Abstract
Background
artificial intelligence (AI) for cellular phenotyping diagnosis of nasal polyps by whole-slide imaging (WSI) is lacking. We aim to establish an AI chronic rhinosinusitis evaluation platform 2.0 (AICEP 2.0) to obtain the proportion of inflammatory cells for cellular phenotyping diagnosis of nasal polyps and to explore the clinical significance of different phenotypes of nasal polyps on the WSI.
Methods
a total of 453 patients were enrolled in our study. For the development of AICEP 2.0, 179 patients (WSIs) were obtained from the Third Affiliated Hospital of Sun Yat-Sen University (3HSYSU) from January 2008 to December 2018. A total of 24,625 patches were automatically extracted from the regions of interest under a 400× HPF by Openslide and the number of inflammatory cells in these patches was counted by two pathologists. For the application of AICEP 2.0 in a prospective cohort, 158 patients aged 14–70 years old with chronic rhinosinusitis with nasal polyps (CRSwNP) who had undergone endoscopic sinus surgery at 3HSYSU from June 2020 to December 2020 were included for preoperative demographic characteristics. For the application of AICEP 2.0 in a retrospective cohort, 116 patients with CRSwNP who had undergone endoscopic sinus surgery from May 2016 to June 2017 were enrolled for the recurrence rate. The proportion of inflammatory cells of these patients on WSI was calculated by our AICEP 2.0.
Findings
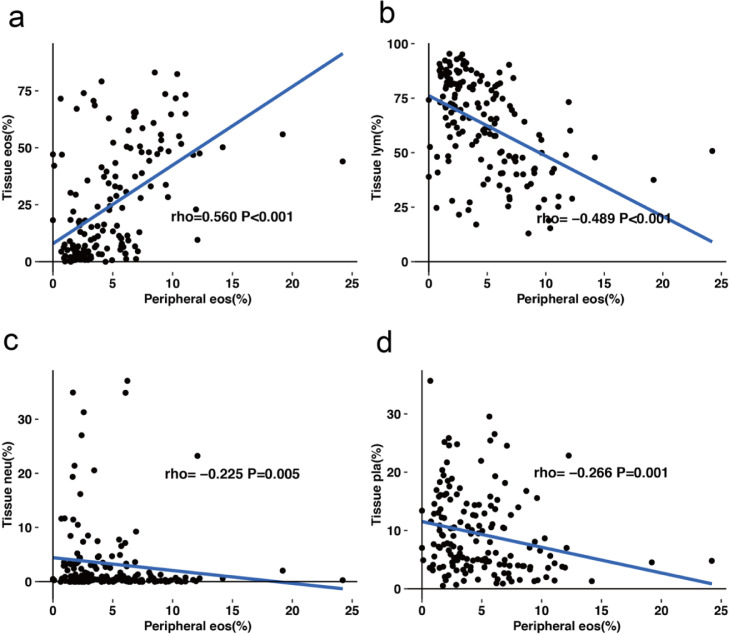
for AICEP 2.0, the mean absolute errors of the ratios of eosinophils, lymphocytes, neutrophils, and plasma cells were 1.64%, 2.13%, 1.06%, and 1.22%, respectively. The four phenotypes of nasal polyps were significantly different in clinical characteristics (including asthma, itching, sneezing, total IgE, peripheral eosinophils%, tissue eosinophils%, tissue neutrophils%, tissue lymphocytes%, tissue plasma cells%, and recurrence rate; P <0.05), but there were no significant differences in age distribution, onset time, total VAS score, Lund-Kennedy score, or Lund-Mackay score. The percentage of peripheral eosinophils was positively correlated with the percentage of tissue eosinophils (r = 0.560, P <0.001) and negatively correlated with tissue lymphocytes% (r = -0.489, P <0.001), tissue neutrophils% (r = -0.225, P = 0.005), and tissue plasma cells% (r = -0.266, P = 0.001) in WSIs.
Keywords: Chronic rhinosinusitis, Artificial intelligence, Nasal polyps, Deep learning, Cellular phenotype, Whole-slide imaging
Research in context.
Evidence before this study
Chronic rhinosinusitis (CRS) is a complex disease consisting of several diseases. Defining the phenotypes of CRS with nasal polyps (CRSwNP) with prognosis may lead to the delivery of personalized treatment. However, artificial intelligence (AI) for cellular phenotyping diagnosis of nasal polyps based on whole-slide imaging (WSI) is lacking. Literature searches were conducted separately using PubMed on 8 September 2020 with the terms (“artificial intelligence” OR “deep learning” OR “machine learning” OR “convolutional neural network”) AND (“nasal polyps” OR “chronic rhinosinusitis”) AND (“whole-slide imaging”), without date or language restrictions. There was only one study found, our previous study. The artificial intelligence (AI) CRS evaluation platform 1.0 (AICEP 1.0), which was established in our previous study, can only distinguish between eosinophilic chronic rhinosinusitis with nasal polyps (eCRSwNP) and non-eCRSwNP, but it failed to obtain the proportion of each inflammatory cell type.
Added value of this study
In this study, we established AICEP 2.0 to obtain the proportion of inflammatory cells for cellular phenotyping diagnosis of nasal polyps and explored the clinical significance of different phenotypes of nasal polyps on WSI.
Implications of the available evidence
The AICEP 2.0 platform extended the previous AICEP 1.0 by further analysing the cellular phenotypes of nasal polyps and can show the distribution concentration of four kinds of inflammatory cells in WSI by hot maps. Furthermore, this platform can help doctors make diagnoses and the decision-making process more easily and accurately.
Alt-text: Unlabelled box
Interpretation AICEP 2.0 was the first AI used for cellular phenotyping diagnosis of nasal polyps based on the proportions of inflammatory cells and WSI. The four phenotypes of nasal polyps based on WSI had different clinical characteristics, and this platform can predict different prognoses and lead to the delivery of personalized treatment. Furthermore, this method was the first confirmation that the percentage of peripheral blood eosinophils was positively correlated with the percentage of eosinophils in polyp tissue on WSI, and it could predict whether patients were eCRSwNP or not.
1. Introduction
Chronic rhinosinusitis (CRS) is a common condition in most of the world and affects 5–12% of the general population, leading to a significant burden on society in terms of healthcare consumption and productivity loss [[1], [2], [3]]. Chronic rhinosinusitis has traditionally been classified into chronic rhinosinusitis with nasal polyps (CRSwNP) and without nasal polyps (CRSsNP). The clinical phenotypes are predominantly eosinophilic CRS (eCRS) and non-eCRS, determined by the histologic quantification of the numbers of eosinophilic, i.e., number/high-power field, which the European position paper on rhinosinusitis and nasal polyps 2020 (EPOS 2020) agreed to be 10 per high-power field (HPF; 400×) or higher [4].
CRS is a complex disease consisting of several disease variants with different underlying pathophysiologies [5,6] Endotyping, based on the pathogenic mechanism, provides a precise picture that is more appropriate for use in clinical practice. However, endotyping CRSwNP is still a challenge for rhinologists [7]. Therefore, some studies defining the phenotypes of CRSwNP with prognosis may lead to the delivery of personalized treatment. Chinese CRSwNP patients may be classified into five phenotypes with different polyp recurrence rates based on the presence of predominantly plasma cells, lymphocytes, neutrophils, eosinophils, or mixed inflammatory cells in 10 random HPFs in the diagnosis of nasal polyps [8]. However, artificial intelligence (AI) for cellular phenotyping diagnosis of nasal polyps based on whole-slide imaging (WSI) is lacking.
Our studies have shown sampling errors among the estimates based on 10 random HPFs in the diagnosis of CRSwNP [9]. Therefore, we considered the ratio of eosinophils (RE) on the WSI as the criterion standard for assessing eCRS with nasal polyps (eCRSwNP) because of its lack of sampling error. The areas under the receiver operating characteristic curve from the internal validation and external test data sets of our artificial intelligence (AI) CRS evaluation platform 1.0 (AICEP 1.0) were 0.974 and 0.957, respectively [9].
However, AICEP 1.0 can only distinguish between eCRSwNP and non-eCRSwNP and it failed to obtain the proportion of each inflammatory cell on the WSI. Therefore, we aim to establish AICEP 2.0 to obtain the proportion of each inflammatory cell for cellular phenotyping diagnosis of nasal polyps and to explore the clinical significance of different phenotypes of nasal polyps on WSI.
2. Methods
2.1. Datasets
First, 179 patients were obtained from the Department of Otolaryngology in the Third Affiliated Hospital of Sun Yat-Sen University (3HSYSU) in China from January 2008 to December 2018 after screening for staining, size, and quality of specimens. The patients (one patient corresponding to one specimen) were randomly divided into 2 groups: 167 patients in the training dataset and 12 patients in the testing dataset. After that, all specimens were scanned through an automatic digital slide scanner (Panoramic 250 FLASH, 3DHISTECH Ltd, Budapest, Hungary), and 179 digital whole-slide imaging (WSIs) were obtained. The lamina propria of the mucosa was sketched, excluding large glands, through automated slide analysis platform software (ASAP, version 1.9, Radboud University Medical Center, The Netherlands) to yield regions of interest (ROIs). Finally, 24,625 patches were automatically extracted from the ROI of WSIs under a 400× HPF by Openslide (version 3.4.1, University of Pittsburgh, Pittsburgh, PA).
2.3. Labelling
For all 24,625 extracted patches, two competent pathologists identified and counted the number of eosinophils (n1), number of lymphocytes (n2), number of neutrophils (n3), and number of plasma cells (n4) of each patch. The number of infiltrating inflammatory cells was regarded as the sum (t = n1 + n2 + n3 + n4). Therefore, we defined the ratios of eosinophils (REpatch-actual), lymphocytes (RLpatch-actual), neutrophils (RNpatch-actual), and plasma cells (RPpatch-actual) as n1/t, n2/t, n3/t, and n4/t, respectively. When the two pathologists’ assessments of Rpatch-actual differed by less than 5%, the average value was used. If the difference was greater than 5%, the patch was rechecked by an expert pathologist who had more than 30 years of experience, and the value was corrected as necessary.
2.4. Development of the AICEP 2.0
A convolutional neural network (CNN) base model was used to extract the features of regions of interest (ROIs) generated from the WSI so that important information could be obtained for regression-fitting tasks.
In our previous study, we compared the counts from AICEP 1.0 with the counts done by the two pathologists [9]. The results showed that compared with Xception and Resnet50, InceptionV3 [10] had the highest AUC and was selected as the final model in AICEP 1.0. With the development of CNN models, we noticed that EfficientNet series models had significant advantages over any previous CNN models, such as Inception, ResNet, and DenseNet, in terms of accuracy and parameter quantity [11].
State-of-the-art EfficientNets are a new baseline network designed by neural architecture search and consist of a family of models. From B0 to B7, the corresponding accuracy increases, but the number of parameters also increases, which leads to a decrease in training and deployment efficiency. The EfficientNet B5 model was considered to be the most suitable base model because of the trade-off between fewer parameters and sufficiently high ImageNet Top-1 accuracy [11]. Thus, we chose the EfficientNet B5 to update AICEP1.0 to AICEP 2.0, which can predict the proportions of inflammatory cells. In our AICEP 2.0, the last layer of EfficientNet B5 was first removed, and then a new layer containing only four neurons was added following the penultimate layer to predict the four ratios of cells. The four neurons in the last layer did not exert any activation function so that the model could output unlimited predicted values.
Then, the datasets were randomly divided into a training dataset, validation dataset, and independent test dataset at a ratio of 3:1:1. The validation data were used to identify the best hyperparameters, such as learning rate (LR) and batch size. Next, the validation data were combined with the training data to form a larger training dataset to retrain AICEP 2.0 and to evaluate its performance on the independent test dataset.
In our study, AICEP 2.0 adopted the Adam optimizer with a learning rate of 8e-4 and batch size of 32. The best model parameter was saved when the mean absolute error (MAE) on the independent testing dataset was the lowest in 500 epochs. The deep learning architecture was Keras 2.2, and the programming language was Python 3.6. Our AICEP 2.0 was trained and tested on one Nvidia Tesla V100 GPU with 32 GB memory with the help of Matplotlib, Numpy, and Scikit-learn.
After the training, AICEP 2.0 can predict the proportions of inflammatory cells in each patch and show the distribution concentration of four kinds of inflammatory cells in WSI by hot maps.
2.5. Performance of AICEP 2.0
To evaluate the performance of AICEP 2.0, MAE, root mean square error (RMSE), R-squared, and explained variance score (EVS) were estimated. MAE was the mean value of the absolute value of error between the real value and the predicted value. RMSE was the square of the difference between the real value and the predicted value; then, the sum was averaged and the square root was obtained. R-squared and EVS represent the degree of the fitting, ranging from 0 to 1; the higher the value is, the better the fitting.
2.6. Application of AICEP 2.0 in the prospective study (Fig. 1)
Fig. 1.
A graphical abstract of the classification process for nasal polyps. (a), Patients with four different phenotypes of nasal polyps. (b), Digitization of the nasal polyp slide into the whole-slide imaging (WSI) by a scanner. (c), WSIs are split into small patches automatically. (d), Patches input into AICEP 2.0, which was trained by EfficientNet B5. (e), Output result with four different groups. (f), Management for different groups.
In our prospective study, 158 patients (117 male and 41 female), aged 14–70 years old with CRSwNP, who had undergone endoscopic sinus surgery (ESS) at 3HSYSU from June 2020 to December 2020 were included for preoperative demographic characteristics. In all, 13 patients (8.2%) had concomitant asthma and none were treated with corticosteroids within 4 weeks before surgery. The patients’ histories of asthma and allergies including acetylsalicylic acid intolerance were obtained from their medical records. For allergic patients, data were also collected from their preoperative clinical reports regarding their total and specific IgE, including for Dermatophagoides pteronyssinus and Dermatophagoides farinae, birch pollen, pellitory, grass mix, and cat and dog dander. The diagnosis of asthma was based on patients’ symptoms of episodic cough, wheezing and/or dyspnoea, and an accumulated dosage of methacholine provoking a 20% fall of forced expiratory volume in the first second (FEV1) <2.505 mg and/or ≥12% increase in FEV1 following inhalation of 200 μg salbutamol.
Prior to surgery, nasal symptoms (including rhinorrhea, nasal obstruction, olfactory dysfunction, and headache/facial pain) were assessed using a visual analogue scale (VAS) of 0 to 10, and a complete peripheral blood cell count and total IgE were performed. The nasal polyps were also scored according to the Lund-Kennedy and Lund-Mackay score staging system before surgery [12,13]. The proportion of each kind of inflammatory cell on the WSI was calculated by AICEP 2.0.
Plasma cell-dominant phenotypes and lymphocyte-dominant phenotypes belong to chronic inflammation and their prognoses are similar [8], so we classified them into plasma cells and lymphocyte-dominant (LP) phenotypes in our study. According to the ratios of inflammatory cells and the different recurrence rates of nasal polyps reported by Lou's study [8], we proposed four phenotypes of CRSwNP based on WSI. (1) Eosinophil-dominant (E) phenotypes, where the rate of eosinophils accounts for more than 60% of inflammatory cells. (2) LP phenotypes, where the rate of lymphocytes and plasma cells accounts for more than 60% of inflammatory cells. (3) E + LP phenotypes, where both the rate of eosinophils and the rate of lymphocytes and plasma cells account for more than 30% of inflammatory cells. (4) Neutrophils and LP-dominant (N + LP) phenotypes, where both the rate of neutrophils and the rate of lymphocytes and plasma cells account for more than 30% of inflammatory cells. In our study, the primary endpoints were the Lund-Kennedy score [12] and Lund-Mackay score [13] at month 36. Secondary endpoints included the total VAS score (including rhinorrhea, nasal obstruction, olfactory dysfunction, and headache/facial pain) at month 36. A score of 0 is given when a symptom is not present, and numbers up to 10 are given when a symptom is present, with 10 indicating the greatest severity [14].
2.7. Application of AICEP 2.0 in a retrospective cohort
116 patients with CRSwNP who had undergone endoscopic sinus surgery at 3HSYSU from May 2016 to June 2017 were retrospectively enrolled for the recurrence rate of the four phenotypes of nasal polyps. Recurrence of nasal polyps was characterized after surgery by nasal endoscopy as the presence of nasal polyps, together with bothersome symptoms persisting for at least one month [15]. All patients were followed-up postoperative for 37.58 ± 2.09 months.
2.8. Ethics statement
This study was approved by the Research Ethics Committee of the Institute of Basic Research in Clinical Medicine, Third Affiliated Hospital of Sun Yat-Sen University ([2020]02-001-01). The research was registered at the Chinese Clinical Trials Registry (http://www.chictr.org.cn/index.aspx) with the number ChiCTR2000033779. Written informed consent was obtained from all patients in this study.
2.9. Statistical analysis
All statistical analyses were performed by SPSS 17.0. Categorical variables are described as frequencies, and the chi-square test or Kruskal-Wallis method was performed to test group differences when appropriate. Continual variables are described as medians and interquartile ranges because of nonnormal distribution, and differences among different types were tested by the Kruskal-Wallis method. The correlation between the percentage of peripheral eosinophils and the percentage of polyp tissue cells in the WSI was evaluated by the Spearman correlation coefficient. All tests were two-sided, and P <0.05 was considered statistically significant.
2.10. Role of funding source
The Funders didn't have any role in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
-
(1)
Comparison between AICEP 2.0 and AICEP 1.0
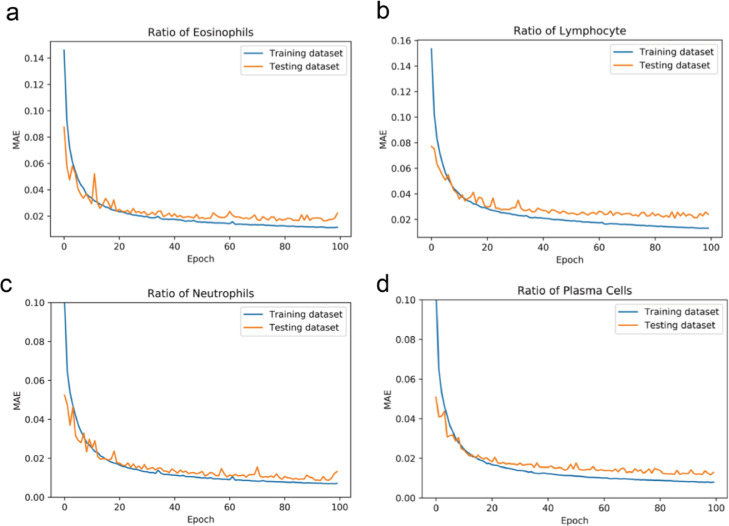
The functional differences and performance differences between AICEP 2.0 and AICEP 1.0 are shown in Tables 1 and 2, respectively. For AICEP 2.0, the MAEs of the ratio of eosinophils (RE), lymphocytes (RL), neutrophils (RN), and plasma cells (RP) were 1.64%, 2.13%, 1.06%, and 1.22%, respectively (Fig. 2).
-
(1)
Panoramic display of phenotypes of nasal polyps based on WSI
Table 1.
The functional differences between AICEP 2.0 and AICEP 1.0.
| Version | Neural Network | Panoramic Display | Quantification |
|---|---|---|---|
| AICEP 2.0 | EfficientNet B5 | Yes | four kinds of inflammatory cells |
| AICEP 1.0 | Inception V3 | No | eosinophils |
Table 2.
The performance differences between AICEP 2.0 and AICEP 1.0.
| Performance | Version | RE | RL | RN | RP |
|---|---|---|---|---|---|
| MAE (%) | AICEP 2.0 | 1.64 | 2.13 | 1.06 | 1.22 |
| AICEP 1.0 | 4.30 | -* | - | - | |
| RMSE (%) | AICEP 2.0 | 2.79 | 3.51 | 1.81 | 1.62 |
| AICEP 1.0 | 7.16 | - | - | - | |
| R2 | AICEP 2.0 | 0.98 | 0.97 | 0.96 | 0.97 |
| AICEP 1.0 | 0.92 | - | - | - | |
| EVS | AICEP 2.0 | 0.99 | 0.98 | 0.97 | 0.98 |
| AICEP 1.0 | 0.94 | - | - | - |
Fig. 2.
Performance of AICEP 2.0. (a), Mean absolute errors (MAE) of the ratio of eosinophils. (b), MAE of the ratio of lymphocytes. (c), MAE of the ratio of neutrophils. (d), MAE of the ratio of plasma cells.
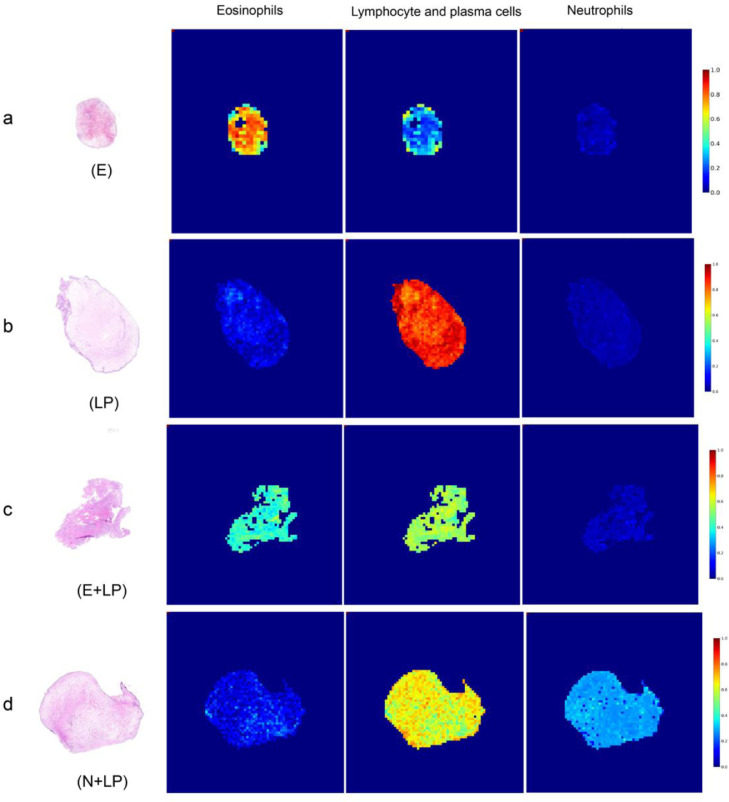
Fig. 3 a shows eosinophils -dominant phenotypes (E). The proportions of eosinophils, lymphocytes and plasma cells (LPs) and neutrophils were 68.8%, 28.8%, and 2.3%, respectively. Fig. 3 b shows lymphocyte- and plasma cell-dominant phenotypes (LPs). The proportions of eosinophils, LP, and neutrophils were 22.3%, 73.1%, and 4.7%, respectively. Fig. 3 c shows eosinophils, lymphocytes and plasma cell-dominant phenotypes (E + LP). The proportions of eosinophils, LP and neutrophils were 40.9%, 53.8% and 5.3%, respectively. Fig. 3 d shows neutrophil-, lymphocyte- and plasma cell-dominant phenotypes (N + LP). The proportions of eosinophils, LP, and neutrophils were 10.0%, 58.9%, and 31.1%, respectively.
-
(1)
Clinical characteristics of phenotypes of CRSwNP on WSI
Fig. 3.
Phenotypes of nasal polyp based on the WSI. (a), Eosinophils-dominant phenotypes (E). (b), Lymphocyte- and plasma cell-dominant phenotypes (LP). (c), Eosinophil-, lymphocyte- and plasma cell-dominant phenotypes (E+LP). (d), Neutrophil-, and lymphocyte- and plasma cell-dominant phenotypes (N+LP).
To investigate whether the patients in these subgroups were representative of distinct phenotypes of CRSwNP on WSI, the clinical and histological characteristics of these four phenotypes were compared (Tables 3 and 4).
Table 3.
Clinical characteristics of CRS subgroups.
| E (n =19) | LP (n =104) | E+LP (n=31) | N+LP (n=4) | χ2 | P | |
|---|---|---|---|---|---|---|
| Age | 47 (36,59) | 37 (29.25,52.75) | 37 (29,48) | 44.5 (22.25,64.5) | 2.971 | 0.396 |
| Gender (M/F) | 14/5 | 77/27 | 23/8 | 3/1 | 0.222 | 1.000 |
| Onset time (months) | 60 (24,120) | 48 (12,120) | 36(12,120) | 39 (4.5,288) | 1.142 | 0.767 |
| Itching (Y/N) | 8/11 | 15/89 | 20/11 | 0/4 | 31.306 | <0.001* |
| Sneezing (Y/N) | 8/11 | 17/87 | 21/10 | 0/4 | 31.334 | 0.013* |
| Asthma (Y/N) | 4/15 | 4/100 | 5/26 | 0/4 | 9.106 | 0.016* |
| Rhinorrhea score | 6 (5,7) | 6 (5,7) | 6 (5,7) | 6 (3,6.75) | 1.298 | 0.730 |
| Nasal obstruction score | 6 (6,7) | 6 (5,6.75) | 6 (5,6) | 6 (5.25,6.75) | 3.578 | 0.311 |
| Olfactory dysfunction score | 6 (5,7) | 5 (2.25,6) | 6 (3,8) | 6 (2.25,6.75) | 7.058 | 0.070 |
| Headache/facial pain score | 2 (1,6) | 2 (1,4.75) | 3 (1,5) | 3 (0.25,5) | 1.473 | 0.689 |
| Total VAS score | 21 (19,24) | 19 (15,21) | 21 (15,24) | 21.5 (10.75,24.75) | 6.397 | 0.094 |
| Lund-Kennedy score | 8 (6,10) | 8 (5,10) | 6 (4,10) | 6.5 (4.5,10.75) | 1.737 | 0.629 |
| Lund-Mackay score | 11 (9,16) | 11 (8,18) | 10 (8,14) | 16 (8,19.5) | 1.360 | 0.715 |
| Total IgE (IU/ml) (1^/2#/3&) | 5/3/11 | 59/12/33 | 5/14/12 | 4/0/0 | 15.878 | 0.001* |
| Peripheral lym% | 31.9 (29.09,34.32) | 32.7 (28.45,39.05) | 31.72 (27.48,36.92) | 31.49 (27.89,32.07) | 2.865 | 0.413 |
| Peripheral neu% | 53.17 (49.25,57.06) | 54.85 (48.24,59.92) | 49.16 (45.56,56.31) | 55.29 (51.1,61.55) | 5.452 | 0.142 |
| Peripheral eos% | 6.9 (3.5,9.87) | 2.9 (1.84,5) | 7.48 (5.23,9.66) | 4.32 (1.9,6.18) | 40.377 | <0.001* |
| Peripheral bas% | 0.46 (0.33,0.96) | 0.58 (0.34,0.66) | 0.71 (0.38,0.96) | 0.48 (0.28,1.13) | 3.355 | 0.340 |
| Recurrence$ (Y/N) | 12/4 | 5/66 | 15/11 | 1/2 | 44.459 | <0.001* |
E= eosinophil-dominant phenotypes; LP: plasma cells and lymphocyte-dominant phenotypes;
E+LP= eosinophils, plasma cells and lymphocyte-dominant phenotypes;
N+LP= neutrophils, plasma cells and lymphocyte-dominant phenotypes;
1^, Total IgE<100 IU/ml; 2#, 100 IU/ml <Total IgE<200 IU/ml; 3&, Total IgE>200 IU/ml.
$data from a retrospective cohort.
*P<0.05.
Table 4.
Histological characteristics of CRS subgroups.
| E(n=19) | LP(n=104) | E+LP(n=31) | N+LP(n=4) | χ2 | P | |
|---|---|---|---|---|---|---|
| Tissue eos% | 68.61 (64.92,73.6) | 6.39 (1.8,15.96) | 48.05 (43.42,51.77) | 6.41 (3.98,10.41) | 105.209 | <0.001* |
| Tissue neu% | 0.32 (0.06,0.71) | 1.06 (0.23,3.63) | 0.25 (0,0.6) | 34.92 (32.21,36.6) | 29.753 | <0.001* |
| Tissue lym% | 25.37 (21.65,29.66) | 76.55 (65.98,85.22) | 43.42 (40.48,48.5) | 49.45 (38.4,56.62) | 106.332 | <0.001* |
| Tissue pla% | 3.89 (1.48,4.94) | 10.05 (5.11,15.76) | 4.9 (3.58,11.54) | 6.91 (1.85,22.24) | 26.826 | <0.001* |
E= eosinophil-dominant phenotypes; LP: plasma cells and lymphocyte-dominant phenotypes;
E+LP= eosinophils, plasma cells and lymphocyte-dominant phenotypes;
N+LP= neutrophils, plasma cells and lymphocyte-dominant phenotypes;
*P<0.05.
The four phenotypes of CRSwNP on WSI were not significantly different in age distribution, onset time, total VAS score, Lund-Kennedy score, or Lund-Mackay score. However, the proportions of inflammatory cells (including eosinophils, neutrophils, lymphocytes, and plasma cells) in nasal polyps in these subgroups, were significantly different, all with P <0.05 (Kruskal-Wallis method). Phenotypes of E and E + LP comprised a significantly high proportion of allergy symptoms (itching and sneezing) and peripheral eosinophils, high total IgE and recurrence rate.
The E phenotype contained 12.0% (n =19) of the total patients, with 73.7% (14/19) being male. It had a higher rate of allergy symptoms (42.1%), the worst subjective symptoms (total VAS score), and the highest proportion of tissue eosinophils, rate of comorbid asthma (21.0%) and recurrence rate (75%).
The LP phenotype occurred in 65.8% (n =104) of the total patients was the most common phenotype, and 74.0% (77/104) were male. It had the highest proportion of peripheral lymphocytes and tissue lymphocytes, and the lowest recurrence rate (7%).
The E + LP phenotype contained 12.1% (n = 31) of the total patients, with 74.2% (23/31) being male. It had the highest rate of allergy symptoms (itching 64.5% and sneezing 67.7%), the highest proportion of peripheral eosinophils and basophils, and a higher rate of comorbid asthma (19.2%) and recurrence rate (57.7%).
The N+LP phenotype comprised only 2.5% (n = 4) of the total patients, and 75% (3/4) were male. It had the highest proportion of tissue neutrophils, without allergy symptoms.
-
(1)
Correlation between percentage of peripheral eosinophils (peripheral eos%) and percentage of inflammatory cells on WSI in polyp tissue
Peripheral eos% was positively correlated with tissue eosinophils% (r = 0.560, P <0.001) and negatively correlated with tissue lymphocytes% (r = -0.489, P <0.001), tissue neutrophils% (r = -0.225, P = 0.005) and tissue plasma cells% (r = -0.266, P = 0.001; Spearman correlation coefficient) in the WSI (Fig. 4).
Fig. 4.
Correlation between percentage of peripheral eosinophils (peripheral eos%) and percentage of inflammatory cells in polyp tissue in WSI. (a), Peripheral eos% and tissue eosinophils%. (b), Peripheral eos% and tissue lymphocyte%. (c), Peripheral eos% and tissue neutrophils%. (d), Peripheral eos% and tissue plasma cells%.
4. Discussion
To the best of our knowledge, AICEP 2.0 was the first AI used for cellular phenotyping diagnosis of nasal polyps on WSI. The four phenotypes of nasal polyps on WSI have different clinical characteristics, and they can predict different prognoses and lead to the delivery of personalized treatment.
Our previous study showed sampling errors among the estimates based on 10 random HPFs in the diagnosis of CRSwNP. Because the total count of inflammatory cells in WSI is time-consuming and laborious, the load of inflammatory cells has not received attention. Our AICEP 1.0 addressed the quantification of eosinophils, and its MAE for eosinophils in internal validation data was 4.3%. However, this method cannot quantify other inflammatory cells and cannot identify the local distribution of various inflammatory cells and the identification of cell accumulation areas [9]. In this study, AICEP 2.0 is the first AI used to quantify all inflammatory cells in the WSI and to visualize the density of each inflammatory cell distribution. Compared with AICEP 1.0, the state-of-the-art EfficientNet B5 model was employing in AICEP 2.0. The MAE of AICEP 2.0 for eosinophils was 1.62% which was lower than that of AICEP 1.0. Moreover, compared with the 2–3 h of manual diagnosis, AICEP 2.0 only takes a few minutes to ten minutes to diagnose a patient. Understanding the number of inflammatory cells in nasal polyps and their distribution in WSI may further improve our diagnosis and provide more personalized treatment. In summary, AICEP 2.0 can help doctors make diagnoses and decisions more easily and accurately. Furthermore, to a certain extent, the use of AICEP 2.0 can save some manpower and material resources in China, which has a large population but not enough doctors.
A new clinical classification for CRS based on the disease being localized or diffuse is proposed by EPOS 2020 [4]. Both of these groups can be further divided into type 2 or no-type 2 disease. However, recent studies with monoclonal antibodies directed at type 2 endotypes have not found reliable biomarkers to predict reaction to medication [16,17]. Currently, the combination of phenotype (e.g., CRSwNP), response to treatment and markers such as eosinophils in either blood or tissue leads us to the best estimation of the endotype and reaction to treatment. Brescia's study showed that cluster analysis can identify different histotypes among CRSwNP patients [18], and some studies identified clusters of CRS based on eosinophilic inflammation and clinical presentation in Asian patients or white patients using cluster analysis [19,20]. However, their study did not consider the prognosis of CRSwNP as a characterization factor. Thus, Lou and his colleagues used cluster analysis to generate 5 clusters with the recurrence rate of nasal polyps based on clinical and pathological data from a large cohort of Chinese patients with CRSwNP [8].
Based on Lou's study [8], the prognosis of plasma cell-dominant phenotypes and lymphocyte-dominant phenotypes was similar, and both lymphocytes and plasma cells are chronic inflammatory cells with similar morphology. Therefore, to facilitate clinical work, we classified cases as plasma cell and lymphocyte-dominant (LP) phenotypes and changed its five phenotypes into four phenotypes. The cut-off values of our four types of cells are similar to Lou's study [8], but our classification was based on the WSI and combined with AICEP 2.0 is more convenient for the clinic.
Our results showed that the four phenotypes of nasal polyps had significant differences in asthma, itching, sneezing, total IgE, peripheral eos%, tissue eos%, tissue lym%, and recurrence rate but no significant differences in polyp scores or severity of other symptoms. These results were consistent with the findings of other studies [5,8,21].
Our study demonstrated that the E phenotype and E + LP phenotype had a higher proportion of eosinophils in tissues and peripheral blood and a higher rate of comorbid asthma and recurrence rate than the other two phenotypes, which was similar to cluster 5 (eosinophil-dominant phenotype; a highest recurrence rates of 98.5%) and cluster 3 (mixed inflammatory cell phenotype; a higher recurrence rates of 75%) by Lou [8]. A tissue eosinophil proportion of 27% of total cells or a tissue eosinophil absolute count of 55 eosinophils/HPF may act as a reliable prognostic indicator for nasal polyp recurrence within 2 years after surgery [22]. Another study demonstrated that when linked to comorbid asthma, eosinophilic nasal polyps often represented a form of severe eosinophilic airway inflammation and patients experienced a high incidence of recurrence within 5 years [23]. Moreover, the higher-risk CRSwNP groups could be identified better by a three-variable panel (age<65 years, serum basophil percentage, and eosinophilic type) which had an area under the receiver operating characteristic curve (AUC) of 0.7028.[24] Additionally, our study also showed that the E+LP phenotype had the highest proportion of peripheral blood eosinophils and basophils. It is known that basophils are related to the severity of eosinophilic inflammation [25,26].
Moreover, the LP phenotype of our study modified from clusters1 and 2 had the highest proportion of peripheral lymphocytes and tissue lymphocytes. The N + LP phenotype, similar to cluster 4, had the highest proportion of tissue neutrophils, without allergy symptoms [8].
Therefore, our four phenotypes of the CRSwNP on the WSI had different clinical characteristics (including asthma, peripheral eos%, tissue eos%, tissue lym% and recurrence rate) which were consistent with other studies [5,8,21]. These phenotypes do help to diagnose and may predict different prognoses. The higher the proportion of eosinophils is, the worse the prognosis is [8,27]. This early prediction was conducive to the early use of biological agents (which can reduce eosinophilia), reduce the recurrence of nasal polyps and improve the prognosis of patients [28].
It is well known that high eosinophilic infiltration in polyps predicts worse outcomes and a higher risk for polyp recurrence after surgical treatment [27]. The identification of an eosinophilic phenotype is therefore important in guiding the management of these patients. Some studies showed that blood eosinophils were significantly correlated with eCRSwNP [29,30], and blood eosinophil number (109/L) was positively correlated with polyp tissue eosinophil number/HPF [31]. However, there is a lack of studies that investigate the relationship between blood eosinophils and polyp tissue eosinophils in WSI. Our study demonstrated that the percentage of peripheral blood eosinophils was positively correlated with the percentage of eosinophils in polyp tissue in the WSI.
In this study, we also found a negative correlation between peripheral eos% and tissue lymphocytes%, tissue neutrophils% and tissue plasma cells%. No reports discuss the relationship of peripheral eos% with lymphocytes%, neutrophils% and plasma cells% of nasal polyps. However, the numbers of neutrophils and the mucous gland area were increased and the numbers of eosinophils were reduced in transverse sections of large and small airways from patients with asthma [32]. The numbers of lymphocytes correlated with the numbers of eosinophils in the fatal asthma group (r = 0.60; p <0.0001) and to a lesser extent in the nonfatal (r = 0.34; p = 0.001) and control groups (r = 0.32; p = 0.001) [33]. Therefore, more studies are needed to reveal the relationship of peripheral eos% with lymphocytes%, neutrophils% and plasma cells% of nasal polyps.
The strengths in our study are as follows. (1) The AICEP 2.0 platform extended the previous AICEP 1.0 by further analysing the cellular phenotypes of nasal polyps and can show the distribution concentration of four kinds of inflammatory cells in WSI by hot maps. (2) AICEP 2.0 achieved better performance than version 1.0 under four different metrics. (3) The four phenotypes of the CRSwNP on the WSI had different clinical characteristics and can predict different prognoses and lead to the delivery of personalized treatment.
However, there are some limitations in our study that should be mentioned. First, the real-world diagnostic accuracy of AI was lower than that reported in a previous study conducted with screening data sets [34]. Our study showed a similar result: AICEP performed better in the internal validation data set than in the external test data set. In our study, the internal training data set and the data validation set came from a similar process regarding slicing, staining, and WSI scanning, whereas these aspects may differ in the external test data set [9]. Thus, to make AICEP 2.0 more generalisable to other populations of patients with CRSwNP, we are trying to use tissue samples from different areas of China and samples from non-Asian populations and to include more pathologists from multiple centers to construct a cloud-based multi-institutional AI platform in the follow-up experiment. Second, each WSI is approximately 2–5 GB, which poses a major challenge to the transmission between countries and regions. With the 5G and cloud platforms widely used, these problems will be solved in the future. Finally, because patients with nasal polyps often need to be followed up for 36 months and our patients in the prospective study were included from June 2020 to December 2020, the primary and secondary endpoints and the prognosis of our four phenotypes of CRSwNP in our study are currently unclear. Therefore, our next step will be to investigate, in a larger series and a long time for regular follow-up, the clinical (e.g., prognostic) implications of identifying four phenotypes of CRSwNP on WSI.
In conclusion, AICEP 2.0 was the first AI for cellular phenotyping diagnosis of nasal polyps by WSI based on the proportions of inflammatory cells. The four phenotypes of nasal polyps have different clinical characteristics, and they may predict different prognoses and lead to the delivery of personalized treatment. Furthermore, the percentage of peripheral blood eosinophils was positively correlated with the percentage of eosinophils in polyp tissue on WSI and may predict whether patients were eCRSwNP.
Contributors
Conception and design: L. Han and Q. Yang.
Collection and assembly of data: Q. Wu, J. Chen, Y. Ren and H. Qiu.
Data analysis and interpretation: L. Yuan, H. Deng, Y. Zhang, R. Zheng, H. Hong and Y. Sun.
Manuscript writing: Q. Wu, X. Wang, X. Huang and C. Shao.
Revised advisor: H. Lin.
Final approval of manuscript: All authors.
Data sharing statement
As the study involved human participants, the data cannot be made freely available in the manuscript, nor a public repository because of ethical restrictions. However, the data are available from the Third Affiliated Hospital of Sun Yat-Sen University for researchers who meet the criteria for access to confidential data. Interested researchers can send data access requests to the corresponding author (yangqint@mail.sysu.edu.cn).
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
The authors would like to thank their colleagues (Department of Pathology, The Third Affiliated Hospital of Sun Yat-Sen University) for their help. This work was supported by the National Natural Science Foundation of China (Grant No. U20A20399 and 81870704), the Key-area Research and Development Program of Guangdong Province (No.2020B0101130015), Sun Yat-Sen University Clinical Research 5010 Programme (No.2019006) and The Third Affiliated Hospital of Sun Yat-Sen University, Clinical Research Programme (No. QHJH201901).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103336.
Contributor Information
Lanqing Han, Email: hanlance@tsinghua-gd.org.
Qintai Yang, Email: yangqint@mail.sysu.edu.cn.
Appendix. Supplementary materials
References
- 1.Hastan D, Fokkens WJ, Bachert C. Chronic rhinosinusitis in Europe–an underestimated disease. A GA²LEN study. Allergy. 2011;66:1216–1223. doi: 10.1111/j.1398-9995.2011.02646.x. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch AG, Stewart WF, Sundaresan AS. Nasal and sinus symptoms and chronic rhinosinusitis in a population-based sample. Allergy. 2017;72:274–281. doi: 10.1111/all.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundaresan AS, Hirsch AG, Storm M. Occupational and environmental risk factors for chronic rhinosinusitis: a systematic review. Int Forum Allergy Rhinol. 2015;5 doi: 10.1002/alr.21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fokkens WJ, Lund VJ, Hopkins C. European Position paper on rhinosinusitis and nasal polyps 2020. Rhinology. 2020;58 doi: 10.4193/Rhin20.600. [DOI] [PubMed] [Google Scholar]
- 5.Wei B, Liu F, Zhang J. Multivariate analysis of inflammatory endotypes in recurrent nasal polyposis in a Chinese population. Rhinology. 2018;56:216–226. doi: 10.4193/Rhin17.240. [DOI] [PubMed] [Google Scholar]
- 6.Tomassen P, Vandeplas G, Van Zele T. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137 doi: 10.1016/j.jaci.2015.12.1324. [DOI] [PubMed] [Google Scholar]
- 7.Brescia G, Zanotti C, Parrino D. Nasal polyposis pathophysiology: endotype and phenotype open issues. Am J Otolaryngol. 2018;39:441–444. doi: 10.1016/j.amjoto.2018.03.020. 2018/03/20. [DOI] [PubMed] [Google Scholar]
- 8.Lou H, Meng Y, Piao Y. Cellular phenotyping of chronic rhinosinusitis with nasal polyps. Rhinology. 2016;54:150–159. doi: 10.4193/Rhin15.271. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Chen J, Deng H. Expert-level diagnosis of nasal polyps using deep learning on whole-slide imaging. J Allergy Clin Immunol. 2020;145 doi: 10.1016/j.jaci.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Christian Szegedy, Vincent Vanhoucke, Sergey Ioffe, Jonathon Shlens, Zbigniew Wojna. Rethinking the inception architecture for computer vision. arXiv Preprint arXiv:1512.00567.
- 11.Mingxing Tan, Quoc.V. Le. EfficientNet: rethinking model scaling for convolutional neural networks. arXiv Preprint arXiv:1905.11946.
- 12.Lund V.J., Mackay I.S. Staging in rhinosinusitus. Rhinology. 1993;31:183–184. 1993/12/01. [PubMed] [Google Scholar]
- 13.Lund V.J., Kennedy D.W. Staging for rhinosinusitis. Otolaryngol Head Neck Surg. 1997;117 doi: 10.1016/s0194-5998(97)70005-6. S35-40. 1997/10/23. [DOI] [PubMed] [Google Scholar]
- 14.Lund V.J., Kennedy D.W. Quantification for staging sinusitis. Ann Otol Rhinol Laryngol Suppl. 1995;167:17–21. [PubMed] [Google Scholar]
- 15.Vlaminck S., Vauterin T., Hellings P.W. The importance of local eosinophilia in the surgical outcome of chronic rhinosinusitis: a 3-year prospective observational study. Am J Rhinol Allergy. 2014;28:260–264. doi: 10.2500/ajra.2014.28.4024. [DOI] [PubMed] [Google Scholar]
- 16.Bachert C., Sousa A.R., Lund V.J. Reduced need for surgery in severe nasal polyposis with mepolizumab: randomized trial. J Allergy Clin Immunol. 2017;140 doi: 10.1016/j.jaci.2017.05.044. [DOI] [PubMed] [Google Scholar]
- 17.Bachert C, Han JK, Desrosiers M. Efficacy and safety of dupilumab in patients with severe chronic rhinosinusitis with nasal polyps (LIBERTY NP SINUS-24 and LIBERTY NP SINUS-52): results from two multicenter, randomized, double-blind, placebo-controlled, parallel-group phase 3 trials. Lancet. 2019;394:1638–1650. doi: 10.1016/S0140-6736(19)31881-1. [DOI] [PubMed] [Google Scholar]
- 18.Brescia G, Alessandrini L, Giacomelli L. A classification of chronic rhinosinusitis with nasal polyps based on structured histopathology. Histopathology. 2020;76:296–307. doi: 10.1111/his.13969. 2019/08/14. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama T, Asaka D, Yoshikawa M. Identification of chronic rhinosinusitis phenotypes using cluster analysis. Am J Rhinol Allergy. 2012;26:172–176. doi: 10.2500/ajra.2012.26.3749. [DOI] [PubMed] [Google Scholar]
- 20.Soler ZM, Hyer JM, Ramakrishnan V. Identification of chronic rhinosinusitis phenotypes using cluster analysis. Int Forum Allergy Rhinol. 2015;5:399–407. doi: 10.1002/alr.21496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wynn R., Har-El G. Recurrence rates after endoscopic sinus surgery for massive sinus polyposis. Laryngoscope. 2004;114:811–813. doi: 10.1097/00005537-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Lou H, Meng Y, Piao Y. Predictive significance of tissue eosinophilia for nasal polyp recurrence in the Chinese population. Am J Rhinol Allergy. 2015;29:350–356. doi: 10.2500/ajra.2015.29.4231. 2015/07/30. [DOI] [PubMed] [Google Scholar]
- 23.Matsuwaki Y, Ookushi T, Asaka D. Chronic rhinosinusitis: risk factors for the recurrence of chronic rhinosinusitis based on 5-year follow-up after endoscopic sinus surgery. Int Arch Allergy Immunol. 2008;146(Suppl 1):77–81. doi: 10.1159/000126066. 2008/06/25. [DOI] [PubMed] [Google Scholar]
- 24.Brescia G, Marioni G, Franchella S. Can a panel of clinical, laboratory, and pathological variables pinpoint patients with sinonasal polyposis at higher risk of recurrence after surgery? Am J Otolaryngol. 2015;36:554–558. doi: 10.1016/j.amjoto.2015.01.019. 2015/02/24. [DOI] [PubMed] [Google Scholar]
- 25.Kagoya R, Kondo K, Baba S. Correlation of basophil infiltration in nasal polyps with the severity of chronic rhinosinusitis. Ann Allergy Asthma Immunol. 2015;114:30–35. doi: 10.1016/j.anai.2014.09.017. 2014/12/03. [DOI] [PubMed] [Google Scholar]
- 26.Pawankar R, Lee KH, Nonaka M. Role of mast cells and basophils in chronic rhinosinusitis. Clin Allergy Immunol. 2007;20:93–101. 2007/05/31. [PubMed] [Google Scholar]
- 27.Grgić MV, Ćupić H, Kalogjera L. Surgical treatment for nasal polyposis: predictors of outcome. Eur Arch Otorhinolaryngol. 2015;272:3735–3743. doi: 10.1007/s00405-015-3519-7. 2015/01/31. [DOI] [PubMed] [Google Scholar]
- 28.Massanari M, Holgate ST, Busse WW. Effect of omalizumab on peripheral blood eosinophilia in allergic asthma. Respir Med. 2010;104:188–196. doi: 10.1016/j.rmed.2009.09.011. 2009/10/23. [DOI] [PubMed] [Google Scholar]
- 29.Xu M, Zhang W, Chen D. Diagnostic significance of serum periostin in eosinophilic chronic sinusitis with nasal polyps. Acta Otolaryngol. 2018;138:387–391. doi: 10.1080/00016489.2017.1388540. 2017/10/31. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y, Cao P-P, Liang G-T. Diagnostic significance of blood eosinophil count in eosinophilic chronic rhinosinusitis with nasal polyps in Chinese adults. The Laryngoscope. 2012;122:498–503. doi: 10.1002/lary.22507. [DOI] [PubMed] [Google Scholar]
- 31.Zuo K, Guo J, Chen F. Clinical characteristics and surrogate markers of eosinophilic chronic rhinosinusitis in Southern China. European archives of oto-rhino-laryngology. 2014;271:2461–2468. doi: 10.1007/s00405-014-2910-0. [DOI] [PubMed] [Google Scholar]
- 32.Carroll N, Carello S, Cooke C. Airway structure and inflammatory cells in fatal attacks of asthma. Eur Respir J. 1996;9:709–715. doi: 10.1183/09031936.96.09040709. [DOI] [PubMed] [Google Scholar]
- 33.Carroll N, Cooke C, James A. The distribution of eosinophils and lymphocytes in the large and small airways of asthmatics. Eur Respir J. 1997;10:292–300. doi: 10.1183/09031936.97.10020292. [DOI] [PubMed] [Google Scholar]
- 34.Long E, Lin H, Liu Z. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nat Biomed Eng. 2017;1 doi: 10.1038/s41551-016-0024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.