Abstract
Oxycodone is a semisynthetic μ- and κ-opioid receptor with agonist with a broad scope of use including postoperative analgesia as well as control of neuropathic and cancer pain. Advantages over other opioids include prolonged duration of action, greater potency than morphine and lack of histamine release or ceiling effect. Individual responses to oxycodone can vary due to genetic differences. This review article aims to summarize the oxycodone literature and provide context on its pharmacogenomics and pharmacokinetics. The evidence for clinical effect of genetic polymorphisms on oxycodone is conflicting. There is stronger evidence linking polymorphic genetic enzymes CYP2D6 and CYP3A with therapeutic outcomes. Further, research is needed to discern all of oxycodone’s metabolites and their contribution to the overall analgesic effect.
Keywords: : ABCB1, analgesia, CYP2D6, CYP3A, cytochrome P450, genetic, opioid, OPRM1, oxycodone, pharmacodynamics, pharmacogenomics, pharmacokinetics, polymorphism, variant
Oxycodone is a semisynthetic opioid analgesic that was first derived from the opium alkaloid thebaine in 1916 and introduced into clinical practice in Germany in 1917 [1–3]. In the USA, the oxycodone market share grew from 10% in 1996 to 53% in 2000, making it a leading opioid [4]. Oxycodone is a selective μ-opioid receptor agonist and at higher doses it binds to κ-opioid receptors in the central, peripheral and autonomic nervous systems [3]. Oxycodone is a selective μ-opioid receptor agonist (with potential κ-opioid receptor agonism). It is widely used in clinical practice for control of postoperative pain, neuropathic pain and cancer pain [5]. Routes of administration include intravenous (IV), intramuscular (IM), intranasal (IN), subcutaneous (SC), rectal, epidural and oral using immediate-release solutions and immediate controlled-release tablets [3].
Oxycodone is used for pain of moderate-to-severe intensity; its potency is greater than that of morphine when used for postoperative pain relief [6]. In postoperative pain management, oxycodone has shown analgesic and oral administration advantages over other strong opioids [7,8]. The duration of action is longer and it does not cause histamine release when compared with morphine which makes it more suitable for individuals with underlying disease states that exacerbate with increase histamine release [9].
In patient controlled intravenous analgesia (PCIA) following abdominal surgery, demand-only oxycodone provided comparable overall pain relief, lower pain on movement, less analgesic dose requirement and fewer adverse effects compared with continuous sufentanil infusion [10] and fentanyl [11].
Individual genetic variations can alter drug effects; hence an understanding of genetic factors is necessary. An individuals’ genetic data can be used to design a targeted approach and personalized therapy [12]. This can apply to dosing guidelines, annotated drug label, potential adverse effects and the risk for hypersensitivity reactions. SNPs are responsible for these genetic variants and variants influencing oxycodone pharmacodynamics and pharmacokinetic have been described. This review article will provide an overview of the literature related to the pharmacogenetics of oxycodone.
Materials & methods
Pubmed and Google Scholar search was done for related articles. MeSH terms such as oxycodone, pharmacogenetics, pharmacodynamics, pharmacokinetics, gene variants, single nucleotide polymorphism, genetic polymorphism was used to build a search. Studies that genotyped patients on oxycodone with data such as potency, adverse effects, antinociceptive effect, drug consumption and dose were selected. Citations were added to the citation manager EndNote X9. Pharmacogenetics of oxycodone affecting its pharmacokinetics and pharmacodynamics are reviewed below.
Pharmacokinetics of oxycodone
Oxycodone is available for IM, IV, SC, IN, rectal or oral (capsule or solution) administration. It has also been used epidurally in a few studies. The pharmacokinetics variability with different formulations of oxycodone has been summarized in Table 1. The bioavailability of the orally administered drug is about 50–90% [13,14] and the time to peak concentration is about 1–1.5 h. Fatty food appears to increase the bioavailability of the drug, while prolonging the time to peak concentration [15]. The bioavailability of oxycodone given rectally is similar to that of the orally administered drug (∼60%) [16]. Oxycodone has a volume of distribution of 2–3.5 l/kg and a total plasma clearance of 45–48 l/h. The half-life of the drug is about 3–5 h [13,15,17–19]. Although IV oxycodone has a shorter half-life (~3 h), the duration of action is similar between these formulations [17]. The short half-life requires the drug to be given every 4–6 h. However, the controlled release oral formulation of the drug has an extended half-life of about 8 h, facilitating 12-h dosing [20].
Table 1. . Pharmacokinetic parameters of various formulations of oxycodone.
| Formulations | Bioavailability | Time to peak concentration | Half-life |
|---|---|---|---|
| IV | 100% | – | 2–3 h |
| IM | 100% | ∼1 h | ∼4.9 h |
| Oral | 50–90% | 1–1.5 h | 3–5 h |
| Oral (extended release) | 60–87% | ∼2.6 h | ∼8 h |
| Rectal | ∼62% | ∼2.8 h | ∼3.7 h |
Oxycodone is primarily bound to albumin (44–46%) and is not bound to α1-acid glycoprotein. It has a liposolubility close to that of morphine, with partition coefficients of 0.7 and 0.5, respectively [21]. The volume of distribution of oxycodone ranges from 211 to 249 l [17]. Oxycodone is extensively metabolized in the liver primarily by the CYP enzymes and in a small amount by the uridine diphosphate glucuronosyltransferases (UGT). Human intestinal microsomes have a much lower N-demethylation activity than liver microsomes, suggesting that there is minimal first pass metabolism of the drug in the gut [22]. The primary enzymes involved in metabolism are CYP2D6, CYP3A4/5, UGT1A3, UGT1A6 and UGT2B7, as demonstrated by Romand et al. with an in vitro study [23]. About 45% of the drug undergoes N-demethylation to noroxycodone noroxymorphone and α- and β-noroxycodol, about 11% of the drug undergoes O-demethylation to oxymorphone and α- and β-oxymorphol and about 8% of the drug undergoes 6-keto-reduction to α- and β-oxycodol. Noroxymorphone is a metabolite produced from O-methylation or N-methylation of noroxycodone or oxymorphone, respectively.
Oxycodone, oxymorphone and noroxymorphone also undergo conjugation with the help of UGT [23]. The minimally formed conjugated products are readily eliminated in the urine and therefore found in very low concentrations in the body. CYP3A4/5 and CYP2D6 are the primary enzymes involved in N-demethylation and O-demethylation of oxycodone, respectively [22,24]. Oxymorphone is ten-times more potent than the parent drug, but is formed in clinically negligible quantities and has a relatively lesser penetration across the blood–brain barrier [24]. Noroxymorphone, although less potent than oxymorphone, is present in a higher concentration than oxymorphone and it also minimally crosses the blood–brain barrier. Consequently, both of these potent metabolites have been shown to cause no significant clinical effect in a pharmacokinetic–pharmacodynamic modelling study [24].
The metabolic pathways of oxycodone are described in Figure 1 [19,24–28]. Metabolites of oxycodone have a longer half-life (about 5.8, 8.8 and 9 h for noroxycodone, oxymorphone and noroxymorphone, respectively) [24]. Consequently, with chronic therapy, the levels of the primary metabolite noroxycodone can exceed the concentrations of the parent drug [29]. About 10–19% of the drug is excreted unchanged by the kidneys [13,15,24]. The conjugated products of the drug and the metabolites (oxymorphone and noroxymorphone) are also renally excreted.
Figure 1. . Oxycodone primarily gets metabolized to noroxycodone and oxymorphone and later to noroxymorphone.
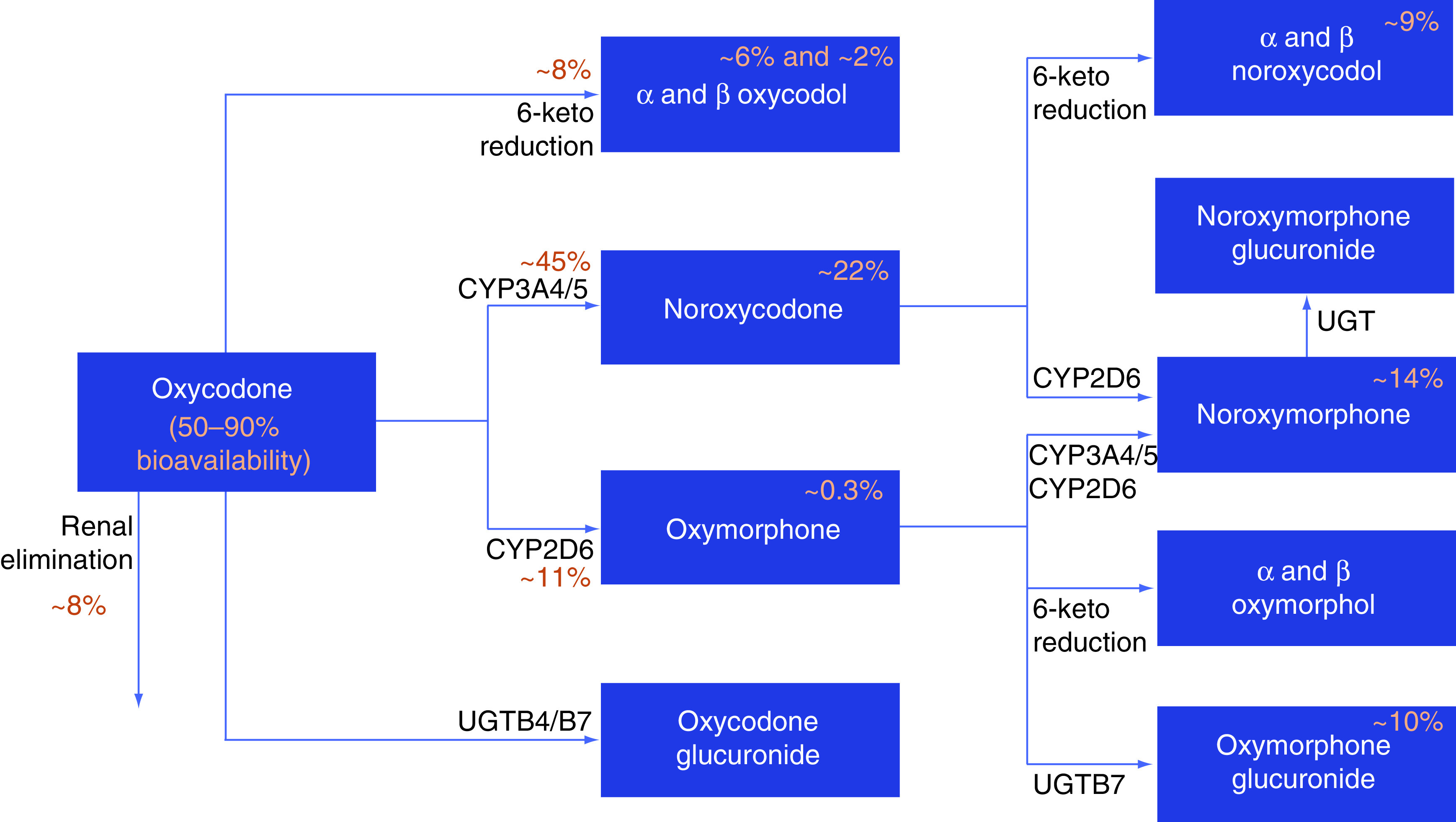
The drug and it metabolites can also undergo glucuronidation before elimination. Oxycodol, noroxycodol and oxymorphol are reductive metabolites of oxycodone, noroxycodone and oxymorphone respectively, with each of them having two stereoisomers (α and β). % mentioned next to an arrow represents the percentage of drug eliminated in that specific pathway and % mentioned in the boxes represent the percentage of eliminated metabolite compared with the parent drug. Pathways with no % mentioned have very minimal contribution to the drug’s elimination.
CYP: Cytochrome P450; UGT: Uridine 5′-diphospho-glucuronosyltransferase or UDP-glucuronosyltransferase.
Given the high amount of hepatic metabolism, hepatic dysfunction increases the maximum concentration (Cmax) and area under curve (AUC) of oxycodone. In a study of controlled-release oxycodone pharmacokinetics with 24 subjects, hepatically-impaired patients had a 40% increase in Cmax, a 90% increase in AUC and a longer half-life (7.7 vs 5.4 h) compared with healthy patients [30]. In a Phase I study of 18 patients with hepatic dysfunction, Cmax and AUC showed a steady increase with decreasing hepatic function. The AUCs were 105.7, 134.7 and 218 for normal, mild and moderately impaired hepatic function, respectively [31]. Tallgren et al. studied the pharmacokinetics of oxycodone in six end-stage cirrhosis patients before and after liver transplantation. The elimination half-life reduced from 13.9 to 3.4 h soon after transplant, which highlights the danger of using it in severely impaired hepatic patients [32].
Although only 10–14% is eliminated renally, patients with severely impaired renal function have increased half-life, increasing the exposure of the drug. Elimination of its metabolites, including the more potent metabolite oxymorphone, is severely impaired. The volume of distribution is increased in uremic patients due to the increase in fat-tissue index caused by the decrease in lean tissue mass index [33,34]. Malhotra et al. showed that the mean AUCs were 210.7, 271.6, 299.5 and 493.5 in individuals with normal, mild, moderate and severely impaired renal function [31]. A dose reduction by 50% is advised if a patient’s glomerular filtration rate is less than 50 ml/min [15,35]. The physicochemical properties of oxycodone suggest it to be dialyzable, although there is no available data to confirm this [34].
The CYP3A4 enzyme is a major contributor to the metabolism of oxycodone – about sevenfold higher than the CYP2D6-related metabolism [24]. CYP3A4 could be a target for drug interactions, as inhibition or induction of the enzyme could change the levels of the drug. In a drug interaction study conducted by Samer et al., administering oxycodone with ketoconazole (a strong CYP3A4 inhibitor) led to a tripling of oxymorphone concentration and reduction of noroxycodone and noroxymorphone AUCs by 80% [36]. Similar results were obtained in other studies where telithromycin and itraconazole increased the AUC of oxycodone by 80 and 144%, respectively, by inhibiting CYP3A4 [37,38]. In a placebo-controlled randomized crossover drug interaction study with rifampicin, the AUC of oxycodone was reduced by two- and sevenfold when administered IV and orally, respectively. As expected, the AUC of noroxycodone increased [39].
Quinidine, an inhibitor of CYP2D6, was investigated with oxycodone in a drug interaction study by Samer et al. Results showed a 40% decrease in Cmax and AUC of oxymorphone and 80% decrease in the Cmax and AUC of noroxymorphone, along with an increase in the AUC in oxycodone and noroxycodone. Although the available evidence is conflicting, use of an alternative drug in CYP2D6 poor or ultra-rapid metabolizers is recommended [36].
Women metabolize oxycodone faster than men, with higher levels of metabolites compared with men [25]. This could be due to the increased CYP3A4 expression and activity generally seen in women [40]. However, in one of the older studies, women consistently showed a higher level of oxycodone AUC compared with men across age groups [30]. This study failed to adjust for body weight, which might be responsible for the observed difference.
Race does not have any effect on the pharmacokinetics of oxycodone [15]; however, oxycodone pharmacokinetics seem to differ with the extreme ages. Kokki et al. found that the elimination half-lives across extremely preterm neonates, preterm neonates, older neonates and infants aged 6–24 months were 8.8, 7.4, 4.1 and 2 h, respectively [41]. Similar results were shown in a population pharmacokinetic study, where the clearance increased with age and the volume of distribution reduced with age in children less than 1 year of age [42]. Children more than 6 months of age reach adult metabolic activity fairly quickly [43]. One in vitro study using cryopreserved hepatocytes from different age groups showed that change in CYP3A activity across various age groups is the major determinant of their metabolic clearance [44].
The AUC indicative of exposure of oxycodone is greatly increased in the elderly. Patients above 70 years of age were found to have 50–80% higher exposure to oxycodone [45]. In a population pharmacokinetic study by Saari et al. with 77 individuals ranging from 19–89 years of age, lean body mass and age were found to be significant covariates on the clearance of the drug. Age was associated with a proportional decrease in clearance [46].
Genetic factors affecting the pharmacokinetics of oxycodone
All known genetic factors affecting the pharmacokinetics of oxycodone are related to the metabolism of the drug involving genetic polymorphism in the related enzymes. While the genetic variants of CYP3A and CYP2D6 are well characterized, the genetic variations of UGTB7 are not. The pharmacological effects of the various genes and their variants are summarized in Table 2.
Table 2. . Genetic variants affecting the pharmacodynamics and the pharmacokinetics of oxycodone.
| Gene | Variant | Effect | Study (year) | Ref. |
|---|---|---|---|---|
| OPRM1 | ||||
| rs1799971 (118A>G) | i. Reduced antinociceptive effect exposed to experimental pain ii. Reduced ability to keep focus |
Zwisler et al. (2010) | [47] | |
| No difference in any of the pain measurements or adverse drug reactions in the first 24 h after surgery | Zwisler et al. (2012) | [48] | ||
| Reduced analgesic response to experimental visceral pressure | Olesen et al. (2015) | [49] | ||
| Gene dosage effect with the GG genotype requiring the highest dose | Cajanus et al. (2014) | [50] | ||
| Higher dose in GG compared with the AA and the AG genotypes in patients with cancer pain | Lin et al. (2018) | [51] | ||
| rs589046 (C>T) | Improved analgesic response to experimental skin heat and visceral pressure | Olesen et al. (2015) | [49] | |
| rs563649 (C>T) | Improved analgesic response to experimental skin heat | Olesen et al. (2015) | [49] | |
| rs9479757 (G>A) | Improved pain tolerance threshold to experimental visceral pressure | Olesen et al. (2015) | [49] | |
| rs533586 (C>A/T) | Poor analgesic response to experimental visceral pressure | Olesen et al. (2015) | [49] | |
| OPRD1 | ||||
| rs419335 (A>G) | Poor analgesic response to experimental visceral heat | Olesen et al. (2015) | [49] | |
| rs2234918 (C>G/T) | Improved analgesic response to experimental muscle pressure | Olesen et al. (2015) | [49] | |
| ABCB1 | ||||
| C3435T | Less pronounced adverse drug reactions: nausea/vomiting, tiredness/drowsiness, itching and for the total sum of adverse drug reactions | Zwisler et al. (2010) | [47] | |
| No difference in any of the pain measurements or adverse drug reactions in the first 24 h after surgery | Zwisler et al. (2012) | [48] | ||
| G2677T/A | i. Variant T allele had improved antinociceptive effect of oxycodone ii. Lesser adverse drug reactions like nausea/vomiting, dizziness, itching and increased adverse drug reactions like urine retention, headaches iii. 3435CC-2677GG was associated with reduced antinociceptive effect of oxycodone |
Zwisler et al. (2010) | [47] | |
| No difference in any of the pain measurements or adverse drug reactions in the first 24 h after surgery | Zwisler et al. (2012) | [48] | ||
| Increased risk of sedation in breastfeeding mothers. No association with oxycodone-induced depression in neonates. | Lam et al. (2012) | [52] | ||
| COMT | ||||
| Multiple SNPs | 22 SNPs within or nearby the COMT gene showed i. 3 SNPs, rs4646312, rs2239393 and rs4818 associated with pain intensity during motion ii. rs887200 (recessive model) and rs1544325 (dominant model) associated with cold pain iii. No association with total oxycodone consumption after multiple testing correction. |
Kambur et al. (2013) | [53] | |
| CYP2D6 | ||||
| PM and EM | i. PM had an increase in pain tolerance thresholds ii. Plasma oxymorphone/oxycodone ratio was lower in PM compared with EM |
Zwisler et al. (2009) | [100] | |
| IM/PM, EM and UM | AUC of oxymorphone were increased while that of noroxycodone were reduced in UM compared with others | Samer et al. (2010) | [36] | |
| IM/PM, EM and UM | i. CYP2D6 ultra-rapid metabolizers experienced increased pain tolerance thresholds ii. Inverse relation between activity and sedation iii. Psychomotor testing by mean of the digit symbol substitution test should gene dose effect |
Samer et al. (2010) | [54] | |
| PM, EM and UM | i. Significant increase in serum concentrations of oxymorphone and noroxymorphone from PM to EM and from PM to UM ii. No difference between the groups with regard to pain, tiredness and nausea |
Andreassen et al. (2012) | [55] | |
| *4/*4 | Trend toward decreased clearance but the effect could not be observed in population pharmacokinetic analysis | Saari et al. (2012) | [46] | |
| PM, IM and UM | Oxymorphone trough concentrations were higher in EM than in IM but did not affect dose escalation | Naito et al. (2013) | [56] | |
| PM, IM, EM and UM | Cmax and AUC of oxymorphone and oxymorphone/oxycodone ratio, was increased with EM compared with PM and IM | Balyan et al. (2017) | [57] | |
| PM, IM, EM and UM | i. *6 and *9 alleles were associated with therapeutic failure in chronic low back pain ii. UM (CYP2D6*2N) associated with an increased risk of side effects |
Dagostino et al. (2018) | [58] | |
| CYP3A5 | ||||
| *3 | i. Daily dose escalation rate was increased ii. Noroxycodone trough concentrations were increased with *1 |
Naito et al. (2013) | [56] | |
All effects are in relation to the variant allele as notated in the variant column unless otherwise specified.
AUC: Area under curve; EM: Extensive metabolizer; IM: Intermediate metabolizer; PM: Poor metabolizer; UM: Ultra-rapid metabolizer.
CYP3A
CYP3A is the major metabolizing enzyme for more than 50% of the drugs administered in clinical care. Generally, variants in CYP3A4 occur in less than 5% of the population as a heterozygous allele. Also, there is no evidence of a null allele (nonfunctional allele) known with CYP3A4 as seen with the other CYP genes. CYP3A4 has many genetic variants, yet few of them (*1b, *2, *3 and *22) result in clinically relevant phenotypes [59]. CYP3A5*3, a nonfunctional variant, is the most common variant of CYP3A5 – seen in 85% of Caucasians and 65% of Asians. It is commonly linked with CYP3A4*1b. CYP3A7 is the major CYP3A isoform in the fetal liver. Although genetic variation will be important in the metabolism of oxycodone, it has not been well studied under in vivo or in vitro conditions.
There is very sparse information available on the effect of CYP3A genetic variation on the metabolism of oxycodone. In a study by Naito et al., CYP3A5*3 decreased the 12 h trough concentration of noroxycodone and was associated with increased incidence of dose escalation in a multivariate analysis [56]. This could have been due to decreased synthesis of the active metabolite noroxymorphone from noroxycodone.
CYP2D6
Genetic variations seen with CYP2D6 include gene duplications, tandem arrangements, gene deletion and extensive regular allelic variants. This complex variation in the genotype gives rise to the poor metabolizers (PM; homozygous for reduced-function variant allele), intermediate metabolizers (IM; heterozygous for nonfunctional allele with a normal function allele) and extensive or normal metabolizers (EM/NM; homozygous wild-type alleles) phenotypes. The most commonly studied variants are CYP2D6*2, *3, *4, *5, *10, *17 and *41. The *1 and *2 alleles have normal activity while the *10, *17 and *41 alleles have reduced activity and *3, *4 and *5 alleles have null activity [60] Duplications of the normal activity alleles result in the UM phenotype. The incidence of each of these variants differs greatly between ethnic groups. All of these factors contribute to the high variation in enzymatic activity of CYP2D6 seen between individuals.
Balyan et al. studied the effect of CYP2D6 genetic polymorphism on oxycodone pharmacokinetics in 30 children. Though there were no changes in pharmacokinetic outcomes of oxycodone, oxymorphone Cmax, AUC and oxymorphone/oxycodone ratio, were increased in children with EM genotype and phenotype compared with PM and IM phenotypes [57]. Similar results were observed in a much smaller study proving the significance of CYP2D6 in the formation of oxymorphone. Further, this study showed that the UM patients have reduced noroxycodone concentrations [36]. In a population pharmacokinetic modelling study, although univariate analysis showed lower clearance in individuals with CYP2D6*4/*4, it did not improve the final fit of the model and, therefore, was omitted [46]. A large genotyping study of 450 cancer patients revealed that patients with PM phenotypes had lower concentrations of oxymorphone and noroxymorphone, while the levels of oxycodone and noroxycodone were unaffected between PM, EM and UM phenotypes. The daily intake of doses did not differ between the groups [55]. Similar results with decreased oxymorphone and unaffected oxycodone levels were also seen in other studies [56,61]. None of these studies found a difference in responder rates despite the high degree of genetic variation. Liukas et al. found no significant correlation between the oxycodone concentrations and genotype, although there was a trend toward higher plasma oxycodone and noroxycodone concentrations and lower oxymorphone and noroxymorphone concentrations in PM and IM compared with the EM and UM phenotypes, signifying the possibility of such an effect [45].
Pharmacodynamics of oxycodone
Oxycodone is an opioid receptor agonist with a weak affinity for the μ-opioid receptor and minimal or no affinity for the delta-opioid receptor and a subtype of the κ-opioid receptor [62,63]. The exact mechanism of action of oxycodone is not fully known. Oxycodone and its active metabolites can selectively bind to the μ-opioid receptor, at higher doses the κ- and δ-opioid receptors in the CNS and periphery and induce a G-protein-coupled receptor signaling pathway. Oxycodone has a lower binding affinity for the μ-opioid receptor than methadone and morphine, although its active metabolite (oxymorphone) has greater affinity that the parent drug [3,64].
The active metabolite contributes largely to the pharmacodynamic characteristics of oxycodone [3,65]. Oxymorphone formed from 3-O-demethylation of oxycodone has two- to five-times the μ-opioid receptor affinity and analgesic potency than morphine [66] while it is 14-fold more potent with 40-times higher μ-opioid receptor affinity than its parent drug oxycodone [67]. A larger primary active metabolite, noroxycodone, produced from N-demethylation of oxycodone has four-times lower μ-opioid receptor-binding affinity [66] and a poor antinociceptive potency in rats. However, noroxymorphine, a further metabolite of noroxycodone, is a more potent shifter of D-ala2,N-MePhe4,Gly-ol (DAMGO)-enkephalin from the μ-opioid receptor in rat brain when compared with oxycodone [64,66,68]. The opioid receptor-binding attributes are yet to be characterized for other metabolites obtained via reduction of oxycodone to α- and β-oxycodol, noroxycodone to α- and β-noroxycodol and oxymorphone to α- and β-oxymorphol [22,66]. However, α- and β-oxymorphol have been shown to be two- to threefold more potent than oxycodone, despite having low plasma concentration after administration of oxycodone [66]. Klimas et al. [69] described the contribution of oxycodone and its metabolites to the general analgesic effect. Their results showed that, even though oxymorphone and noroxymorphone have much higher μ-receptor affinity than oxycodone, oxycodone is accountable for 83 and 95% of analgesic effect during oral and IV administration, respectively.
When oxycodone is compared with morphine, several studies [3,64,66,70,71] have shown its lower affinity for the μ-opioid receptor. Likewise, the potency of oxycodone in the μ-opioid receptor activation of intracellular G proteins is lower than that of morphine [66]. However, Silvasti et al. [72] showed both drugs to be equipotent in a study of 50 patients that compared the analgesic efficacy of oxycodone and morphine in postoperative PCIA. Equal analgesia was also demonstrated following PCIA titration in the management of cancer pain [73].
Oxycodone & breastfeeding
Pain can hinder a woman’s ability to care for herself and her newborn; however, opioid use has been linked to birth defects [74,75]. Opioid pain medication intervention is one of the reasons for late breastfeeding initiation after birth [76]. Niklasson et al. [77] evaluated the efficacy and safety of oral oxycodone for postoperative analgesia in 80 women following cesarean section, showing reduced pain intensity, provoked pain and total opioid consumption with oxycodone compared with IV morphine/codeine. They also reported no serious adverse events in mothers and newborns. Conversely, Lam et al. [52] showed a 20% incidence of CNS depression in neonates breastfed by mothers receiving oxycodone compared with 16.7% on codeine and 0.5% on acetaminophen. A case reported by Timm also described oxycodone intoxication of a breastfed neonate [78].
Oxycodone use disorder insights from animal studies
Oxycodone is a leading contributor to the global opioid abuse crisis. Opioid reward is generally believed to be mediated by activation of μ-opioid receptors located on GABAergic interneurons or afferents in the ventral tegmental area [79] and central dopaminergic system [80]. A highly selective dopamine (DA) D3 receptor antagonist, VK4-40 was examined on the rewarding and analgesic effects of oxycodone in rodents; VK4-40 produced a dose-dependent reduction in oxycodone self-administration and blocked the oxycodone brain stimulation reward pathway [80]. In addition, VK4-40 showed antinociceptive properties as efficacious as oxycodone, possibly by augmenting opioid analgesia via DA-dependent mechanisms [80]. The observation that dopamine antagonists inhibit oxycodone self-administration is supported by other studies including heroin self-administration in wildtype, but not in D3R-knockout mice [81] and oxycodone self-administration in rats [82].
OrxR1 and OrxR2 have been associated with regulation of motivation, arousal and stress. After rats were taught to self-administer oxycodone, blockade of OrxR1 was shown to be associated with decreased oxycodone self-administrations and oxycodone-seeking behavior whereas blockade of OrxR2 had no such effects [83,84] The investigators then examined if immunochemotherapy directed against oxycodone would alter antinociception action or intravenous self-administration rate. In those rats trained to self-administer oxycodone, administration of an immunoconjugate vaccine (Oxy-TT) reduced oxycodone concentrations in brain by 50% and vaccination decreased the rat's oxycodone intake and provided protection against the reinforcing effects of oxycodone. The exact neuronal pathways that lead to opioid addiction and abuse need further investigating. However, promising new research and drugs are providing light on potential areas that can be targeted to help decrease opioid use, treat those already addicted and help combat relapse.
Genetic factors affecting the pharmacodynamics of oxycodone
Genetic variations influence the response to opioids including oxycodone. Most of the studies related to genetic variability influencing the pharmacodynamics of oxycodone have focused on the antinociceptive, pupil, sedation and adverse effects of oxycodone. The best-characterized genetic variants involve the opioid receptors and P-glycoprotein transporter. This section will compare different study outcomes in relation to the pharmacogenomics of oxycodone.
Oxycodone & clinical outcomes
Opioid receptors
Opioid receptor polymorphisms mediate various analgesic and adverse effects by impacting the expression and function of binding sites. Approximately 100 variants of the μ-opioid receptor gene (OPRM1) have been identified [85]. A small number of OPRM1 mutations based on allelic frequency affect opioid therapy [86]. OPRM1 polymorphism is an important factor influencing opioid pharmacodynamics.
The A118G SNP of OPRM1 was found to have a variant allele frequency of 10–15% in the Caucasian population [86]. The SNP 118A>G (rs1799971) on chromosome 6 in OPRM1 leads to substitution of aspartate for asparagine, changing the N-glycosylation of the receptor protein [87]. This change can affect the response to opioid analgesia. Olesen et al. [49] evaluated 19 opioid receptor genetic polymorphisms and found that variability in oxycodone response to skin heat was associated with OPRM1 SNPs rs589046 and rs563649 (p < 0.0001). The OPRM1 SNPs rs589046 (p = 0.015), rs1799971 (p = 0.045), rs9479757 (p = 0.009) and rs533586 (p = 0.046) were associated with variability in oxycodone response to visceral pressure. Delta opioid receptor (OPRD1) SNP rs419335 reached oxycodone visceral heat threshold (p = 0.015) while OPRD1 SNP rs2234918 (p = 0.041) was associated with muscle pressure. However, no association was found between κ-opioid receptor (OPRK1) SNPs and oxycodone response for any of the pain modalities.
Zwisler et al. [47] studied the association between the variant allele of the OPRM1 SNP A118G and altered antinociceptive effect and adverse drug reactions of oxycodone. In 33 healthy subjects exposed to experimental pain (including electrical stimulation and cold pressor test), the authors found that the variant G allele was associated with reduced antinociceptive effect when measured by pain tolerance threshold to single electrical nerve stimulation (8% increase vs 25% for the wild-type carriers, p = 0.007).
In contrast, Zwisler et al. [48] later showed no association between OPRM1 SNP A118G and the analgesic effect of IV oxycodone for clinical postoperative pain in 268 patients. Similarly, Skarke et al. [88] showed no effect of A118G genotype on the response to transcutaneous electrical stimulation for morphine and morphine-6-glucuronide (M6G). Carriers of the G allele had less nausea and vomiting after receiving M6G but not morphine.
The association between OPRM1 SNPs and postoperative analgesic requirement in surgical patients remains controversial. Hayashida et al. [89] studied 138 Japanese patients following major open abdominal surgery and found that the 118G homozygous (GG) patients required 24-h postoperative analgesics more than 118A homozygous (AA) and heterozygous (AG) patients. This relationship was verified between OPRM1 118A/G gene polymorphism and oxycodone analgesic dose in patients with cancer pain when investigated by Lin et al. [51]. The authors genotyped 203 patients with moderate-to-severe cancer pain and showed that the dosage of oxycodone in GG genotype was significantly higher than that in AA genotype and AG genotype (15.44 ± 10.19 for GG vs 10.25 ± 4.53 for AA and 10.49 ± 5.26 for AG on day 3; 89.15 ± 27.69 for GG vs 43.59 ± 12.19 for AA and 48.27 ± 18.79 for AG on day 30 after treatment; p < 0.05). In another similar study with a group of 1000 women received oxycodone after breast cancer surgery [50], a similar conclusion was obtained when they demonstrated that the 118A>G variant was associated with the amount of oxycodone requested for adequate analgesia. The highest oxycodone consumption was shown in patients with GG genotype (0.16 mg/kg), moderate for AG genotype (0.13 mg/kg) and lowest for AA genotype (0.12 mg/kg). Conversely, Naito et al. [90], studied 62 cancer patients and reported that OPRM1 A118G SNP had no effect on dose escalation for patients receiving oxycodone. Likewise, Kim et al. [91] did not find any association between postoperative fentanyl consumption and genetic polymorphisms of OPRM1 in 196 Korean gynecologic patients.
ATP-binding cassette subfamily B member 1 (ABCB1)
Zwisler et al. [47] studied the association between the variant allele of the SNPs C3435T and G2677T/A in the ABCB1 gene and altered antinociceptive effect and adverse drug reactions of oxycodone. In 33 healthy subjects exposed to experimental pain (electrical stimulation and cold pressor test), they showed that carriers of the variant T allele in the SNP C3435T had less adverse drug reactions on oxycodone when compared with carriers of the wild-type genotype. Likewise, for SNP G2677T/A, carriers of the variant T allele had a better antinociceptive effect of oxycodone than the wild-type genotype in the cold pressor test (25% reduction vs 15%, p = 0.015) and less adverse drug reactions. The combined wild-type genotype 3435CC-2677GG had less antinociceptive effect in the cold pressor test (13% reduction vs 23%, p = 0.019) and more severe adverse drug reactions than carriers of the variant alleles. However, Zwisler et al. [48] did not find any association between SNPs C3435T and G2677T/A in ABCB1 and the analgesic effect of IV oxycodone in clinical postoperative pain in 268 patients. This finding was attributed to the fact that patients were in an uncontrolled environment and were also co-medicated with inhibitors and substrates of P-glycoprotein. In cancer patients receiving oxycodone, no significant difference was found between oxycodone consumption and ABCB1 SNPs [90].
Catechol-O-methyltransferase
The effect of SNPs in catechol-O-methyltransferase (COMT) was studied by Kambur et al. [53]. Although there were multiple SNPs identified to be related with pain during motion and cold pain, there was no association between the SNPs and postoperative oxycodone consumption.
Genetics related to nursing mothers & neonates
Opioids are prescribed to >80% of women following Caesarean section and up to 30% of women following vaginal delivery and millions of women breastfeed while receiving opioids each year [92]. The US FDA’s 2017 updated warning recommends against using codeine and tramadol in breastfeeding women due to excess sleepiness and serious breathing problems including death in breastfed infants [93]. Codeine and tramadol are metabolized in the liver to active/more potent metabolites by an enzyme called cytochrome P450 isoenzyme 2D6 (CYP2D6). CYP2D6 ultra-rapid metabolizers convert codeine to morphine and tramadol to an active metabolite, M1, faster and to a greater extent than in other people. In such mothers, unsafe levels of morphine and M1 are found in blood and breast milk [93] and most are unaware of their CYP2D6 metabolic status as an ultra-rapid metabolizer and the associated risks [93].
Lam et al. [94] investigated the effect of maternal CYP2D6, CYP345, ABCB1 and OPRM1 polymorphisms in relation to both neonatal and maternal CNS depression after oxycodone use during lactation. No association was found between the maternal genetic variants and oxycodone induced depression in neonates. However, the authors did note that mothers of symptomatic infants used oxycodone longer than asymptomatic infants and carriers of at least one copy of the ABCB1 2677 T variant experienced an increased risk of sedation.
Both oxycodone and hydrocodone are also metabolized by CYP2D6 and have the same risk for life-threatening respiratory depression in an infant breastfeeding from an ultra-rapid metabolizer mother. It took >20 years to recognize the life-threatening complications and deaths associated with codeine from CYP2D6 genetic variations in children undergoing tonsillectomy and nursing mothers. If we are not proactive in identifying those at risk and modifying their management, oxycodone (and hydrocodone) that are more potent and addictive than codeine (morphine) would unfortunately pose greater risks to these vulnerable patient populations.
Genetics related to oxycodone abuse
There has been an increase in abuse of prescription opioids including oxycodone, which contributes significantly to opioid overdose deaths. Wightman et al. [95] compared the likeability and/or abuse potential of hydrocodone, oxycodone and morphine and reported that oxycodone demonstrated high abuse liability based on its high likability scores and a relative lack of negative subjective effects. Jones et al. [96] analyzed variations in the genes that encode for μ-, k- and δ-opioid receptors and the dopamine metabolizing enzyme, COMT. Genetic variants in the μ- (rs6848893) and δ-opioid receptor (rs581111) influenced the responses to oxycodone administration [96]. Furthermore, self-reported ‘stimulated’ effects of oxycodone varied significantly as a function of COMT rs4680 genotype [96].
OPRM1 gene encodes the primary receptor target for most opioids. The largest and recent genome-wide association study in adults identified OPRM1 rs1799971 as associated with opioid use disorder, genome-wide significance; p = 1.51 × 10-8 [97], DRD2 gene encodes central dopamine receptor 2. DRD2 genetic variants, rs1076560 and rs1800497 (TaqA1 of ANKK1), have been associated with OD, opioid addiction [98,99].
Cytochrome P450 (CYP) 2D6, CYP3A genetic polymorphism & oxycodone pharmacodynamics
Oxycodone pharmacodynamics are largely dependent on metabolic activity of the polymorphic enzymes CYP2D6 and CYP3A. This was demonstrated by Dagostino et al. [58] when they CYP2D6-genotyped 224 patients with chronic low back pain (CLBP). Carriers of CYP2D6*6 and *9, with reduced and absent enzymatic activity, respectively, were associated with therapeutic failure. CYP2D6 ultra-rapid metabolizers (UM) (CYP2D6*2N patients) had increased risk of side effects and rare alleles like CYP2D6*1/*11, *4/*6 and *41/*2N demonstrated strong associations of efficacy and side effects with chronic opioid treatment.
Samer et al. [54] also corroborated these effects in a study of ten healthy volunteers genotyped for CYP2D6 in which experimental pain (cold pressor test, electrical stimulation, or thermode), pupil size, psychomotor effects and toxicity were assessed after oral oxycodone. The CYP2D6 UM demonstrated an increased pharmacodynamic effect with 1.5- to sixfold increase of analgesic effect as compared with extensive metabolizers (EM) (subjective pain threshold) after electrical stimulation, tolerance to cold pressor test, thermal pain threshold). Conversely, poor metabolizers (PM) had a two- to 20-fold reduction of analgesic effects but cold pressor test and pupil size were unchanged relative to EM. In a different experimental pain study of 33 healthy volunteers, Zwisler et al. [100] found less reduction in pain area under curve on oxycodone for PM compared with EM (14 vs 26%, p = 0.012) following cold pressure. In the same study [54], the pharmacodynamic effect of drug–drug interaction after CYP2D6 blockade with quinidine showed reduced subjective pain threshold for oxycodone by 30%, while blockade of CYP3A4 with ketoconazole increased the analgesic efficacy and toxicity of oxycodone in CYP2D6 UM. Similarly, Hagelberg et al. [101] also demonstrated increased pharmacodynamic effects of oxycodone when administered with voriconazole to 12 healthy subjects. However, Zwisler et al. [61] found no difference in postoperative analgesic effect of oxycodone between the PM and EM of CYP2D6 in 270 patients.
CYP2D6 & Opioids Clinical Pharmacogenetics Implementation guidelines & translational implications
The Clinical Pharmacogenetics Implementation guidelines recommend against or caution using opioids that are metabolized by CYP2D6 (codeine, tramadol, hydrocodone and oxycodone) in ultra-rapid and poor metabolizers [102,103]. Unfortunately, stronger opioids such as oxycodone and hydrocodone are now increasingly prescribed as alternatives to codeine and tramadol in children and nursing mothers [92,104]. The CYP2D6 metabolites of oxycodone (oxymorphone) and hydrocodone (hydromorphone) are five- to 14-fold stronger than codeine’s and tramadol’s CYP2D6 metabolites. This shift in clinical practice to stronger opioids such as oxycodone places new mothers and their breastfed infants at greater, currently unknown, risk of adverse effects (life-threatening respiratory depression, persistent opioid use, addiction and accidental ingestion by young children) [105]. The practice of prescribing oxycodone as an alternative to codeine without considering CYP2D6 genotype is reported to be dangerous for breastfed newborns [52,78] and nursing mothers [94] as oxycodone caused significant CNS depression [52,94], infantile life-threatening respiratory depression [78] and death [106].
Conclusion
Oxycodone has several favorable pharmacodynamic and pharmacokinetic properties which account for its use in treating postoperative and cancer pain. However, its metabolism and effects are influenced by several genetic polymorphisms that can cause considerable variability in its efficacy and safety. There is growing evidence linking polymorphic genetic enzymes CYP2D6 and CYP3A with clinical outcomes. Further research is needed to study the effects of these genetic polymorphisms, the contribution of oxycodone’s metabolites to the overall clinical effects and impact on inter-individual variations in clinical effects. Understanding of the effect of genes related to opioid use disorder is warranted to aid in safe use of this commonly prescribed opioid.
Future perspective
Due to altered CYP2D6 metabolism, severe adverse effects such as life-threatening respiratory depression from codeine [107–110] and tramadol [111,112] were reported, resulting in the FDA’s strongest black box warning contraindicating the use of codeine for the treatment of pain following tonsillectomy and adenoidectomy in children [113] and in nursing mothers. CYP2D6 genetic factors are also known to affect the metabolism of oxycodone and increase the risk in vulnerable populations. With the FDA warnings on codeine and tramadol in vulnerable patients, despite higher abuse potential than codeine and tramadol and metabolism by CYP2D6 pathway, oxycodone is used currently as a preferred opioid analgesic. Therefore, large and robust studies exploring pharmacokinetics, clinical outcomes and pharmacogenetics are needed to improve understandings of inter-individual variabilities in metabolism and clinical responses and to minimize risks of immediate and long-term adverse outcomes risks associated with oxycodone.
Executive summary.
Oxycodone is a semisynthetic opioid analgesic. It acts as a selective μ-opioid agonist and potential kappa receptor agonist, with little to no affinity for the delta receptor.
The μ-opioid receptor’s binding affinity is greater for methadone and morphine than for oxycodone; however, oxymorphone, the active metabolite of oxycodone, has a greater binding affinity than oxycodone.
Oxymorphone is 14-times more potent and has a 40-fold higher μ-opioid receptor affinity than its parent drug oxycodone. Noroxycodone has four-times less μ-opioid receptor-binding affinity and poor antinociceptive potency in rats. Noroxymorphone, a further metabolite of noroxycodone, is a more potent shifter of DAMGO enkephalin from the μ-opioid receptor in rat brains.
One study analyzed the general analgesic effects of oxycodone and its metabolites, revealing that even though oxymorphone and noroxymorphone have much higher μ-receptor affinities than oxycodone, oxycodone is accountable for 83 and 95% of the analgesic effect during oral and intravenous (IV) administration, respectively. This may be attributed to limited crossing of the blood–brain barrier for oxymorphone and noroxymorphone.
Oxycodone has lower μ-opioid receptor affinity and G-protein activation than morphine. However, through the utilization of patient-controlled IV analgesia, equal analgesic efficacy was found for treating postoperative and cancer pain.
Breastfeeding mothers taking oxycodone showed reduced pain and total opioid consumption as compared with breastfeeding mothers using IV morphine/codeine. No serious adverse events were found in mothers or newborns. Rates of CNS depression in newborns were 20.1% with oxycodone, 16.7% with codeine and 0.5% in acetaminophen groups.
OPRM1 polymorphism is an essential factor influencing the pharmacodynamics of opioids. About 100 variants of the OPRM1 gene have been identified. SNP 118>G (rs1799971) variant (G-allele) showed a strong association with the amount of oxycodone for sufficient analgesia.
Variability was also found regarding skin heat with genetic polymorphisms OPRM1 SNPs rs589046 and rs563649. Variability was found with visceral pain rs589046, rs1799971, rs9479757 and rs533586. No association was found for the κ-opioid receptor (OPRK1) SNPs for any pain modalities.
SNP A118G in the OPRM1 gene was linked with reduced antinociceptive effect. C3435T (ABCB1 gene) T allele variant had less adverse drug reactions to oxycodone compared with wild-type alleles. However, G2677T/A (ABCB1 gene) carriers of T allele had better antinociceptive effect of oxycodone than the wild-type genotype and less adverse drug reactions. The combined 3435CC-2677GG had reduced antinociceptive and more severe adverse drug reactions than carriers of the variant alleles.
Conversely, a different study did not find any association between SNPs A118G, C3435T and G2677T and IV analgesic effect in postoperative pain. Confounding factors included an uncontrolled environment and inhibitors of P-glycoprotein.
One study found that, following major open abdominal surgery, patients with 118A homozygous (GG) required more 24-h postoperative analgesics than 118A homozygous (AA) and heterozygous (AG) patients. This relationship was verified in another study regarding cancer pain. This relationship was further strengthened by a study that looked at oxycodone with extensive research on adequate analgesia.
No effect of polymorphisms in catechol-O-methyltransferase was found to be associated with postoperative oxycodone consumption. The pharmacodynamics of oxycodone is mostly dependent on polymorphic enzymes CYP2D6 and CP3A. CYP2D6 PM phenotype shows decreased efficacy, while UM shows strong efficacy and increased side effects. The blockade of CYP3A4 increased the analgesic efficacy and toxicity of oxycodone.
The primary enzymes involved in the metabolism of oxycodone are CYP2D6, CYP3A4/5, UGT1A6, UGT1A3 and UGT2B7. Oxycodone is comprehensively highly metabolized in the liver by CYP enzymes (and in a limited amount by uridine diphosphate and UGT). Oxycodone is weakly metabolized by intestinal microsomes. A total of 45% of the drug is converted to noroxycodone, 11% to oxymorphone and 8% to α- and β-oxycodol.
Many studies show that inducing the CYP3A4/5 pathway reduces area under curve (AUC) of oxycodone, while inhibiting the pathway increases AUC. However, inducing the CYP2D6 pathway increases the AUC.
Women were found to have higher CYP3A4 expression and metabolize oxycodone faster than men. Clearance is lowest in preterm neonates and increases as term neonates transition to infants; however, increasing age was associated with a proportional decrease in clearance. Children older than 6 months of age reach peak metabolic activity quickly. Changes in CYP3A activity seems to be responsible for these differences. One study also found that lean body mass and age were significant covariates on clearance of oxycodone.
Sparse information is available regarding CYP3A genetic variation and its outcomes on oxycodone. One study found CYP3A5*3 decreased 12 h concentration and increased incidence of dose escalation.
CYP2D6 genetic variations give rise to poor metabolizers (homozygous for reduced function allele), intermediate metabolizer (heterozygous) and extensive or normal metabolizer (homozygous for wild-type allele) Most common variants include CYP2D6 *2, 3, 4, 5, 10, 17 and 41. CYP2D6 *1 and *2 have normal activity; *10, *17 and *41 have reduced activity and *3, *4 and *5 have null activity. Duplications of normal alleles result in ultra-rapid metabolizers. Incidence differs between ethnic groups.
Oxycodone has high hepatic metabolism and care must be taken when given to patients with hepatic dysfunction. Although only 10–14% of oxycodone is renally eliminated, a dose reduction is advised when glomerular filtration rate is less than 50 ml/min.
Footnotes
Author contributions
All authors contributed to the drafting of the work and revising it critically for content. All authors have reviewed and approve of this manuscript for submission. All authors agree to be accountable for all aspects of the work.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial & competing interests disclosure
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the NIH funded under Award Numbers, R01HD089458 (PI: S Sadhasivam), R21HD094311 (PI: S Sadhasivam) and R01HD096800 (PI: S Sadhasivam). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The departments of Anesthesia and Clinical Pharmacology at Indiana University School of Medicine (IUSM) provided salary support for the authors. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest
- 1.Lenz GR, Evans SM, Walter DE, Hopfinger AJ. Opiates. Academic Press, Orlando, FL, USA: (1986). [Google Scholar]
- 2.Falk E. Eukodal, ein neues Narkotikum. MMW Munch. Med. Wochenschr. 20, 381–384 (1917). [Google Scholar]
- 3.Kalso E. Oxycodone. J. Pain Symptom Manage 29(Suppl. 5), S47–S56 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Coluzzi F, Mattia C. Oxycodone. Pharmacological profile and clinical data in chronic pain management. Minerva Anestesiol. 71(7–8), 451–460 (2005). [PubMed] [Google Scholar]
- 5.Lei F, Ye J, Wang J, Xia Z. A Bibliometric Analysis of Publications on Oxycodone from 1998 to 2017. BioMed Res. Int. 2019, 9096201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalso E, Pöyhiä R, Onnela P, Linko K, Tigerstedt I, Tammisto T. Intravenous morphine and oxycodone for pain after abdominal surgery. Acta Anaesthesiol. Scand. 35(7), 642–646 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Raff M, Belbachir A, El-Tallawy S et al. Intravenous oxycodone versus other intravenous strong opioids for acute postoperative pain control: a systematic review of randomized controlled trials. Pain Ther. 8(1), 19–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MK, Ahn SE, Shin E, Park SW, Choi JH, Kang HY. Comparison of analgesic efficacy of oxycodone and fentanyl after total hip replacement surgery: a randomized controlled trial. Medicine (Baltimore) 97(49), e13385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ennis M, Schneider C, Nehring E, Lorenz W. Histamine release induced by opioid analgesics: a comparative study using porcine mast cells. Agents Actions 33(1–2), 20–22 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Han L, Su Y, Xiong H et al. Oxycodone versus sufentanil in adult patient-controlled intravenous analgesia after abdominal surgery: a prospective, randomized, double-blinded, multiple-center clinical trial. Medicine (Baltimore) 97(31), e11552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang BY, Kwon JY, Kim E, Lee DW, Kim TK, Kim HK. Oxycodone vs. fentanyl patient-controlled analgesia after laparoscopic cholecystectomy. Int. J. Med. Sci. 11(7), 658–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimmesgern E, Benediktsson I, Norstedt I. Personalized medicine in Europe. Clin. Transl. Sci. 10(2), 61–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pöyhiä R, Seppälä T, Olkkola KT, Kalso E. The pharmacokinetics and metabolism of oxycodone after intramuscular and oral administration to healthy subjects. Br. J. Clin. Pharmacol. 33(6), 617–621 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cone EJ, Heltsley R, Black DL, Mitchell JM, LoDico CP, Flegel RR. Prescription opioids. I. Metabolism and excretion patterns of oxycodone in urine following controlled single dose administration. J. Anal. Toxicol. 37(5), 255–264 (2013). [DOI] [PubMed] [Google Scholar]
- 15.US FDA. Roxicodone drug label (2008). https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021011s002lbl.pdf
- 16.Leow KP, Cramond T, Smith MT. Pharmacokinetics and pharmacodynamics of oxycodone when given intravenously and rectally to adult patients with cancer pain. Anesth Analg. 80(2), 296–302 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Leow KP, Smith MT, Williams B, Cramond T. Single-dose and steady-state pharmacokinetics and pharmacodynamics of oxycodone in patients with cancer. Clin. Pharmacol. Ther. 52(5), 487–495 (1992). [DOI] [PubMed] [Google Scholar]
- 18.Söderberg Löfdal KC andersson ML, Gustafsson LL. Cytochrome P450-mediated changes in oxycodone pharmacokinetics/pharmacodynamics and their clinical implications. Drugs 73(6), 533–543 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Kinnunen M, Piirainen P, Kokki H, Lammi P, Kokki M. Updated clinical pharmacokinetics and pharmacodynamics of oxycodone. Clin. Pharmacokinet. 58(6), 705–725 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Kalso E. Oxycodone. J. Pain Symptom Manage 29(5), 47–56 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Pöyhiä R, Seppälä T. Liposolubility and protein binding of oxycodone in vitro. Pharmacol. Toxicol. 74(1), 23–27 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Lalovic B, Phillips B, Risler LL, Howald W, Shen DD. Quantitative contribution of CYP2D6 and CYP3A to oxycodone metabolism in human liver and intestinal microsomes. Drug Metab. Dispos. 32(4), 447–454 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Romand S, Spaggiari D, Marsousi N et al. Characterization of oxycodone in vitro metabolism by human cytochromes P450 and UDP-glucuronosyltransferases. J. Pharm. Biomed. Anal. 144, 129–137 (2017). [DOI] [PubMed] [Google Scholar]; • By far the best attempt in characterizing the in vitro metabolism of oxycodone through the Phase I and II pathways. It also studied the effect of oxycodone on the relevant CYP and UGT enzymes.
- 24.Lalovic B, Kharasch E, Hoffer C, Risler L, Liu-Chen L-Y, Shen DD. Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin. Pharmacol. Ther. 79(5), 461–479 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Andreassen TN, Klepstad P, Davies A et al. Influences on the pharmacokinetics of oxycodone: a multicentre cross-sectional study in 439 adult cancer patients. Eur. J. Clin. Pharmacol. 67(5), 493–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korjamo T, Tolonen A, Ranta V-P, Turpeinen M, Kokki H. Metabolism of oxycodone in human hepatocytes from different age groups and prediction of hepatic plasma clearance. Front. Pharmacol. 2(87), 1–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePriest AZ, Puet BL, Holt AC, Roberts A, Cone EJ. Metabolism and disposition of prescription opioids: a review. Forensic Sci. Rev. 27(2), 115–145 (2015). [PubMed] [Google Scholar]
- 28.Huddart R, Clarke M, Altman RB, Klein TE. PharmGKB summary: oxycodone pathway, pharmacokinetics. Pharmacogenet. Genomics 28(10), 230–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy J, Norris R, Anderson H, O'Shea A, Charles B. Is saliva a valid substitute for plasma in pharmacokinetic studies of oxycodone and its metabolites in patients with cancer? Support. Care Cancer 20(4), 767–772 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Kaiko RF. Pharmacokinetics and pharmacodynamics of controlled-release opioids. Acta Anaesthesiol. Scand. 41(1 Pt 2), 166–174 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Malhotra BK, Schoenhard GL, de Kater AW, Friedmann N. The pharmacokinetics of oxycodone and its metabolites following single oral doses of Remoxy®, an abuse-deterrent formulation of extended-release oxycodone, in patients with hepatic or renal impairment. J. Opioid Manag. 11(2), 157–169 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Tallgren M, Olkkola KT, Seppälä T, Höckerstedt K, Lindgren L. Pharmacokinetics and ventilatory effects of oxycodone before and after liver transplantation. Clin. Pharmacol. Ther. 61(6), 655–661 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Kirvela M, Lindgren L, Seppala T, Olkkola KT. The pharmacokinetics of oxycodone in uremic patients undergoing renal transplantation. J. Clin. Anesth. 8(1), 13–18 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Dean M. Opioids in renal failure and dialysis patients. J. Pain Symptom Manage 28(5), 497–504 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Pham PC, Khaing K, Sievers TM et al. 2017 update on pain management in patients with chronic kidney disease. Clin. Kidney J. 10(5), 688–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samer C, Daali Y, Wagner M et al. The effects of CYP2D6 and CYP3A activities on the pharmacokinetics of immediate release oxycodone. Br. J. Pharmacol. 160(4), 907–918 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grönlund J, Saari T, Hagelberg N et al. Effect of telithromycin on the pharmacokinetics and pharmacodynamics of oral oxycodone. J. Clin. Pharmacol. 50(1), 101–108 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Saari TI, Grönlund J, Hagelberg NM et al. Effects of itraconazole on the pharmacokinetics and pharmacodynamics of intravenously and orally administered oxycodone. Eur. J. Clin. Pharmacol. 66(4), 387–397 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Nieminen TH, Hagelberg NM, Saari TI et al. Rifampin greatly reduces the plasma concentrations of intravenous and oral oxycodone. Anesthesiology 110(6), 1371–1378 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Soldin OP, Mattison DR. Sex differences in pharmacokinetics and pharmacodynamics. Clin. Pharmacokinet. 48(3), 143–157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kokki M, Heikkinen M, Välitalo P et al. Maturation of oxycodone pharmacokinetics in neonates and infants: oxycodone and its metabolites in plasma and urine. Br. J. Clin. Pharmacol. 83(4), 791–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Välitalo P, Kokki M, Ranta V-P, Olkkola KT, Hooker AC, Kokki H. Maturation of oxycodone pharmacokinetics in neonates and infants: a population pharmacokinetic model of three clinical trials. Pharm. Res. 34(5), 1125–1133 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Kokki H, Rasanen I, Reinikainen M, Suhonen P, Vanamo K, Ojanperä I. Pharmacokinetics of oxycodone after intravenous, buccal, intramuscular and gastric administration in children. Clin. Pharmacokinet. 43(9), 613–622 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Korjamo T, Tolonen A, Ranta V-P, Turpeinen M, Kokki H. Metabolism of oxycodone in human hepatocytes from different age groups and prediction of hepatic plasma clearance. Front. Pharmacol. 2, 87–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liukas A, Kuusniemi K, Aantaa R et al. Plasma concentrations of oral oxycodone are greatly increased in the elderly. Clin. Pharmacol. Ther. 84(4), 462–467 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Saari TI, Ihmsen H, Neuvonen PJ, Olkkola KT, Schwilden H. Oxycodone clearance is markedly reduced with advancing age: a population pharmacokinetic study. Br. J. Anaesth. 108(3), 491–498 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Zwisler ST, Enggaard TP, Noehr-Jensen L et al. The antinociceptive effect and adverse drug reactions of oxycodone in human experimental pain in relation to genetic variations in the OPRM1 and ABCB1 genes. Fundam. Clin. Pharmacol. 24(4), 517–524 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Zwisler ST, Enggaard TP, Mikkelsen S et al. Lack of association of OPRM1 and ABCB1 single-nucleotide polymorphisms to oxycodone response in postoperative pain. The J. Clin. Pharmacol. 52(2), 234–242 (2012). [DOI] [PubMed] [Google Scholar]; • The authors do not find any association between the variant G allele in the OPRM1 gene and the variant Tallele in the ABCB1 gene with alteration in analgesic effect of oxycodone. This study contrasts most reviewed articles that found significant associations between SNPs of OPRM1 and ABCB1 and altered antinociceptive of oxycodone.
- 49.Olesen AE, Sato H, Nielsen LM et al. The genetic influences on oxycodone response characteristics in human experimental pain. Fundam. Clin. Pharmacol. 29(4), 417–425 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Cajanus K, Kaunisto MA, Tallgren M, Jokela R, Kalso E. How much oxycodone is needed for adequate analgesia after breast cancer surgery: effect of the OPRM1 118A>G polymorphism. J. Pain. 15(12), 1248–1256 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Lin T, Li X, Song J, Zhang C, Bie M. [Relationship of OPRM1 118A/G gene polymorphism and oxycodone analygesic dose in paitents with cancer pain]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 35(6), 887–890 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Lam J, Kelly L, Ciszkowski C et al. Central nervous system depression of neonates breastfed by mothers receiving oxycodone for postpartum analgesia. J. Pediatr. 160(1), 33–37 e32 (2012). [DOI] [PubMed] [Google Scholar]; • Compares the incidence of CNS depression in neonates breastfed by mothers taking with oxycodone as compared with neonates whose breastfeeding mothers used codeine or acetaminophen only and concludes that oxycodone is not a safer alternative to codeine in breastfed infants.
- 53.Kambur O, Kaunisto MA, Tikkanen E, Leal SM, Ripatti S, Kalso EA. Effect of catechol-o-methyltransferase-gene (COMT) variants on experimental and acute postoperative pain in 1,000 women undergoing surgery for breast cancer. Anesthesiology 119(6), 1422–1433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samer CF, Daali Y, Wagner M et al. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br. J. Pharmacol. 160(4), 919–930 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andreassen TN, Eftedal I, Klepstad P et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross-sectional multicentre study. Eur. J. Clin. Pharmacol. 68(1), 55–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naito T, Takashina Y, Yamamoto K et al. CYP3A5*3 affects plasma disposition of noroxycodone and dose escalation in cancer patients receiving oxycodone. J. Clin. Pharmacol. 51(11), 1529–1538 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Balyan R, Mecoli M, Venkatasubramanian R et al. CYP2D6 pharmacogenetic and oxycodone pharmacokinetic association study in pediatric surgical patients. Pharmacogenomics 18(4), 337–348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Aims to associate CYP2D6 genotype and oxycodone’s metabolism in children who had inpatient surgery and reported that CYP2D6 phenotypes explain metabolism of oxycodone and oxymorphone exposure is higher in CYP2D6 extensive metabolizers compared with poor metabolizers.
- 58.Dagostino C, Allegri M, Napolioni V et al. CYP2D6 genotype can help to predict effectiveness and safety during opioid treatment for chronic low back pain: results from a retrospective study in an Italian cohort. Pharmgenomics Pers Med. 11, 179–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klein K, Zanger UM. Pharmacogenomics of cytochrome P450 3A4: recent progress toward the “missing heritability” problem. Front. Genet. 4, 12–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin. Pharmacokinet. 48(11), 689–723 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Zwisler ST, Enggaard TP, Mikkelsen S, Brosen K, Sindrup SH. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol. Scand. 54(2), 232–240 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Raffa RB, Pergolizzi JV, Segarnick DJ, Tallarida RJ. Oxycodone combinations for pain relief. Drugs Today (Barc.) 46(6), 379–398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monory K, Greiner E, Sartania N et al. Opioid binding profiles of new hydrazone, oxime, carbazone and semicarbazone derivatives of 14-alkoxymorphinans. Life Sci. 64(22), 2011–2020 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Chen ZR, Irvine RJ, Somogyi AA, Bochner F. Mu receptor binding of some commonly used opioids and their metabolites. Life Sci. 48(22), 2165–2171 (1991). [DOI] [PubMed] [Google Scholar]
- 65.Lemberg KK, Kontinen VK, Siiskonen AO et al. Antinociception by spinal and systemic oxycodone: why does the route make a difference? In vitro and in vivo studies in rats. Anesthesiology 105(4), 801–812 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Lalovic B, Kharasch E, Hoffer C, Risler L, Liu-Chen LY, Shen DD. Pharmacokinetics and pharmacodynamics of oral oxycodone in healthy human subjects: role of circulating active metabolites. Clin. Pharmacol. Ther. 79(5), 461–479 (2006). [DOI] [PubMed] [Google Scholar]; • Gives a detailed description of oxycodone’s metabolites and their impact to oxycodone’s pharmacodynamic was evaluated by pharmacokinetic–pharmacodynamic modeling.
- 67.Childers SR, Creese I, Snowman AM, Synder SH. Opiate receptor binding affected differentially by opiates and opioid peptides. Eur. J. Pharmacol. 55(1), 11–18 (1979). [DOI] [PubMed] [Google Scholar]
- 68.Leow KP, Smith MT. The antinociceptive potencies of oxycodone, noroxycodone and morphine after intracerebroventricular administration to rats. Life Sci. 54(17), 1229–1236 (1994). [DOI] [PubMed] [Google Scholar]
- 69.Klimas R, Witticke D, El Fallah S, Mikus G. Contribution of oxycodone and its metabolites to the overall analgesic effect after oxycodone administration. Expert Opin. Drug Metab. Toxicol. 9(5), 517–528 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Lemberg K, Heiskanen T, Kontinen V, Kalso E. Pharmacology of oxycodone: does it explain why oxycodone has become a bestselling strong opioid? Scand. J. Pain 1, S18–S23 (2009). [Google Scholar]
- 71.Peckham EM, Traynor JR. Comparison of the antinociceptive response to morphine and morphine-like compounds in male and female Sprague-Dawley rats. J. Pharmacol. Exp. Ther. 316(3), 1195–1201 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Silvasti M, Rosenberg P, Seppälä T, Svartling N, Pitkänen M. Comparison of analgesic efficacy of oxycodone and morphine in postoperative intravenous patient-controlled analgesia. Acta Anaesthesiol. Scand. 42(5), 576–580 (1998). [DOI] [PubMed] [Google Scholar]
- 73.Kalso E, Vainio A. Morphine and oxycodone hydrochloride in the management of cancer pain. Clin. Pharmacol. Ther. 47(5), 639–646 (1990). [DOI] [PubMed] [Google Scholar]
- 74.Yazdy MM, Mitchell AA, Tinker SC, Parker SE, Werler MM. Periconceptional use of opioids and the risk of neural tube defects. Obstet. Gynecol. 122(4), 838–844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Broussard CS, Rasmussen SA, Reefhuis J et al. Maternal treatment with opioid analgesics and risk for birth defects. Am. J. Obstet. Gynecol. 204(4), 314.e31–314.e311 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Fan HSL, Wong JYH, Fong DYT, Lok KYW, Tarrant M. Association between intrapartum factors and the time to breastfeeding initiation. Breastfeed Med. 15(6), 394–400 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Niklasson B, Arnelo C, Georgsson Öhman S, Segerdahl M, Blanck A. Oral oxycodone for pain after caesarean section: a randomized comparison with nurse-administered IV morphine in a pragmatic study. Scand. J. Pain 7(1), 17–24 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Timm NL. Maternal use of oxycodone resulting in opioid intoxication in her breastfed neonate. J. Pediatr. 162(2), 421–422 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Fields HL, Margolis EB. Understanding opioid reward. Trends Neurosci. 38(4), 217–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jordan CJ, Humburg B, Rice M et al. The highly selective dopamine D(3)R antagonist, R-VK4-40 attenuates oxycodone reward and augments analgesia in rodents. Neuropharmacology 158, 107597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boateng CA, Bakare OM, Zhan J et al. High affinity dopamine D3 receptor (D3R)-selective antagonists attenuate heroin self-administration in wild-type but not D3R knockout mice. J. Med. Chem. 58(15), 6195–6213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You ZB, Gao JT, Bi GH et al. The novel dopamine D3 receptor antagonists/partial agonists CAB2-015 and BAK4-54 inhibit oxycodone-taking and oxycodone-seeking behavior in rats. Neuropharmacology 126, 190–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matzeu A, Martin-Fardon R. Targeting the orexin system for prescription opioid use disorder: orexin-1 receptor blockade prevents oxycodone taking and seeking in rats. Neuropharmacology 164, 107906 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen JD, Hwang CS, Grant Y, Janda KD, Taffe MA. Prophylactic vaccination protects against the development of oxycodone self-administration. Neuropharmacology 138, 292–303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ikeda K, Ide S, Han W, Hayashida M, Uhl GR, Sora I. How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol. Sci. 26(6), 311–317 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Stamer UM, Stüber F. The pharmacogenetics of analgesia. Expert Opin. Pharmacother. 8(14), 2235–2245 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Mura E, Govoni S, Racchi M et al. Consequences of the 118A>G polymorphism in the OPRM1 gene: translation from bench to bedside? J. Pain Res. 6, 331–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skarke C, Darimont J, Schmidt H, Geisslinger G, Lötsch J. Analgesic effects of morphine and morphine-6-glucuronide in a transcutaneous electrical pain model in healthy volunteers. Clin. Pharmacol. Ther. 73(1), 107–121 (2003). [DOI] [PubMed] [Google Scholar]
- 89.Hayashida M, Nagashima M, Satoh Y et al. Analgesic requirements after major abdominal surgery are associated with OPRM1 gene polymorphism genotype and haplotype. Pharmacogenomics 9(11), 1605–1616 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Naito T, Takashina Y, Yamamoto K et al. CYP3A53 affects plasma disposition of noroxycodone and dose escalation in cancer patients receiving oxycodone. J. Clin. Pharmacol. 51(11), 1529–1538 (2011). [DOI] [PubMed] [Google Scholar]
- 91.Kim KM, Kim HS, Lim SH et al. Effects of genetic polymorphisms of OPRM1, ABCB1, CYP3A4/5 on postoperative fentanyl consumption in Korean gynecologic patients. Int. J. Clin. Pharmacol. Ther. 51(5), 383–392 (2013). [DOI] [PubMed] [Google Scholar]
- 92.Badreldin N, Grobman WA, Chang KT, Yee LM. Opioid prescribing patterns among postpartum women. Am. J. Obstet. Gynecol. 219(1), 103.e1–103.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Communication FDS. FDA restricts use of prescription codeine pain and cough medicines and tramadol pain medicines in children; recommends against use in breastfeeding women (2017). https://wwwfdagov/media/104268/download
- 94.Lam J, Kelly L, Matok I et al. Putative association of ABCB1 2677G>T/A with oxycodone-induced central nervous system depression in breastfeeding mothers. Ther. Drug Monit. 35(4), 466–472 (2013). [DOI] [PubMed] [Google Scholar]
- 95.Wightman R, Perrone J, Portelli I, Nelson L. Likeability and abuse liability of commonly prescribed opioids. J. Med. Toxicol. 8(4), 335–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jones JD, Mumtaz M, Manubay JM et al. Assessing the contribution of opioid- and dopamine-related genetic polymorphisms to the abuse liability of oxycodone. Pharmacol. Biochem. Behav. 186, 172778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou H, Rentsch CT, Cheng Z et al. Association of OPRM1 functional coding variant with opioid use disorder: a genome-wide association study. JAMA Psychiatry. 77(10), 1072–1080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Doehring A, Hentig N, Graff J et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenet. Genomics 19(6), 407–414 (2009). [DOI] [PubMed] [Google Scholar]
- 99.Clarke TK, Weiss AR, Ferarro TN et al. The dopamine receptor D2 (DRD2) SNP rs1076560 is associated with opioid addiction. Ann. Hum. Genet. 78(1), 33–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zwisler ST, Enggaard TP, Noehr-Jensen L et al. The hypoalgesic effect of oxycodone in human experimental pain models in relation to the CYP2D6 oxidation polymorphism. Basic Clin. Pharmacol. Toxicol. 104(4), 335–344 (2009). [DOI] [PubMed] [Google Scholar]
- 101.Hagelberg NM, Nieminen TH, Saari TI et al. Voriconazole drastically increases exposure to oral oxycodone. Eur. J. Clin. Pharmacol. 65(3), 263–271 (2009). [DOI] [PubMed] [Google Scholar]
- 102.Crews KR, Gaedigk A, Dunnenberger HM et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95(4), 376–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Summarizes evidence from the literature and provide therapeutic recommendations for opioid use on CYP2D6 genotype.
- 103.Crews KR, Caudle KE, Dunnenberger HM, Sadhasivam S, Skaar TC. Considerations for the utility of the CPIC guideline for CYP2D6 genotype and codeine therapy. Clin. Chem. 61(5), 775–776 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Smolina K, Weymann D, Morgan S, Ross C, Carleton B. Association between regulatory advisories and codeine prescribing to postpartum women. JAMA 313(18), 1861–1862 (2015). [DOI] [PubMed] [Google Scholar]
- 105.Finkelstein Y, Macdonald EM, Gonzalez A et al. Overdose risk in young children of women prescribed opioids. Pediatrics 139(3), e20162887 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Levine B, Moore KA, Aronica-Pollak P, Fowler DF. Oxycodone intoxication in an infant: accidental or intentional exposure? J. Forensic Sci. 49(6), 1358–1360 (2004). [PubMed] [Google Scholar]
- 107.Gasche Y, Daali Y, Fathi M et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N. Engl. J. Med. 351(27), 2827–2831 (2004). [DOI] [PubMed] [Google Scholar]
- 108.Voronov P, Przybylo HJ, Jagannathan N. Apnea in a child after oral codeine: a genetic variant – an ultra-rapid metabolizer. Paediatr. Anaesth. 17(7), 684–687 (2007). [DOI] [PubMed] [Google Scholar]
- 109.Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet 368(9536), 704 (2006). [DOI] [PubMed] [Google Scholar]
- 110.Madadi P, Ross CJ, Hayden MR et al. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin. Pharmacol. Ther. 85(1), 31–35 (2009). [DOI] [PubMed] [Google Scholar]
- 111.Stamer UM, Stüber F, Muders T, Musshoff F. Respiratory depression with tramadol in a patient with renal impairment and CYP2D6 gene duplication. Anesth. Analg. 107(3), 926–929 (2008). [DOI] [PubMed] [Google Scholar]
- 112.Orliaguet G, Hamza J, Couloigner V et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics 135(3), e753–755 (2015). [DOI] [PubMed] [Google Scholar]
- 113.Kuehn BM. FDA: no codeine after tonsillectomy for children. JAMA 309(11), 1100–1100 (2013). [DOI] [PubMed] [Google Scholar]


