Abstract
Now a days, more than 200 countries faces the health crisis due to epidemiological disease COVID-19 caused by SARS-CoV-2 virus. It will cause a very high impact on world’s economy and global health sector. Earlier the structure of main protease (Mpro) protein was deposited in the RCSB protein repository. Hydroxychloroquine (HCQ) and remdesivir were found to effective in treatment of COVID-19 patients. Here we have performed docking and molecule dynamic (MD) simulation study of HCQ and remdesivir with Mpro protein which gave promising results to inhibit Mpro protein in SARS-CoV-2. On the basis of results obtained we designed structurally modified 18 novel derivatives of HCQ, remdesivir and tetrahydrocannabinol (THC) and carried out docking studies of all the derivatives. From the docking studies six molecules DK4, DK7, DK10, DK16, DK17 and DK19 gave promising results and can be use as inhibitor for Mpro of SARS-CoV-2 to control COVID-19 very effectively. Further, molecular dynamics simulation of one derivative of HCQ and one derivative of tetrahydrocannabinol showing excellent docking score was performed along with the respective parent molecules. The two derivatives gave excellent docking score and higher stability than the parent molecule as validated with molecular dynamics (MD) simulation for the binding affinities towards Mpro of SARS-CoV-2 thus represented as strong inhibitors at very low concentration.
Keywords: COVID-19, SARS-CoV-2, Hydroxychloroquine, Remdesivir, THC, Docking, MD simulation
1. Introduction
In the Wuhan city, the capital of Hubei province (China), since late December of 2019, firstly a case of pneumonia like viral infection attracted the attention of scientists, academicians and pharmaceuticals worldwide [1], [2], [3], [4]. It was an outbreak of pneumonia like symptoms with abnormal lungs, due to the anonymous reasons with an epidemiological among humans [2]. Earlier the terms like “the new coronavirus” and “Wuhan coronavirus” were used in a very common manner by January 2020. On the 11th February 2020, this virus strain, earlier termed as ‘2019-nCoV & Wuhan virus’ was officially announced a taxonomic name “severe acute respiratory syndrome coronavirus-2” (SARS-CoV-2) [5] due to the resemblance of the RNA genome of the previously identified coronavirus (SARS-CoV). On the same day, finally the world health organization (WHO) referred the name of this disease as COVID-19 [6]. This virus represents a pandemic threat to global public health and has already infected millions of the people and leads to more than hundred thousands of deaths globally in more than 200 countries. These numbers are increasing rapidly as of this manuscript writing. If the rapid spread of this virus- SARS-CoV-2 will be not controlled, it will probably bring a foremost challenge for global health system and world’s economy [7], [8], [9].
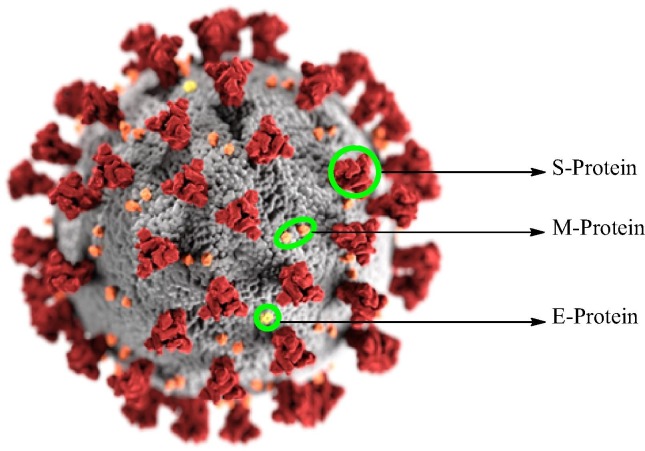
Coronaviruses (CoVs) are comparatively larger in size than other viruses and shared a greater than 79% sequence identity to SARS-CoV [10]. Coronaviruses (CoVs) are enveloped single stranded positive sense RNA viruses and their family can be subdivided into four groups- alpha-, beta-, gamma and delta- CoV [11], [12]. Before the identification of SARS-CoV-2, only six of the previously known CoVs were cause mild to severe illnesses in humans. The alpha coronavirous (HCoV-229E and HCoV-NL63), cause milder respiratory disease in the upper respiratory tract. However the beta form of CoVs (HCoV-OC43, HKU1, and SARS-CoV are responsible for severe acute respiratory syndrome [12], [13]. While it’s another form MERS-CoV (Middle east resipartory syndrome coronavirus) is responsible for infection in the lower respiratory tract [13]. The coronavirus has its appearance like crown shape (Fig. 1 ) and its structure is quite complex which contains four structural proteins [14] spike proteins (S), envelope protein (E), membrane protein (M) and nucleocapsid proteins (N) and several unidentified nonstructural protein. Recent studies shows that the spike protein and chymotrypsin-like protease (3CLpro) also called as main protease (Mpro) plays an important role in mediating viral replication and transcription [15], [16], [17], [18] and hence is an attractive target for drug development for SARS-CoV-2. The 3CLprocatalyze the cleavage of polyprotein at 11 distinct sites and produces a various nonstructural protein which is responsible for viral replications. Therefore by the inhibiting the activity of this enzyme the researchers can cure the viral replication [17], [18].
Fig. 1.
Structure of the virus caused COVID-19 i.e. SARS-CoV-2.
The coronavirus can infect both animals as well as humans. Few animals like bat, hosts various types of coronaviruses appears to be immune to coronavirus prompted sickness [19]. Latest establishment shows that SARS-CoV-2 infects the lower respiratory system and results in viral pneumonia. Along with viral pneumonia SARS-CoV-2 may also attacks at kidney, heart, liver, gastrointestinal and central nervous system causing multiple organ failure [20], [21]. Current advancement about its transmission suggests that this new coronavirus strain, SARS-CoV-2 is more contagious than another coronavirus family [22]. Therefore structure-based virtual computational studies will be used to investigate the best drugs and is a fundamental approach to design highly potent inhibitor which can cure the viral replication.
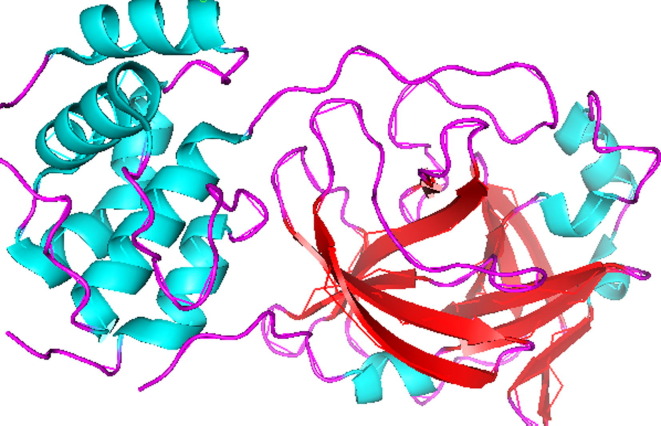
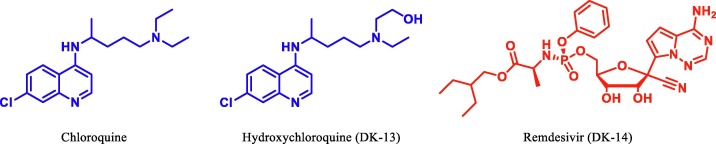
The crystal structure of main protease (Mpro) of SARS-CoV-2 is determined and deposited in RCSB with PDB ID: 6LU7 [23] Fig. 2 . According to our recent literature studies, most of the potential drugs contains heterocyclic moieties and was found to be active against SARS-CoV-2 Mpro. Recently antiviral drug Remdesivir [24], [25] and antimalerial drug hydroxychloroquine (HCQ)/chloroquine [26], [27], [28], [29] were found to inhibit effectively to control SARS-CoV-2 in vitro and are currently being used in the treatment of patients with SARS-CoV-2 infection (Fig. 3 ).
Fig. 2.
The crystal structure of Mpro (PDB id: 6LU7) of SARS-CoV-2.
Fig. 3.
Chemical structure of Chloroquine, HCQ and Remdesivir.
HCQ is less toxic than chloroquine and also found to be a potent inhibitor against the SARS-CoV-2 in vitro [30]. Herein, in order to manage/control this pandemic situation we have suggested few modified chloroquine, remdesivir and tetrahydrocannabinol (THC) and studied their inhibition abilities towards Mpro using molecular docking and were further validated by molecular dynamics (MD) simulation study. In order to understand the properties of molecular assemblies in terms of their structure and the microscopic interactions between them computer simulations plays a significant role. Molecular dynamics simulation provides a route to dynamical properties of a system; transport coefficients, time-dependent responses to perturbations, spectra, rheological properties [31]. Molecular dynamics simulation comprised of numerical, step-by-step solution of the classical equations of motion that for a simple atomic system can be written as Eq. (1).
| (1) |
For this we need to calculate the forces fi, acting on the atoms, and these are derived from a potential energy μ (rN), where rN = (r1, r2, r3…..rN) representing the complete set of 3 N atomic coordinates.
Molecular dynamics simulations in drug discovery play an important role as they provide insight into the protein motion. The static models provided by homology modelling, NMR, X-ray crystallography gives insights about the macromolecular structure while drug binding and molecular recognition are very dynamic processes that can be studied using molecular dynamics simulations [32]. MD simulations are often used to explore the conformation space of molecules, especially the large molecules such as proteins. In MD simulation the energy surface is studied using the newton’s law of motion for system. In the docked structures MD simulation is useful to confirm whether the major contacts found in docking are maintained during the MD simulation hence presenting the more reliable results. In other words the static view presented by docking is verified by the MD simulation. Generally by combining the two in-silico techniques, docking and MD simulation will provide the more reliable results.
In the current prevailing situation where HCQ, remdesivir are being used by the clinical practitioners in the treatment of patients with SARS-CoV-2 infection, then in the current study we proposed the structural modifications of these drugs on the basis of strong theoretical chemistry concepts. Then, computationally assessed the binding affinity of the designed molecules by docking studies and were further validated by the molecular dynamics simulation study (MD simulation) of the two potential derivatives showing the binding affinity more than the parent compounds.
2. Calculations
The step-wise methodology followed for in-silico interaction pattern study of hydroxychloroquine (HCQ), remdesivir, and tetrahydrocannabinol (THC) and their derivatives for characterizing the potent drug molecules targeting Mpro/3CLpro protein of SARS-CoV-2 is described as follow.
2.1. Protein, hydroxychloroquine, remdesivir and tetrahydrocannabinol derivatives designing and their geometry optimization
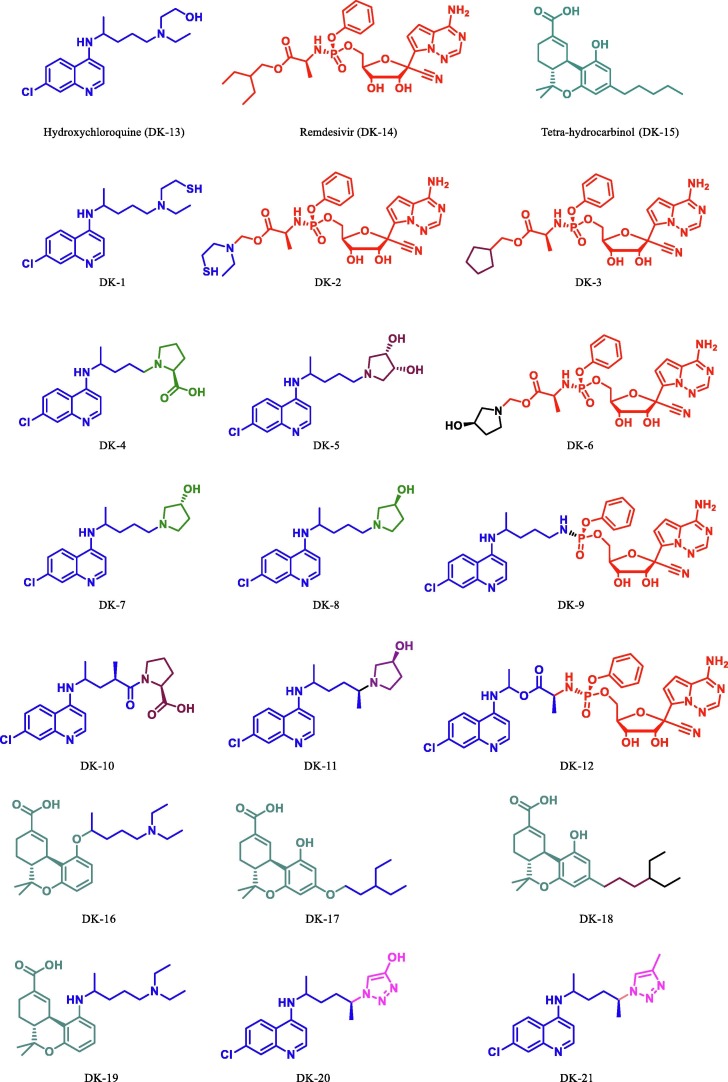
A dataset of eighteen derivatives of hydroxychloroquine, remdesivir, and tetrahydrocannabinol were designed (Fig. 4 ). The initial structures of all the derivatives were drawn in Gauss view 3.0 and all the molecular structures of derivatives along with the parent drugs were optimized at the level of parametrization method 6 (PM6) semi-empirical method in Gaussian 09, quantum chemistry software [33], [34], [35].
Fig. 4.
Derivatives of HCQ/Chloroquine, Ramdesivir and THC.
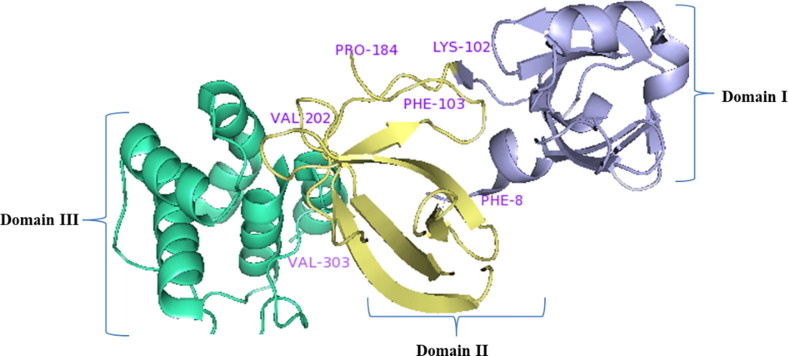
Main protease protein (Mpro/3CLpro) of SARS-CoV-2 is a homodimer with two chains, A and B [36]. The two chains are associated by crystallographic 2-fold symmetry to form dimer. Each protomer is comprises of three domains. Domain I (residues 8–101) and domain II (residues 102–184) forms an antiparallel beta-barrel structure and domain III (residues 201–303) comprised five alpha-helices those are arranged into a largely into an antiparallel globular structure and is connected to domain II by means of long loop region (residues 185–200) (Shown in Fig. 5 ). Mpro/3CLpro protein of SARS-CoV-2 has a catalytic dyad comprises of Cys-His and substrate binding site is located in the cleft between domain I and II [37].
Fig. 5.
Structure of main protease protein (Mpro/3CLpro) of SARS-CoV-2.
The functional role of Mpro in the life-cycle of virus and also no closely related homologue of this protein in humans identify it as an attractive target for the anti-viral drugs. The three-dimensional structure of chain A from Mpro/3CLpro (PDB ID: 6LU7, 2.16 Å) of SARS-CoV-2 was taken from PDB (http://www.rcsb.org/) [23]. In the current study chain A was taken for macromolecule preparation. Chimera 1.13.1 software was used for carrying out the minimization of protein structure implementing Amber ff99Sb force field [38].
2.2. Interpretation of binding site
The Mpro active site mainly contains sub-sites commonly named as S1, S2, S1′, S2′, S4. The amino acid residue Phe140, Leu141, His163, Met165, Gln166 and His172 are present in S1 site, while Met49 and Asp187, Glu189 are present in bulky hydrophobic subsite S2 [39]. The amino acid residue Thr25, Leu27, Cys38, Pro-39, Val42, and Cys145 present in S1′ subsite and play an important role in catalysis [18]. The Thr-26, Asn-28, Tyr-118, Asn-119, and Gly-143 are found in S2,’while the flexible S4 site contains Leu167, Gln192 [39], [40]. The active site was predicted by CastP software active site prediction tool [41].
2.3. Molecular docking study
Dataset of optimized molecular structures of hydroxychloroquine, remdesivir, THC and its derivatives were subjected to molecular docking against the Mpro of SARS-CoV-2 in the above defined active site using AutoDock 4.2.2 [42] and AutoDock Vina [43]. AutoDock implies 3-dimemsional potential grids to calculate the potential energy of docking molecules. Grid preparation module of AutoDock was used for the generation of a 3-dimensional grid box encompassing the active-site residues and a grid box with grid points dimensionality of X = 100, Y = 126, Z = 96 having grid spacing of 0.375 Å was generated. Lamarckian genetic algorithm and empirical scoring function of the docking software was employed for carrying out molecular docking and for ranking the docked molecules respectively. Binding conformation of the protein–ligand complex was used to calculate the intermolecular energy taking the unbound protein–ligand conformation as the initial conformation. For the docking process all the molecules were set with the population size of 150, number of maximum generations 27,000, maximum evaluations of 2,50,000 with a gene mutation rate of 0.02, and crossover rate of 0.8. All the molecules were employed for the generation of 100 binding poses for the respective docking.
2.4. Molecular dynamics simulations
To study the stability of protein–ligand complex i.e., The molecular dynamics (MD) simulation study [44], we have used the Gromacs 5.1.2 software which is widely used. The MD simulation of the hydroxychloroquine and tetrahydrocannabinol and their derivative DK7 and DK16 respectively those were showing the best binding affinity from the docking study against Mpro was carried out. All the four systems were employed for 100 ns time scale simulation. Protein topology was generated by GROMOS 96 54a7 forcefield [45] in Gromacs while the ligands topology was obtained using automated topology builder (ATB) server [46]. Solvation of protein–ligand complex was carried out in a cubic box of size 9.73 × 9.73 × 9.73 nm using SPC water model. Systems were neutralized by the addition of four Cl− ions and then the energy minimization of the systems was carried out using the steepest descent algorithm. Particle-Mesh Ewald method [47] was used for the calculation of electrostatic interactions of the systems. A normal cut-off of 0.9 nm was implied for Van der Waals interaction. Constraints imposed by the covalent bonds including hydrogen atoms of the system were calculated by a linear constraint solver for molecular simulations (LINCS) algorithm. NVT (Number of particles, Volume and Temperature) and NPT (Number of particles, Pressure and Temperature) simulations were run on the system for 1 ns using Berendsen thermostat method [48] to control the temperature at 300 K with a coupling time of 0.1 ps and pressure was restrained at 1 bar [49]. The co-ordinates were saved in 0.002 fs for all the systems [50]. Different analytical tools such as gmx rms, gmxrmsf, gmx gyration, gmxh_bond, and gmxsasa were implied for the calculation of RMSD, RMSF, Rg, Hydrogen bonds and SASA respectively. The RMSD and RMSF trajectories were also resolved with the help of visualization software like Visual Molecular Dynamics (VMD) [51] and Chimera 1.13.1 [38] visualization tools and the graphs were plotted using the Origin software.
2.5. Principal component analysis
Analysis of eigenvectors and eigenvalues was performed using Principal component analysis (PCA) or Essential dynamics method in Gromacs for obtaining the projection of first two Principal Components (PCs). Protein function is majorly defined by the associated motion of the protein which was obtained by PCA [52]. All the rotational and translational movements were removed and co-variance matrix was formed. The atomic coordinates from the positional covariance C and its eigenvectors were used in the following equation for the calculation of elements of the positional covariance matrix C:
| (2) |
The ith Cα atom was represented by the cartesian coordinate qi and Cα atom was denoted by N in the protein–ligand bound systems. Complete translational and rotational movements using ‘Least square method’ of the equilibrated trajectory was removed that was further superimposed on a reference structure. For the prediction of rest of eigenvectors and eigenvalues λi, all the matrices were diagonalized using orthogonal coordinate transformation matrix Λ.
| (3) |
In the equation, eigenvectors corresponding to the direction of motion relative to <qi> were represented in the columns and each eigenvector associated with the eigenvalue represents the total mean-square fluctuation of the system ahead the corresponding eigenvector. gmxcovar and gmxanaeig tools of Gromacs were used for the calculation of eigenvector and eigenvalues for the obtained last 20 ns trajectories.
3. Results and discussion
3.1. Molecular docking
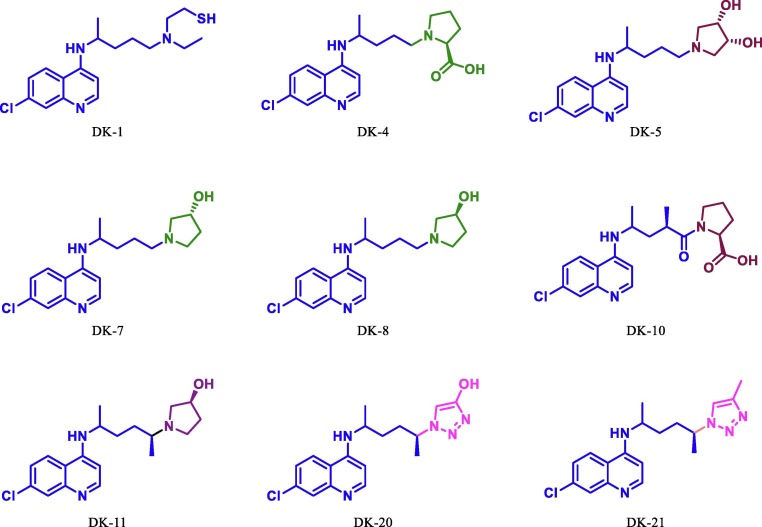
Since chloroquine based antimalarial drug HCQ and remdesivir, the antiviral drug is in use for the treatment of COVID-19. Few reports suggest that cannabis and their derivatives have many therapeutic applications including antiviral activity [53], [54], [55], [56], [57]. Herein, we have done structural modification in the HCQ/chloroquine, remdesivir and THC (Fig. 4) and perform virtual screening on Mpro. Dataset of optimized molecular structures of hydroxychloroquine, remdesivir, THC and its derivatives were subjected to molecular docking against the Mpro of SARS-CoV-2 (PDB Id: 6LU7) in the defined active site using AutoDock 4.2.2 [42] and AutoDock Vina [43].The free binding energy and inhibitory constant (Ki) of all molecules are sown in table 1 . During our initial studies for the designing of potent COVID-19 inhibitor first we put modification at chain of the tertiary nitrogen atom of hydroxychloroquine/chloroquine derivative. This can be done by substituting a variety of aliphatic open chain and different derivatives of pyrrolidine. By using this concept, we have studied and compare the free binding energy along with inhibition constant of nine novel derivatives of hydroxychloroquine/chloroquine, these are DK1, DK4, DK5, DK7, DK8, DK10, DK11, DK20 and DK21 (Fig. 6 ). The effect of the substituent was further compared with the standard drug DK13 (hydroxychloroquine).
Table 1.
Docking study result of the derivatives of hydroxychloroquine, remdesivir, and tetrahydrocannabinol with PM6 optimized geometry against main protease (Mpro) showing the binding affinity and the interacting residues.
| S No. | Molecule | aFree binding energy(autodock)(kcal.mol−1) | Inhibiton Constant Ki ((µM) autodock |
bFree binding energy (autodock vina) (kcal.mol−1) | Interacting residues |
|---|---|---|---|---|---|
| DK1 | −5.37 | 115.27 | −5.30 | Phe3, Arg4, Lys5, Trp207, Leu282, Leu286, Leu287, Glu288, Asp289, Glu290, Phe291 | |
| DK2 | −6.45 | 16.38 | −6.60 | Gly2, Phe3, Arg4, Lys5, Trp207, Leu282, Phe291 | |
| DK3 | −6.52 | 16.52 | −7.02 | Phe3,Arg4, Lys5, Met6, Ala7, Val125, Tyr126, Gln127, Trp207, Leu282, Phe291 | |
| DK4 | −7.11 | 6.12 | −6.90 | Phe8, Pro9, Asn151, Ile152, Asp153, Tyr154, Phe294, Arg298 | |
| DK5 | −6.01 | 39.13 | −6.50 | Asp33, Tyr37, Pro99, Lys100, Tyr101, Lys102, Phe103 | |
| DK6 | −5.92 | 45.72 | −6.12 | Phe3, Arg4, Lys5, Tyr126, Gln127, Lys137, Gly138, Ser139, Trp207, Ser284, Glu288, Phe291 | |
| DK7 | −7.56 | 2.72 | −7.21 | His41, Phe140, Leu141, Ser144, His163, Met165, Glu166, Asp187, Arg188, Gln189 | |
| DK8 | −6.78 | 10.76 | −6.42 | Thr198, Thr199, Tyr237, Asn238, Tyr239, Leu271, Leu272, Gly275, Met276, Leu286, Leu287 | |
| DK9 | −5.59 | 79.38 | −6.14 | Phe3, Arg4, Lys5, Val125, Tyr126, Gln127, Trp207, Leu282, Gly283, Ser284, Leu286, Glu288, Phe291 | |
| DK10 | −7.14 | 5.87 | −7.23 | Phe3, Arg4, Lys5, Trp207, Leu282, Glu288, Phe291 | |
| DK11 | −6.86 | 9.33 | −6.98 | Pro108, Gly109, Gln110, Pro132, Ile200, Val202, Asn203, Glu240, His246, Thr292, Pro293, Phe294 | |
| DK12 | −5.57 | 83.11 | −5.22 | Phe8, Pro9, Gln110, Thr111, Asn151, Ile152, Asp153, Tyr154, Phe294, Val297, Arg298 | |
| DK13* | −5.41 | 108.7 | −5.78 | Glu14, Gly15, Cys16, Met17, Gln19, Trp31, Gln69, Ala70, Gly71,Asn95,Lys97, Asn119, Gly120, Ser121, Pro122 | |
| DK14** | −5.30 | 129.48 | −5.60 | Phe3, Arg4, Lys5, Met6, Ala7, Val125, Tyr126, Gln127, Phe291 | |
| DK15*** | −6.99 | 7.56 | −6.58 | Glu14, Gly15, Met17, Val18,Gln19, Gln69, Ala70, Gly71, Lys97, Asn119, Gly120, Pro122 | |
| DK16 | −7.64 | 2.49 | −7.92 | Lys102, Val104 Ile106, Gln107, Gln110, Thr111, Asn151, Ile152, Asp153, Ser158, Phe294, | |
| DK17 | −7.62 | 2.90 | −7.46 | Thr26, Leu27, His41, Met49, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Pro168, Gln189, | |
| DK18 | −6.88 | 9.09 | −7.04 | Lys5, Met6, Ala7, Val125,Tyr126, Gln127, Cys128, Lys137, Gly138, Ser139, Glu290 | |
| DK19 | −7.41 | 3.66 | −7.67 | Phe8,Lys102, Val104, Ile106, Gln107, Gln110, Thr111, Asn151, Ile152, Asp153,Ser158, Phe294 | |
| DK20 | −6.16 | 30.28 | −6.29 | His41, Phe140, Leu141, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Gln189 | |
| 21. | DK21 | −6.13 | 32.27 | −6.32 | Phe8, Asn151, Ile152, Asp153, Tyr154, Phe294, Arg298, |
Hydroxychloroquine.
Remdesivir.
Tetrahydrocannabinol.
Data using Autodock.
Data using Autodock; Residues in bold are representing hydrogen bond forming residues.
Fig. 6.
Structurally modified HCQ/Chloroquine.
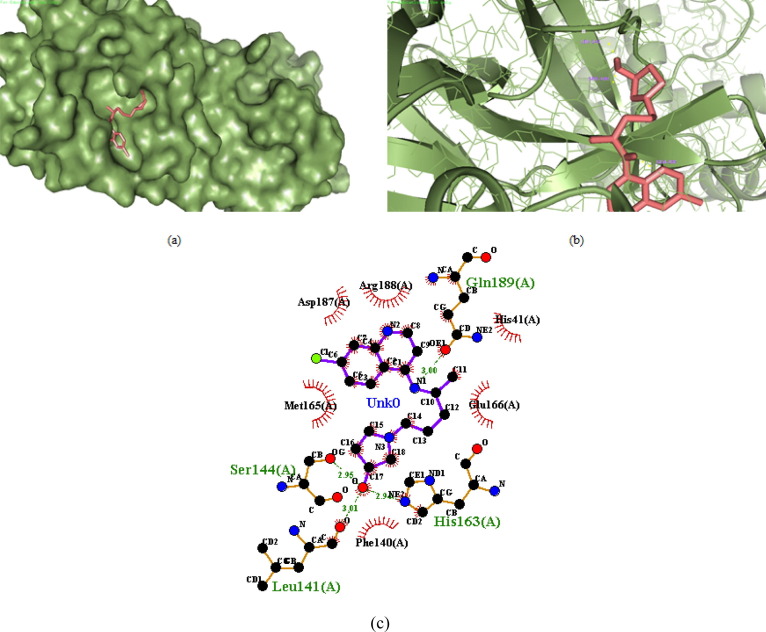
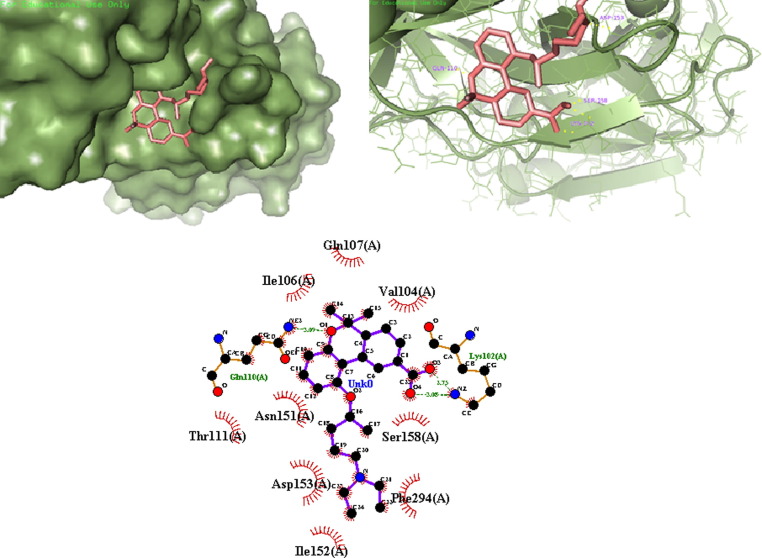
Out of the nine designed derivatives DK4, DK7 and DK10 were found to be more potent inhibitor against Mpro as compared to the standard drug hydroxychloroquine. The free binding energy for hydroxychloroquine (DK13) was found to be −5.65 kcal mol−1 while it was −7.11 kcal mol−1,-7.56 kcal mol−1, −7.14 kcal mol−1, for DK4, DK7 and DK10 respectively (Table1). The molecular Docking of DK7 suggested that the hydroxy substituted pyrrolidine ring makes a hydrogen bond with the Leu141, Ser144 and His163 of the active site of Mpro. The anilinic NH makes a hydrogen bond with the oxygen atom of Gln189, while His41, Phe140, Glu166, Asp187, Arg188 are involve in the hydrophobic interaction with the ring residue of the active site respectively (Shown in Fig. 7 , 2D and 3D images of DK7).
Fig. 7.
(a) 3D surface image of protein with DK7 represented in sticks model showing the interactions in the binding pocket of protein. (b) Protein is represented in cartoon model and DK7 is represented in sticks model showing the polar interaction in 3D with the bond lengths represented by yellow dashed lines and interacting residues of the proteins are labelled with residue name using the PyMOL interactive visualization tool. (c) 2D representation of DK7 with Mpro by Ligplot+.
We have also compared the free binding energy and inhibition activity of DK7 −7.56 kcal mol−1 with its isomer DK8 (−6.78 kcal mol−1), surprisingly, it was a more than three-fold change in inhibition constant against Mpro. We further investigate the effect of more polar —COOH functional group of pyrrolidine DK10. The molecular docking data reveals that the activity of DK10 (−7.13 kcal mol−1) is more than that of DK8 but less than that of DK7. We have also compared the effect of introducing one more OH group in pyrrolidine ring (adjacent and syn to –OH group in DK7), in DK5 (free binding energy −6.01 kcal mol−1, and it was found to be less active than DK7. It was also observed that the replacement of pyrrolidine moiety with planar 1,2,3 triazole derivative (DK20 and DK21, see Table 1) has no significant effect on the activity of the molecule against Mpro of SARS-CoV-2.
3.2. Molecular dynamics simulation HCQ-Mpro and DK7-Mpro
Further, molecular dynamics (MD) simulation which is a computerized method for studying the physical movements of the atoms while binding [58] was carried out for HCQ-Mpro and DK7-Mpro complexes. MD simulation of the docked complexes for 100 nano second (ns) was carried out and equilibration state for both the systems was obtained. The trajectories were observed for RMSF, Rg, SASA, and H-bonds for last 20 ns equilibrated trajectories including the binding free energy analysis also for last 20 ns.
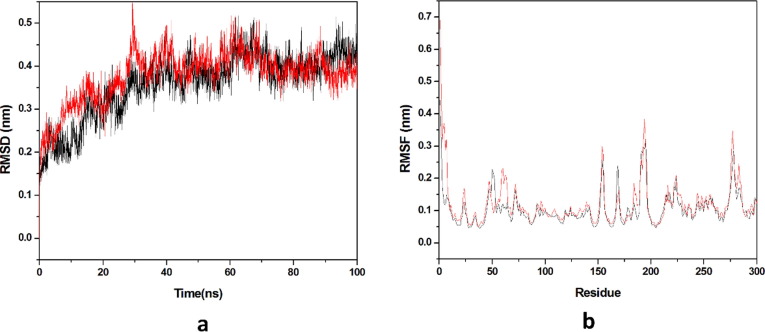
3.2.1. Root mean square deviation (RMSD) and root mean square fluctuation (RMSF)
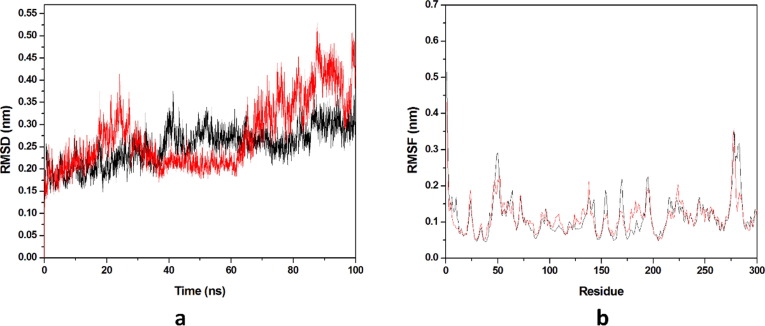
Dynamic stability of all the systems is described by the RMSD, it calculates the changes in the protein’s backbone framework during the simulation time scale. Two systems occupied for the calculation of RMSD values and the trajectories of all the systems were observed for last 20 ns as shown in (Fig. 8 a). Average values of RMSD for HCQ-Mpro & DK7-Mpro were 0.3582 nm & 0.3777 nm respectively. RMSD pattern observed for DK7 is comparable to HCQ representing the stable complex. Residue mobility calculation for the last 20 ns equilibrated trajectory was done after the RMSD calculation as illustrated in (Fig. 8b). Residue mobility analysis describes how ligand binding induces the conformational changes at residue level, as for the protein function an exact geometry is required. The average RMSF values obtained for HCQ-Mpro was 0.1088 nm while DK7-Mpro had 0.1226 nm. DK7 showed average RMSF value comparable to the parent drug.
Fig. 8.
Molecular dynamics simulation. (a) RMSD of the Cα backbone for 100 ns MD simulation at 300 K, (b) RMSF value of Cα atoms for last 20 ns. In all panels the color code is: HCQ-Mpro (black) and the derivativeDK7-Mpro (red).
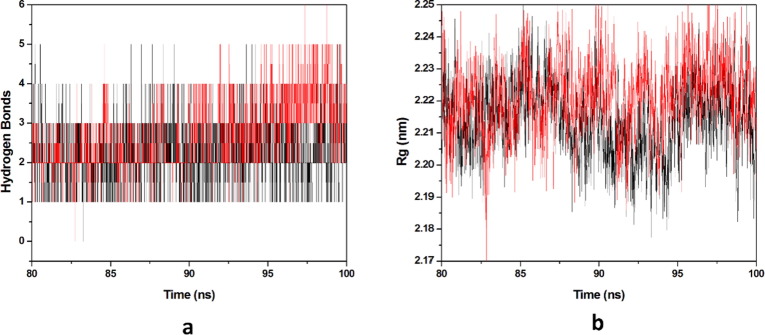
3.2.2. Hydrogen bonds and compactness analysis
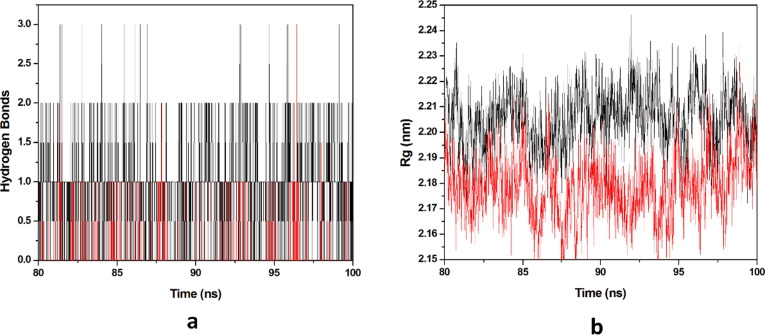
Protein-ligand stability is majorly defined by the hydrogen bonds as they contribute to transient interactions in the complex formation providing stability to the docked complex. In this study, we calculated the number of hydrogen bonds formed in the complexes while binding. The number of hydrogen bonds formed in the complexes in the time scale is plotted in (Fig. 9 a). Number of hydrogen bonds formed for HCQ-Mpro & DK7-Mpro were 0–5 & 0–5 respectively. Derivative showed the number of hydrogen bonds as equal as the parent molecule in formation of complex representing derivative stability comparable to the parent molecule. Whereas, radius of gyration (Rg) factor that defines the compactness of the protein–ligand complexes during the simulation time-scale. The distance measure between centers of mass of protein atoms with its terminal atoms in a particular time frame is called radius of gyration. Compact protein shows a lesser amount of variation in the gyration value while higher Rg value represents diffused structure. In this study, we plotted the Rg values vs. time for all the complexes (Fig. 9b). The average Rg values obtained for HCQ-Mpro & DK7-Mpro were 2.2141 & 2.2214 nm respectively. From this analysis it has been predicted that the derivative is as good as the parent molecule.
Fig. 9.
Molecular dynamics simulation. (a) Number of hydrogen bonds interaction between protein and ligand during simulation time scale for HCQ and DK7 (b) Rg vs time for HCQ and DK7 during MD simulation. In all panels the color code is: HCQ-Mpro (black) and the derivative DK7-Mpro (red).
3.2.3. Solvent accessible surface area
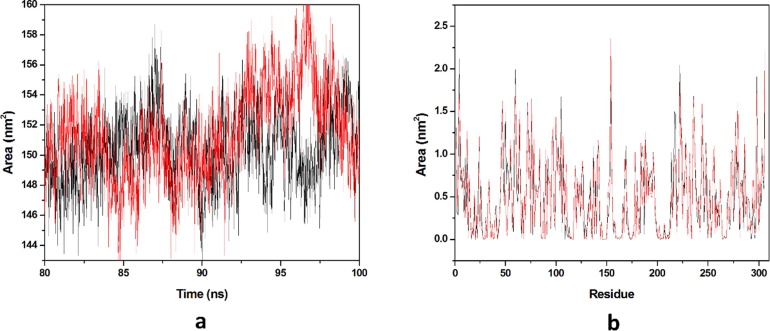
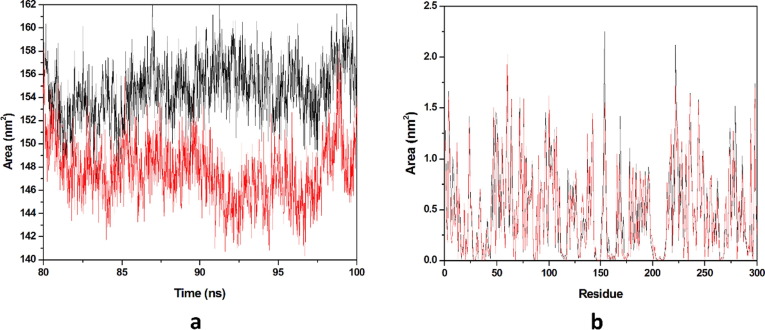
Solvation free energy of a protein is the result of interaction among polar and non-polar residues of the protein. SASA is that surface area of the protein which is monitored by the solvent molecule’s probe when it seeks the van der Waals surface of the protein. Hydrophobic residues are most responsible for the increment of the SASA value. The SASA value of each complex was calculated from the last 20 ns equilibrated trajectory and shown in (Fig. 10 a). We got the average SASA values of 150.443 & 151.448 nm2 for HCQ-Mpro & DK7-Mpro respectively. An important parameter, residue SASA value that gives understanding of conformational change per residue contribution. A graph has been plotted between residues and SASA value (Fig. 10b). The average residue SASA values gained HCQ-Mpro & DK7-Mpro were 0.4916 & 0.4949 nm2 respectively. The SASA analysis represents that the derivative is as good as the parent molecule.
Fig. 10.
Solvent accessible surface area. (a) The SASA plot for HCQ and derivative complex in water with respect to time, (b) The Residue SASA plot for HCQ and other derivative complex. In all panels, the color code is HCQ-Mpro (black) and the derivativeDK7-Mpro (red).
3.2.4. Principal component analysis
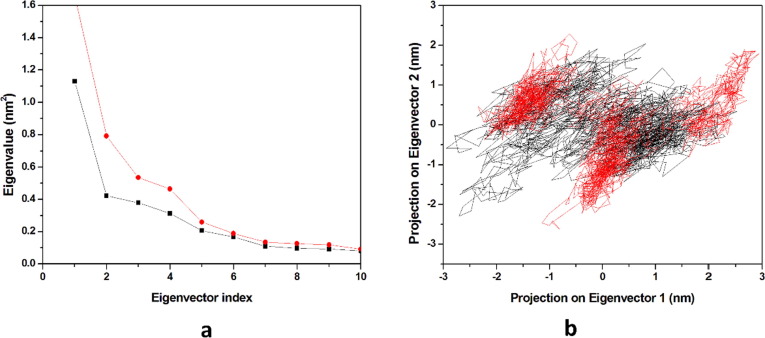
In the molecular dynamics simulation, the principal component analysis predicts the correlated motion of the complexes, in the calculation of the Principal Components (PCs) the overall motions in the proteins is described by a few key eigenvectors. Hence in the study, we had plotted the eigenvalues with the eigenvectors for the 10 eigenvectors in (Fig. 11 a). The first ten eigenvectors of HCQ-Mpro, & DK7-Mpro explained 29.97% & 43.72% of the motions respectively for the last 20 ns equilibrated trajectory. It was observed that the derivative of HCQ showed comparable motions as compared to HCQ. As previously stated, we had taken 10 eigenvectors for predicting the correlated motion of the complexes and had observed that the first five eigenvectors are the key for studying overall protein dynamics. The first two eigenvectors were plotted opposite to each other in a phase space where each one of the spectrum represents the correlated motions (Fig. 11b). The denser cluster characterizes the stability of the complex while the dispersed cluster represents the less stable cluster. It was seen from the results obtained that DK7 and HCQ showed the similar cluster pattern towards Mpro .
Fig. 11.
Principal component analysis. (a) The plot of eigenvalues vs. eigenvector index. First 10 eigenvectors were considered (b) Projection of the motion of the protein in phase space along the PC1 and PC2. In all panels the color code is: HCQ-Mpro (black) and the derivativeDK7-Mpro (red).
3.3. Molecular docking of conjugate of hydroxychloroquine with remdesivir and derivatives of tetrahydrocannabinol
In our second studies we have designed a conjugate of hydroxychloroquine with remdesivir. Several report suggest that remdesivir is effectively in use with HCQ for the treatment of COVID-19 patient. So, we planned to design a hybrid of HCQ and remdesivir. These hybrid molecules (DK2, DK3, DK6, DK9 and DK12) were docked with active site of the Mpro of SARS-CoV-2. The docking score obtained from the molecular docking studies shows, among these conjugates DK3, DK6, DK9 and DK12 are little better but not that much active as compared to HCQ alone while DK2 (free binding energy −4.22 kcal.mol−1), was found to have lower free binding energy and poor inhibition with respect to HCQ against Mpro of SARS-CoV-2 (Table 1).
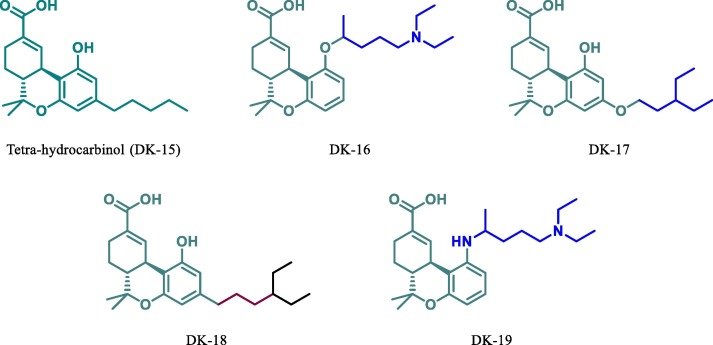
Since tetrahydrocannabinol is useful in the cure of various disease [53], [54], [55], [56], [57]. Hence, taking THC as a parent moiety (DK15, Fig. 12 ) we have designed its four derivatives (DK16, DK17, DK18 and DK19, see Fig. 12) for docking study against Mpro of SARS-CoV-2. The molecular docking data shows that parent molecule (DK15, free binding energy −6.99 kcal.mol−1 see table 1) is itself more potent inhibitor than the HCQ (DK13) itself. This attracted our attention towards modification in this moiety and designs a more potent inhibitor. Now based on the docking result of DK15 we have designed and perform their docking against Mpro of SARS-CoV-2. Based on this docking results of these derivatives (DK16),
Fig. 12.
Chemical structure of THC and their derivatives.
DK17, DK18 and DK19) it was found that all derivatives are more active than the parent molecule (DK15). Among all these the best result was obtained by DK16, DK17 and DK19 (-7.64 kcal mol−1, −7.62 kcal mol−1, −7.42 kcal mol−1 respectively). It has also been found that DK16 is more potent inhibitor for Mpro of SARS-CoV-2 than the HCQ’s derivative DK7. The molecular docking of DK16 suggests that the carboxylic acid group present at parent moiety makes a hydrogen bond with Lys102 and oxygen atom of tetrahydropyran ring is involved in hydrogen bond formation with Gln110. While the other amino acid residue Val104, Ile106, Gln107, Thr111, Asn151, Ile152, Asp153, Ser 158, Phe294 are involved in hydrophobic interaction with the different residue of the moiety (Fig. 13 ).
Fig. 13.
(a) 3D surface image of protein with DK16 represented in sticks model showing the interactions in the binding pocket of protein. (b) Protein is represented in cartoon model and DK16 is represented in sticks model showing the polar interaction in 3D with the bond lengths represented by yellow dashed lines and interacting residues of the proteins are labelled with residue name using the PyMOL interactive visualization tool. (c). 2D representation of DK16 with PDB6LU7 by Ligplot+.
3.4. Molecular dynamics simulation complex of Mpro with THC and DK16
Hence molecular dynamics simulation for 100 ns of the docked complex of Mpro with THC and DK16 was carried out to further validate the results obtained from the docking studies. In the study, equilibration state for both the systems were obtained and last 20 ns equilibrated trajectories were observed for RMSF, Rg and SASA H-bonds including binding free energy analysis for last 20 ns.
3.4.1. Root mean square deviation (RMSD) and root mean square fluctuation (RMSF)
Average values of RMSD for THC-Mpro& DK16-Mpro were 0.1863 & 0.1516 nm respectively. RMSD pattern observed for DK16 showed less average RMSD value than THC, representing a stable complex formation with Mpro . Also, average RMSF values obtained for THC-Mpro & DK16-Mpro were 0.1185& 0.1178 nm respectively. An optimized geometry of the protein is required for the functioning of protein and ligand binding disrupts the original conformation of the protein. DK16 showed average RMSF value less than the THC representing that ligand complex is a stable complex as compared to the parent drug (Fig. 14 a and b).
Fig. 14.
Molecular dynamics simulation. (a) RMSD of the Cα backbone for 100 ns MD simulation at 300 K, (b) RMSF value of Cα atoms for last 20 ns. In all panels the color code is: HCQ-Mpro (black) and the derivativeDK7-Mpro (red).
3.4.2. Hydrogen bonds and compactness analysis
Number of hydrogen bonds formed THC-Mpro & DK16-Mpro complexes was 0–3 & 0–2 respectively in the time scale, plotted (Fig. 15 a). Derivatives showed the number of hydrogen bonds as equally as the parent molecule in formation of complex representing derivatives is stability as compared to the parent molecules. The average Rg values obtained for THC-Mpro & DK16-Mpro were 2.2055 & 2.1787 nm respectively (Fig. 15b). DK16 derivative showed less Rg value than the parent molecule. This analysis indicates that the derivative is good as compared to the parent molecule.
Fig. 15.
Molecular dynamics simulation. (a) Number of hydrogen bonds interaction between protein and ligand during simulation time scale for THC and DK16 (b) Rg vs time for THC and DK16 during MD simulation. In all panels the color code is: THC-Mpro (black) and the derivative DK16-Mpro (red).
3.4.3. Solvent accessible surface area
Average SASA values of 154.761 & 147.616 nm2 for THC-Mpro & DK16-Mpro. The average SASA value for DK16 is less than the parent molecule. SASA analysis reveals that the derivative is comparably good than the THC. The SASA value of each complex was calculated from the last 20 ns equilibrated trajectory and shown in (Fig. 16 a). Residue SASA value that gives understanding of conformational change per residue contribution. A graph has been plotted between residues and SASA value (Fig. 16b). The average residue SASA values gained for THC-Mpro & DK16-Mpro were 0.5057 & 0.4824, nm2 respectively. This analysis also in favor that the derivative is good as that the parent drug.
Fig. 16.
Solvent accessible surface area. (a) The SASA plot for THC and derivative complex in water with respect to time, (b) The Residue SASA plot for THC and other derivative complex. In all panels, the color code is THC-Mpro (black) and the derivative DK16-Mpro (red).
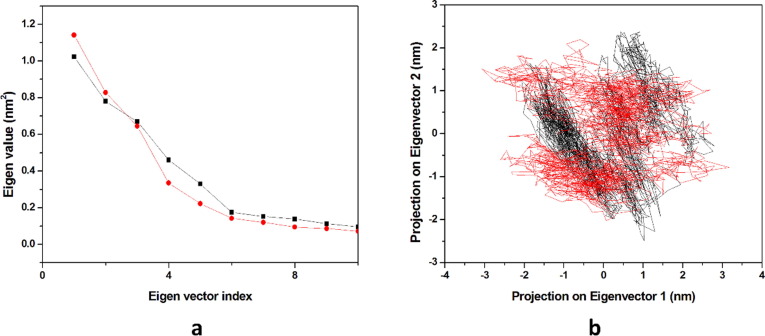
3.4.4. Principal component analysis
In the MD simulation, the principal component analysis predicts the correlated motion of the complexes. The first five eigenvectors of THC-Mpro & DK16-Mpro explained 39.37% & 36.84% of the motions respectively for the last 20 ns equilibrated trajectory. It was observed that DK16 showed less motion as compared to parent molecule. Hence, the modeled results are suggesting the stability of DK16 towards binding with Mpro protein, (Fig. 17 a). The first two eigenvectors were plotted opposite to each other in a phase space where each one of the spectrum represents the correlated motions (Fig. 17b). The denser cluster characterizes the stability of the complex while the dispersed cluster represents the less stable cluster. It was seen from the results obtained that DK16 showed the dense cluster representing stability towards Mpro.
Fig. 17.
Principal component analysis. (a) The plot of eigenvalues vs. eigenvector index. First 10 eigenvectors were considered (b) Projection of the motion of the protein in phase space along the PC1 and PC2. In all panels the color code is: THC-Mpro (black) and the derivative DK16-Mpro (red).
3.5. MMPBSA binding free energy analysis
The binding ability of the ligands to the protein is assessed with the help of binding free energy calculation which is the sum of non-bonded interaction energies of the complex. MMPBSA tool [59] that is directed by the Gromacs, was adopted for the calculation of binding free energy. All the binding free energies of the complexes are summarized in Table 2 , showing the stability of the complex formation with the derivatives as compared to the parent molecule.
Table 2.
Table represents the Van der Waals, electrostatic, polar solvation, SASA and binding energy in kJ.mol−1 for molecules.
| S. No | Compound | Van der Waals energy | Electrostatic Energy | Polar solvation energy | SASA energy | Binding energy |
|---|---|---|---|---|---|---|
| 1 | Hydroxy-chloroquine | −172.01 ± 13.53 | −75.44 ± 10.74 | 158.32 ± 13.58 | −19.56 ± 0.87 | −108.69 ± 12.87 |
| 2 | DK7 | −37.40 ± 49.55 | −34.93 ± 46.08 | 73.17 ± 77.72 | −5.24 ± 7.69 | −4.40 ± 57.28 |
| 3 | Tetrahydro-cannabinol | −94.88 ± 50.87 | −16.58 ± 10.40 | 62.87 ± 38.50 | −10.49 ± 5.96 | −59.09 ± 47.98 |
| 4 | DK16 | −9.25 ± 29.79 | −2.37 ± 7.56 | 8.40 ± 58.88 | −1.45 ± 5.26 | −4.68 ± 56.47 |
4. Conclusion
Since COVID-19 is epidemiological and created a globally challenged health crisis, till date there is no any particular vaccine or drug have been developed to control it. Initially it was found that hydroxychloroquine and remdesivir are little bit active and are using as a drug for management of COVID-19. Keeping it in mind we have designed derivatives of these drugs and studied about their inhibition and binding with Mpro of SARS-CoV-2. In our docking studies we have concluded that DK4, DK7, DK10, DK16, DK17 and DK19 can be use as inhibitor for Mpro of SARS-CoV-2 to control COVID-19 very effectively. Also, MD simulation for HCQ, DK7, THC, and DK16 those were highest scoring molecules obtained in the docking studies was carried out. RMSD, RMSF, hbonds, Rg, SASA, PCA parameters were calculated to validate the stability of the protein ligand complexes and found that DK16 showed higher stability as compared to the parent molecule as well as from DK7. DK7 molecule showed comparable stability as the HCQ with Mpro protein of SARS-CoV-2. It is anticipated that both these derivatives will be potent drug candidates and can be further validated experimentally.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are thankful to Scfbio, IIT Delhi for providing the facility of Gaussian software to optimize the structures and also supporting in carrying out molecular dynamics simulation studies.
References
- 1.Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown etiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel Med. 2020:1–3. doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D.S. Hui, E.I. Azhar, T.A. Madani, F. Ntoumi, R. Kock, G. Dar Ippolito, T. D. Mchugh, Z.A. Memish, C. Drosten, A. Zumla, A. Petersen, The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 91 (2020) 264–266. [DOI] [PMC free article] [PubMed]
- 3.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.P. Zhou, X-L. Yang, X-G.Wang, B. Hu, L. Zhang, W. Zhang, H-R. Si, Y. Zhu, B. Li, C- L.Huang, H-D. Chen, J. Chen, Y. Luo, H. Guo, R-D. Jiang, M-Q. Liu, Y. Chen, X-R. Shen, X. Wang, Z-L. Shi, A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, 579 (2020) 270–273. [DOI] [PMC free article] [PubMed]
- 5.Gorbalenya A.-E., Baker S.-C., Baric R.-S., de Groot R.-J., Drosten C., Gulyaeva A.-A., Haagmans B.-L., Lauber C., Leontovich A.-M., Neuman B.-W., Penzar D., Perlman S., Poom L.-L.-M., Samborskiy D.-V., Sidorov I.-A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baig A.-M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 7.A-E. Gorbalenya, S-C. Baker, R. Baric, R-J. Groot, C. de Drosten, A-A. Gulyaeva, b-L. Haagmans, C. Lauber, A-M. Leontovich, B-W. Neuman, D. Penzar, Severe acute respiratory syndrome-related coronavirus: The species and its viruses–a statement of the Coronavirus Study Group, 2020.
- 8.Kupferschmidt K., Cohen J. Will novel virus go pandemic or be contained? Science. 2020;367:610–611. doi: 10.1126/science.367.6478.610. [DOI] [PubMed] [Google Scholar]
- 9.S. Klebnikov, Coronavirus is now expected to curb global economic growth by 0.3% in. https://www.forbes.com/sites/sergeiklebnikov/2020/02/11/coronavirus-is-now-expected-to-curb-global-economic-growthby-03-in-2020/#5de149ad16da (accessed on 20-Nov-2020).
- 10.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repur-posing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr A.-R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong Y., Ren X. Coronavirus entry and release in polarized epithelial cells: a review. Rev. Med. Virol. 2014;24:308–315. doi: 10.1002/rmv.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutictargets for SARS-CoV-2 and discovery of potential drugs bycomputational methods. Acta Pharmaceutica Sinica B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Zhang J., Wang N., Li H., Shi Y., Guo G., Liu K., Zeng H., Zou Q. Therapeutic drugs targeting 2019-nCoV main protease byhigh-throughput screening. BioRxiv. 2020 [Google Scholar]
- 16.Xu Z., Peng C., Shi Y., Zhu Z., Mu K., Wang X., Zhu W. Nelfinavir was predicted to be a potential inhibitor of 2019-nCovmain protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 2020 [Google Scholar]
- 17.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang H. Structure of Mpro from SARSCoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 18.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.-C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: Implications for substratebinding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S.-J. Anthony, C.-K. Johnson, D.-J. Greig, S. Kramer, X. Che, H. Wells, A.-L. Hicks, D.-O. Joly, N.-D. Wolfe, P. Daszak, W. Karesh, W.-I. Lipkin, S.-S. Morse, J.-A.-K. Mazet, T. Goldstein, Global patterns in coronavirus diversity, Virus Evol. 3 (2017) vex012. [DOI] [PMC free article] [PubMed]
- 20.Su S., Wong G., Shi W., Liu J., Lai A.-C., Zhou J., Liu W., Bi Y., Gao G.-F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.B. Tang, N.L. Bragazzi, Q. Li, S. Tang, Y. Xiao, J. Wu, An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov), Infect. Dis. Model. 5 (2020) 248–255. [DOI] [PMC free article] [PubMed]
- 23.M. Bzowka, K. Mitusinska, A. Raczynska, A. Samol, J.A. Tuszynski A. Gora, Structural and evolutionary analysis indicate that theSARS-CoV-2 Mpro is a challenging target for small-molecule inhibitordesign, Int. J. Mol. Sci. 21 (2020) 3099. [DOI] [PMC free article] [PubMed]
- 24.Scavone C., Brusco S., Bertini M., Sportiello L., Rafaniello C., Zoccoli A., Berrino L., Racagni G., Rossi F., Capuano A. Currentpharmacological treatments for COVID-19: What’s next? Br. J. Pharmacol. 2020;177:4813–4824. doi: 10.1111/bph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug againstlife-threatening RNA virus infections. Pharmacol. & Therap. 2020 doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for thetreatment of COVID-19. J. Criti. Care. 2020;57:279–283. doi: 10.1016/j.jcrc.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallikarjuna N., Vasudharani D., Ambrish S. Screening ofchloroquine, hydroxychloroquine and its derivatives for their bindingaffinity to multiple SARS-CoV-2 protein drug targets. J. Biomol. Struct. Dyn. 2020;1–13 [Google Scholar]
- 28.P. Gautret, J.-C. Lagier, P. Parola, V.T. Hoang, L. Meddeb, M. Mailhe, B. Doudier, J. Courjon, V. Giordanengo, V.E. Vieira, H. Tissot Dupont, S. Honore, P. Colson, E. Chabriere, B. La Scola, J.M. Rolain, P. Brouqui, D. Raoult, Hydroxychloroquine and azithromycinas atreatment of COVID-19: Results of an open-label non-randomizedclinical trial, Int. J. Antimicrob. Agents 56 (2020)105949. [DOI] [PMC free article] [PubMed]
- 29.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Liu, R. Cao, M. Xu, Xi. Wang, H. Zhang, H. Hu, Y. Li, Z. Hu, W. Zhong, M. Wang, Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discov. 6 (2020) 16. [DOI] [PMC free article] [PubMed]
- 31.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durrant J.D., McCammon J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M.J. Frish, Gaussian 09 (Reversion B. 01), Gaussian. Inc. Pittsburgh, 2009.
- 34.C. Tabti, N. Benhalima, Molecular structure, vibrational assignments and non-linear optical properties of 4, 4’Dimethylaminocyanobiphenyl (DMACB) by DFT and ab Initio HF Calculations, A. M. P. C. 5 (2015) 221–228.
- 35.Kwiecien R.A., Rostkowski M., Dybała-Defratyka A., Paneth P. Validation of semiempirical methods for modeling of corrinoid systems. J. Inorg. Biochem. 2004;98:1078–1086. doi: 10.1016/j.jinorgbio.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 36.Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, Structure of Mpro from COVID-19 virus and discovery of its inhibitors, Bio. Rxiv. (2020). [DOI] [PubMed]
- 37.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. E. M. B. O. J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Wang F., Chen C., Tan W., Yang K., Yang H. Structure of mainprotease fromhuman coronavirus NL63: Insights for wide spectrumanti-coronavirus drug design. Sci. Rep. 2016;6:22677. doi: 10.1038/srep22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal L., Kumari A., Srivastava M., Singh M., Asthana S. Identification of potential molecules against COVID-19 main proteasethrough structure-guided virtual screening approach. J. Biomole. Struct. Dyn. 2020:1–26. doi: 10.1080/07391102.2020.1768151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.W. Tian, C. Chen, X. Lei, J. Zhao, J.Liang, CASTp 3.0: computed atlas of surface topography of proteins, Nucl. Acids. Res. 46 (2018) W363–W367. [DOI] [PMC free article] [PubMed]
- 42.Goodsell D.S., Morris G.M., Olson A.J. Automated Docking of Flexible Ligands: Applications of AutoDock. J. Mol. Recog. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 43.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Der Spoel D., Lindalh E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 45.N. Schmid, A.P. Eichenberg, A. Choutko, S. Riniker, M. Winker, A.E. Mark, W.F.van,Gunsteren Definition and testing of the GROMOS force-field versions 54A7 and 54B7, Eur. Biophys. J. 40 (2011)843. [DOI] [PubMed]
- 46.A.K. Malde, L. Zuo, M. Breeze, M. Stroet, D. Poger, P.C. Nair, C. Oostenbrink, A.E.Mark, An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0, J. Chem. Theory Comput. 7 (2011)4026–4037. [DOI] [PubMed]
- 47.Petersen H.G. Accuracy and efficiency of the particle Mesh Ewald method. J. Chem. Phys. 1995;103:3668–3679. [Google Scholar]
- 48.Berendsen H.J., Postma J.V., van Gunsteren W.F., Dinola A.R.H.J., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 49.Andersen H.C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 1980;72:2384–2393. [Google Scholar]
- 50.Van Gunsteren W.F., Berendsen H.J. A leap-frog algorithm for stochasticdynamics. Mol. Simulat. 1998;1:173–185. [Google Scholar]
- 51.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 52.Lever J., Krzywinski M., Altman N. Points of significance: Principal component analysis. Nat. Methods. 2017;14:641–642. [Google Scholar]
- 53.Fernández-Ruiz J., Galve-Roperh I., Sagredo O., Guzmán M. Possible therapeutic applications of cannabis in the neuropsychopharmacology field. Eur. Neuropsychopharmacol. 2020;1:18. doi: 10.1016/j.euroneuro.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Sarris J., Sinclair J., Karamacoska D., Davidson M., Firth J. Medicinal cannabis for psychiatric disorders: a clinically-focused systematic review. BMC Psychiatry. 2020;20:24. doi: 10.1186/s12888-019-2409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo E.B. History of cannabis and its preparations in saga, science, and sobriquet. Chem. Biodivers. 2007;4:1614–1648. doi: 10.1002/cbdv.200790144. [DOI] [PubMed] [Google Scholar]
- 56.Touw M. The religious and medicinal uses of Cannabis in China, India and Tibet. J. Psychoact. Drugs. 1981;13:23–34. doi: 10.1080/02791072.1981.10471447. [DOI] [PubMed] [Google Scholar]
- 57.Lowe H.I., Toyang N.J., McLaughlin W. Potential of cannabidiol for the treatment of viral hepatitis. Phcog. Res. 2017;9:116–118. doi: 10.4103/0974-8490.199780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M.J. Abraham, T. Murtola, R. Schulz, S. Pall, J.C. Smith, B. Hess, E.L. Lindalh GROMACS: High performance molecular simulations through multi-level parallelism from laptopsto supercomputers, SoftwareX 1 (2015) 19–25.
- 59.R. Kumari, R. Kumar, Open Source Drug Discovery Consortium, & Lynn, A.g_mmpbsa--A GROMACS tool for high-throughput MM-PBSA calculations, J. Chem. Inf. Model. 54 (2014) 1951–1962. [DOI] [PubMed]