Abstract
Seeing the global emergence and the lack of a definitive cure for COVID-19, it is essential to find the most sensitive and specific detection method to identify infected patients in a timely manner.
Our paper aims to compare the clinical sensitivity of different commercial RT-qPCR (Genesig, 1copy, DNA-Techonolgy and Charité primer-probe sets), isothermal PCR (Ustar Isothermal Amplification-Real Time Fluorescent Assay) and immunochromatographic antigen detection (BIOCREDIT COVID-19 Ag) assays developed to use in laboratory diagnosis of COVID-19.
A total of 119 nasopharyngeal swab specimens were collected from symptomatic patients. A subset of samples, positive with two RT-qPCR assays were then tested with isothermal PCR and rapid antigen tests.
Of the 119 specimens, 65 were positive by at least two PCR assays. All PCR assays showed substantial or perfect match, although some variations in the clinical performance was observed. Of the 37 and 32 remnant nasopharyngeal samples positive by RT-qPCR, respectively, three were positive by the BIOCREDIT COVID-19 Ag and 14 were detected by the isothermal amplification assay.
In conclusion, in the clinical settings we recorded that each of the RT-qPCR assays was superior to other test formats, in particular, the routine use of the DNA-technology assay is recommended. Although alternative recommendations exist, we belive that the use of isothermal amplifiaction assays and antigen rapid tests for COVID-19 diagnosis can only serve as adjuncts while awaiting the PCR result because of their high false-negative rate.
Keywords: COVID-19, SARS CoV-2, RT-qPCR, Laboratory diagnostics
1. Introduction
At the end of 2019, a novel coronavirus (severe acute respiratory syndrome coronavirus-2, SARS-CoV-2), the causative agent of the Coronavirus Disease 2019 (COVID-19) outbreak, emerged from Wuhan, China (Zhou et al., 2020). As of 25 March 2021, the SARS-CoV-2 has been confirmed in more than 120 million cases and has been responsible for ∼ 2,7 million deaths (ECDC, 2020). As a result, the rapid spread of the virus has placed a great burden on the healthcare system globally (Nicola et al., 2020).
Despite intensive research, a definite cure for COVID-19 is still an unmet clinical need. Therefore, the key to controlling the pandemic currently lies in early diagnosis, supportive treatment and the isolation of infected patients and their contacts, necessitating an accurate and fast diagnosis of SARS-CoV-2 (Cascella M and Cuomo, 2021). In the laboratory diagnosis of SARS-CoV-2, reverse transcriptase real-time PCR (RT-qPCR) has played a pivotal role (Lau et al., 2005). Since the start of the COVID-19 outbreak, various RT-qPCR assays have been developed and made available on the market (finddx.org, 2020).
SARS-CoV-2 is an enveloped, positive-stranded RNA virus that belongs to the genus Betacoronavirus in the Coronaviridae family (Venter and Richter, 2020). Its extraordinarily large genome encodes 16 non-structural proteins, including the RNA-dependent RNA polymerase (RdRp) from an open reading frame termed ORF1a/b at the 5′ end of the viral genome, as well as structural proteins, including the Spike (S), Envelope (E) and Nucleocapsid (N) encoded by ORFs at the 3′ end (Su et al., 2016). In the laboratory diagnosis of SARS-CoV-2, RT-qPCR assays most frequently target the RdRp, E, N and S genes (van Kasteren et al., 2020). For example, both 1copy and the Charité protocol targets the E and RdRp genes, DNA-technology targets the SARS like coronaviruses sequence, the N and E genes, while according to the manufacturer, the Genesig assay primer/probe is specific for the SARS-CoV-2 (the manufacturer has not made the target gene public).
In consequence of many countries going into lockdown, a great shortage in the supply of SARS-CoV-2 RT-qPCR assay kits have occurred, forcing many laboratories to use different RT-qPCR assays that are acutely available on the market. Moreover, with the escalation of the pandemic, manufacturers have been rushed to develop more rapid detection assays, such as lateral flow assays, as well as isothermal amplification assays. However, in comparison to the amount of assays available on the market, the number of studies comparing their clinical performance are scarce. Therefore, in the present paper, we provide a comparison of four widely used RT-qPCR assays, an immunochromatographic rapid detection assay and an isothermal cross priming amplification (ICPA) assay, each developed to use in the acute diagnosis of SARS-CoV-2 infection
2. Materials and methods
2.1. Sample collection and handling
Nasopharyngeal swabs were collected from patients that were admitted to hospital with presumable SARS-CoV-2 infection between April and July of 2020 Hungary. Nasopharyngeal swabs were immersed into viral transport medium (VTM) immediately after sample collection. Samples were stored at 4 °C until testing.
2.2. Nucleic acid extraction
Total nucleic acid was extracted from VTM using the Ribo-Virus Viral RNA/DNA extraction kit (Sacace Biotechnologies, Como, Italy) according to the manufacturer’s instructions. Aliquoted nucleic acid and residual samples were stored at -70 C° until further testing with RT-qPCR tests. All samples were tested after one initial freeze-thaw cycle. Commercially available PCR assays were performed according to the manufacturers’ instruction using the Quantstudio 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, United States)
2.3. Genesig coronavirus (COVID-19) real-time PCR
Testing samples with the Genesig COVID-19 assay was carried out according to the manufacturer’s instructions. The reaction mix consists of 12 μL master mix and 8 μL extracted nucleic acid. The assay detects a specific, however, unnamed segment of the SARS-CoV-2 RNA using a hydrolysis probe system, labelled with the FAM fluorophore. According to the the product’s manual, the limit of detection (LoD) value of the kit is 0.58 copies/μL. The kit utilizes an internal control, which should be added during nucleic acid extraction.
2.4. 1copy COVID-19 qPCR multi kit
This simplex assay detects the non-SARS-CoV-2 specific E gene and the SARS-CoV-2 specific RdRp gene of the SARS-CoV-2 virus, in separate reactions/separate master mixes. The reaction mix consists of 15 μL master mix and 5 μL template. According to the user manual, the LoD value of the kit is 200 copies/mL (= 0.2 copies/μL).
2.5. DNA-technology SARS-CoV-2 multiplex real-time PCR detection kit
The assay detects different genome segments, including the “SARS-CoV-like Coronaviruses” gene, the E gene and the N gene of SARS-CoV-2. The reaction mix utilizes 10 μL template and 15 μL master mix. The detection limit of the kit is 10 copies per 25 μL reaction mix (= 0.4 copies/μL).
2.6. Charité (Universitätsmedizin berlin institute of virology, Germany) primer-probe sets
In our comparison, we adapted the Charité protocol, which was published on 17 January 2020 (Christian Drosten et al., 2020). We used the E_Sarbeco_F1 and E_Sarbeco_R2 primers with the E_Sarbeco_P1 probe for the detection of the E gene, and the RdRP_SARSr-F2 and RdRP_SARSr-R1 primers with the RdRP_SARSr-P2 probe for detection of the RdRp gene. All primers and probes were used at the concentrations recommended by the protocol. For the PCR reactions, we used the GoTaq® Probe 1-Step RT-qPCR System (Promega Corporation, 2800 Woods Hollow Road, Madison, U.S.).
2.7. Isothermal amplification-real time fluorescent assay
The Novel-Coronavirus (2019-nCoV) RNA Diagnostic Kit, and the fully automated nucleic acid analysis platform (Ustar Biotechnologies Ltd. Hangzhou, Zhejiang, China) This kit utilizes cartridges that are equipped with multiple hydrophobic separation layers to isolate the lysate, the cleaning solution and the reaction solution in the cartridge. Briefly, following thorough vortexing, 1000 μL of the magnetic bead solution and 500 μL of the sample (from either viral transport medium or sputum) is pipetted into the cartridge. Thus, the assay requires minimal hands-on time. The reaction time is 79 min, and the instrument is capable of running 2 separate samples at the same time. The kit uses Crossing Priming Amplification (CPA) (Xu et al., 2012) to detect the ORF1ab and the N gene sequences of the SARS-CoV-2.
2.8. Antigen assay
BIOCREDIT COVID 19 Ag test is an immunochromatographic rapid test developed for the rapid detection of SARS-CoV2. Sample processing was carried out according to the manufacturer instructions. Briefly, following vortexing of the VTM, 100 μL of the sample was added directly into the assay’s lysis buffer and the mixture was homogenized. The assay’s dilution tube was sealed securely with a filter cap and 3–4 drops of the reagent was added into the sample well of the device. Results were interpreted following a 15 min incubation.
2.9. Interpretation of results
All results were interpreted based on the manufacturers’ instructions. Samples positive for only one specific SARS CoV-2 gene were considered marginal. Here, marginal samples were interpreted as negative results.
2.10. Evaluation of workflow
Workflow was evaluated by using stopwatches to measure the time needed for each performed step of the assay. Hands-on time (HoT) and assay runtime were determined. The parameters were calculated based on the throughput of samples per run.
2.11. Statistical methods
Samples that were positive by at least two of the four PCR diagnostic assays were deemed PCR-positive. Positive percentage agreement (PPA), negative percentage agreement (NPA), Kappa values, linear regression and PCR efficiency, were calculated using GraphPad Prism 8. Agreement between various kits were characterized by Cohen’ kappa values, and were categorized as almost-perfect (>0.90), strong (0.80 to 0.90), moderate 180 (0.60 to 0.79), weak (0.40 to 0.59), minimal (0.21 to 0.39), or none (0 to 0.20) (Landis and Koch, 1977).
3. Results
3.1. Genesig coronavirus (COVID-19) real-time PCR
Among the 119 tested samples, 64 were identified as positive by the Genesig Coronavirus (COVID-19) Real-time PCR. The Ct values varied between 16.21 and 39.9, with a mean Ct value of 28.9.
3.2. 1copy COVID-19 qPCR multi kit
Among the 119 tested samples, 59 were positive with this assay. The Ct values varied between 12.8 and 37.66, with a mean Ct value of 28.43 for the E gene, and varied between 12.14 and 39.94 with a mean Ct value of 30.07 for the RdRp gene.
3.3. DNA-technology SARS-CoV-2 multiplex real-time PCR detection kit
Out of the 119 tested samples, the DNA-Technology SARS-CoV-2 Multiplex real-time PCR Detection Kit identified 60 as positive. The Ct values for the SARS like Coronaviruses specific probe, for the N gene and the E gene varied between 13.28 and 41.46 (mean: 32.00); 17.6 and 42.7 (mean: 32.33); 18.3 and 43.00 (mean: 32.20), respectively.
3.4. Charité (Universitätsmedizin berlin institute of virology, Germany) primer-probe sets
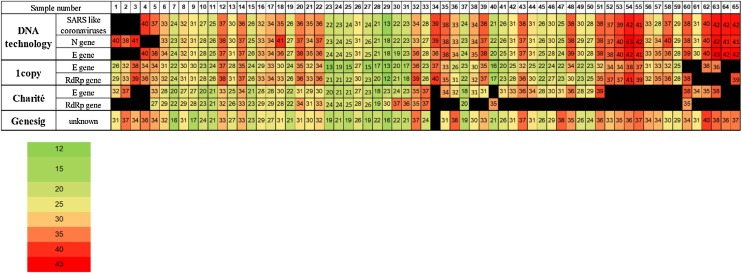
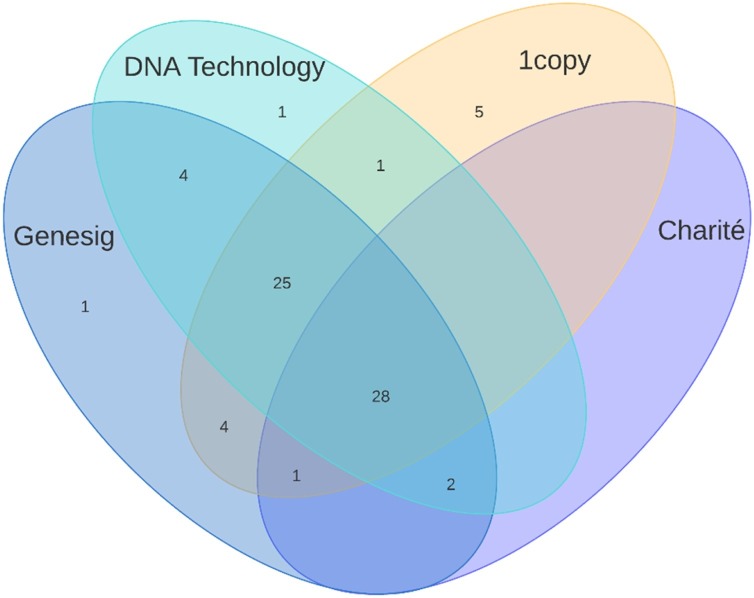
Of the 119 samples, 31 were positive with the Charité primer probe set. The Ct values ranged from 17.70 to 39.30 (mean: 28.76) for the E gene, and from 18.9–37.1 (mean: 28.48) for the RdRp gene. The results of each PCR kit, their positive percentage agreement and negative percentage agreement values are shown in Table 1 . Kappa values between the different PCR assays are highlighted in Table 2 . Further details of the positive samples using the different PCR assays can be seen on Fig. 1, Fig. 2 .
Table 1.
Clinical performance of the compared four PCR kits.
| Molecular assay | Final result1 |
PPA | NPA | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Charité | Positive | 31 | 0 | 47.69 % | 100 % |
| Negative | 34 | 54 | |||
| 1copy | Positive | 59 | 5 | 90.77 % | 90.74 % |
| Negative | 6 | 49 | |||
| Genesig | Positive | 64 | 1 | 98.46 % | 98.15 % |
| Negative | 1 | 53 | |||
| DNA Technology | Positive | 60 | 1 | 92.31 % | 98.15 % |
| Negative | 5 | 53 | |||
The final result of a sample was positive when at least two of the four kits yielded a positive result.
Table 2.
Inter-rater agreement defined by Cohen’s kappa values.
| Genesig | 1Copy | DNA-Technology | Charité | |
|---|---|---|---|---|
| Genesig | – | κ = 0.795 | κ = 0865 | κ = 0453 |
| St. error 0.056 | St. error = 0046 | St. error = 0066 | ||
| CI = 0.685−0.905 | CI = 0,775−0,995 | CI = 0,323−0,583 | ||
| 1Copy | – | – | κ = 0714 | κ = 0,4 |
| St. error = 0064 | St. error = 0069 | |||
| CI = 0,588−0,840 | CI = 0,264−0,536 | |||
| DNA-Technology | – | – | – | κ = 0469 |
| St. error = 007 | ||||
| CI = 0,332−0,605 |
Fig. 1.
Heatmap of Ct values by PCR assay.
Of the 119 nasopharyngeal samples, 65 were positive by at least two PCR assays. The diagram shows the Ct values of the screened genes by the different RT-qPCR assays. Black bars represent undeterminable Ct values. Five results of DNA technology, four results of 1copy and 21 results of Charité were considered marginal.
Fig. 2.
Number of positive samples with each kit, depicted on a Venn-diagram.
3.5. Antigen testing
To assess the sensitivity and reliability of the BIOCREDIT COVID-19 Ag test in comparison to RT-qPCR, of the 65 PCR positive samples, we randomly selected 37 samples. Of the 37 PCR-positive samples, 3 (8.11 %) were identified as positive with the rapid antigen test.
3.6. Isothermal amplification
Additionally, we also evaluated the clinical sensitivity of a PCR assay, which is based on isothermal amplification. An Ustar Isothermal Amplification-Real Time Fluorescent Assay was carried out with 32 samples, which were previously deemed positive with at least 2 PCR kits out of the 4. Among the 32 samples, 14 (43.75 %) were detected by the ICPA assay as positive.
3.7. Workflow evaluation
In the clinical decision making, it is particularly important to receive accurate and fast diagnosis. Therefore, workflow was determined based on the assessment of HoT, and the assay runtime for each assay compared in the paper. Summary of the workflow results are shown in Table 3 . Of the RT-qPCR assays, 1copy had both the longest HoT, followed by the the rest of the assays, which had a comparable values. All the assays had a similar runtime with an average of 90 min. (Table 3)
Table 3.
Hands-on time and assay runtime with the examined assays.
| Method | HoT (minutes)1 | Assay runtime (minutes:seconds) |
|---|---|---|
| Genesig Coronavirus (COVID-19) Real-time PCR | Average = 26.05 | 94:00 |
| 95% CI = 24.17–27.92 | ||
| 1Copy | Average = 52.1 | 95:41 |
| 95% CI = 48.34–55.85 | ||
| DNA-Technology | Average = 23.5 | 96:43 |
| 95% CI = 22.27–24.72 | ||
| Charité Primer-Probe sets | Average = 26.625 | 91:07 |
| 95% CI = 25.4–27.85 | ||
| Ustar Isothermal Amplification | < 1 | 79:00 |
| BIOCREDIT COVID-19 Ag rapid test | < 1 | 10−15 |
In the case of PCR assays, the hands-on-time was measured for the preparation of 96 samples. In the case of the isothermal amplification and the antigen rapid test, HoT is measured for 1 sample.
4. Discussion
Here we demonstrate a comparison of four RT-qPCR assays used in the detection of SARS-CoV-2. Additionally, samples that were positive based on two commercially available assays were further tested with two point-of care tests (POCTs): an immunochromatographic, lateral flow-based antigen rapid test and an ICPA assay.
With the escalation of the pandemic, authorities were forced to grant emergency use authorization (EUA) to RT-qPCR detection assays with a limited number of experiments required; thus, their sensitivity and specificity might be questionable (FDA, 2020). Our data suggest that there was discordance between the four PCR methods, as the kappa values varied notably between the different assays. The highest kappa value (0.865) yielded between Genesig and DNA-technology, the lowest (0.4) was seen between 1copy and the Charité protocol. The clinical performance also showed a variation between the different assays. The Genesig assay had the highest sensitivity (PPA: 98.46 %, NPA: 98.15 %), while the Charité protocol had the lowest performance (PPA: 47.69 %, NPA: 100 %), missing 34 positive samples out of 65. Both 1copy (PPA: 90.77 %, NPA: 90.74 %) and DNA-Technology (PPA: 92.31 %, NPA: 98.15 %) have acceptable analytical sensitivity for SARS-CoV-2, with 6 and 5 missed positive samples, respectively. Ideally, RT-qPCR assays used for the laboratory detection of SARS-CoV-2 should have, firstly, a high PPA value; secondly, a high NPA value; and finally, should require a short period of time for detection. However, integrating all these parameters into one assay is challenging. Assays with the highest number of detected positive samples should be used for diagnostic purposes since it has previously been highlighted that a significant number of false-negative results occur by the use of RT-qPCR assays (Suo et al., 2020). The average basic reproduction number (R0) of the SARS-CoV-2 was previously estimated to be 2.2; therefore, false-negative results are especially concerning, as it may be a driver of the pandemic (Zhen et al., 2020).
Interestingly, one of the first protocols published was the Charité (Universitätsmedizin Berlin Institute of Virology) primer-probe sets, which had the lowest sensitivity in our experimental settings. This poor performance may be due to a mutation in the site where primers bind and initiate amplification, as it has been previously demonstrated by Artesi et. al that a C to U transition in the E gene of the SARS-CoV-2 resulted in the failure of the Cobas SARS-CoV-2 assay identifying the gene (Artesi et al., 2020). Moreover, at the time when these primer-probes were published, a relatively small number of complete genome sequences were available, making the designing more difficult. In line with these findings, PCR assays that screen more SARS-CoV-2 genes should be preferred, as similar mutations may decrease the sensitivity of detection assays, increasing the number of false negative results. Therefore, the routine monitoring of SARS-CoV-2 for mutations that may adversely affect PCR assays is recommended.
Regarding HoT and assay runtimes, all RT-qPCR assays had similar HoT except for 1copy (Table 3). In the case of 1copy, PCR setup requires two distinct PCR tubes, as the master mix is singleplex and the E and RdRp genes are detected in separate PCR tubes, almost doubling HoT.
According to the manual of the kits, DNA Technology screens the highest number of SARS-CoV-2 specific genes. By screening both N and E genes, false-negative results occurring in consequence of a mutation may be minimized. Although there were five false negative results with this assay, these samples weren’t completely lost, as all five samples were positive for at least one specific SARS-CoV-2 gene. In the case of marginal samples, according to the manufacturer, samples likely contain a low copy number of SARS-CoV2 and resampling or retesting should be performed.
In addition, we also compared the performance of PCR assays to two POCTs. ICPA assays compared to RT-qPCR assays have a significantly shorter HoT (∼25-minutes vs less than 1 min). However, in our experimental settings, the sensitivity of the ICPA assay was only 43 %; thus, this method of detection is not able to completely replace RT-qPCR kits in the first-line diagnosis of SARS-CoV-2. Despite these shortcomings, in the case of laboratory overload, the application of this method at first-line treatment centers may be beneficial while awaiting the RT-qPCR results. It should also be noted that since the finalization of the conducted experiments in this study, the manufacturer has produced a novel kit available on the market, which may have higher sensitivity based on our preliminary testing.
The other POCT compared in this study was an immunochromatographic antigen assay, whose main advantage is its short HoT and runtime, requiring 10−15 minutes for the detection of SARS-CoV-2 antigens. Based on our data, BIOCREDIT COVID-19 Ag tests are not reliable for SARS-CoV-2 antigen detection from virus transport medium, as only 3 positive samples were identified out of the 37 PCR-positive samples. Since nasopharyngeal swabs were immersed into VTM and only a small amount of VTM is required for the assays lysis buffer, this poor clinical performance may result from a significant dilution (a total of 20-fold dilution) in the kit’s dilution tube. This is further supported by the findings of CK Mak et al., who reported that rapid antigen tests had a much higher clinical sensitivity when nasopharyngeal swabs were directly transferred into the dilution buffer (Mak et al., 2021).
In summary, multiplex RT-qPCR assays are the cornerstone of the laboratory diagnosis of SARS-CoV-2; however, “all-in-one” kits based on amplification that significantly reduces HoT and speeds up the clinical decision-making process might serve as an alternative method.
Author contributions
Bence Kenyeres: Conceptualization, methodology, investigation, writing- original draft preparation, visualization.
Noel Ánosi: Conceptualization, methodology, software, investigation, writing- original draft preparation, visualization.
Andrea Kiss: Investigation.
Mária Mátyus: Visualization, methodology, data curation.
László Orosz: Conceptualization, writing- original draft preparation.
Beatrix Kele: methodology, data curation, formal analysis.
Katalin Burián: Conceptualization, methodology, writing- reviewing and editing, supervision.
Krisztián Bányai: Writing- original draft preparation, writing- reviewing and editing, supervision.
György Lengyel: Conceptualization, methodology, writing- original draft preparation, writing- reviewing and editing, supervision, validation, data curation, project administration, resources.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Dr. Péter Kisfali and Amplikon Kft. for providing the Ustar Isothermal Amplification platform and the reagents. We would like to thank Fanni Rapcsák, Gábor Deli and Edina Takács for their technical assistance.
References
- Artesi M., Bontems S., Gobbels P., Franckh M., Maes P., Boreux R., Meex C., Melin P., Hayette M.P., Bours V., Durkin K. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the e gene quantitative reverse Transcription-PCR utilized in a commercial dual-target diagnostic assay. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M R.M., Cuomo A., et al. Features, evaluation, and treatment of coronavirus (COVID-19) StatPearls. 2021 [PubMed] [Google Scholar]
- Christian Drosten V.C., Bleicker Tobias, Brünink Sebastian. 2020. Diagnostic Detection of 2019-nCoV by Real-time RT-PCR.https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf?sfvrsn=a9ef618c_2 Accessed at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . 2020. COVID-19 Situation Update Worldwide, As of 30 March 2021. Accessed at: https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases 2020. [Google Scholar]
- FDA . 2020. Coronavirus Disease 2019 (COVID-19) Emergency Use Authorizations for Medical Devices.https://www.fda.gov/medical-devices/emergency-use-authorizations-medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices Accessed at. [Google Scholar]
- finddx.org . 2020. SARS-CoV-2 Diagnostic Pipeline. [Google Scholar]
- Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Lau S.K., Che X.Y., Woo P.C., Wong B.H., Cheng V.C., Woo G.K., Hung I.F., Poon R.W., Chan K.H., Peiris J.S., Yuen K.Y. SARS coronavirus detection methods. Emerg Infect Dis. 2005;11:1108–1111. doi: 10.3201/eid1107.041045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak G.C.K., Lau S.S.Y., Wong K.K.Y., Chow N.L.S., Lau C.S., Lam E.T.K., Chan R.C.W., Tsang D.N.C. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J. Clin. Virol. 2021;134 doi: 10.1016/j.jcv.2020.104712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola M., Alsafi Z., Sohrabi C., Kerwan A., Al-Jabir A., Iosifidis C., Agha M., Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int. J. Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020;9:1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M., Richter K. Towards effective diagnostic assays for COVID-19: a review. J. Clin. Pathol. 2020;73:370–377. doi: 10.1136/jclinpath-2020-206685. [DOI] [PubMed] [Google Scholar]
- Xu G., Hu L., Zhong H., Wang H., Yusa S., Weiss T.C., Romaniuk P.J., Pickerill S., You Q. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci. Rep. 2012;2:246. doi: 10.1038/srep00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Manji R., Smith E., Berry G.J. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J. Clin. Microbiol. 2020:58. doi: 10.1128/JCM.00743-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]