Murine hepatitis virus (MHV) strain 3, one of the most important inducers of viral hepatitis, has been extensively studied as an organism to gain a better understanding of coronavirus biology and pathogenesis. Only one sequence is currently available.
ABSTRACT
Murine hepatitis virus (MHV) strain 3, one of the most important inducers of viral hepatitis, has been extensively studied as an organism to gain a better understanding of coronavirus biology and pathogenesis. Only one sequence is currently available. Another representative isolate has now been sequenced and added to the arsenal of MHV-3 variants.
ANNOUNCEMENT
The group of coronaviruses (CoV) includes several species that infect a variety of hosts (1). Murine hepatitis virus (MHV) belongs to the family Coronaviridae, subfamily Orthocoronavirinae, and genus Betacoronavirus and contains a positive-sense single-stranded RNA genome sequence approximately 25 to 31 kb in size (2). With Mus musculus as the main host, MHV encompasses a set of well-described more virulent (MHV-2, MHV-3, MHV-A59, and MHV-JHM) and less virulent (MHV-1, MHV-S, MHV-Y, and MHV-Nu) strains (3). It is noteworthy that, since MHV shares the same genus (β-coronavirus) as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), MHV together with mouse models could offer, through a translational approach, mechanistic insight into the SARS-CoV-2 biology and pathogenesis (3, 4).
MHV-3, stored at the Laboratory of Virology, University of Campinas (UNICAMP), Brazil, originally from the Culture Collection Laboratory of Animal Quality Control, Multidisciplinary Center for Biological Investigation on Laboratory Animal Science (CEMIB), UNICAMP, was grown on cell line L-929 (ATCC CCL-1). RNA was extracted from the supernatant (centrifugation at 2,000 rpm for 10 min) using a viral RNA minikit (Qiagen), and cDNA synthesis was performed using random primers with the high-capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer’s instructions. PCR was performed with primers designed from the primalscheme tool (ZiBRA project), aiming to produce 36 overlapping amplicons (Table 1), and their products were purified and quantified using the ExoSAP-IT Express PCR product cleanup reagent and Qubit v.2.0 fluorometer (Invitrogen), respectively. Next, the MHV-3 coding genome was Sanger sequenced at both ends using a primer walking approach. Genome assembly and full annotation (based on GenBank accession numbers FJ647224.1 and NC_048217.1) were performed using the Unipro UGENE v.37.1, Geneious v.9.1, and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) platforms.
TABLE 1.
Primers used for whole-genome sequencing of MHV-3 from UNICAMP
| Primer | Sequence | Size (nucleotides) | Primer positiona |
|---|---|---|---|
| MHV-3_1_LEFT | 5′-ACTTTATAAACGGCACTTCCTGC-3′ | 23 | 47–1048 |
| MHV-3_1_RIGHT | 5′-GCCACACAGGGAATAACTCCTT-3′ | 22 | |
| MHV-3_2_LEFT | 5′-GCTATCGCGGTGTTAATCCCAT-3′ | 22 | 917–1935 |
| MHV-3_2_RIGHT | 5′-GCGGGGCACTAAACCATCTAAT-3′ | 22 | |
| MHV-3_3_LEFT | 5′-AATAGGGGCGACTACAGTCTCC-3′ | 22 | 1804–2808 |
| MHV-3_3_RIGHT | 5′-AAACTCTGAACACGCCTTCGAA-3′ | 22 | |
| MHV-3_4_LEFT | 5′-TCAATGCTGGAGGTTTCCTTGC-3′ | 22 | 2658–3694 |
| MHV-3_4_RIGHT | 5′-TAATTGACTTGGGCACGCTCTT-3′ | 22 | |
| MHV-3_5_LEFT | 5′-GCTGGCTGCGTTCTACTTTGAT-3′ | 22 | 3548–4568 |
| MHV-3_5_RIGHT | 5′-ACATTCTTTGTCACAACACCAAGT-3′ | 24 | |
| MHV-3_6_LEFT | 5′-ACTTTTAGAACGTGCCTATCAGCA-3′ | 24 | 4437–5447 |
| MHV-3_6_RIGHT | 5′-GACACAAACCTTAGTGGCTTGC-3′ | 22 | |
| MHV-3_7_LEFT | 5′-AAGCAGGCGAATAACAATTGCT-3′ | 22 | 5314–6305 |
| MHV-3_7_RIGHT | 5′-CTCAAAGATGCATCACCATGGC-3′ | 22 | |
| MHV-3_8_LEFT | 5′-TTACAGAATGGCCAGCAGCTAC-3′ | 22 | 6173–7198 |
| MHV-3_8_RIGHT | 5′-TTCTATAGCCCACATCCGATGC-3′ | 22 | |
| MHV-3_9_LEFT | 5′-TCTGCCTAATATTGGATTCTTCCCT-3′ | 25 | 7080–8087 |
| MHV-3_9_RIGHT | 5′-TCCACCATCAGCATCGGTCTAT-3′ | 22 | |
| MHV-3_10_LEFT | 5′-GTGTGCCTGAAACCCATGTAGT-3′ | 22 | 7976–8944 |
| MHV-3_10_RIGHT | 5′-TAACAGCAACCACAACAGGACA-3′ | 22 | |
| MHV-3_11_LEFT | 5′-TAATGGTGTGCTACGGGATGTG-3′ | 22 | 8805–9825 |
| MHV-3_11_RIGHT | 5′-GTTTGAAACAACGACTGCTGCA-3′ | 22 | |
| MHV-3_12_LEFT | 5′-CAACCCTTTATTTCCCATCGGAGA-3′ | 24 | 9707–10672 |
| MHV-3_12_RIGHT | 5′-CGGTGTGACAACCAGTACTCAA-3′ | 22 | |
| MHV-3_13_LEFT | 5′-CCCAAGGAGCCTTCCATGTTAC-3′ | 22 | 10517–11521 |
| MHV-3_13_RIGHT | 5′-AGTACCACATAGACACCAGGCT-3′ | 22 | |
| MHV-3_14_LEFT | 5′-GGTGTCGTGTTGCTAGTTGCTA-3′ | 22 | 11407–12377 |
| MHV-3_14_RIGHT | 5′-ATACGTTCGAGCTTACGTGCAA-3′ | 22 | |
| MHV-3_15_LEFT | 5′-AGGCTAGTGGCTCTGCTAATCA-3′ | 22 | 12266–13232 |
| MHV-3_15_RIGHT | 5′-CAATTAGTGACAGGAGCGCCAC-3′ | 22 | |
| MHV-3_16_LEFT | 5′-ATCGTCGACGGTAAGATTGCAG-3′ | 22 | 13089–14088 |
| MHV-3_16_RIGHT | 5′-CAGTGTTAAGCAGGGCCCTATT-3′ | 22 | |
| MHV-3_17_LEFT | 5′-CCTATGCTGAGTGTGAAGAGTCC-3′ | 23 | 13963–14969 |
| MHV-3_17_RIGHT | 5′-ACACCCACCCTCATAGATCTCA-3′ | 22 | |
| MHV-3_18_LEFT | 5′-AGGATGGTAATGCTGCTATTACTGA-3′ | 25 | 14839–15828 |
| MHV-3_18_RIGHT | 5′-CCTTGGACGCGAACTCTGAATT-3′ | 22 | |
| MHV-3_19_LEFT | 5′-TGACCCCGCATTTGTTAGTGAG-3′ | 22 | 15719–16701 |
| MHV-3_19_RIGHT | 5′-ACAATTTAAGGCGCTCGGTACA-3′ | 22 | |
| MHV-3_20_LEFT | 5′-ACAATCTTGTACTGGTTCGCCC-3′ | 22 | 16586–17548 |
| MHV-3_20_RIGHT | 5′-GTTCCTTTATTCAGCAGCACACG-3′ | 23 | |
| MHV-3_21_LEFT | 5′-CCCGAGTTGGTGACTGACATTA-3′ | 22 | 17391–18407 |
| MHV-3_21_RIGHT | 5′-CCCAATGCTATCACGTATCGCA-3′ | 22 | |
| MHV-3_22_LEFT | 5′-GCTTGACTTGACCCTTGATGGT-3′ | 22 | 18284–19295 |
| MHV-3_22_RIGHT | 5′-CGTGTCGAACCTACACACAACT-3′ | 22 | |
| MHV-3_23_LEFT | 5′-ATGATGCCTCGCCTGTTGTTAA-3′ | 22 | 19147–20149 |
| MHV-3_23_RIGHT | 5′-ACAACTACGCCATTTAGCTCGG-3′ | 22 | |
| MHV-3_24_LEFT | 5′-TGATGGTCGTGATAATGGTGCT-3′ | 22 | 20018–21018 |
| MHV-3_24_RIGHT | 5′-CCTTATCTGACCCTGCACCAAG-3′ | 22 | |
| MHV-3_25_LEFT | 5′-CTCTGGAATTATGGCAAGCCGA-3′ | 22 | 20868–21902 |
| MHV-3_25_RIGHT | 5′-TCCACCTGTTTGTATTGTTCTGCT-3′ | 24 | |
| MHV-3_26_LEFT | 5′-TCCATTGGCCCAATTTAGTGGC-3′ | 22 | 21753–22775 |
| MHV-3_26_RIGHT | 5′-ACCTGAATGGCACAACTCTTGG-3′ | 22 | |
| MHV-3_27_LEFT | 5′-TGATGACTGGTTCCTCTTTGGC-3′ | 22 | 22664–23636 |
| MHV-3_27_RIGHT | 5′-CGTGCAAGGAAATACACTGCAC-3′ | 22 | |
| MHV-3_28_LEFT | 5′-TGTCAGCCGCCATATTGTTTCT-3′ | 22 | 23519–24528 |
| MHV-3_28_RIGHT | 5′-ACGCATAAAAAGTACCACCCTGT-3′ | 23 | |
| MHV-3_29_LEFT | 5′-GCTTTTGGCACACAGATGTCAA-3′ | 22 | 24409–25420 |
| MHV-3_29_RIGHT | 5′-CTCCTAGAACATTCCCGATGCG-3′ | 22 | |
| MHV-3_30_LEFT | 5′-TGCCTATGCCCAGCAATGTTTT-3′ | 22 | 25277–26275 |
| MHV-3_30_RIGHT | 5′-CCATCAATGGCTTGAACACTATCA-3′ | 24 | |
| MHV-3_31_LEFT | 5′-GGTGCTGGACTATGCGTTGATT-3′ | 22 | 26145–27119 |
| MHV-3_31_RIGHT | 5′-AGAAGCACTAATAGCGCCAAAC-3′ | 22 | |
| MHV-3_32_LEFT | 5′-TGGGTTCGATGCAACCAATTCT-3′ | 22 | 26999–27977 |
| MHV-3_32_RIGHT | 5′-ATCCTGGTGTCCTCCATACTCA-3′ | 22 | |
| MHV-3_33_LEFT | 5′-GGCCTTGGTACATTTGGTTGCT-3′ | 22 | 27832–28823 |
| MHV-3_33_RIGHT | 5′-TTTCGAGCAACAAGGCCCTAAA-3′ | 22 | |
| MHV-3_34_LEFT | 5′-CGAAAAATCAGGCCACCCAAAA-3′ | 22 | 28696–29689 |
| MHV-3_34_RIGHT | 5′-AAAACCGCTAACACCGTCTACC-3′ | 22 | |
| MHV-3_35_LEFT | 5′-GTGGCCACCTCTATATGCAAGG-3′ | 22 | 29544–30579 |
| MHV-3_35_RIGHT | 5′-GACTTCTTTGGCGCTTTGCTTT-3′ | 22 | |
| MHV-3_36_LEFT | 5′-CGTGGGCCAAATAATCGCTCTA-3′ | 22 | 30418–31400 |
| MHV-3_36_RIGHT | 5′-GGGCATTGCAGGAATAGTACCC-3′ | 22 |
Based on the MHV-3 sequence found under GenBank accession number FJ647224.1.
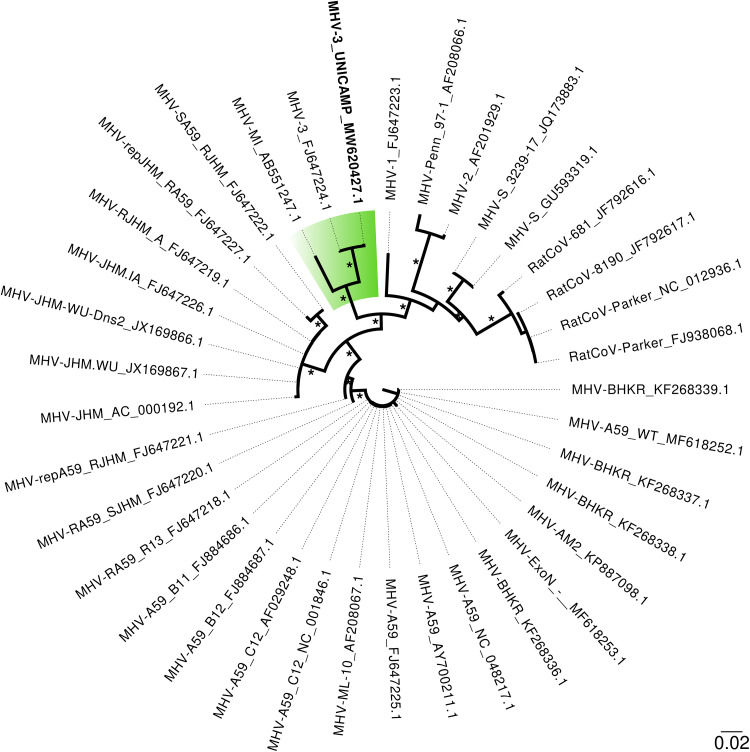
The final consensus of the MHV-3 genome sequence (length, 31,218 nucleotides; GC content, 41.97%) obtained from UNICAMP shares 99.81% nucleotide identity with the only available MHV-3 full-genome sequence, from the J. Craig Venter Institute in Rockville, MD (GenBank accession number FJ647224.1). All murine CoV whole-genome sequences (n = 36) were retrieved from the GenBank database, aligned using MAFFT v.7 (Web version), and manually edited (Unipro UGENE v.37.1). Phylogenetic analysis using the maximum likelihood method (IQ-TREE) indicated that it groups (100% of SH-aLRT/aBayes/ultrafast bootstrap support values) (5) with MHV-MI (AB551247.1), a virus closely related to the highly virulent strain MHV-JHM (6) (Fig. 1). All tools were run using default parameters unless otherwise specified. Lastly, with this MHV-3 strain from UNICAMP sequenced, the scientific community now has a new standard for comparison for further studies involving murine CoV-induced hepatitis.
FIG 1.
Maximum likelihood phylogenetic tree based on the 36 representative genome sequences of murine coronavirus strains. MHV-3, the strain from this study, is highlighted and in bold. The labels include GenBank accession numbers. The asterisks indicate 100% of SH-aLTR/aBayes/ultrafast bootstrap support values. The branch lengths are drawn to scale of nucleotide substitutions per site according to the bar in the figure.
Data availability.
The sequence of MHV-3 has been deposited in the GenBank database under accession number MW620427.
ACKNOWLEDGMENTS
Amanda Barbosa Garcia is the recipient of an institutional scholarship from the Coordination for the Improvement of Higher Education Personnel (CAPES), Brazil (grant number 88887.505801/2020-00). Clarice Weis Arns and Ricardo Durães-Carvalho are supported by the National Council for Scientific and Technological Development (CNPq; grant number 403761/2020-4) and the São Paulo Research Foundation (FAPESP), Brazil (grant number 2019/01255-9), respectively.
REFERENCES
- 1.Tizaoui K, Zidi I, Lee KH, Ghayda RA, Hong SH, Li H, Smith L, Koyanagi A, Jacob L, Kronbichler A, Shin JI. 2020. Update of the current knowledge on genetics, evolution, immunopathogenesis, and transmission for coronavirus disease 19 (COVID-19). Int J Biol Sci 16:2906–2923. doi: 10.7150/ijbs.48812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artika IM, Dewantari AK, Wiyatno A. 2020. Molecular biology of coronaviruses: current knowledge. Heliyon 6:e04743. doi: 10.1016/j.heliyon.2020.e04743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korner RW, Majjouti M, Alcazar MAA, Mahabir E. 2020. Of mice and men: the coronavirus MHV and mouse models as a translational approach to understand SARS-CoV-2. Viruses 12:880. doi: 10.3390/v12080880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Albuquerque N, Baig E, Ma X, Zhang J, He W, Rowe A, Habal M, Liu M, Shalev I, Downey GP, Gorczynski R, Butany J, Leibowitz J, Weiss SR, McGilvray ID, Phillips MJ, Fish EN, Levy GA. 2006. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. J Virol 80:10382–10394. doi: 10.1128/JVI.00747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Islam MM, Toohey B, Purcell DF004A, Kannourakis G. 2015. Suppression subtractive hybridization method for the identification of a new strain of murine hepatitis virus from xenografted SCID mice. Arch Virol 160:2945–2955. doi: 10.1007/s00705-015-2592-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of MHV-3 has been deposited in the GenBank database under accession number MW620427.