with micronucleus (Fig. S3 D). These results were corroborated by immunofluorescence experiments where we detected a significant increase in the percentage of multipolar mitosis in iTERT hPSCs with short telomeres (long telomere = 0.5%; short telomere = 14%; Fig. 3, D and E). Most of these multipolar mitoses had three to four centrosomes (Fig. 3, D and E).
Figure 3.
Loss of p53 exacerbates mitotic abnormalities induced by telomere (Tel.) attrition in hPSCs. (A) Illustrative time-lapse images of a regular mitosis, slippage, death during a multipolar mitosis, and a multinucleated cell undergoing multipolar mitosis in iTERT hPSCs expressing H2B-mApple. (B) Percentage of mitotic cells undergoing slippage. (C) Percentage of multinucleated mitotic cells. (D) Immunofluorescence illustrative images of normal and multipolar mitosis. Mitotic chromosomes are stained in green (pHistone H3Ser10), centrosomes in magenta (Aurora A), and DNA in blue (DAPI). (E and F) Distribution of the number of Aurora A foci per mitotic cell (average ± SD) in iTERT hPSCs and iTERT_TP53 KO hPSCs, respectively, with long or short telomeres. (G) Percentage of death during multipolar mitosis. (H) Percentage of abnormal karyotypes in iTERT and iTERT_TP53KO hPSCs with long and short telomeres. PDLs for cells with short telomeres: 149–159 (iTERT hPSCs); 205–211 (iTERT_TP53 KO hPSCs). Statistical analysis: one-way ANOVA followed by Bonferroni’s test. *, P < 0.05; **, P < 0.01. n = 3 independent experiments.
The ablation of p53 significantly increased the frequency of slippage and multinucleation in hPSCs with short telomeres (Fig. 3, B and C). It also increased the occurrence and severity of multipolar mitosis: >40% of mitoses in p53-null hPSCs with short telomeres were multipolar (Fig. 3 F), with ∼15% of them displaying five or more centrosomes (up to 30 centrosomes and 10 nuclei per cell were also detected; Fig. S3, E and F).
Although iTERT and iTERT_TP53 KO hPSCs with short telomeres displayed similar mitotic death rates (11.5% and 12.2%, respectively; Fig. 2 F), p53-null cells showed significantly more mitotic instability (Fig. 3, B, C, E, and F). Accordingly, G-band analysis confirmed that telomere shortening led to karyotype abnormalities in iTERT hPSCs (long telomere = 0%; short telomere = 45% of abnormal karyotypes), which was significantly potentiated by p53 loss (long telomere = 0%; short telomere = 80%; Figs. 3 H and S3 G; and Table S1). Combined, these data indicate that loss of p53 increases the formation of chromosomal abnormalities and resistance to death during aberrant mitosis in hPSCs with short telomeres. Accordingly, we observed that ∼60% of the iTERT hPSCs die during multipolar mitosis, while in iTERT_TP53 KO hPSCs, this value is significantly lower (∼20%; Fig. 3 G).
ATR is the main driver of dysfunction induced by telomere attrition in hPSCs
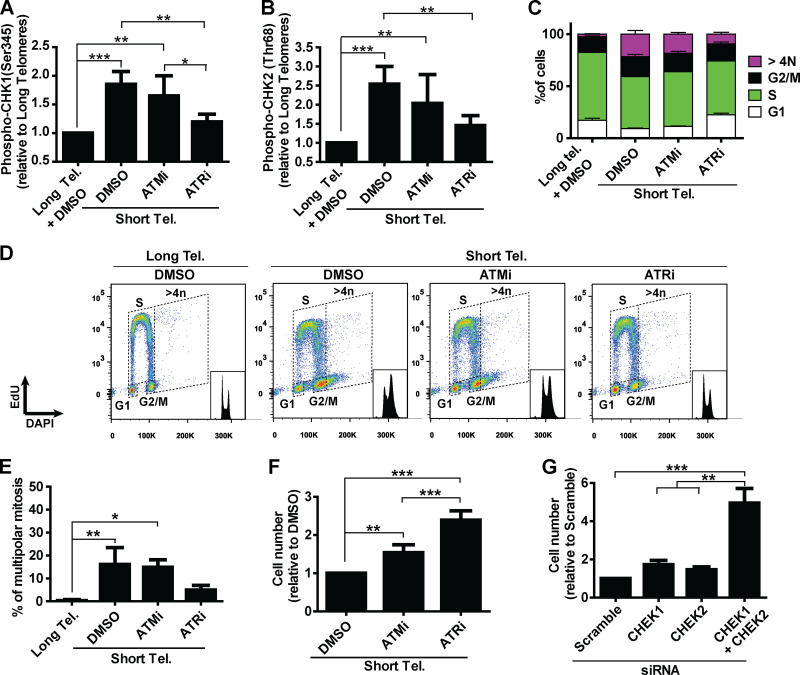
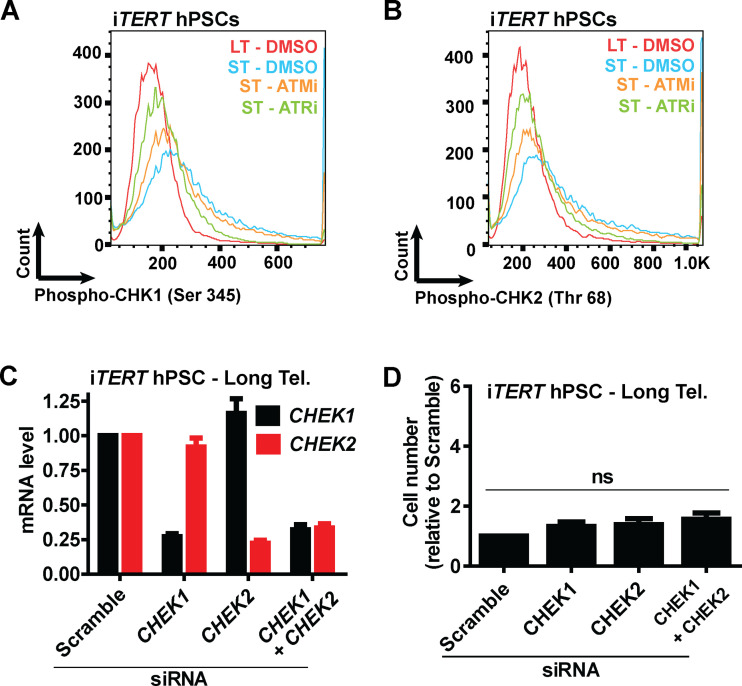
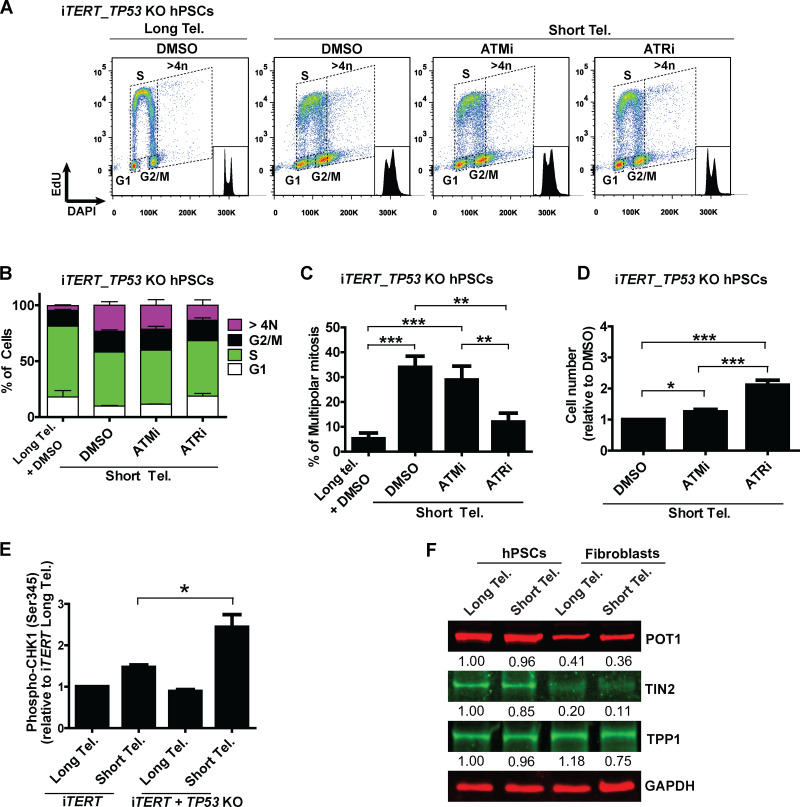
Next, we asked what connects telomere dysfunction to cell cycle/mitotic abnormalities in hPSCs. We focused on ATM and ATR, known regulators of DDR that act upstream of p53 in the response of fibroblasts to telomere dysfunction (Abraham, 2001; Davoli et al., 2010). We confirmed activation of DDR in hPSCs with short telomeres, with increased phosphorylation of CHK1 (Ser345) and CHK2 (Thr68) by flow cytometry (Fig. 4, A and B; and Fig. S4, A and B). We then tested the impact of the ATM inhibitor (ATMi) KU-55933 (Hickson et al., 2004) and the ATR inhibitor (ATRi) VE-821 (Prevo et al., 2012) on G2/M accumulation, aneuploidy (>4n), multipolar mitosis, and cell proliferation in hPSCs with short telomeres (Fig. 4, C–F). The inhibition of ATR induced a more significant rescue of these phenotypes than ATMi in hPSCs with short telomeres (Fig. 4, C–F). This result was unexpected, as ATM is generally considered the dominant pathway in the response of human cells to telomere attrition (Herbig et al., 2004). Also, ATRi was able to reduce CHK1 and CHK2 phosphorylation to a greater extent than ATMi in hPSCs with short telomeres (Fig. 4, A and B; and Fig. S4, A and B). To investigate the involvement of CHK1 and CHK2 in more detail, we depleted CHEK1 and/or CHEK2 (Fig. S4 C). Codepletion of CHEK1 and CHEK2 significantly increased proliferation of iTERT hPSCs with short telomeres (Fig. 4 G), but not of iTERT hPSCs with long telomeres (Fig. S4 D). Our results show that CHK1 and CHK2 are both relevant in the response of hPSCs to telomere dysfunction and demonstrate a major role of ATR in the DDR of these cells.
Figure 4.
A determinant role for ATR after telomere (Tel.) attrition in hPSCs. (A and B) Analysis of phosphoCHK1 (Ser345) and phosphoCHK2 (Thr68), respectively, by flow cytometry in iTERT hPSCs with short telomeres in the presence of ATMi (10 µm) or ATRi (1 µm). Average ± SD of five independent experiments. (C and D) Cell cycle distribution in iTERT hPSCs with short telomeres treated with ATMi or ATRi. Average ± SD of three independent experiments are shown in C and representative images in D. (E) Multipolar mitosis analysis of iTERT hPSCs with short telomeres in the presence of ATMi or ATRi by immunofluorescence. (F) Cell proliferation analysis in iTERT hPSCs with short telomeres in the presence of ATMi or ATRi. (G) Cell proliferation analysis in iTERT hPSCs with short telomeres 7 d after transfection with the indicated siRNAs. In E–G, average ± SD of three independent experiments. Statistical analysis: one-way ANOVA followed by Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. PDLs for iTERT hPSCs with short telomeres: 149–159.
Figure S4.
Analysis of the effects of ATM or ATRis or CHEK1 and/or CHEK2 silencing on iTERT hPSCs with short telomeres (Tel.). (A and B) Representative histograms of phosphorylated CHK1 (Ser 345) and phosphorylated CHK2 (Thr 68), respectively, in iTERT hPSCs with short telomeres treated with ATMi (10 µm) or ATRi (1 µm) for four consecutive days. (C) mRNA levels of CHEK1 and CHEK2 72 h after transfection with the indicated siRNAs. Bar graphs represent average ± SD of three biological replicates, normalized to housekeeping gene ACTB. (D) Cell proliferation analysis of iTERT hPSCs with long telomeres 7 d after transfection with the indicated siRNAs. Average ± SD of three independent experiments. PDL for iTERT hPSCs with short telomeres: 159.
As G2/M accumulation, aneuploidy, and mitotic catastrophe were also observed in p53-null hPSCs with short telomeres (Figs. 1 and 3), we tested whether ATR acts independently of p53 to induce those phenotypes. Indeed, ATR inhibition rescued all phenotypes induced by short telomeres in hPSCs, regardless of p53 status (Fig. S5, A–D). Additionally, as our results indicate that ablation of p53 increases the resistance of hPSCs to death induced by telomere attrition (Fig. 2 H), we tested if these cells would show increased ATR activation. Accordingly, we observed increased CHK1 phosphorylation levels in p53-null iTERT hPSCs with short telomeres (Fig. S5 E).
Figure S5.
Analysis of the effects of ATM or ATRis on iTERT_TP53 KO hPSCs with short telomeres (Tel.), shelterin components between isogenic iTERT hPSCs and fibroblasts. (A and B) Cell cycle distribution as detected by EdU and DAPI staining by flow cytometry in iTERT_TP53 KO hPSCs with short telomeres in the presence of ATMi or ATRi. Representative analyses are shown in A, and quantification of three independent experiments (average ± SD) are depicted in B. (C) Multipolar mitosis analysis in iTERT_TP53 KO hPSCs with short telomeres in the presence of ATMi or ATRi by immunofluorescence. Average ± SD of three independent experiments. (D) Cell proliferation analysis of iTERT_TP53 KO hPSCs with short telomeres in the presence of ATMi or ATRi. (E) Analysis of phosphorylated CHK1 (Ser 345) between iTERT and iTERT_TP53 KO hPSCs. (F) Analysis of shelterin components in isogenic iTERT hPSCs and fibroblasts. Average ± SD of three independent experiments. Statistics: one-way ANOVA followed by Bonferroni’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. PDL for cells with short telomeres: 149–159 (iTERT hPSCs); 205–210 (iTERT_TP53 KO hPSCs); 45–47 (iTERT fibroblasts).
Human stem cells with short telomeres accumulate ssDNA at the telomeric G-strand
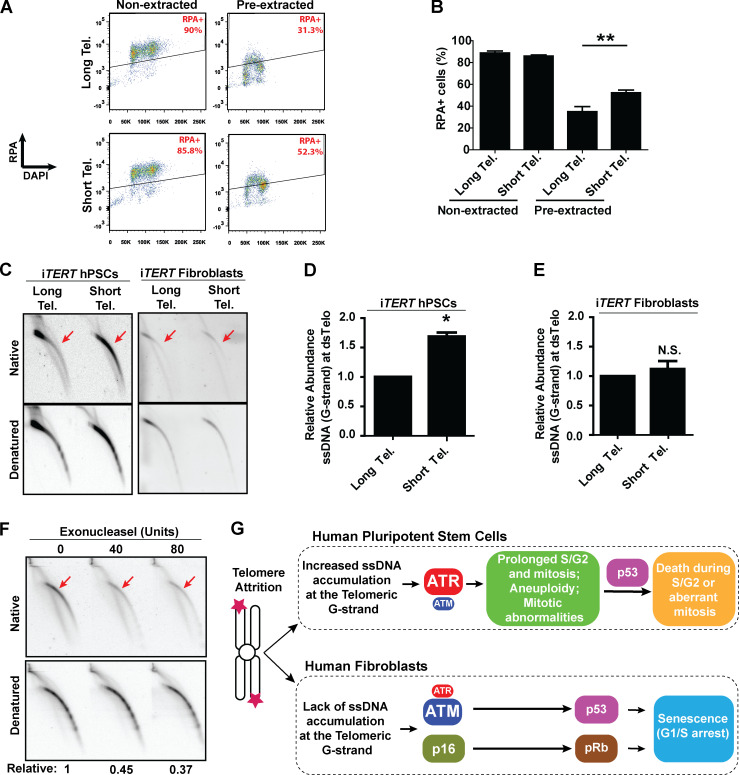
Our next step was to determine the mechanism behind the predominant role of ATR in the response of hPSCs with short telomeres. As ATR responds to accumulation of ssDNA coated by replication protein A (RPA; Saldivar et al., 2017), we analyzed chromatin-bound RPA by flow cytometry (Forment et al., 2012). Our results indicate that telomere shortening significantly increases the fraction of hPSCs positive for chromatin-bound RPA (Fig. 5, A and B). To explore this response further, we investigated the formation of ssDNA specifically at telomeres of hPSCs. Using 2D gel analysis, we observed that hPSCs with short telomeres have a significant increase in the relative abundance of ssDNA at the telomeric G-strand relative to hPSCs with longer telomeres (Fig. 5, C and D). Interestingly, this was not observed in their isogenic fibroblast counterparts (Fig. 5, C and E), indicating that this response is specific to pluripotent cells. Digestion of our DNA samples with Exonuclease I before running 2D gels degraded most of the observed signal, confirming it arises from single-stranded telomeric DNA overhang (Fig. 5 F). As shelterin binds both double-stranded and single-stranded telomeric DNA, we decided to test if the reduced expression of shelterin components (when compared with fibroblasts) could be a trigger for the accumulation of unprotected ssDNA in hPSCs with short telomeres. However, our analysis of the expression of POT1, TPP1, and TIN2 shows that, if anything, hPSCs have higher levels of expression of shelterin components when compared with their differentiated progeny (Fig. S5 F). Therefore, while at this moment our data do not provide clarity on the trigger for ssDNA accumulation in hPSCs with short telomeres, they indicate that the accumulation of ssDNA at telomeric overhangs provides the molecular culprit that activates ATR in hPSCs with short telomeres. A model including our main findings is shown in Fig. 5 G.
Figure 5.
ssDNA accumulates at telomeric G-strand (overhang) of hPSCs with short telomeres (Tel.). (A and B) Detection of chromatin-bound RPA by flow cytometry in iTERT hPSCs. Illustrative images are shown in A, and average ± SD of three independent experiments shown in B. Statistical analysis: paired t test; **, P < 0.01. (C–E) G-overhang detection in iTERT hPSCs and iTERT fibroblasts by 2D gel analysis. A representative image is shown in C, and quantification of three independent experiments (average ± SD; *, P < 0.05) is shown in D and E. (D and E) PDLs for cells with short telomeres: 149–159 (iTERT hPSCs), 45–47 (iTERT fibroblasts). N.S., not significant. (F) Exonuclease I–mediated degradation of Telomeric ssDNA in iTERT hPSCs with short telomeres. Units of enzyme are shown on top; relative indicates native/denatured signal. (G) Model: Telomere attrition in hPSCs leads to accumulation of ssDNA at telomeric overhangs and an ATR-mediated response that results in prolonged S/G2 and mitosis, mitotic abnormalities, and death. In fibroblasts, ATM has a dominant role in the activation of p53 upon telomere attrition and, together with p16/pRb, induces senescence.
While most data generated to date detailing human cells’ response to genetic instability has been derived from terminally differentiated cells, patients with reduced DNA repair capability or reduced telomere maintenance potential commonly show defects in stem or progenitor cells (Armanios and Blackburn, 2012; Fok et al., 2017; Munroe et al., 2020; Shukla et al., 2020). Successful approaches to restore tissue function in these patients must therefore derive from studies that take into consideration the intrinsic differences between stem cells and their differentiated progenies. In support of this theory, it has recently been demonstrated that TRF2, a shelterin component essential for telomere protection in differentiated cells, is dispensable in mouse embryonic stem cells, which are viable in its absence (Markiewicz-Potoczny et al., 2021; Ruis et al., 2021). Our work adds to this concept of a stem cell–specific reaction to telomere dysfunction and shows that hPSCs have a unique response to telomere shortening that greatly differs from fibroblasts. We describe that ATR activation is the main driver of loss of function of hPSCs with short telomeres that show accumulation of genetic abnormalities, prolonged S/G2 and mitosis phases of the cell cycle, and induction of death preferentially during S/G2 and mitosis as well. We also establish that induction of chromosomal aberrations and survival are significantly up-regulated in the absence of p53 in hPSCs with short telomeres.
It is known that ATR activation can induce a G2/M checkpoint (Abraham, 2001; Liu et al., 2020) and that a prolonged G2 arrest can induce both centrosome amplification and multipolar mitosis (Churikov et al., 2006; Dodson et al., 2004; Sato et al., 2000). Thus, it is tempting to speculate that ATR activation in stem cells with short telomeres prolongs the G2 phase of the cell cycle, leading to centrosome duplication and mitotic catastrophe. Cells that avoid mitotic death become aneuploid and might be eliminated by p53 during the upcoming S/G2 phase of the cell cycle. In the absence of p53, aneuploid stem cells would then proceed to additional rounds of abnormal mitosis, increasing karyotypic abnormalities. It is therefore tempting to speculate that death during multipolar mitosis in p53-ablated hPSCs with short telomeres results from the mechanical disruption induced by centrosome overduplication (Fragkos and Beard, 2011), a possibility that needs to be tested further.
As mutations in telomerase cause significant chromosomal aberrations, loss of tissue function, and elevated cancer predisposition (Alter et al., 2009; Dokal and Luzzatto, 1994), it will be important to determine the role of ATR activation in the adult stem cell compartments of patients with telomeropathies and determine the potential of modulation of ATR, or its downstream targets, to mitigate some of the deleterious phenotypes observed in these patients.
Materials and methods
Culture of hPSCs
hPSCs were grown in mTeSRTM1 (#85850; Stemcell Technologies) with 1% penicillin-streptomycin (#15140122; Gibco), in the presence of 5% O2, 5% CO2 at 37°C. hPSCs were grown on cell culture plates coated with Matrigel (see below; #08-774-552; Thermo Fisher Scientific), and medium was replaced daily. For single-cell passaging, cells were washed once with DPBS 1X (#14190136, Gibco) and incubated with Accutase (#07920; Stemcell Technologies) for 5 min at 37°C, followed by dissociation by pipetting and centrifugation (1,500 rpm, 3 min). Cells were then resuspended in mTeSR-1 with 10 µM of Y-27632 (#1293823; Peprotech) and plated at a density of 105 per well of a six-well plate. For passaging of cells as small aggregates, cells were washed once with DPBS 1X and incubated with ReLeSR (#05872; Stemcell Technologies) for 30 s at room temperature, followed by aspiration of ReLeSR and incubation of cells for an additional 5 min at 37°C. Then, hPSCs were resuspended in mTeSR1, gently dissociated into small clumps using a 5-ml pipette, and plated. For freezing, small aggregates of hPSCs were resuspended in Bambanker (#NC9582225; Wako Chemicals), transferred to Cryovials (Greiner Bio-One), placed in a Mr. Frosty Freezing container (#5100-0001; Thermo Fisher Scientific), and stored at −80°C overnight. For long-term storage, frozen cells were transferred to a liquid nitrogen container. To coat culture plates with Matrigel: A stock bottle was thawed overnight in an ice bucket placed inside a refrigerator. Using cold pipettes, 5 ml of ice-cold DMEM and penicillin-streptomycin (1% final) were mixed with the Matrigel stock, aliquoted into ice-cold 1.5-ml Eppendorf tubes (1 ml per tube), and stored at −20°C. To coat plates, 1 aliquot was thawed overnight in an ice bucket inside a refrigerator, mixed with 14 ml of DMEM and penicillin-streptomycin (1%) to make a Matrigel use solution, and aliquoted into refrigerated cell culture plates. These plates were immediately moved to the refrigerator and incubated for at least 6 h. Immediately before plating hPSCs, Matrigel use solution was transferred to another refrigerated cell culture plate, and the coated plate incubated for 30 min at 37°C. The Matrigel use solution was reused up to eight times.
Genome editing of hPSCs
Construction of iTERT hPSCs
A Flag–human (h)TERT cDNA containing silent mutations in the Exon 1 region (details below) was synthesized by Genewiz and cloned into the AAVS1-SA-2A-PURO-TRE-eGFP donor plasmid (#22074; Addgene) using the NcoI and SalI restriction enzyme sites (AAVS1-SA-2A-PURO-TRE-FLAG-TERT donor plasmid). On the day of transfection, a transfection mixture was prepared by combining 100 µl Iscove's Modified Dulbecco's Medium (#12440046; Gibco) with 1.5 µg AAVS1-SA-2A-PURO-TRE-FLAG-TERT donor plasmid, 1.5 µg AVS1-SA-2A-NEO-CAG-RTTA3 (#60431; Addgene), 0.2 µg PGK-AAVS1-ZFN-Right (#60915; Addgene), and 0.2 µg GK-AAVS1-ZFN-Left (#60916; Addgene; Sim et al., 2016). Then, 3 µl X-tremeGENE 9 DNA Transfection Reagent (#06 365 779 001; Roche) was added to this mixture (3:1 ratio), mixed by gently flicking the tube, and incubated at room temperature for 20 min. This mixture was carefully pipetted dropwise on one well of a six-well plate containing BJ-hiPSCs (BJ human-induced PSCs) colonies at ∼40% confluence growing in mTeSR-1 without antibiotics. Medium was replaced 16 h later, and when cells reached ∼90% confluence, they were passaged (1:6 split ratio using ReLeSR). Selection with Puromycin (0.5 µg/ml in mTeSR-1 medium; #P8833; Sigma-Aldrich) started the next day. Once kill control was dead (usually 2–3 d), selection with Geneticin/Neomycin (40 µg/ml; #10131035; Life Technologies) was started and proceeded for ∼7–10 d. A surviving clone was expanded in regular hPSC culture medium, and correct integration of the PURO-TRE-FLAG-TERT and of the NEO-CAG-RTTA3 constructs was checked by PCR, as described in detail in Sim et al. (2016). This clone was then submitted to endogenous hTERT KO using CRISPR/Cas9, and the resulting cell line was named inducible hTERT, or simply iTERT, since expression of the exogenous hTERT (hence telomerase activity) is induced by DOX. These cells were grown in the presence of 75 ng/ml DOX during the entire hTERT KO step to ensure that telomerase activity was not lost at any moment. The gRNAs 5′-CCCCAGGCGCCGCACGAACG-NGG-3′ and 5′-CCACGTTCGTGCGGCGCCTG-NGG-3′ were used to generate a double-strand break in the endogenous hTERT gene. Importantly, the hTERT cDNA inserted into the AAVS1 safe harbor locus is not cleaved by these gRNAs, as it contains silent mutations on the protospacer adjacent motif sequence sites. Nucleofection of the gRNAs, clonal expansion, next-generation sequencing (NGS) to confirm successful KO of the endogenous hTERT, mycoplasma testing, and karyotype analysis were performed by the Genome Engineering and iPSC Center of Washington University in St. Louis (WUSTL).
TP53 and Rb1 gene KO
iTERT hPSCs were dissociated into single cells with Accutase, and 2 × 105 cells were resuspended in a mix of P4 Primary Cell Solution (16.4 µl) with Supplement-1 (3.6 µl; #PBP4-02250; Lonza). 200 ng of the gRNA vector (see below) and 900 ng of the p3s-Cas9HC vector (#43945; Addgene) were added to this mix, immediately transferred to a 16-well Nucleocuvette, and nucleofected in a 4D Nucleofector System (Lonza) using the CA-137 program. 15 min after nucleofection, cells were gently resuspended in 100 µl mTeSR-1, transferred to a 15-ml conical tube, and plated at low density in a six-well plate (in the presence of 10 µM Y-27632 without antibiotics). The day after, medium was replaced (without Y-27632), and cells were grown for ∼8 d, when colonies were ready to be manually picked under a stereoscope. NGS to confirm successful KO of the target gene and mycoplasma tests were performed by the Genome Engineering and iPSC Center. Karyotype analysis were performed by Cell Line Genetics.
To knock out TP53, the gRNA sequence 5′-TCGACGCTAGGATCTGACTG-NGG-3′ was cloned into the vector MLM3636 (#43860; Addgene) using the BsmBI restriction site (Fok et al., 2017). KO of RB1 was performed on iTERT_TP53 KO hPSCs using the same methodology described above using the RB1 gRNA sequence 5′-GGTGGCGGCCGTTTTTCGGG-NGG-3′.
Culture of hPSC-derived fibroblasts
Fibroblasts were grown in fibroblast medium comprising DMEM (#11995073; Gibco), 20% FBS (#100-500; GeminiBio), and 1% penicillin-streptomycin, in the presence of 5% O2, 5% CO2 at 37°C. Fibroblasts were grown on cell culture plates coated with gelatin 0.1% (see below), and medium was replaced every 4 days. For passaging (usually once a week), cells were washed once with DPBS 1X and incubated with Trypsin-EDTA 0.05% (#25300054; Gibco) for 5 min at 5% CO2 at 37°C and dissociated by pipetting, and trypsin was neutralized with growth medium. Cells were then centrifuged (1,500 rpm, 3 min), resuspended in growth medium, and plated at a density of 8 × 104 cells per well of a six-well plate. To freeze fibroblasts, single cells were resuspended in freezing medium (90% FBS and 10% DMSO), transferred to cryovials, placed in a Mr. Frosty Freezing container, and stored at −80°C overnight. For long-term storage, frozen vials were transferred to a liquid nitrogen container. To coat culture plates with gelatin, a 0.1% stock solution of gelatin (#G9391; Sigma-Aldrich) was prepared by dissolving 1 g into 1 liter of DPBS 1X and autoclaving, followed by filtration on a 0.2-µm PES 500-ml filter unit (#569-0020; Nalgene). Solution was stored at 4°C. An aliquot of the stock solution was added to empty culture wells, incubated for 10 min at room temperature, and aspirated, and cells were immediately plated.
Differentiation of hPSCs into fibroblasts
This protocol was adapted from Xu et al. (2004). hPSC cultures that were ∼90% confluent were dissociated with ReleSR and resuspended in differentiation medium comprising KO DMEM (#10829018; Gibco), 1 mM L-glutamine (#25030081; Gibco), 0.1 mM β-mercaptoethanol (#ES-007; Millipore), 20% FBS, and 1% nonessential amino acids (#11140050; Gibco). Cells were cultured in nonadherent cell culture plates for 4 d, in the presence of 5% O2, 5% CO2 at 37°C, to form embryoid bodies. Medium was carefully replaced on day 2. Then, embryoid bodies were plated on gelatin-coated plates in the presence of differentiation medium and grown for an additional 9 d. Medium was replaced 4 d after plating. Then, the outgrowth culture was passaged by incubating cells with 2 mg/ml Collagenase II (#07418; Stemcell Technologies) in DPBS 1× for 30 min at 37°C. Cells were carefully dissociated by pipetting, centrifuged (1,500 rpm, 3 min), resuspended in differentiation medium, and plated on gelatin-coated plates at a density of 4 × 104 cells per well of a 12-well plate. Cells were grown for 7 d (medium was replaced 4 d after plating) before they were detached with Trypsin 0.05%, resuspended in fibroblast medium (DMEM supplemented with 20% FBS and 1% penicillin-streptomycin), plated on gelatin-coated plates at a density of 8 × 104 cells per well of a six-well plate, and grown as described in Culture of fibroblasts.
Karyotype analysis
Karyotype analysis was performed by Cell Line Genetics (Madison, WI).
Drug treatment
Stock solutions of ATMi KU-55933 (#SML1109; Sigma-Aldrich) and ATRi VE-821 (#SML1415; Sigma-Aldrich) were prepared by dissolving these drugs in DMSO to a concentration of 10 mM (ATMi) or 1 mM (ATRi) and stored as small aliquots at −80°C. Before use, an aliquot was thawed and diluted in the culture medium to a final concentration of 10 µM (ATMi) or 1 µM (ATRi). Aliquots were thawed up to three times and then discarded. ATMi or ATRi was added daily to fresh hPSC medium during inhibitor treatment.
A stock solution of etoposide (#E1383; Sigma-Aldrich) was prepared by dissolving it in DMSO to a concentration of 300 µM and stored as small aliquots at −80°C. Aliquots were thawed up to three times and then discarded. Drug treatment with etoposide was performed by culturing cells with the desired drug concentration for 24 h, followed by a recovery period of 3 d.
Cell viability using DRAQ5 and Sapphire700 staining
5 × 103 hPSCs were seeded as single cells per well of 96-well plate (#655180; Greiner Bio-One). The next day, they were treated with etoposide for 24 h, when medium was replaced and cells were grown for an additional 3 d. Then, cells were washed once with DPBS 1X, fixed with 4% PFA in DPBS 1X for 10 min at room temperature, washed twice with DPBS 1X, and stored at 4°C. Before staining, fixed samples were permeabilized with 0.1% Triton X-100 (#X100; Sigma-Aldrich) for 15 min at room temperature, washed once with DPBS 1X, and incubated with staining solution of DRAQ5 + Sapphire700 (prepared according to manufacturer’s instruction) for 30 min at room temperature, protected from light. Cells were washed three times with DPBS 1X and dried, and data were acquired on an Odyssey CLx Imager (LI-COR Biosciences) using a focus offset of 3.0 mm. The signal intensity of each well was analyzed using Image Studio Lite software (LI-COR).
PDLs
To calculate PDL, the formula PDL = X + 3.322 (log Y – log I) was used, where X = initial PDL, I = initial cell number seeded, and Y = number of cells at the end of the growth period.
Quantitative RT-PCR
RNA was obtained from frozen cell pellets using Trizol (#15596018; Ambion) following the manufacturer’s instruction. DNase treatment was performed on 4 µg isolated RNA from each sample with the TURBO DNA-free kit (#AM1907; Ambion) for 25 min at 37°C. Approximately 0.75 µg DNase-treated RNA was then reverse transcribed using SuperScript III reverse transcription (#18080044; Invitrogen) following the manufacturer’s instruction. cDNA was diluted 10-fold in Nuclease-Free Water (#AM9937; Ambion). Quantitative RT-PCR was performed on a StepOnePlus Real-Time PCR System (#4376600; Applied BioSystems). Reaction: 3 µl diluted cDNA, 3 µl forward + reverse primer mix (2.4 µM each in the primer mix), and 6 µl EvaGreen 2x Master Mix R (#MX-R; Lamda Biotech). Primers were designed using PrimerQuest software (IDT) and ordered from IDT. Primer sequences used were CHEK1 forward, 5′-TCTTCTGTTGGATGAAAGGGATAA-3′, and reverse 5′- TGGTAAAGTACCACACATCTTGT-3′; CHEK2 forward, 5′-AAGCTAAATCATCCTTGCATCATC-3′, and reverse, 5-GCTTCTTTCAGGCGTTTATTCC-3′; and ACTB forward, 5′-TTCCTTCCTGGGCATGGAGT-3′, and reverse, 5′-AATGCCAGGGTACATGGTGG-3′.
Western blotting
Protein was extracted from frozen cell pellets using NP-40 buffer (25 mM Hepes, 150 mM KCl, 1.5 mM MgCl2, 0.5% IGEPAL CA-630, and 10% glycerol, pH 7.5, in RNase-free water) supplemented with Pierce Protease inhibitor (#A32955; Thermo Fisher Scientific) and PhosSTOP (#04906837001; Roche) following the manufacturer’s instructions. Samples were incubated on ice for 15 min and centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was transferred to a clean tube placed on ice. Protein quantification was performed using the Pierce BCA Protein Assay Kit (#23225; Thermo Fisher Scientific) according to the manufacturer’s instruction. 20 µg of protein per sample was resolved in a gradient polyacrylamide gel in 1× Tris/glycine/SDS buffer (#1610732; Bio-Rad) and transferred onto 0.2-µm nitrocellulose membrane (#1620112; Bio-Rad) at 400 mA for 1:15 h in 1× Tris/glycine/SDS buffer with 20% methanol (#322415; Sigma-Aldrich). Membranes were blocked for 1 h in blocking solution consisting either of 5% BSA (#A-420-250; GoldBio) in TBS buffer (20 mM Tris and 150 mM NaCl in water) or 5% nonfat milk in TBS buffer, depending on primary antibody. Primary antibody was diluted in blocking solution + 0.1% Tween20 (#P1379; Sigma-Aldrich) and incubated with gentle shaking overnight at 4°C. Membranes were washed four times for 5 min with TBS-Tween buffer, and secondary antibody was incubated in blocking solution + 0.1% Tween20 for 1 h with gentle shaking at room temperature, protected from light. Membranes were washed four times for 5 min with TBS-Tween buffer and scanned on an Odyssey CLx Imager. Antibodies and dilutions: mouse p53 (DO-1; 1:5,000, #SC-126; Santa Cruz Biotechnology); rabbit GAPDH (D16H11) XP (1:5,000, #5174), mouse Rb (4H1; 1:2,000, #9309; Cell Signaling); mouse Flag M2 (1:5,000, #F3165; Sigma-Aldrich); mouse TPP1 (1:1,000, #AB54685), rabbit POT1 (1:1,000, #AB21382; Abcam); and mouse TIN2 (1:1,000, #NB600-1522; Novus).
Telomere restriction fragment (TRF) analysis
Genomic DNA was extracted from frozen cell pellets by incubating samples on 500 µl tail lysis buffer (100 mM Tris, 5 mM EDTA, 0.2% SDS, and 200 mM NaCl, pH 8.5, in water) supplemented with 400 µg/ml Proteinase K (#AM2546; Ambion) on a thermocycler overnight at 55°C with shaking (500 rpm). Then, 600 µl isopropanol (#437522; Sigma-Aldrich) was added, and samples were vigorously shaken and centrifuged (13,000 rpm, 5 min, room temperature). The supernatant was removed, and the pellet was washed with 1 ml of 70% ethanol, centrifuged (13,000 rpm, 5 min, room temperature), and resuspended in TE (10 mM Tris and 1 mM EDTA, pH 8.0). 10 µg extracted DNA was digested overnight with 30 U each RsaI (#R0167; New England Biolabs) and HinfI (#R0155; New England Biolabs) restriction enzymes and resolved on a 0.8% agarose gel for 16 h at 85 V in Tris/borate/EDTA (TBE) buffer (89 mM Tris, 89 mM boric acid, and 2 mM EDTA in water). Gel was then soaked in denaturing solution (1.5 M NaCl and 0.5 M NaOH in water) for 45 min with gentle shaking, briefly washed with deionized water, soaked in neutralizing buffer (1.5 M NaCl and 1 M Tris, pH 7.4, in water) for 1 h with gentle shaking, and transferred to a Hybond-N+ nylon membrane (#RPN303B; GE Healthcare) by capillarity for 48 h in 10× SSC buffer (1.5 mM NaCl and 150 mM sodium citrate, pH 7.0, in water). Membrane was cross-linked with 700 J/m2 UVC on a Hoefer UVC 500 Crosslinker and hybridized with a 32P-labeled telomere probe. To label the telomere probe with 32P, a mix was prepared in water (final volume 20 µl) containing 10 µM of the oligonucleotide (CCCTAA)4; 10 U T4 polynucleotide kinase (#M0201; NEB); 1× T4 polynucleotide kinase buffer (#B0201; NEB); and 5 µl ATP γ-32P (#BLU502A250UC; PerkinElmer). The reaction was incubated at 37°C for 30 min and 95°C for 5 min, and then put in ice. To label the telomeres with the radioactive probe, the cross-linked membrane was preincubated with 15 ml of Rapid-hyb buffer (#RPN1636; GE Healthcare) placed inside a hybridization bottle (#25000-010; VWR) at 42°C for 20 min in a hybridization oven with slow rotation. Then, the radiolabeled probe mix was added and incubated for an additional 2 h. Membrane was washed once with 5× SSC + 0.1% SDS for 20 min, and then washed twice with 1× SSC + 0.1% SDS at 42°C in the hybridization oven at 42°C with rotation. The hybridized membrane was exposed (overnight up to 48 h) to a Kodak BioMax MR film (#8701302; Carestream) at −80°C and then developed.
Telomeric repeat amplification protocol (TRAP) analysis
Protein extraction and quantification were performed as described in the Western blotting topic of this section with addition of RNase OUT (#P/N 51535; Invitrogen) to a final concentration of 40 U/ml in the NP-40 protein extraction buffer. Telomere extension reactions were performed using 2.0, 0.5, and 0.125 µg of protein extract per sample, and the resulting products were amplified by PCR following a two-step TRAP protocol from the manufacturer (TRAPeze Telomerase Detection Kit, #S7700; Millipore). PCR products were resolved on a 9% polyacrylamide 37:5:1 gel at 250 V for 3 h. Gel was dried on a gel dryer (#583; Bio-Rad) at 80°C for 30 min, exposed for 15 min to a Kodak BioMax MR film (#8701302; Carestream) at −80°C, and developed.
Lentivirus generation and hPSC transduction
Lentiviruses were generated by transfecting 293T cells that were ∼80% confluent with third-generation lentivirus packaging plasmids (pMDL [Gag/Pol], pVSVG, and pREV) together with the transfer vector (DNA proportion was 3.97:2.58:1.39:1, respectively). Lipofectamine 2000 (#11668019; Invitrogen) was used as transfection reagent. The DNA/Lipofectamine 2000 mix was prepared according to the manufacturer’s instruction in Opti-MEM (#31985062; Gibco). Medium was replaced 6 h after transfection with 293T growth medium (10% FBS in DMEM). Medium containing virus was collected 24 and 48 h after transfection, filtered through a cellulose acetate 0.45-µm filter, concentrated using Lenti-X Concentrator (#PT4421-2; Clontech), resuspended in mTeSR-1, aliquoted, and frozen at −80°C using a Mr. Frosty container. hPSCs were transduced by adding lentivirus preparation to growth medium and incubating cells for 24 h in the presence of 5% O2, 5% CO2 at 37°C. Lentivirus packaging vectors (#VB160226-10009, #VB160226-10010, and #VB160226-10011) were purchased from Vector Builder. The pLV-EF1α_H2B-EGFP_TagBFP_hCdt1_mCherry_hGeminin vector was adapted from Sato et al. (2019) and synthesized by Vector Builder. The pBOB-EF1α_H2B_mApple vector was a kind gift from Dr. Sheila Stewart (WUSTL).
Immunofluorescence
hPSCs were grown on 12-mm Circle Microscope Cover Glass (#22-293232; Thermo Fisher Scientific) coated with Matrigel and fixed with 4% PFA (#28908; Thermo Fisher Scientific) in DPBS 1X at room temperature for 10 min. Fixed cells were washed twice with DPBS 1X, permeabilized with 0.2% Triton X-100 for 10 min at room temperature and incubated for 1 h with blocking solution (2% BSA in DPBS 1X). Samples were incubated with primary antibodies overnight at 4°C, washed three times with DPBS 1X, and incubated with secondary antibodies for 1 h at room temperature, protected from light. After three washes with DPBS 1X, the cover glass was lowered on top of a 6-µl drop of ProLong Gold reagent (#P-36931; Life Technologies) placed on top of a microscope slide (#2949-75X25; Corning). Cells were imaged 4–24 h after mounting using a Leica DM6B upright digital research microscope and a Leica DFC7000 T camera (Leica Microsystems). Images were acquired using Leica Las X Core software. The objective used was a PL APO 40×/0.95 CORR, Apochromatic corrected objective with high numerical aperture, FWD: 0.17 mm, CORR: 0.11.0,23 mm. Primary antibodies and dilutions used were phospho-histone H3 (Ser10; 6G3; 1:300, #9706) and Aurora A/AIK (1G4; 1:300, #4718). Secondary antibodies and dilutions used were: goat anti-rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 (1:1,000, #A32733), and goat anti-mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:1,000, #A-11001; Thermo Fisher Scientific).
DNA replication analysis by immunofluorescence
For DNA replication analysis, we used the Click-iT EdU Alexa Fluor 488 Kit (#C10337; Life Technologies) according to the manufacturer’s instruction. Briefly, EdU was added to the culture medium of hPSCs or fibroblasts grown on top of coverslips to a final concentration of 10 µM EdU and incubated for 1 h in the presence of 5% O2, 5% CO2 at 37°C. Then, cells were immediately washed twice with DPBS 1X and fixed with PFA 4% in DPBS 1X for 10 min at room temperature. Fixed samples were then washed twice with 3% BSA in DPBS 1X, permeabilized with 0.2% Triton X-100 0.2% for 20 min at room temperature, and washed twice with 3% BSA in DPBS 1X, and the cocktail reaction (prepared according to the manufacturer’s instruction) was added to the samples. Samples were incubated for 30 min protected from light, and then immediately washed twice with 3% BSA in DPBS 1X. The cover glass was lowered on top of a 6-µl drop of ProLong Gold reagent (#P-36931; Life Technologies) placed on top of a microscope slide (#2949-75X25; Corning). Cells were imaged 4–24 h after mounting using a Leica DM6B upright digital research microscope (Leica Microsystems).
Cell cycle analysis by flow cytometry
Cell cycle analysis was performed using the Click-iT EdU Alexa Fluor 488 Kit (#C10337; Life Technologies). EdU was added to the culture medium of hPSCs or fibroblasts to a final concentration of 10 µM EdU, and cells were incubated for 1 h in the presence of 5% O2, 5% CO2 at 37°C. Cells were washed twice with DPBS 1X, dissociated into single cells using Accutase (hPSCs) or trypsin 0.05% (fibroblasts), centrifuged (1,500 rpm, 3 min), resuspended in fixing solution (PFA 2% in DPBS 1X), and incubated at room temperature for 15 min. Then, cells were centrifuged (3,000 rpm, 3 min from now on for all centrifugation steps in this section), washed twice with DPBS 1X, and stored (in DPBS 1X) at 4°C for up to 2 mo. On the day of the analysis, cells were centrifuged, resuspended in 1% BSA in DPBS 1X, incubated for 10 min at room temperature, centrifuged, resuspended in permeabilizing solution (1% BSA, 0.5% saponin [#8047-15-2; Sigma-Aldrich] in DPBS 1X), and incubated for 30 min. Cells were then centrifuged and resuspended in 200 µl EdU detection solution (prepared according to the manufacturer’s instruction) and incubated for 30 min, protected from light. Cells were then centrifuged, washed with 3% BSA in DPBS 1X, centrifuged, and resuspended in 1% BSA solution containing 2 µg/ml DAPI (#62248; Thermo Fisher Scientific) in DPBS 1X. Cells were incubated for 15–30 min protected from light and immediately analyzed in a FACS-Canto II (BD) in the Flow Cytometry & Fluorescence Activated Cell Sorting Core of WUSTL. Cell doublets were detected and excluded based on DAPI width versus DAPI area fluorescence signal. 10,000–20,000 events were acquired per sample.
Protein detection by flow cytometry
hPSCs were dissociated with Accutase, centrifuged (1,500 rpm, 3 min), resuspended in fixing solution (PFA 2% in DPBS 1X), and incubated at room temperature for 10 min. Then, cells were centrifuged (4,000 rpm, 3 min from now on for all centrifugation steps in this section), washed twice with DPBS 1X, and stored (in DPBS 1X) at 4°C for ≤2 mo. Cells were centrifuged, resuspended in blocking buffer (1% BSA and 0.5% saponin in DPBS 1X), incubated for 10 min at room temperature, centrifuged, resuspended in blocking buffer containing primary antibody, and incubated overnight at 4°C. 1 ml blocking buffer was added to each sample, centrifuged, resuspended in blocking solution containing secondary antibody, and incubated for 1 h protected from light. Then, 1 ml blocking buffer was added to each sample, centrifuged, and resuspended in 0.4 ml DAPI solution (1% BSA and 2 µg/ml DAPI in DPBS 1X). Cells were incubated for 15 min at room temperature protected from light and immediately analyzed in a FACS-Canto II (BD) in the Flow Cytometry & Fluorescence Activated Cell Sorting Core. Cell doublets were detected and excluded based on DAPI width versus DAPI area fluorescence signal. 20,000 events were acquired per sample. Primary antibodies and dilutions used were rabbit phospho-Chk1 (Ser345; 133D3; 1:200, #2348) and rabbit phospho-Chk2 (Thr68; 1:200, #2661; Cell Signaling). Secondary antibodies and dilutions used were goat anti-rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 647 (1:1,000, #A32733), and goat anti-mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (1:1,000, # A-21235; Thermo Fisher Scientific).
Chromatin-bound RPA detection by flow cytometry
The assay was adapted from Forment et al. (2012). hPSCs were dissociated with Accutase, centrifuged (1,500 rpm, 3 min), and washed with DPBS 1X, and the pellet was gently resuspended in ice-cold permeabilization buffer (0.1% Triton X-100 in cytoskeletal buffer). Cells were incubated for 10 min on ice. 1 ml of 0.1% BSA in DPBS 1X was added to the permeabilized samples and centrifuged (2,000 rpm, 5 min). The pellet was resuspended in fixing solution (2% PFA in DPBS 1X), incubated for 10 min at room temperature, and washed twice with DPBS 1X. Cell permeabilization and antibody staining was performed exactly as described in Protein detection by flow cytometry. RPA antibody used was mouse RPA32/RPA2 antibody [9H8] (#ab2175; Abcam; 1:300).
Cell death analysis by flow cytometry using PI and Annexin V staining
The assay was performed using the FITC Annexin V Apoptosis Detection Kit (#556547; BD) according to the manufacturer’s instruction. Detached (supernatant) and attached hPSCs (dissociated with Accutase) were combined, centrifuged (1,500 rpm, 3 min), resuspended in ice-cold DPBS 1X, counted, aliquoted (105 cells per 1.5-ml Eppendorf tube), centrifuged again, and resuspended in 100 µl of 1× binding buffer(100 mM Hepes/NaOH, pH 7.5, containing 1.4 M NaCl and 25 mM CaCl2). 5 µl FITC Annexin V and 5 µl PI solutions were added to each sample, gently vortexed to mix, and incubated at room temperature for 15 min, protected from light. Then, 400 µl of 1× binding buffer was added to each sample, and cells were immediately analyzed in a FACS-Canto II (BD) in the Flow Cytometry & Fluorescence Activated Cell Sorting Core. 10,000 events were acquired per sample. Live cells are double negative; dead cells are positive for PI or for both PI and Annexin V; cells at early stages of apoptosis are Annexin V positive and PI negative. Cells treated with etoposide (75 nM) for 24 h were used as positive control in every assay. Unstained cells, or etoposide-treated cells stained either with PI or with Annexin V, were used to set up compensation and quadrants.
Reverse transfection of siRNA
A 10-µM stock solution of siRNA was prepared by resuspending the lyophilized duplexes in RNase-free 1× siRNA buffer (#B-002000-UB-100; Horizon Discovery) and stored as small aliquots at −80°C. On the day of transfection, 0.34 µl Lipofectamine RNAiMAX Transfection Reagent (#13778100; Invitrogen) was added to 25 µl Opti-MEM (tube 1). In a separate tube (tube 2), 1.8 µl of the 10-µM stock solution of siRNA was added to 25 µl Opti-MEM. After 5 min, tubes 1 and 2 were mixed and incubated for 20 min at room temperature. In the meantime, hPSCs were dissociated into single cells using Accutase, and 5 × 104 cells were resuspended in 450 µl mTeSR-1 medium containing 10 µM Y-27632 and no antibiotics. 50 µl RNAiMAX/siRNA mix was added to the cells (final volume = 500 µl) and plated on a well of a 24-well plate. Medium was changed 24 h after reverse transfection. siRNAs used were siGENOME Human CHEK1 (1111) siRNA–SMARTpool, 5 nmol (#M-003255-04-0005), siGENOME Human CHEK2 (11200) siRNA–SMARTpool, 5 nmol (#M-003256-06-0005), and siGENOME Human Non-Targeting siRNA–SMARTpool, 5 nmol (#D-001206-14-20), from Dharmacon.
Time-lapse imaging
Cells were grown in Matrigel-coated Chambered 1.5 Borosilicate Cover Glass System (#155382PK; Nunc Lab-Tek II; Thermo Fisher Scientific) in mTeSR-1 medium, and time-lapse imaging was performed using a Yokogawa CSU-X1 spinning-disk confocal microscope on a Nikon Ti:Eclipse inverted microscope frame (Washington University Center for Cellular Imaging) using a 20× objective and the PerfectFocus stability mechanism. The camera used was a Hamamatsu ORCA-Fusion Gen-III sCMOS, and acquisition software was Nikon NIS Elements AR v5.21.03. Cells were maintained in the presence of 5% CO2 at 37°C during entire live imaging. For mitosis analysis, 3D images from hPSCs stably expressing H2B-mApple (lentivirus) were obtained every 4 min, for a total of 24 h; each z-stack image was created by compiling approximately eight confocal images. At least 50 mitoses were analyzed per sample in each independent experiment. For FUCCI analysis, single-plane images from hPSCs stably expressing H2B-EGFP_TagBFP-hCdt1_mCherry-hGeminin (lentivirus) were obtained every 20 min, for a total of 36 h; >200 individual cells were analyzed per experimental group in order to visualize at least 100 complete G1 events, 100 complete S/G2 events, and 100 complete mitoses. During the recording of the hPSCs with short telomeres, we observed a small percentage of cells in which the S/G2 phase lasted for the entire 36-h recording period. These cells were not taken into consideration during our analysis, since we could determine neither the beginning nor the terminus of S/G2. Imaging processing and data analysis were performed using Fiji software (Schindelin et al., 2012).
Neutral-neutral 2D gel electrophoresis and in-gel hybridization
Neutral-neutral 2D agarose gel electrophoresis was performed as described (Zhang et al., 2017). Briefly, 10 µg genomic DNA was extracted (Qiagen) and digested with restriction enzymes RsaI (NEB) and HinfI (NEB), to which Exonuclease I (NEB) was added 2 h before restriction enzyme treatment to remove 3′ overhang when appropriate. Digested genomic DNA was subjected to first dimensional gel using 0.4% agarose gel in 1× TBE buffer and electrophoresed at 1 V/cm for 12 h at room temperature. Sample lanes were excised and immersed in 0.3 µg/ml ethidium bromide (Sigma-Aldrich) for 30 min and casted into 1% agarose gel with 0.3 µg/ml ethidium bromide in 1× TBE. The secondary dimensional gel was electrophoresed at 3 V/cm for 6 h at 4°C. Afterward, the gel was pumped to dry using gel dryer (Bio-Rad) and subjected to native and denatured in gel hybridization with 32P-labeled C-probe. The gel was washed and exposed to PhosphorImager screen, which was then scanned by Storm and analyzed with Fiji.
Statistical analysis
The software GraphPad Prism was used to perform statistical significance analysis. The number of independent experiments analyzed, statistical tests performed, and statistical significance are indicated in the figure legends.
Online supplemental material
Fig. S1 shows the engineering strategy and characterization of iTERT hPSCs with long and short telomeres. Fig. S2 shows the engineering strategy and characterization of iTERT hPSCs lacking TP53 or TP53 and Rb1 and PDL curve of hPSCs and isogenic fibroblasts. Fig. S3 shows analysis of live imaging and karyotype analysis of iTERT and iTERT_TP53 KO hPSCs with short telomeres. Fig. S4 shows histograms of phosphoCHK1 (Ser345) and phosphoCHK2 (Thr68) analysis, CHEK1/CHEK2 knockdown levels, and impact on proliferation of iTERT hPSCs with long telomeres. Fig. S5 shows multiple analysis in iTERT_TP53 KO hPSCs with short telomeres in the presence of ATMi or ATRi, pCHK1 analysis in iTERT vs iTERT_TP53 KO hPSCs with short telomeres, as well as shelterin analysis by Western blotting in isogenic iTERT hPSCs and fibroblasts. Table S1 shows G-band karyotype analysis in iTERT and iTERT_TP53 KO hPSCs with long or short telomeres.
Supplementary Material
shows karyotype analysis in iTERT and iTERT_TP53 KO hPSCs with long and short telomeres.
Acknowledgments
We thank J. Fitzpatrick, M. Shih, and P. Bayguinov (WUSTL Center for Cellular Imaging) for technical support; S. Stewart (WUSTL) for the EF1α-H2B-mApple lentivirus vector; and Genome Engineering and iPSC Center (GEIC-WUSTL) for assistance with gene editing.
A.T. Vessoni was supported by the Philip W. Majerus, MD, Fellowship. M. Munroe is supported by the National Heart, Lung, and Blood Institute T32 Training Grant (HL007088-41). This research was supported by the National Cancer Institute (R01CA237263) to A. Vindigni and by the National Heart, Lung, and Blood Institute (1R01HL137793), the Alvin J. Siteman Cancer Center at Washington University in St. Louis, the American Cancer Society, the American Federation for Aging Research, the Longer Life Foundation, and the Center of Regenerative Medicine at Washington University in St. Louis to L.F.Z. Batista.
The authors declare no competing financial interests.
Author contributions: A.T. Vessoni and L.F.Z. Batista conceptualized the study, administered the project, and wrote the manuscript. A.T. Vessoni conceptualized experiments and was involved in the direct investigation of most of the data. T. Zhang performed and analyzed 2D agarose electrophoresis and in-gel hybridization. A. Quinet assisted with conceptualization and data analysis. M. Munroe, H.-C. Jeong, M. Wood, and E. Tedone were involved in investigation and analysis. A. Vindigni, J.W. Shay, and R. Greenberg provided resources and interpreted the data. All authors reviewed and edited the final manuscript. L.F.Z. Batista supervised the project.
References
- Abraham, R.T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177–2196. 10.1101/gad.914401 [DOI] [PubMed] [Google Scholar]
- Alter, B.P., Giri N., Savage S.A., and Rosenberg P.S.. 2009. Cancer in dyskeratosis congenita. Blood. 113:6549–6557. 10.1182/blood-2008-12-192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios, M., and Blackburn E.H.. 2012. The telomere syndromes. Nat. Rev. Genet. 13:693–704. 10.1038/nrg3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios, M., Alder J.K., Parry E.M., Karim B., Strong M.A., and Greider C.W.. 2009. Short telomeres are sufficient to cause the degenerative defects associated with aging. Am. J. Hum. Genet. 85:823–832. 10.1016/j.ajhg.2009.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A.A. 2016. The molecular genetics of the telomere biology disorders. RNA Biol. 13:696–706. 10.1080/15476286.2015.1094596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov, D., Wei C., and Price C.M.. 2006. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol. Cell. Biol. 26:6971–6982. 10.1128/MCB.01011-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna, F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., and Jackson S.P.. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 426:194–198. 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Davoli, T., and de Lange T.. 2012. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 21:765–776. 10.1016/j.ccr.2012.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli, T., Denchi E.L., and de Lange T.. 2010. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 141:81–93. 10.1016/j.cell.2010.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson, H., Bourke E., Jeffers L.J., Vagnarelli P., Sonoda E., Takeda S., Earnshaw W.C., Merdes A., and Morrison C.. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 23:3864–3873. 10.1038/sj.emboj.7600393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal, I., and Luzzatto L.. 1994. Dyskeratosis congenita is a chromosomal instability disorder. Leuk. Lymphoma. 15:1–7. 10.3109/10428199409051671 [DOI] [PubMed] [Google Scholar]
- Fok, W.C., Niero E.L.O., Dege C., Brenner K.A., Sturgeon C.M., and Batista L.F.Z.. 2017. p53 Mediates Failure of Human Definitive Hematopoiesis in Dyskeratosis Congenita. Stem Cell Reports. 9:409–418. 10.1016/j.stemcr.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment, J.V., Walker R.V., and Jackson S.P.. 2012. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 81:922–928. 10.1002/cyto.a.22155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos, M., and Beard P.. 2011. Mitotic catastrophe occurs in the absence of apoptosis in p53-null cells with a defective G1 checkpoint. PLoS One. 6:e22946. 10.1371/journal.pone.0022946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi, A.J., Jensen F.C., and Koprowski H.. 1965. Sv40-Induced Tranformation of Human Diploid Cells: Crisis and Recovery. J. Cell. Comp. Physiol. 65:69–83. 10.1002/jcp.1030650110 [DOI] [PubMed] [Google Scholar]
- Guo, X., Deng Y., Lin Y., Cosme-Blanco W., Chan S., He H., Yuan G., Brown E.J., and Chang S.. 2007. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 26:4709–4719. 10.1038/sj.emboj.7601893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, L.Y., Armanios M., Strong M.A., Karim B., Feldser D.M., Huso D., and Greider C.W.. 2005. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 123:1121–1131. 10.1016/j.cell.2005.11.020 [DOI] [PubMed] [Google Scholar]
- Hara, E., Tsurui H., Shinozaki A., Nakada S., and Oda K.. 1991. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 179:528–534. 10.1016/0006-291X(91)91403-Y [DOI] [PubMed] [Google Scholar]
- Hayashi, M.T., Cesare A.J., Rivera T., and Karlseder J.. 2015. Cell death during crisis is mediated by mitotic telomere deprotection. Nature. 522:492–496. 10.1038/nature14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig, U., Jobling W.A., Chen B.P., Chen D.J., and Sedivy J.M.. 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell. 14:501–513. 10.1016/S1097-2765(04)00256-4 [DOI] [PubMed] [Google Scholar]
- Hickson, I., Zhao Y., Richardson C.J., Green S.J., Martin N.M., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., and Smith G.C.. 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 64:9152–9159. 10.1158/0008-5472.CAN-04-2727 [DOI] [PubMed] [Google Scholar]
- Jacobs, J.J., and de Lange T.. 2004. Significant role for p16INK4a in p53-independent telomere-directed senescence. Curr. Biol. 14:2302–2308. 10.1016/j.cub.2004.12.025 [DOI] [PubMed] [Google Scholar]
- Jaiswal, S.K., Oh J.J., and DePamphilis M.L.. 2020. Cell cycle arrest and apoptosis are not dependent on p53 prior to p53-dependent embryonic stem cell differentiation. Stem Cells. 38:1091–1106. 10.1002/stem.3199 [DOI] [PubMed] [Google Scholar]
- Lee, H.W., Blasco M.A., Gottlieb G.J., Horner J.W. II, Greider C.W., and DePinho R.A.. 1998. Essential role of mouse telomerase in highly proliferative organs. Nature. 392:569–574. 10.1038/33345 [DOI] [PubMed] [Google Scholar]
- Liu, K., Zheng M., Lu R., Du J., Zhao Q., Li Z., Li Y., and Zhang S.. 2020. The role of CDC25C in cell cycle regulation and clinical cancer therapy: a systematic review. Cancer Cell Int. 20:213. 10.1186/s12935-020-01304-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimets, T., Neganova I., Armstrong L., and Lako M.. 2008. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 27:5277–5287. 10.1038/onc.2008.166 [DOI] [PubMed] [Google Scholar]
- Markiewicz-Potoczny, M., Lobanova A., Loeb A.M., Kirak O., Olbrich T., Ruiz S., and Lazzerini Denchi E.. 2021. TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature. 589:110–115. 10.1038/s41586-020-2959-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilović, O., Choi S., Varum S., Bakkenist C., Schatten G., and Navara C.. 2009. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells. 27:1822–1835. 10.1002/stem.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe, M., Niero E.L., Fok W.C., Vessoni A.T., Jeong H.C., Brenner K.A., and Batista L.F.Z.. 2020. Telomere Dysfunction Activates p53 and Represses HNF4α Expression Leading to Impaired Human Hepatocyte Development and Function. Hepatology. 72:1412–1429. 10.1002/hep.31414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neganova, I., Zhang X., Atkinson S., and Lako M.. 2009. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 28:20–30. 10.1038/onc.2008.358 [DOI] [PubMed] [Google Scholar]
- Padgett, J., and Santos S.D.M.. 2020. From clocks to dominoes: lessons on cell cycle remodelling from embryonic stem cells. FEBS Lett. 594:2031–2045. 10.1002/1873-3468.13862 [DOI] [PubMed] [Google Scholar]
- Palm, W., and de Lange T.. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42:301–334. 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Prevo, R., Fokas E., Reaper P.M., Charlton P.A., Pollard J.R., McKenna W.G., Muschel R.J., and Brunner T.B.. 2012. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol. Ther. 13:1072–1081. 10.4161/cbt.21093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis, P., Van Ly D., Borel V., Kafer G.R., McCarthy A., Howell S., Blassberg R., Snijders A.P., Briscoe J., Niakan K.K., et al. 2021. TRF2-independent chromosome end protection during pluripotency. Nature. 589:103–109. 10.1038/s41586-020-2960-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar, J.C., Cortez D., and Cimprich K.A.. 2017. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18:622–636. 10.1038/nrm.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, H., Wu B., Delahaye F., Singer R.H., and Greally J.M.. 2019. Retargeting of macroH2A following mitosis to cytogenetic-scale heterochromatic domains. J. Cell Biol. 218:1810–1823. 10.1083/jcb.201811109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N., Mizumoto K., Nakamura M., and Tanaka M.. 2000. Radiation-induced centrosome overduplication and multiple mitotic spindles in human tumor cells. Exp. Cell Res. 255:321–326. 10.1006/excr.1999.4797 [DOI] [PubMed] [Google Scholar]
- Savage, S.A. 2018. Beginning at the ends: telomeres and human disease. F1000 Res. 7:524. 10.12688/f1000research.14068.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay, J.W. 2016. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 6:584–593. 10.1158/2159-8290.CD-16-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay, J.W., and Wright W.E.. 2019. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 20:299–309. 10.1038/s41576-019-0099-1 [DOI] [PubMed] [Google Scholar]
- Shay, J.W., Pereira-Smith O.M., and Wright W.E.. 1991. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 196:33–39. 10.1016/0014-4827(91)90453-2 [DOI] [PubMed] [Google Scholar]
- Shukla, S., Jeong H.C., Sturgeon C.M., Parker R., and Batista L.F.Z.. 2020. Chemical inhibition of PAPD5/7 rescues telomerase function and hematopoiesis in dyskeratosis congenita. Blood Adv. 4:2717–2722. 10.1182/bloodadvances.2020001848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, X., Cardenas-Diaz F.L., French D.L., and Gadue P.. 2016. A Doxycycline-Inducible System for Genetic Correction of iPSC Disease Models. Methods Mol. Biol. 1353:13–23. 10.1007/7651_2014_179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska, A., and de Lange T.. 2002. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21:4338–4348. 10.1093/emboj/cdf433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri, H., and Benchimol S.. 1996. From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: the telomere loss/DNA damage model of cell aging. Exp. Gerontol. 31:295–301. 10.1016/0531-5565(95)02025-X [DOI] [PubMed] [Google Scholar]
- Xu, C., Jiang J., Sottile V., McWhir J., Lebkowski J., and Carpenter M.K.. 2004. Immortalized fibroblast-like cells derived from human embryonic stem cells support undifferentiated cell growth. Stem Cells. 22:972–980. 10.1634/stemcells.22-6-972 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Hu Q., and Zhao Y.. 2017. Analysis of Telomere-Homologous DNA with Different Conformations Using 2D Agarose Electrophoresis and In-Gel Hybridization. Methods Mol. Biol. 1587:197–204. 10.1007/978-1-4939-6892-3_18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows karyotype analysis in iTERT and iTERT_TP53 KO hPSCs with long and short telomeres.