Summary
Genetic differences are a primary reason for differences in the susceptibility and severity of COVID-19. As induced pluripotent stem (iPS) cells maintain the genetic information of the donor, they can be used to model individual differences in SARS-CoV-2 infection in vitro. We found that human iPS cells expressing the SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2) (ACE2-iPS cells) can be infected w SARS-CoV-2. In infected ACE2-iPS cells, the expression of SARS-CoV-2 nucleocapsid protein, budding of viral particles, and production of progeny virus, double membrane spherules, and double-membrane vesicles were confirmed. We performed SARS-CoV-2 infection experiments on ACE2-iPS/ embryonic stem (ES) cells from eight individuals. Male iPS/ES cells were more capable of producing the virus compared with female iPS/ES cells. These findings suggest that ACE2-iPS cells can not only reproduce individual differences in SARS-CoV-2 infection in vitro but also are a useful resource to clarify the causes of individual differences in COVID-19 due to genetic differences.
Subject areas: Health Sciences, Virology, Cell Biology
Graphical abstract
Highlights
-
•
ACE2 expression is required for SARS-CoV-2 to infect human iPS cells
-
•
SARS-CoV-2 life cycle can be reproduced in the ACE2-iPS cells
-
•
COVID-19 candidate drugs can be evaluated using ACE2-iPS cells
-
•
ACE2-iPS cells can reproduce individual differences in SARS-CoV-2 infection
Health Sciences ; Virology ; Cell Biology
Introduction
The number of patients with coronavirus disease 2019 (COVID-19) and deaths continues to rise. Interestingly, the symptoms of COVID-19 are known to vary widely among individuals and include asymptomatic cases. Genetic differences are one cause for the differences in susceptibility and severity of COVID-19 (Anastassopoulou et al., 2020; Group, 2020; Pairo-Castineira et al., 2021; Zeberg and Pääbo, 2020). Approximately 20% of patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will present severe symptoms (Weiss and Murdoch, 2020). To develop drugs for patients with severe COVID-19, it is necessary to identify the causes of the worsening symptoms. Although several models, including cells such as Vero, Calu-3, and Caco-2 cells; organoids; and animals such as angiotensin-converting enzyme 2 (ACE2)-transgenic mice, hamsters, and ferrets have been used to study SARS-CoV-2 infection (Takayama, 2020), they do not reproduce individual differences well.
Human induced pluripotent stem (iPS) cells can be established from any individual (Takahashi et al., 2007). Furthermore, they are widely used as genetic disease models because they inherit the genetic information of their donors (Park et al., 2008). Our institute (Center for iPS Cell Research and Application, CiRA) has established iPS cells from more than 500 individuals. As this iPS cell panel was established from a human population of diverse genetic backgrounds, it can be a resource for reproducing individual differences in response to pathogens and drugs. Accordingly, in this study, we attempted to reproduce the SARS-CoV-2 infection and its individual differences using this panel.
It has been reported that SARS-CoV-2 infection is dependent on the expression of ACE2 and transmembrane protease, serine 2 (TMPRSS2) in host cells (Hoffmann et al., 2020). Type II alveolar epithelial cells, bronchial ciliated cells, pharyngeal epithelial cells, and intestinal epithelial cells all express high levels of ACE2 (Ziegler et al., 2020) and can be easily infected by SARS-CoV-2. It has also been reported that embryonic stem (ES) cell-derived type II alveolar epithelial cells and intestinal epithelial cells can be infected with SARS-CoV-2 (Han et al., 2021). However, the investigation of infection differences at the individual level and at large scale using iPS cell-derived somatic cells is hindered by the long time (generally more than 3 weeks) to differentiate the iPS cells and the variable differentiation efficiency among iPS cell lines (Kajiwara et al., 2012). SARS-CoV-2 infection experiments in undifferentiated iPS cells would reduce the time.
However, due to the low expression of ACE2 and TMPRSS2, SARS-CoV-2 does not infect iPS cells. Therefore, in this study, we identified SARS-CoV-2-related genes that enable SARS-CoV-2 to infect undifferentiated human iPS cells. To facilitate gene overexpression experiments with multiple iPS cell lines, we used adenovirus (Ad) vectors. Because the gene transfer efficiency of Ad vectors to undifferentiated iPS cells is almost 100%, there was no need to obtain clones expressing SARS-CoV-2-related genes. Next, we investigated whether the life cycle of SARS-CoV-2 can be reproduced in iPS cells expressing these SARS-CoV-2-related genes. We also show the value of ES/iPS cells to test drugs and gender effects. Our findings indicate that iPS cells expressing SARS-CoV-2-related genes are useful for the study of SARS-CoV-2 infection and its individual patient differences.
Results
SARS-CoV-2 does not infect undifferentiated iPS cells
First, we examined whether undifferentiated iPS cells could be infected by SARS-CoV-2 (Figure S1A). Before conducting this experiment, we examined the expression levels of viral receptors and proteases in undifferentiated iPS cells (Figure S1B). The gene expression level of ACE2 was low, but that of CD147 was high. CD147 is reported as a coronavirus receptor (Wang et al., 2020). TMPRSS2, a protease, was also expressed in undifferentiated iPS cells. We thus tried to infect undifferentiated iPS cells with SARS-CoV-2, but the morphology of the iPS cell colonies did not change (Figure S1C). In addition, viral genome in the cell culture supernatant (Figure S1D) and the production of infectious virus (Figure S1E) were not detected. The gene expression levels of undifferentiated markers (Figure S2A) and innate immune response-related markers (Figure S2B) were also unchanged. Furthermore, the expression of SARS-CoV-2 nucleocapsid (N) protein was not detected (Figure S2C). Together, these results indicated that SARS-CoV-2 does not infect undifferentiated iPS cells.
ACE2 expression is required for SARS-CoV-2 to infect human iPS cells
As human ACE2 and TMPRSS2 are known to be important for SARS-CoV-2 to infect cells, we overexpressed human ACE2 and TMPRSS2 in undifferentiated iPS cells by using Ad vectors (Figure 1A). The overexpression of ACE2 in iPS cells (ACE2-iPS cells) caused a large amount of SARS-CoV-2 infection (Figure 1B). Additionally, the amount of viral genome in the cell culture supernatant increased (Figure 1C). This was not the case if only overexpressing TMPRSS2. Furthermore, 2 days after the ACE2-iPS cells were infected with SARS-CoV-2, cell fusion was observed (Figure 1D), and after 4 days many of the cells died. Therefore, these results indicate that ACE2 expression is required for SARS-CoV-2 to infect undifferentiated iPS cells.
Figure 1.
Efficient SARS-CoV-2 infection and replication in ACE2-iPS cells
(A) Undifferentiated human iPS cells (1383D6) were transduced with 600 vector particles (VP)/cell of LacZ-, ACE2-, or TMPRSS2-expressing Ad vectors (Ad-LacZ, Ad-ACE2, or Ad-TMPRSS2, respectively) for 2 h and then cultured with AK02 medium for 2 days. ACE2-expressing human iPS (ACE2-iPS) cells were infected with SARS-CoV-2 (5×104 TCID50/well) for 2 h and then cultured with AK02 medium.
(B) The amount of infectious virus in the supernatant was measured by the TCID50 assay. One-way ANOVA followed by Tukey's post hoc test (∗p < 0.05, ∗∗p < 0.01, compared with Ad-LacZ).
(C) At days 0, 2, 3, and 4 after the SARS-CoV-2 infection, the viral RNA copy number in the cell culture supernatant was measured by qPCR.
(D) At days 2 and 4 after the SARS-CoV-2 infection, phase images of infected ACE2-iPS cells were obtained. Data are represented as means ± SD (n = 3).
See also Figures S1 and S2.
Characterization of SARS-CoV-2-infected ACE2-iPS cells
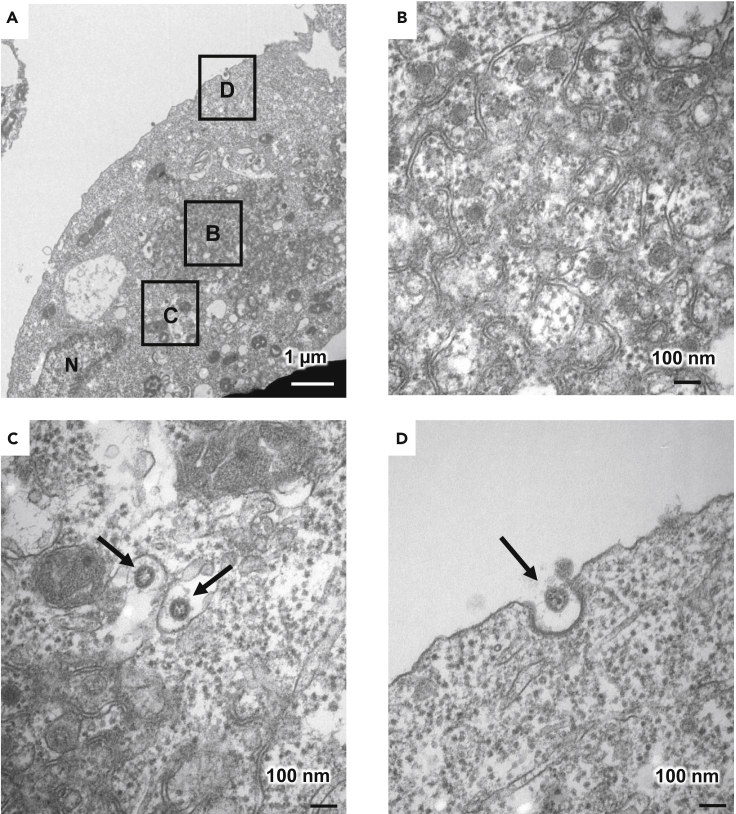
Transmission electron microscopic (TEM) images of ACE2-iPS cells infected with SARS-CoV-2 were obtained (Figures 2 and S3). Zippered endoplasmic reticulum (ER) (Figure 2B), double-membrane spherules (DMS) (Figure 2B) (Maier et al., 2013), and viral particles near the cell membrane (black arrow) (Figure 2D) were observed. So was an ER-Golgi intermediate compartment (ERGIC) containing SARS-CoV-2 particles (black arrows) (Figures 2C and S3). Double-membrane vesicles (DMVs, black arrows) were also observed in infected ACE2-iPS cells (Figure S3). DMVs are the central hubs for viral RNA synthesis (Klein et al., 2020). These structures were not observed in uninfected ACE2-iPS cells. These TEM images show that the life cycle of SARS-CoV-2 can be observed in ACE2-iPS cells.
Figure 2.
TEM images of infected ACE2-iPS cells
(A–D) (A) TEM images of infected ACE2-iPS cells. Zippered ER DMS (B) and virus particles in the ERGIC (black arrows) (C) and near the cell membrane (black arrows) (D) were observed.
See also Figure S3.
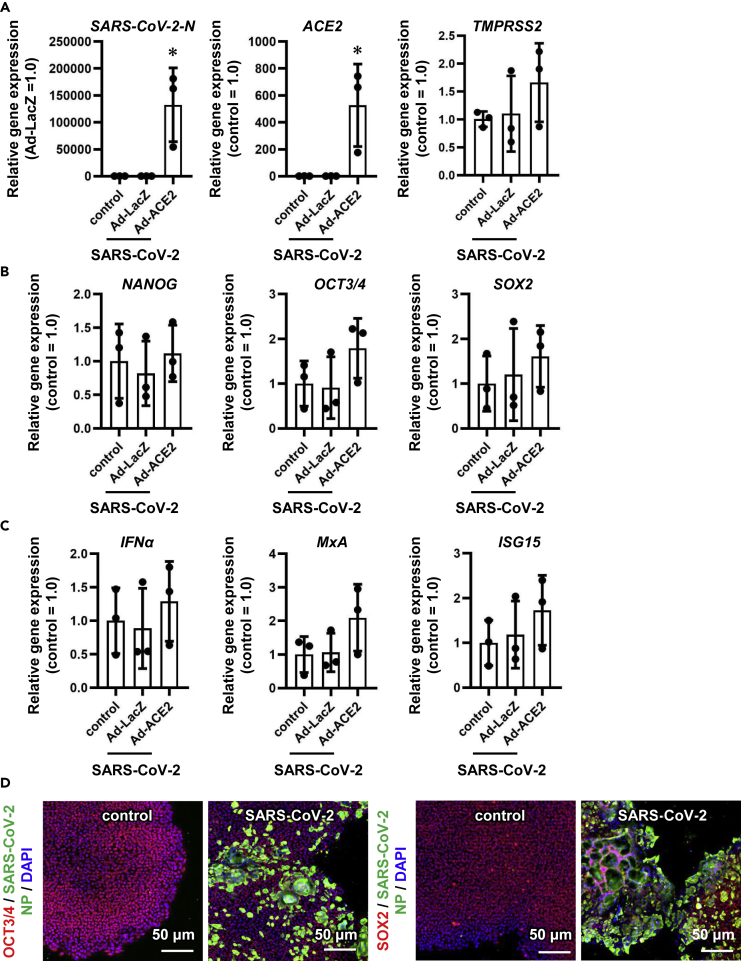
Next, we analyzed gene and protein expressions in uninfected and infected iPS cells 3 days after the viral infection. The intracellular viral genome and ACE2 expression levels in ACE2-iPS cells infected with SARS-CoV-2 were high (Figure 3A). At the same time, ACE2 overexpression and SARS-CoV-2 infection did not alter the gene expression levels of undifferentiated markers (Figure 3B) or innate immune response-related markers (Figure 3C). The gene expression levels of endoderm markers except for CER1 (Figure S4A) and SARS-CoV-2-related genes (CD147, NRP1, and TMPRSS2) (Figures S4B and 3A) were also unchanged. Immunostaining data showed that SARS-CoV-2 N protein was strongly expressed in ACE2-iPS cells 2 days after the infection (Figures 3D and S5).
Figure 3.
The pluripotent state of ACE2-iPS cells is not affected by SARS-CoV-2 infection
LacZ- or ACE2-expressing human iPS cells (LacZ-iPS cells and ACE2-iPS cells, respectively) were infected with SARS-CoV-2 (5×104 TCID50/well) for 2 h and then cultured with AK02 medium for 2 or 3 days. Control human iPS cells were not transduced with Ad vectors.
(A) The gene expression levels of viral genome, ACE2, and TMPRSS2 were examined by qPCR analysis.
(B and C) (B) The gene expression levels of pluripotent markers (NANOG, OCT3/4, and SOX2) and (C) innate immunity-related markers (IFNα, MxA, and ISG15) were examined by qPCR.
(D) Immunofluorescence analysis of SARS-CoV-2 NP (green), OCT3/4 (red), and SOX2 (red) in uninfected and infected ACE2-iPS cells. Nuclei were counterstained with DAPI (blue). One-way ANOVA followed by Tukey's post hoc test (∗p < 0.05, compared with Ad-LacZ). Data are represented as means ± SD (n = 3).
See also Figures S4 and S5, and Tables S2 and S3.
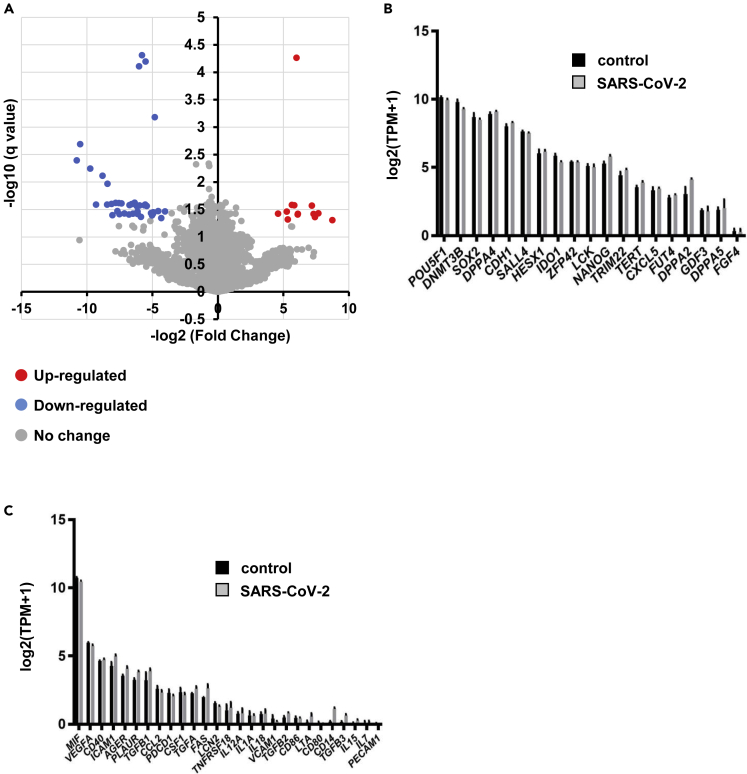
We also performed an RNA sequencing (RNA-seq) analysis of uninfected and infected ACE2-iPS cells. The colored dots in the volcano plot in Figure 4A indicate genes whose expression levels changed significantly more than 4-fold. In total, this change occurred in 6.7% of all genes (Figure S6A). A GO term analysis was performed on these genes (Figures S6B and S6C). None of the genes included undifferentiated markers (Figure 4B) or innate immune-response markers (Figure 4C). The gene expression levels of ectoderm, mesoderm, and endoderm markers were also unchanged after infection with SARS-CoV-2 (Figure S7). Overall, these results suggest that human iPS cells maintain an undifferentiated state even when SARS-CoV-2 replicates in large numbers.
Figure 4.
Global gene expression analysis of infected ACE2-iPS cells
RNA-seq analysis of uninfected or infected ACE2-iPS cells.
(A) A volcano plot of uninfected ACE2-iPS cells versus infected ACE2-iPS cells.
(B and C) Bar plots of pluripotent genes (B) and immune-related genes (C) in uninfected ACE2-iPS cells (control) and infected ACE2-iPS cells (SARS-CoV-2) are shown.
See also Figures S6 and S7.
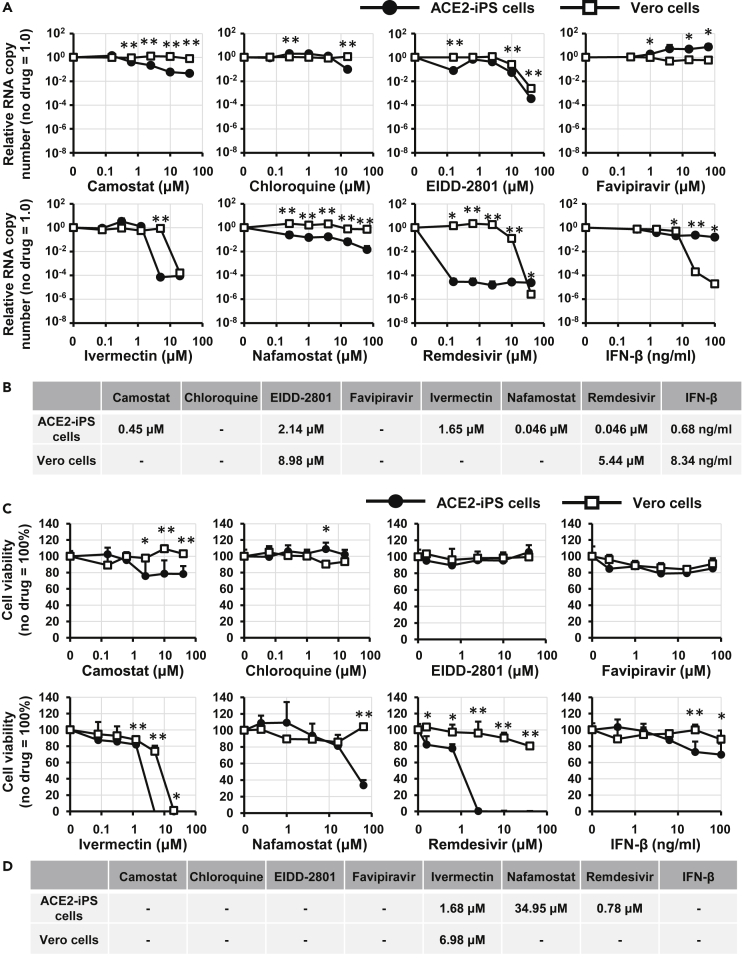
Evaluation of COVID-19 candidate drugs using ACE2-iPS cells
Next, we examined whether ACE2-iPS cells could be used for drug screening. We tried eight drugs used in COVID-19 clinical trials. Vero cells were used as the control. After exposing the cells to various concentrations of a drug, the number of viral RNA copies in the culture supernatant was quantified (Figure 5A). Data fitting resulted in sigmoid curves, and half-maximal effective concentrations (EC50) were calculated (Figure 5B). Among the eight drugs, the antiviral effect of remdesivir was strongest. On the other hand, chloroquine and favipiravir did not inhibit viral replication, and ivermectin was highly cytotoxic (Figure 5C). The EC50 and half-maximal cytotoxic concentration (CC50) values of ivermectin were almost the same between control and infected ACE2-iPS cells (Figure 5D). With the exception of interferon-beta, drug effects were stronger in ACE2-iPS cells than in Vero cells. Last, we confirmed the anti-viral effects of RNA-dependent RNA polymerase (RdRp) inhibitors (remdesivir and EIDD-2801) and TMPRSS2 inhibitors (camostat and nafamostat) in ACE2-iPS cells, indicating that ACE2-iPS cells can be used to evaluate COVID-19 drug candidates.
Figure 5.
Evaluation of anti-COVID-19 drugs in ACE2-iPS cells
(A) ACE2-iPS cells and Vero cells (2.0×104 cells/well) were infected with SARS-CoV-2 (2.0×103 TCID50/well) in the presence or absence of a drug and then cultured with medium for 4 and 2 days, respectively. The viral RNA copy number in the cell culture supernatant was measured by qPCR.
(B) The EC50 values (μM) of anti-COVID-19 drugs in ACE2-iPS cells and Vero cells were calculated using GraphPad Prism8 and are summarized.
(C) Vero cells and ACE2-iPS cells were cultured with medium containing drugs for 4 days. Cell viability was measured by the WST-8 assay.
(D) The CC50 values (μM) of anti-COVID-19 drugs in Vero cells and ACE2-iPS cells were calculated using GraphPad Prism8 and summarized. Data are represented as means ± SD (n = 3). Unpaired two-tailed Student's t test (∗p < 0.05, ∗∗p < 0.01; ACE2-iPS versus Vero cells).
See also Table S1.
SARS-CoV-2 infection experiments in various ACE2-iPS/ES cell lines
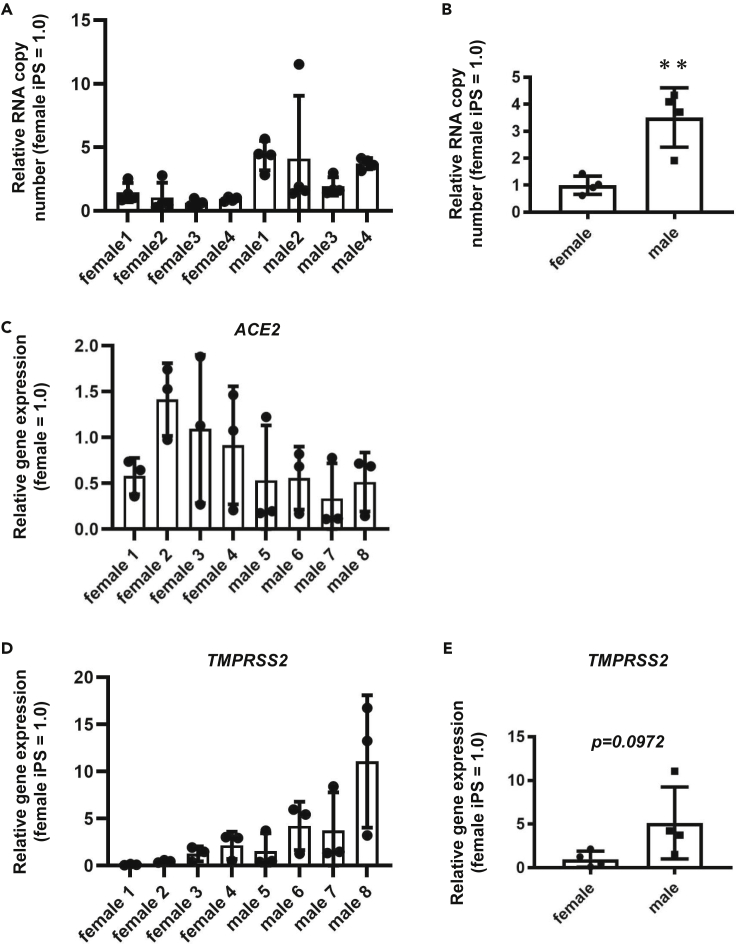
Finally, we performed SARS-CoV-2 infection experiments using human ACE2-iPS/ES cells established from eight donors. The replication efficiency of the virus was different among the ACE2-iPS/ES cell lines (Figure 6A). Interestingly, the viral replication capacity of male ACE2-iPS/ES cells was higher than that of female ACE2-iPS/ES cells (Figure 6B), suggesting that sex differences in the susceptibility to SARS-CoV-2 can be reproduced using ACE2-iPS/ES cells. Note that there was no significant difference in ACE2 expression levels in the ACE2-iPS/ES cell lines (Figure 6C). Recently, it has been speculated that the expression levels of androgen receptor and its target gene, TMPRSS2, are involved in the sex differences in SARS-CoV-2 infection (Wambier et al., 2020). The TMPRSS2 expression levels appeared to be higher in male iPS/ES cells than in female iPS/ES cells (Figure 6D), but there was no significant difference (Figure 6E).
Figure 6.
Sex differences of the SARS-CoV-2 infection rate in ACE2-ES/iPS cells
Four female ES/iPS cell lines and four male ES/iPS cell lines were transduced with 600 VP/cell of ACE2-expressing Ad vectors (Ad-ACE2) for 2 h and then cultured with AK02 medium for 2 days. The cells were then infected with SARS-CoV-2 (5×104 TCID50/well) for 2 h and cultured with AK02 medium.
(A) The viral RNA copy number in the cell culture supernatant was measured by qPCR for each cell line.
(B) The viral RNA copy number in the cell culture supernatant was compared between female iPS/ES cells and male iPS/ES cells.
(C and D) ACE2 (C) and TMPRSS2 (D) expression levels were measured by qPCR for each cell line.
(E) TMPRSS2 expression levels were compared between female iPS/ES cells and male iPS/ES cells. Unpaired two-tailed Student's t test (∗∗p < 0.01). Data are represented as means ± SD (n = 3). Female 1: H9, Female 2: KhES1, Female 3: KhES2, Female 4: 201B7, Male 1: H1, Male 2: KhES3, Male 3: Tic, Male 4: 1383D6.
See also Table S2.
Discussion
In this study, we showed that the life cycle of SARS-CoV-2 can be reproduced in human iPS cells overexpressing ACE2. In addition, we were able to confirm the effects of two TMPRSS2 inhibitors (camostat and nafamostat) and two RdRp inhibitors (remdesivir and EIDD-2801) using these ACE2-iPS cells. Finally, we showed a difference in the efficiency of infection of SARS-CoV-2 among ACE2-iPS/ES cells from eight donors. These results suggest that by using our iPS cell panel, it will be possible to investigate the effects of race and blood type as well as gender on SARS-CoV-2 infection. By conducting SARS-CoV-2 infection experiments using a panel of iPS cells for which genomic information has been obtained, it will also be possible to find genomic mutations that appear with high frequency in susceptible cells.
We observed a difference in the infection efficiency of ACE2-iPS/ES cells between donors, but we do not know if this difference reflects the sensitivity of the original donors to COVID-19, because none were patients with COVID-19. Therefore, we are currently establishing iPS cells from patients with severe and mild COVID-19. We plan to conduct infection experiments in the newly established iPS cells and compare the results with the symptoms of the original patients. These experiments could clarify whether the results of SARS-CoV-2 infection experiments using iPS cells reflect the symptoms of the original donors.
In this study, we confirmed sex differences in the susceptibility to SARS-CoV-2. Testosterone is known as a ligand for the androgen receptor, which regulates the expression of TMPRSS2 (Wambier et al., 2020). By generating iPS cell-derived Leydig cells that are capable of producing testosterone (Li et al., 2019), more prominent sex differences in the susceptibility to SARS-CoV-2 may be confirmed. Thus, ACE2-iPS cells and differentiated cells may have different susceptibility to SARS-CoV-2. In the future, it is expected that more accurate results will be obtained by conducting infection experiments using ACE2-iPS cells and differentiated cells at the same time.
Recently, GWAS analyses were performed on patients with severe and mild COVID-19 (Group, 2020; Pairo-Castineira et al., 2021). The analysis of the severe COVID-19 GWAS group indicated that mutations in the SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1 genes are associated with the severity of the symptoms (Group, 2020). Pairo-Castineira et al. suggested that the mutations and expression levels of the IFNAR2 and TYK2 genes are also associated with the severity of COVID-19 (Pairo-Castineira et al., 2021). However, the function of these genes in COVID-19 is still not fully understood. Further analysis can be performed by examining the relationship between viral infection and gene mutations and expressions in our iPS cell panel. Because single-nucleotide mutations can be easily introduced into iPS cells (Kim et al., 2018), the function of these mutations can be studied using genome-edited iPS cells.
Accordingly, there may be multiple causes of the individual differences in COVID-19 symptoms. The ACE2-iPS cells that we have developed in this study will be one tool to elucidate the cause of these individual differences, which will help identify vulnerable populations and develop new drugs.
Limitations of the study
To infect iPS cells with SARS-CoV-2, we overexpressed ACE2. However, if ACE2 and its related genes are responsible for the individual differences in SARS-CoV-2 infection, our system will not be effective. To analyze the mutation and expression of ACE2 and its related genes, it is essential to use somatic cells expressing ACE2. Also, our system is not effective for studying the non-genetic causes of individual differences in COVID-19 severity. For example, it has been speculated that age-related differences in COVID-19 symptoms are due to impaired cytotoxic CD8+ T cell responses (Westmeier et al., 2020). For such studies, it is more suitable to use blood samples.
Resource availability
Lead contact
Dr. Kazuo Takayama.
Center for iPS Cell Research and Application, Kyoto University, Shogoin Kawaharacho 53, Sakyo-ku, Kyoto 606-8397, Japan.
Phone: +81-75-366-7362, FAX: +81-75-366-7074.
E-mail: kazuo.takayama@cira.kyoto-u.ac.jp.
Materials availability
All unique/stable reagents generated in this study are available from the corresponding authors with a completed Materials Transfer Agreement.
Data and code availability
Raw data of RNA-seq analysis were submitted under Gene Expression Omnibus (GEO) accession number GSE166990.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
The SARS-CoV-2 strain used in this study (SARS-CoV-2/Hu/DP/Kng/19-027) was kindly provided by Dr. Tomohiko Takasaki and Dr. Jun-Ichi Sakuragi (Kanagawa Prefectural Institute of Public Health). Figures 1A and S1A were created using Biorender (https://biorender.com). We thank Dr. Misaki Ouchida (Kyoto University) for creating the graphical abstract, Dr. Peter Karagiannis (Kyoto University) for critical reading of the manuscript; Dr. Masato Nagagawa (Kyoto University) for providing the human iPS cells; Dr. Yoshio Koyanagi and Dr. Kazuya Shimura (Kyoto University) for the setup and operation of the BSL-3 laboratory at Kyoto University; Dr. Toru Okamoto (Osaka University), Dr. Akatsuki Saito (University of Miyazaki), Dr. Hirofumi Ohashi, and Dr. Koichi Watashi (National Institute of Infectious Diseases) for helpful discussion; and Ms. Kazusa Okita and Ms. Satoko Sakurai (Kyoto University) for technical assistance with the RNA-seq experiments. This research was supported by the iPS Cell Research Fund, the COVID-19 Private Fund (to the Shinya Yamanaka laboratory, CiRA, Kyoto University), the Joint Usage/Research Center program of Institute for Frontier Life and Medical Sciences, Kyoto University, the Astellas Foundation for Research on Metabolic Disorders, the Senri Life Science Foundation, the Mitsubishi Foundation, the JST Core Research for Evolutional Science and Technology (JPMJCR20HA), and the Japan Agency for Medical Research and Development (AMED) (20fk0108263s0201, 20fk0108518s0401, 20fk0108511h0201).
Author contributions
E.S. performed the SARS-CoV-2 experiments, analyses, and statistical analysis. S.D. performed the data analysis and statistical analysis. A.S. performed the human cell culture and qPCR analyses. N.M. performed the drug experiments. A.H. obtained the TEM images. Y.M. obtained the TEM images. T.N. obtained the TEM images. T.Y. performed the RNA-seq analysis. K.T. performed the SARS-CoV-2 experiments and analyses and wrote the paper.
Declaration of interests
The authors declare no competing financial interests.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102428.
Supplemental information
References
- Anastassopoulou C., Gkizarioti Z., Patrinos G.P., Tsakris A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum. Genomics. 2020;14:1–8. doi: 10.1186/s40246-020-00290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group S.C.-G. Genomewide association study of severe Covid-19 with respiratory failure. New Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., Endo H., Eto K., Toguchida J., Uemoto S. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2012;109:12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Matsumoto T., Kagawa H., Nakamura M., Hirohata R., Ueno A., Ohishi M., Sakuma T., Soga T., Yamamoto T. Microhomology-assisted scarless genome editing in human iPSCs. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-03044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li Y., Sottas C., Culty M., Fan J., Hu Y., Cheung G., Chemes H.E., Papadopoulos V. Directing differentiation of human induced pluripotent stem cells toward androgen-producing Leydig cells rather than adrenal cells. Proc. Natl. Acad. Sci. U S A. 2019;116:23274–23283. doi: 10.1073/pnas.1908207116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. mBio. 2013;4 doi: 10.1128/mBio.00801-13. e00801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D. Genetic mechanisms of critical illness in Covid-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Park I.-H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takayama K. In vitro and animal models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020;41:513–517. doi: 10.1016/j.tips.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambier C.G., Goren A., Vaño-Galván S., Ramos P.M., Ossimetha A., Nau G., Herrera S., McCoy J. Androgen sensitivity gateway to COVID-19 disease severity. Drug Dev. Res. 2020;81:771–776. doi: 10.1002/ddr.21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss P., Murdoch D.R. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395:1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmeier J., Paniskaki K., Karaköse Z., Werner T., Sutter K., Dolff S., Overbeck M., Limmer A., Liu J., Zheng X. Impaired cytotoxic CD8+ T cell response in elderly COVID-19 patients. mBio. 2020;11 doi: 10.1128/mBio.02243-20. e02243–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- Ziegler C.G., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of RNA-seq analysis were submitted under Gene Expression Omnibus (GEO) accession number GSE166990.