We report a sequencing analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater samples collected in the Frankfurt, Germany, metropolitan area. The majority of the detected mutations have been identified only in clinical genomes outside Frankfurt, indicating that the sequencing of SARS-CoV-2 RNA in wastewater can provide insights into emerging variants in a city.
ABSTRACT
We report a sequencing analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater samples collected in the Frankfurt metropolitan area of Germany. The majority of the detected mutations have been identified only in clinical genomes outside Frankfurt, indicating that the sequencing of SARS-CoV-2 RNA in wastewater can provide insights into emerging variants in a city.
ANNOUNCEMENT
Wastewater-based epidemiology (WBE) can provide complementary information about the dynamics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections at the community level because SARS-CoV-2 (genus Betacoronavirus, family Coronaviridae) RNA has been detected in human urine and feces (1, 2). However, most of the WBE studies have focused primarily on reverse transcription-quantitative PCR analysis (3–5). Since the onset of the coronavirus disease 2019 (COVID-19) pandemic, several clinically relevant mutations have been reported, resulting in various SARS-CoV-2 variants (6–9). A few studies showed that the sequencing of SARS-CoV-2 RNA in wastewater could help to determine which SARS-CoV-2 variants are circulating in the studied areas (10, 11).
We collected 24-h composite samples on 4 December 2020 at three sampling points in the Frankfurt metropolitan area of Germany. The three sampling points were (i) influent of the wastewater treatment plant (WWTP) Niederrad (NR) (50.08N, 8.63E), (ii) influent of the WWTP Sindlingen (SL) (50.09N, 8.51E), and (iii) a sewage sample from Griesheim (GR) (50.09N, 8.60E). One liter of the untreated wastewater was filtered through a 0.45-μm electronegative membrane filter to concentrate the virus-like particles, followed by extraction using the FastRNA Pro Blue kit (MP Biomedicals) according to the manufacturer’s protocol. cDNA synthesis was performed using SuperScript VILO Master Mix (Thermo Fisher Scientific), followed by library preparation using the Ion AmpliSeq SARS-CoV-2 research panel (Thermo Fisher Scientific) according to the manufacturer’s instructions. This panel consists of 237 primer pairs, resulting in an amplicon length range of 125 to 275 bp and covering the nearly full genome of SARS-CoV-2. Libraries were multiplexed and sequenced using an Ion Torrent 530 chip on an Ion S5 sequencer (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Ion Torrent Suite software (v 5.12.2) of the Ion S5 sequencer was used to map the generated reads to a SARS-CoV-2 reference genome (Wuhan-Hu-1 [GenBank accession numbers NC_045512 and MN908947.3]), using TMAP software included in the Torrent Suite. The sequencing data for the samples are summarized in Table 1. For mutation calls, additional Ion Torrent plugins were used. First, all single-nucleotide variants (SNVs) were called using Variant Caller (v5.12.0.4) with “Generic - S5/S5XL (510/520/530) - Somatic - Low Stringency” default parameters. Then, for annotation and determination of the base substitution effect, COVID19AnnotateSnpEff (v1.0.0.1), a plugin developed explicitly for SARS-CoV-2, was used. To determine whether the mutations detected in these samples had already been reported in clinical samples at different locations, we downloaded the clinically reported mutations and their corresponding locations from COVID CG (https://covidcg.org) (12) on 7 March 2021.
TABLE 1.
Summary of sequencing data for the samples
| Sample | Sample location | Total no. of reads | No. of mapped reads | Avg target base coverage depth (×) | Avg read identity to target (%)a | GC content (%) | Accession no. |
|---|---|---|---|---|---|---|---|
| NR | Niederrad | 2,781,914 | 854,804 | 817.9 | 95.68 | 48.3 | SAMN18310569 |
| SL | Sindlingen | 2,143,766 | 675,006 | 680.8 | 94.01 | 46.3 | SAMN18310571 |
| GR | Griesheim | 2,349,476 | 640,702 | 215.1 | 94.84 | 47.4 | SAMN18310570 |
The target sequence was the SARS-CoV-2 reference genome (Wuhan-Hu-1 [GenBank accession numbers NC_045512 and MN908947.3]).
We detected 75 unique mutations across all of the samples, including 5 mutations reported outside Europe, 35 reported in Europe, 13 reported in Germany, 18 reported in the Frankfurt region, and 4 not reported (Fig. 1). In the SL sample, we also observed D614G, a predominant mutation reported around the globe (13) and a key mutation associated with the B.1.1.7, B.1.351, and P.1 variants (14). The 69/70 deletion mutation, a key mutation associated with B.1.1.7 (14), was also found in the SL sample. Due to the plausible presence of SARS-CoV-2 RNA of multiple variants in wastewater, WBE analysis can provide only indirect information about the variants. However, observations from this study suggest that an integration of genomic epidemiology into WBE may support identification of variants that have already been detected in a city, as well as those that have not yet been detected in clinical samples.
FIG 1.
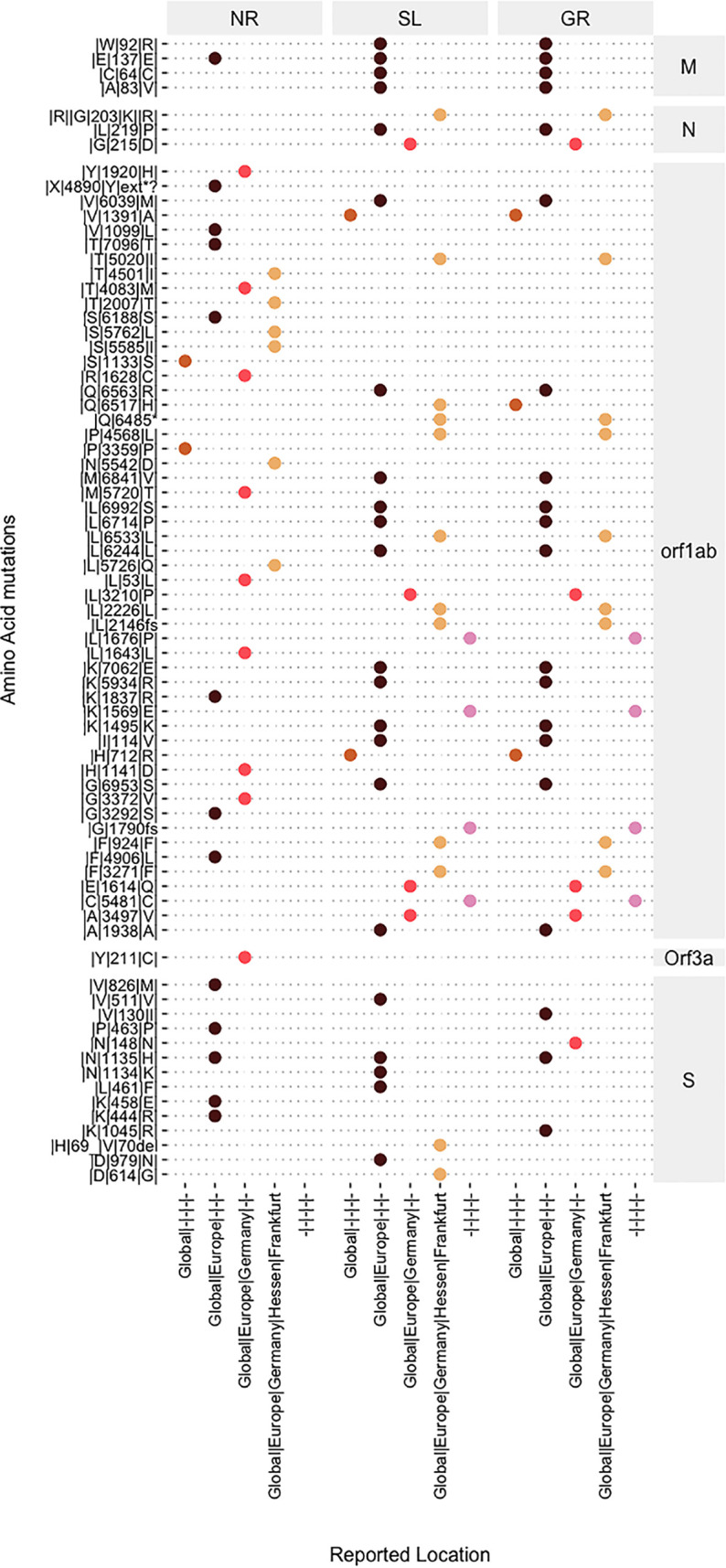
Locations of mutations detected in samples (NR, Niederrad; SL, Sindlingen; GR, Griesheim), corresponding to the locations reporting the mutations in clinical samples. The detected mutations are organized according to the corresponding gene (M, N, open reading frame 1ab [Orf1ab], Orf3a, and S) of the SARS-CoV-2 genome. Points indicate the locations reporting the detected mutations.
Data availability.
The raw metagenomic sequence data are available in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA714504 and SRA accession numbers SRX10340342 (NR), SRX10340344 (SL), and SRX10340343 (GR).
ACKNOWLEDGMENTS
This work was funded by the Hessian Ministry of Economics, Energy, Transport, and Housing within the framework of the IWB-EFRE program for knowledge and technology transfer under grant 20007030.
We thank the employees of the Stadtentwässerung Frankfurt (SEF) and members of our research group for their support with sample collection.
REFERENCES
- 1.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, Yang Y, Liu B, Wang W, Wei C, Yang J, Ye G, Cheng Z. 2020. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol 92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 2.Ren J-G, Li D-Y, Wang C-F, Wu J-H, Wang Y, Sun Y-J, Zhang Q, Wang Y-Y, Chang X-J. 2020. Positive RT-PCR in urine from an asymptomatic patient with novel coronavirus 2019 infection: a case report. Infect Dis (Lond) 52:571–574. doi: 10.1080/23744235.2020.1766105. [DOI] [PubMed] [Google Scholar]
- 3.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. 2020. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ Sci Technol Lett 7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O'Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. 2020. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal S, Orschler L, Lackner S. 2021. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in southern Germany. Sci Rep 11:5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercatelli D, Giorgi FM. 2020. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol 11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfefferle S, Günther T, Kobbe R, Czech-Sioli M, Nörz D, Santer R, Oh J, Kluge S, Oestereich L, Peldschus K, Indenbirken D, Huang J, Grundhoff A, Aepfelbacher M, Knobloch JK, Lütgehetmann M, Fischer N. 2021. SARS coronavirus-2 variant tracing within the first coronavirus disease 19 clusters in northern Germany. Clin Microbiol Infect 27:130.e5–130.e8. doi: 10.1016/j.cmi.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Priesemann V, Balling R, Brinkmann MM, Ciesek S, Czypionka T, Eckerle I, Giordano G, Hanson C, Hel Z, Hotulainen P, Klimek P, Nassehi A, Peichl A, Perc M, Petelos E, Prainsack B, Szczurek E. 2021. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet 397:469–470. doi: 10.1016/S0140-6736(21)00150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eurosurveillance Editorial Team. 2021. Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA: first update. Euro Surveill 26:2101211. doi: 10.2807/1560-7917.ES.2021.26.3.2101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crits-Christoph A, Kantor RS, Olm MR, Whitney ON, Al-Shayeb B, Lou YC, Flamholz A, Kennedy LC, Greenwald H, Hinkle A, Hetzel J, Spitzer S, Koble J, Tan A, Hyde F, Schroth G, Kuersten S, Banfield JF, Nelson KL. 2021. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio 12:e02703-20. doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemudryi A, Nemudraia A, Wiegand T, Surya K, Buyukyoruk M, Cicha C, Vanderwood KK, Wilkinson R, Wiedenheft B. 2020. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep Med 1:100098. doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen AT, Altschuler K, Zhan SH, Chan YA, Deverman BE. 2021. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. Elife 10:e63409. doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isabel S, Graña-Miraglia L, Gutierrez JM, Bundalovic-Torma C, Groves HE, Isabel MR, Eshaghi A, Patel SN, Gubbay JB, Poutanen T, Guttman DS, Poutanen SM. 2020. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci Rep 10:14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong S, Silk BJ, Armstrong GL, Biggerstaff M, Dugan VG. 2021. Emergence of SARS-CoV-2 B.1.1.7 lineage: United States, December 29, 2020–January 12, 2021. MMWR Morb Mortal Wkly Rep 70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw metagenomic sequence data are available in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA714504 and SRA accession numbers SRX10340342 (NR), SRX10340344 (SL), and SRX10340343 (GR).


