Abstract
Background
COVID-19, declared a pandemic in March 2020 by the World Health Organization is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The virus has already killed more than 2.3 million people worldwide.
Object
The principal intent of this work was to investigate lead compounds by screening natural product library (NPASS) for possible treatment of COVID-19.
Methods
Pharmacophore features were used to screen a large database to get a small dataset for structure-based virtual screening of natural product compounds. In the structure-based screening, molecular docking was performed to find a potent inhibitor molecule against the main protease (Mpro) of SARS-CoV-2 (PDB ID: 6Y7M). The predicted lead compound was further subjected to Molecular Dynamics (MD) simulation to check the stability of the leads compound with the evolution of time.
Results
In pharmacophore-based virtual screening, 2,361 compounds were retained out of 30,927. In the structure-based screening, the lead compounds were filtered based on their docking scores. Among the 2,360 compounds, 12 lead compounds were selected based on their docking score. Kazinol T with NPASS ID: NPC474104 showed the highest docking score of −14.355 and passed criteria of Lipinski's drug-like parameters. Monitoring ADMET properties, Kazinol T showed its safety for consumption. Docking of Kazinol T with two Asian mutants (R60C and I152V) showed variations in binding and energy parameters. Normal mode analysis for ligand-bound and unbound form of protease along with its mutants, revealed displacement and correlation parameters for C-alpha atoms. MD simulation results showed that all ligand-protein complexes remained intact and stable in a dynamic environment with negative Gibbs free energy.
Conclusions
The natural product Kazinol T was a predicted lead compound against the main protease of SARS-CoV-2 and will be the possible treatment for COVID-19.
Keywords: Coronavirus, Protease, Mpro, Natural products, Docking
1. Introduction
Tyrrell and colleagues studied several animal viruses using electron microscopy in the 1960s, including infectious bronchitis virus, mouse hepatitis virus and transmissible gastroenteritis virus of swine. These viruses revealed a similar morphology [1,2] and were clustered together as a new virus group. This new set of viruses was termed as ‘coronaviruses’ (corona denoting the crown-like appearance of the surface projections) and was officially accepted as a new genus of viruses [3]. These viruses have a large variety and previously caused several outbreaks. In 2002–2003, a modified strain of coronavirus emerged from southern China, caused severe acute respiratory syndrome (SARS) and spread throughout the world with quantifiable speed [[4], [5], [6]]. In Wuhan, the capital city of Hubei province of China, pneumonia cases of unknown origin were identified in early December 2019 [[7], [8], [9], [10]]. The responsible pathogen was identified as a novel enveloped RNA β-coronavirus [11]. It is now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is phylogenetically similar to SARS-CoV [12]. In recent studies, the severity of some cases of COVID-19 imitated that of SARS-CoV [7,13].
WHO declared the coronavirus disease 2019 (COVID-19) as a public health emergency of international concern on 30th January 2020 (https://www.who.int). According to WHO statistics, there were more than 106 million COVID-19 cases and more than 2.3 million deaths globally from the disease (dated: February 8, 2021; https://www.worldometers.info/coronavirus/). It affected >210 countries and territories around the world. So far, the percentage of severe cases remains high, calling for effective regimes for this highly contagious disease [14,15]. Scientists are striving hard to find specific antiviral drugs. Numerous drugs such as chloroquine, arbidol, remdesivir, and favipiravir were in clinical trials to treat COVID-19, but none provided convincing evidence of safety and efficacy [[8], [9], [10],16]. Therefore, an effective drug against SARS-CoV-2 still needs to be discovered. In the present study, we aimed to evaluate natural product compounds for the inhibition of the main protease of SARS-CoV-2. The comparative binding was also carried out with Asian mutants of Mpro recently reported by Ref. [17]. We believe that our findings will help pharmaceutical chemists to optimize suitable drugs for the clinical management of COVID-19 patients.
2. Methodology
2.1. Data retrieval
The co-crystallized structure of the main protease (ID: 6Y7M) of SARS-CoV-2 was downloaded from PDBJ (https://pdbj.org/) in.pdb format. A natural product compounds library of 30,927 was retrieved from the Natural Product Activity and Species Source database (NPASS) database (http://bidd.group/NPASS/index.php). Protein mutants (R60C and I152V) were created in PyMOL and saved in.pdb format for processing.
2.2. Library screening
NPASS containing 30,927 compounds, was screened using Molecular Operating Environment (MOE) software. Two approaches were adopted for compound screening; (a) pharmacophore-based screening and (b) molecular docking. In pharmacophore-based virtual screening, the pharmacophoric features of the co-crystalized ligand (anionic and cationic atom, H-bond donor and acceptor, aromatic centre, Pi-ring centre and hydrophobic centroid etc) were selected from Compute option of MOE. After ligand feature selection, the software was run to filter compounds based on selected features in the co-crystal ligand. The resulting output file of the compounds was used for molecular docking.
2.3. Molecular docking and downstream analysis
In a molecular docking approach, the pharmacophore-based screened library (2361 compounds) was docked with the three-dimensional structure of the target protein. The protein structure of native and mutant Mpro was prepared in MOE for docking. The 3D protonation and energy minimization of the protein as well as site finder was used to predict the active site residues of the binding pocket [[8], [9], [10]]. Structure preparation was done by adding missing hydrogen atoms and correcting bond order assignments. Furthermore charge states and orientation of various groups and performed restrained minimizations were performed, which allow hydrogen atoms to be freely minimized. The Piper algorithm was used for docking. The resulting structures were grouped into clusters and ranked according to the docking score from the least to the highest. Poisson-Boltzmann Surface Area (PBSA) energies were calculated using the Scientific Vector Language (SVL) script in MOE. The parameters were: Salt = NaCl, molar concentration 0.1, temperature = 300 K, interior dielectic = 1 and exterior dielectic = 80. The top-ranked compound was selected to evaluate its ADMET properties using the SMILE format (C=CC(c1cc(CCCc2cc(O)c3c(c2CC=C(C)C)CC(O3)C(O)(C)C)c(cc1O)O)(C)C) at ADMET SAR server (http://lmmd.ecust.edu.cn/admetsar2/) and SwissADME server (http://www.swissadme.ch/index.php).
2.4. Molecular dynamics simulation
The compound with the best molecular docking results to the viral protein was considered for Molecular Dynamics simulation, in order to understand the interaction and stability of the protein-ligand complex. Maestro-Desmond Interoperability Tools [18] system builder was used for the purpose. The explicit water solvation TIP3P model was used with orthorhombic box shape (size deliberated by box size calculation method) of absolute size with a volume of 412556 Å3. The OPLS3e force field was used to rectify the geometries. The system was then minimized using a maximum of 1000 iterations with a convergence threshold of 1.0 kcal/mol Å3. The in silico simulation of the system was carried out for an interval of 100 ns and with a recording interval of an energy equivalent to 5 and a trajectory of 4.8. The NPT ensemble corresponded to a temperature of 300 K and a pressure of 1.01325 bar [18].
3. Results and discussion
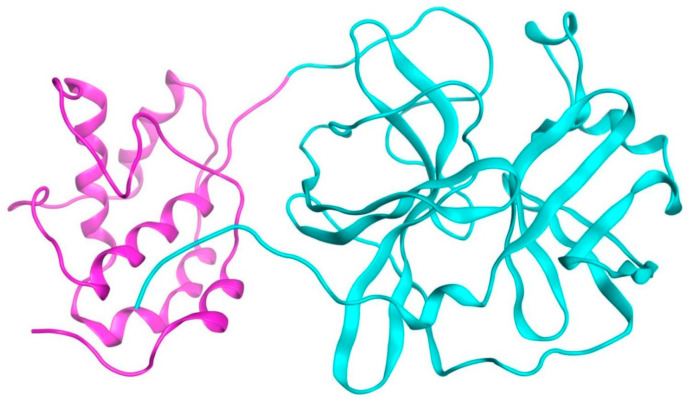
Strong medical support and self-isolation can enhance the cure rate, reduce the mortality rate of infected individuals, and the spread of SARS-CoV-2 (Cheng and Shan, 2019; [19,20]. Still, thousands of severely ill patients are dying every day. The guidelines issued by WHO and National Health Commission for diagnosis and treatment of SARS-CoV-2 suggest that ribavirin, lopinavir/ritonavir antiviral therapy may be used to treat SARS-CoV-2 patients [21], but a permanent and efficient cure is still urgently required. Natural products may be useful in discovering novel therapeutics to ease the current COVID-19 pandemic [22]. The main protease (PDB ID: 6Y7M) is a key protein in the SARS-CoV-2 life cycle. This enzyme cuts the poly-proteins translated from viral RNA to yield functional viral proteins. The first crystal structure of SARS-CoV main protease and its complex with a hexapeptidyl chloromethylketone (CMK) inhibitor has been previously reported [23]. The three-dimensional structure of the protein is shown in Fig. 1 . l-canavanine from Sutherlandia frutescens has been described as potential inhibitor of the SARS-CoV-2 3C-like main protease using informatics based approaches [24].
Fig. 1.
The three-dimensional structure of the SARS-CoV-2 protease protein.
In the present study, the active site of the protein was predicted prior to docking analyses for attaining best docking results. The pocket residues of the protease protein (PDB ID: 6Y7M) were Thr24, Thr25, Thr26, Leu27, His41, Thr45, Met49, Tyr54, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, Met165, Glu166, Pro168, His172, Val186, Asp187, Arg188, Gln189. The binding site of Mpro contains hydrophobic amino acids such as Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168 and His172. They maintain a relatively hydrophobic environment and stabilize its conformation. The identification of a compound, which acts as potent Mpro inhibitor may provide a basis for the development of an anti-corona viral drug [19,20]. Upadhyay et al. [25] screened 51 medicinal plants and found that Camellia sinensis and Terminalia chebula inhibited SARS-COV-2 3CLpro with IC50 values of 8.9 ± 0.5 μg/ml and 8.8 ± 0.5 μg/ml, respectively. Thearubigin, the polymeric phenol, bound to the CYS145 residue of the active site of the protease and might serve as pharmaco-active molecule.
3.1. Inhibitor analysis
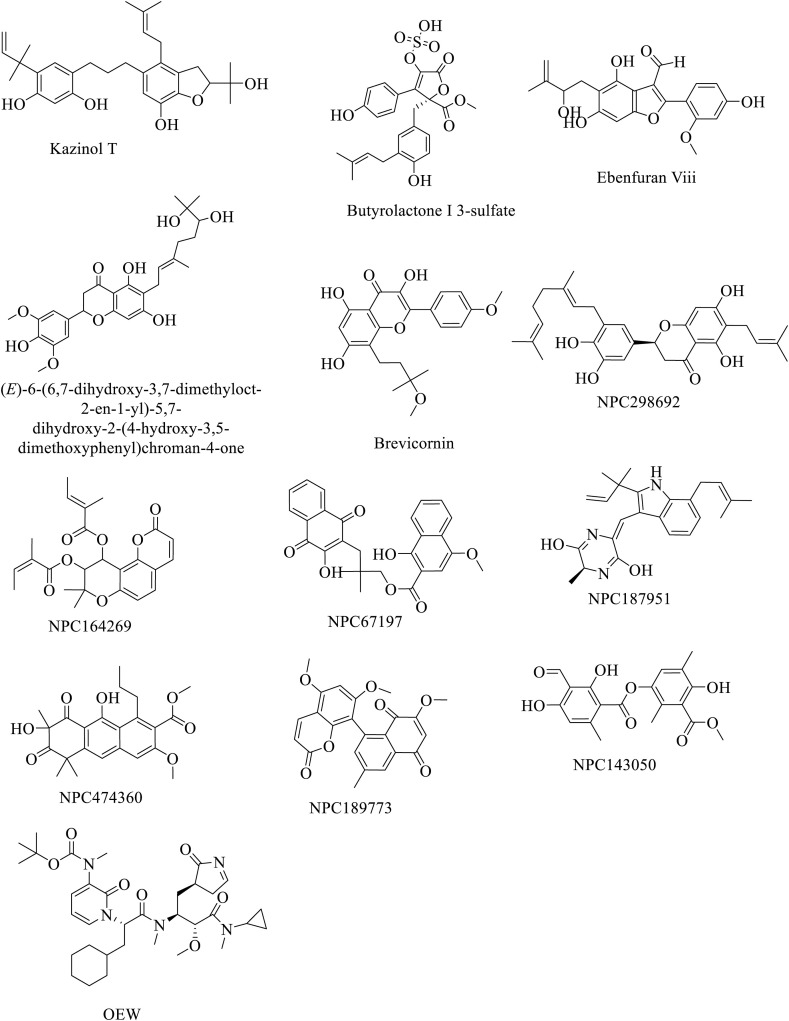
After virtual screening of 30,927 compounds based on pharmacophoric features of the co-crystalized ligand (such as an anionic and cationic atom, H-bond donor and acceptor, aromatic centre, Pi-ring centre and hydrophobic centroid), the dataset was reduced to 2,361 compounds. These compounds were docked to the SARS-COV-2 protease. Twelve compounds derived from various natural sources (e.g. fungi, plants, and microbes) showed docking scores below −12 (Table 1 ). In addition, 18 compounds revealed higher docking score values than the positive control (OEW, docking score −11.5448). However, a cut-off value of −12 was considered in this study. The chemical structures of the compounds and the highest docking scores along with the standard compound (control) are shown in Fig. 2 and Table 1 respectively.
Table 1.
Descriptors and binding affinity values of highest scoring compounds upon receptor docking.
| NPASS ID | Toxicity | Weight (g/mol) | TPSA | logP | S | Binding value (kcal/mol) |
|---|---|---|---|---|---|---|
| NPC474104 | No | 480.64 | 90.15 | 6.03 | −8.00 | −14.355008 |
| NPC173034 | No | 504.51 | 156.6 | 2.28 | −5.98 | −13.853008 |
| NPC476350 | No | 384.38 | 120.3 | 3.52 | −4.54 | −13.556889 |
| NPC472630 | No | 502.56 | 145.9 | 4.02 | −4.38 | −13.469447 |
| NPC124729 | no | 400.43 | 105.4 | 3.9 | −4.58 | −12.732084 |
| NPC298692 | no | 492.61 | 107.2 | 7.05 | −7.5 | −12.71736 |
| NPC164269 | no | 426.46 | 88.13 | 4.3 | −6.10 | −12.695246 |
| NPC67197 | no | 460.48 | 110.1 | 5.0 | −6.56 | −12.586933 |
| NPC187951 | no | 391.51 | 80.97 | 5.8 | −6.10 | −12.577618 |
| NPC474360 | no | 414.45 | 110.13 | 3.09 | −6.2 | −12.227035 |
| NPC189773 | no | 406.39 | 88.13 | 3.52 | −7.1 | −12.054446 |
| NPC143050 | no | 374.34 | 130.36 | 2.55 | −3.6 | −12.021175 |
| Control (Standard compound) | ||||||
| OEW | no | 641.81 | 129.13 | 3.90 | −6.11 | −11.5384 |
Fig. 2.
Chemical structures of the compounds with the highest docking score and standard compound.
Kanjanasirirat et al, [26] tested 122 Thai natural products and found panduratin A from Boesenbergia rotunda as potent anti-SARS-CoV-2 inhibitor. Treatment of Vero E6 cells with B. rotunda extract and panduratin An upon viral infection drastically suppressed SARS-CoV-2 infectivity with IC50 values of 3.62 μg/mL (CC50 = 28.06 μg/mL) and 0.81 μΜ (CC50 = 14.71 μM), respectively.
NPC474104 (Kazinol T) extracted from Broussonetia kazinoki was the top compound identified in our screening. The bioactivity of this compond has been described in different context. It showed inhibitory activities against both the monophenolase and diphenolase activities of tyrosinase [27]. It exhibited the strongest binding affinity score (−14.355008 kcal/mol) compared to all other putative ligands. This was followed by NPC173034 (butyrolactone I 3-sulfate) (docking score −13.8530080 kcal/mol) discovered from Aspergillus terreus (HKI0499). Butyrolactone I 3-sulfate has been previously reported to inhibit CDK1/cyclin B, CDK5/p25, DYRK1A, CK1, and GSK-3R kinases. Sulfate derivatives of fungal origin provided new insights into the inhibitory activity of butyrolactones. It also showed moderate cytotoxicity against HeLa cells [28]. Compound NPC476350 (ebenfuran III; docking score −13.556889 kcal/mol) is a naturally occurring benzofuran derivative isolated from Onobrychis ebenoides (family: Leguminosae). It induced apoptosis and cytotoxicity in DU-145 prostate cancer cells. It also exerted significant cytotoxic effects towards certain other cancer cell lines. This effect is thought to occur via the isoprenyl moiety at the C-5 position of the molecule [29]. Paulowniones A (NPC472630; docking score −13.469447 kcal/mol) is a C-geranylated flavonoid isolated from the fruits of Paulownia tomentosa. It reduced the production of the pro-inflammatory cytokine TNF-α in THP-1 cells [30]. Other top-ranked compounds based on their docking scores are shown in Table 1. Kar et al. [31] found promising inhibitory potentials of the phytochemicals taraxerol, friedelin and stigmasterol by binding to the active binding pockets of viral proteins.
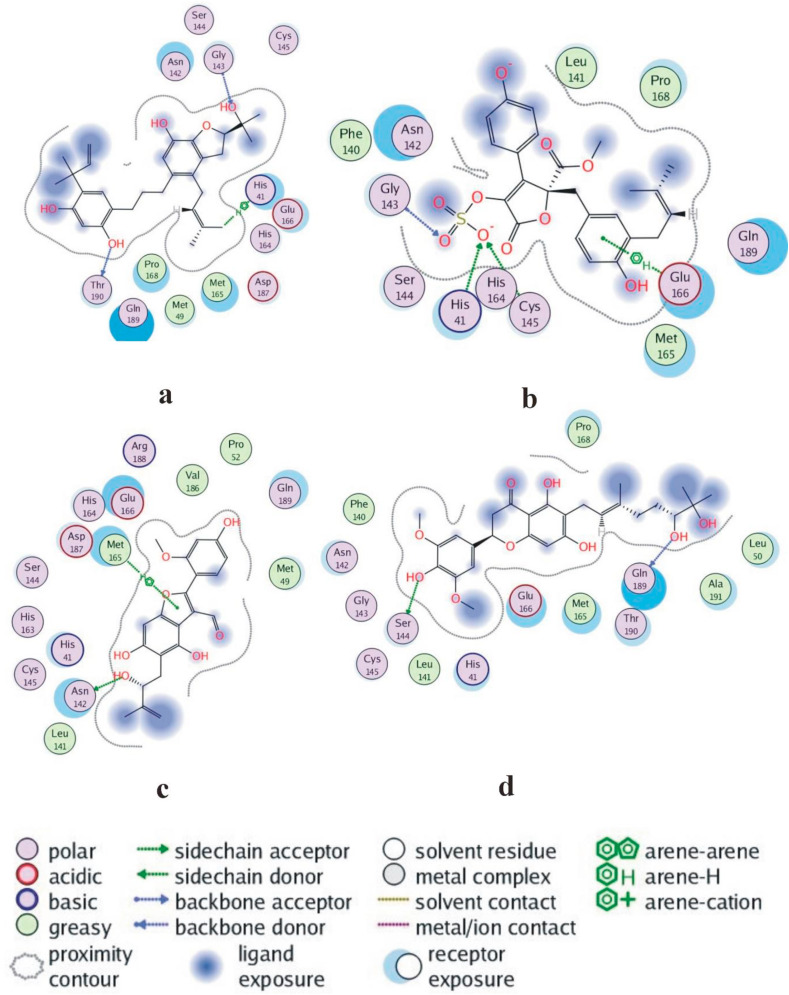
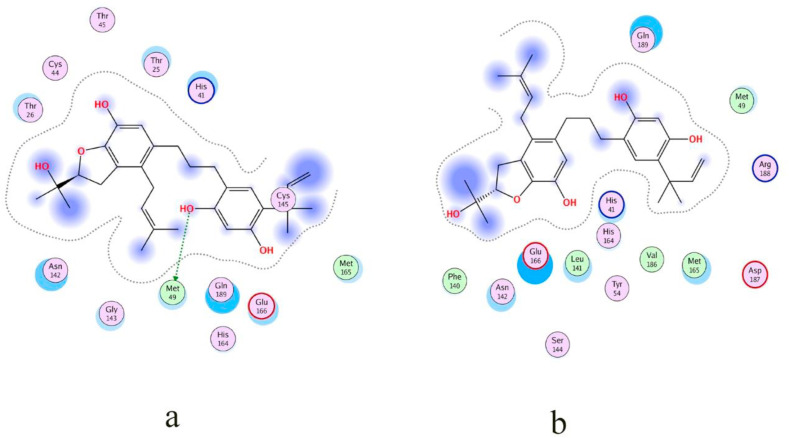
Kazinol T was completely trapped in the predicted binding site of the protein via a complicated network of strong bonding. The hydroxyl group attached to a benzene ring of NPC474104 (kazinol T) exhibited a backbone acceptor (basic) interaction with Gly143 and a backbone donor (greasy) interaction with Thr190. An arene-hydrogen interaction was found between the hydrogen of the ligand and residue His41 of the receptor protein Fig. 3a. The two key residues, Gly143 and His41, were especially noted, because they commonly formed effective interactions. The interaction was mainly influenced by a conformational change of the Gly143 side chain. His41 also formed effective interactions with ligands. Thus, His41 could be utilized as a key residue against a broad range of ligands. Butyrolactone I 3-sulfate and butyrolactone I 4′′-sulfate were the first examples of sulfate bearing secondary metabolites isolated from Aspergillus. His42, Cys145, Gly143 and His164 formed effective interactions with butyrolactone I 3-sulfate, which suggested potential utilization of these key residues for a specific class of drugs. The benzene ring of NPC173034 (butyrolactone I 3-sulfate) exhibited arene-hydrogen interactions with Glu166, His41 and Cys145. It bound by a side-chain acceptor (polar) to the oxygen of the SO4 group. Gly143 bound by backbone acceptor (basic) interaction to another oxygen of the SO4 group of the ligand (Fig. 3b).
Fig. 3.
Two-dimensional interactions of protease with ligands depicting (a) NPC474104 (b) NPC173034 (c) NPC476350 (d) NPC472630 compound.
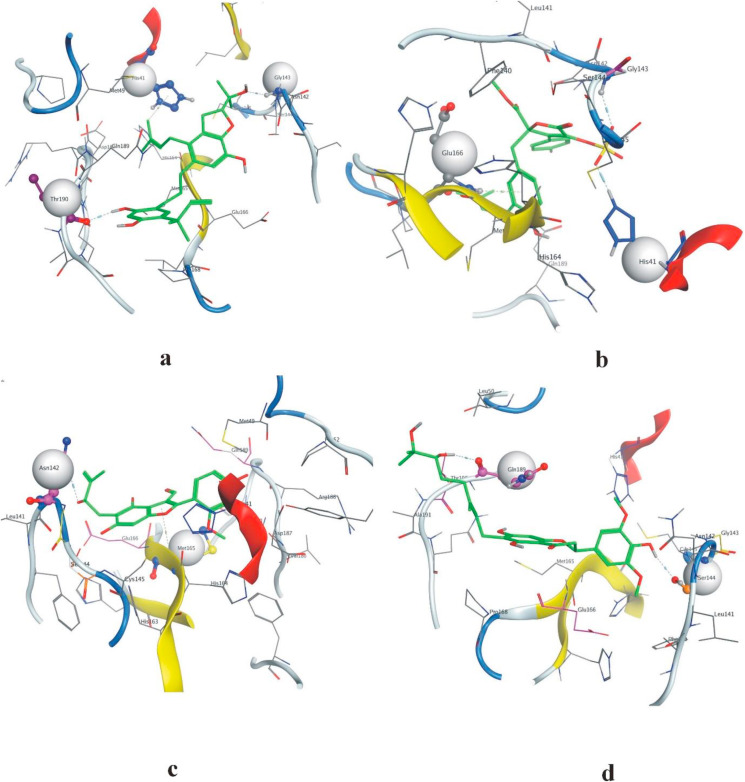
Met165 formed moderate interactions with ebenfuran III, which may be useful for drug development against Mpro. Ebenfuran III may serve as lead molecule to design and synthesize benzofuran derivatives substituted directly at C-2 with different heterocyclic groups. The pentene ring of NPC476350 (ebenfuran III) formed an arene-hydrogen interaction with Met165 and Asn142 and bound to the hydroxyl group of the ligand by the side-chain donor (acidic) (Fig. 3c). Generally, the addition of an isoprenoid chain rendered the derivate pharmacologically more effective than the parent compound, probably because the prenyl group increased the lipophilicity and conferred a strong affinity for biological membranes. Here, the prenyl moiety at position C-5, rather than the formyl group at C-3, was the key determinant of the activity of ebenfuran III. The hydroxyl group of NPC472630 (paulownione A) isolated from Paulownia tomentosa contains a benzene ring. This ring bound to Ser144 by a side-chain donor (acidic) interaction and another hydroxyl group to Gln189 by a backbone donor (greasy) interaction to the receptor protein (Fig. 3d). Gln189 is located in the binding pocket of Mpro and belongs to the loop region (residue 187–192), which is known to be quite flexible. Gln189 could maintain effective interactions by utilizing the flexibilities in its side chain and the backbone of the loop. Because of their location within the loop and the preferred orientation of their side chains, Asp187 and Arg188 (present in the same loop as Gln189) might have relatively less chance to form effective interactions. P. tomentosa prenylated flavonoids showed promising antioxidant, antibacterial, antiphlogistic, cytotoxic, and SARS-CoV papain-like protease (SARS-CoV PLpro) effects. The three-dimensional interactions of top-ranked ligands are shown in Fig. 4 . These results indicate diverse interaction patterns of the respective ligands in the flexible active site of Mpro. The PBSA energy values along with other types of energy changes of four best-docked compounds are mentioned in Table 2 .
Fig. 4.
Three-dimensional interactions of ligands (a) NPC474104 (b) NPC173034 (c) NPC476350 (d) NPC472630 with the receptor protein.
Table 2.
Solvation energies of ligands (kcal/mol) and their SARS-CoV-2 protease bound conformations. Total energies for bond stretching are also mentioned.
| Compound | PBSA | PBSA with bound receptor | Solvation energy | Solvation energy with bound receptor | Bond stretch energy | Bond stretch energy with bound receptor |
|---|---|---|---|---|---|---|
| NPC474104 | −0.8264 | −23.2122 | −92273.2969 | −87545.4531 | 7.2408 | 621.4600 |
| NPC173034 | −0.6643 | −23.3307 | −88473.1172 | −85025.8203 | 10.6801 | 624.8994 |
| NPC476350 | −0.7843 | −23.3050 | −99042.6172 | −94307.0703 | 5.0719 | 619.2912 |
| NPC472630 | −0.8763 | −23.3545 | −86674.2813 | −80113.3750 | 5.3077 | 619.5270 |
The inhibition analysis showed various binding affinities for the mutants R60C and I152V, with decreased S-values of −5.9 and −7.09, respectively (Fig. 5 ). The native protease formed 13 interactions (Fig. 3a), while R60C made 9 common and four new interactions (Thr25, Thr26, Cys44, Thr45). I152V revealed 8 common and six new interactions (Phe140, Leu141, Tyr54, Glu166, Val186, Arg188) compared to the native protease.
Fig. 5.
Interactions of protease mutant (a) R60C (b) I152V with top-scored ligand (NPC474104).
3.2. ADMET analysis of top compound
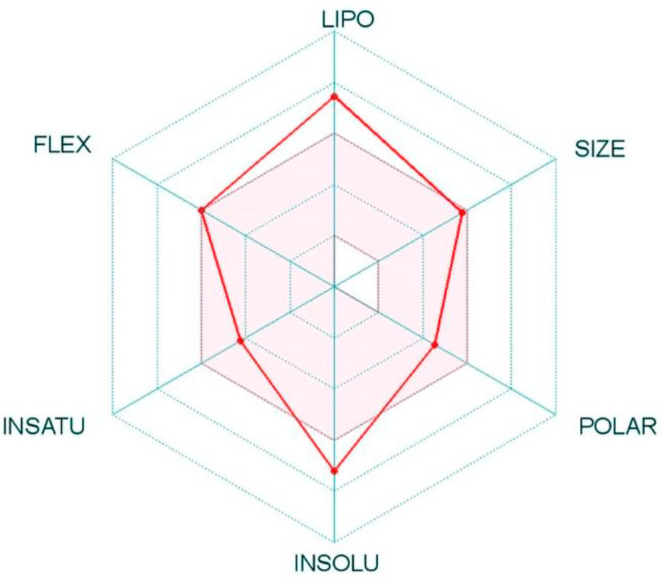
Previously, andrographolide from Andrographis paniculata has been identified in silico as a potential inhibitor of the SARS-COV-2 main protease (Mpro) by molecular docking analysis, toxicity and ADME predictions [32]. We conducted an ADMET analysis of NPC474104 (kazinol T; molecular weight: 480.29 g/ml), which scored the highest inhibition against protease of SARS-CoV-2. A logP value of 6.03, five hydrogen bond acceptors, four hydrogen bond donors and 9 rotatable bonds were obtained. It showed high probability for intestinal absorption (0.99), passing the blood-brain barrier and the potential for inhibition of phase II enzymes (CYP3A4, CYP2C9, CYP2D6, CYP2C19 and CYP1A2) and phase III transporters (P-glycoprotein, BSEP, OATP1B1, OATP1B3, OATP2B1). Kazinol T also bound to other receptors, e.g. receptors for estrogen, androgen, glucocorticoid, aromatase etc. The compound tested negative for hepatotoxicity, AMES mutagenesis and carcinogenicity. Concerning ecotoxicology, it appeared as toxic against honey bees and fishes. The water solubility was −3.565 logS, while the plasma protein binding capability was 1.21%. Acute oral toxicity was recorded as LD50 = 2.825 mg/kg. The Swiss ADME server did not show a violation of the Lipinski's criteria for drug-likeliness and revealed a bioavailability score of 0.55. The synthetic accessibility was 4.87, while the skin permeation was −3.91 cm/s. The physicochemical space for oral bioavailability of this compound is shown in Fig. 6 .
Fig. 6.
Radar plot showing physicochemical space for oral bioavailability, with suitable parameters falling in the coloured zone. POLAR depicts polarity, LIPO depicts lipophilicity (−0.7<XLOGP3<5), INSATU shows insaturation (0.25 <fraction Csp3 < 1) and INSOL shows insolubility value (0< log S (ESOL) < 6). 4 parameters such as polarity, instauration, flexibility and size were in range.
Previously, Chen et al. [33] reported tannic acid and 3-isotheaflavin-3-gallate as SARS-CoV-1 protease inhibitors by screening a natural product library consisting of 720 natural polyphenols from tea. Extracts from pure tea and black tea were also potent inhibitors of SARS-CoV-1 protease activity. We increased the size of the compound library and, hence, the chemical space against the protease of SARS-CoV-2. These compounds could serve as new drug leads for the treatment of viral diseases. Therefore, further research in this area is desirable.
3.3. Molecular dynamic simulation
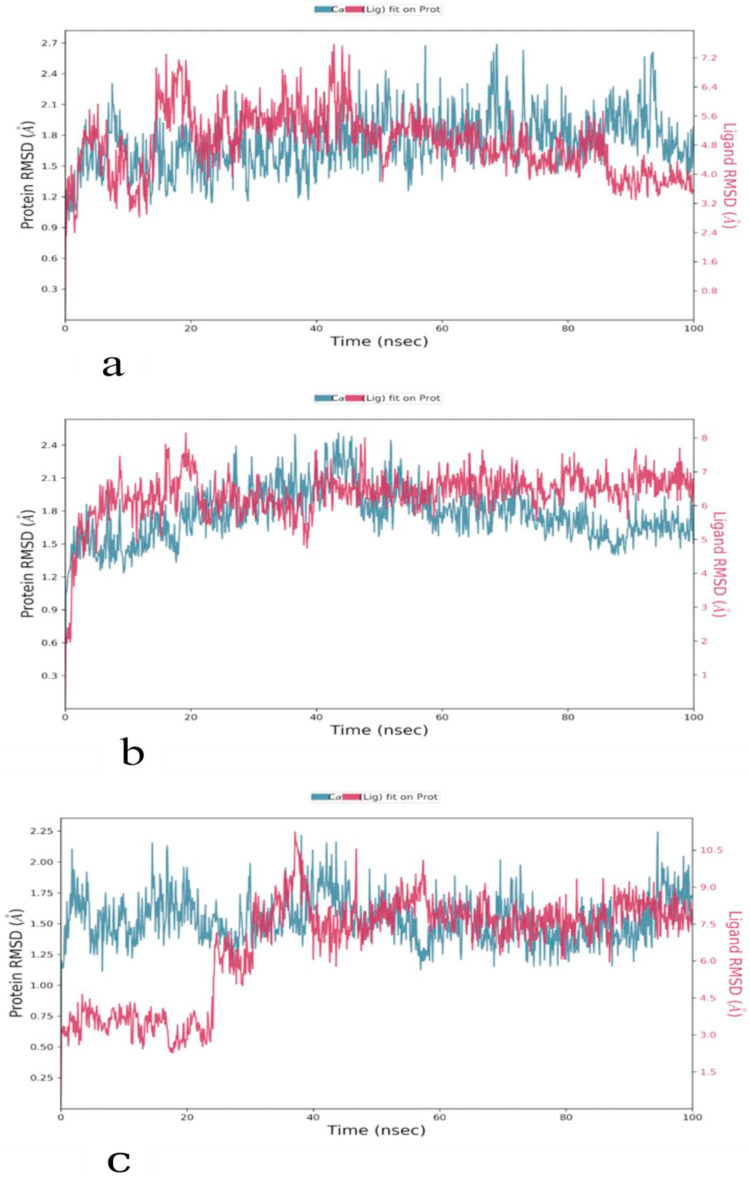
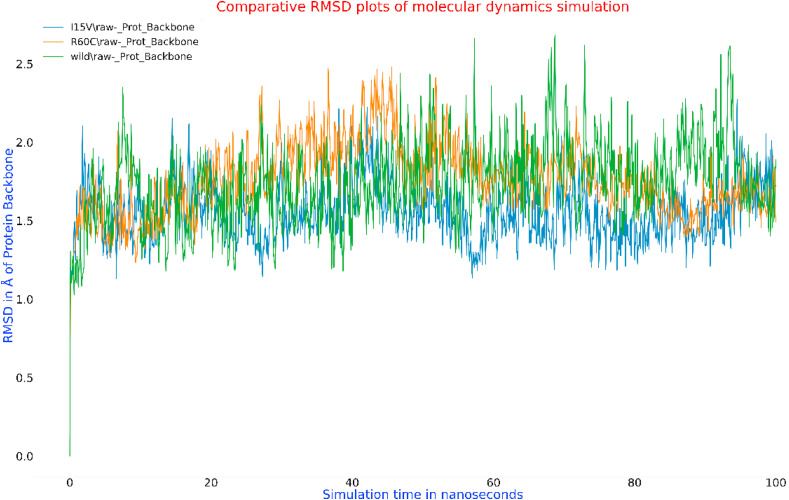
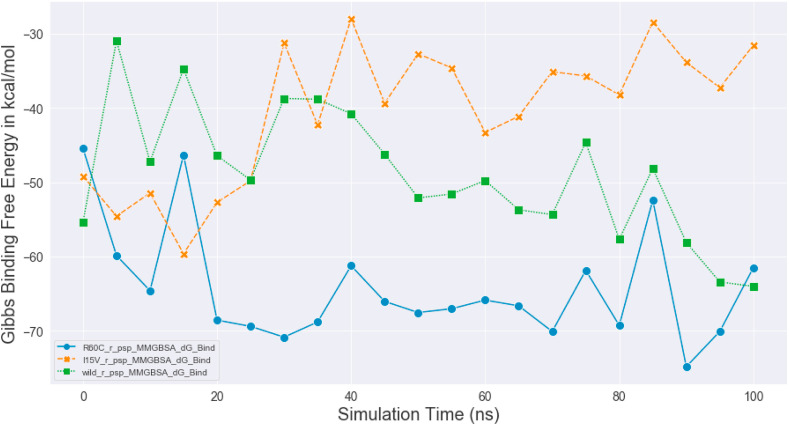
Kar et al. [31] performed molecular dynamics simulation of the SARS-CoV-2 spike protein, RdRp and Mpro with taraxerol. This compound exhibited better binding energy scores with the concerned viral proteins than the drugs that are specifically designed to target them. MD simulations became an important computational tool for understanding the physical basis of the structure and function of biological macromolecules. MD simulations are comparable with an induced-fit model, as it can be used to assess stability, to refine and to rescore docking poses [34,35]. MD simulation was run to monitor the atomic level interaction and stability of the best pose complexes of the main protease and its two mutants (R60C and I152V) with NPC474104 (kazinzol T). The best molecular docking-generated poses of the complexes were subjected to MD simulation by using the Desmond program. The system builder in Desmond uses clear aqueous medium followed by the energy minimization step to lower the energy of the system. The real-time simulation of 100 ns (ns) showed that the kazinol T-protease (wild-type and R60C and I152V mutants) complexes were stable in a dynamic environment at 300 K temperature and 1.01325 bar pressure. H-bonding, hydrophobic interactions, ionic contact and water bridges supported the binding with the protein, resulting in a significant structural change in the viral protein, potentially leading to protein cleavage. In MD simulation, the stability of the protein-ligand complex is represented in terms of Root Mean Square Deviation (RMSD) [36]. The RMSD plots of protein-ligand complexes of wild-type and its two Asian mutants of SARS-CoV-2 are shown in Fig. 7 a, b, and 7c. The RMSD values of proteins and ligand were always on the same trajectory and did not show any deviation. The RMSD pattern averaged around 1.8 Å for the wild-type protease-kazinol T and mutant R60C-kazinol T complexes and 1.5 Å for the mutant I152V-kazinol T complex. The wild-type protease and mutant R60C showed the same RMSD pattern. However, the mutant I152V protease-kazinol T was not a stable complex from 0 to 25 ns, but formed a stable complex from 25 to 100 ns. The comparative protein backbone RMSD plot showed the stability of the protein backbone of the wild-type protease and its two mutants (R60C and I152V) (Fig. 8 ). The RMSD value varied around an average of 1.7 Å. The comparative binding energies of the ligand-protein complexes from 0 to 100 ns were in a range of −30 kcal/mol to −75 kcal/mol for all complexes (Fig. 9 ). These results reflected the stability of our predicted lead compound among the top complexes.
Fig. 7.
The above plot shows the RMSD evolution of a (wild) b (R60C) and c (I152V) proteases (left Y-axis). Ligand RMSD (right Y-axis) indicates how stable the ligand is with respect to the protein and its binding pocket. Time is shown on X-axis.
Fig. 8.
Comparative Root Mean Square Deviation (RMSD) plot of molecular dynamics simulation of the protein's backbone by Desmond with time.
Fig. 9.
Shows the comparative Gibbs binging free energy of the wild (green), mutant R60C (blue) and mutant I152V (orange) proteases with Kazinol T.
4. Conclusion
Computational docking methods are valuable tools aimed to facilitate the costly and time-consuming process of drug development and improvement. Most current approaches assume a rigid receptor structure to allow virtual screening of large numbers of possible ligands and putative binding sites on a receptor molecule. However, it is not possible to mimic and simulate the biological system on a computer.
Natural products are easily available and can be usually extracted at low costs in large quantities. Still, it is a daunting job to narrow down pharmacologically useful compounds from the entire plethora of compounds. In the current study, we used a swift computational strategy to identify inhibitors of SARS-CoV-2 protease from a natural product database with a large number of compounds. Along with several other compounds, kazinol T (NPC474104) was identified as a potent inhibitor, showing a docking score of −14.355008 kcal/mol and no considerable toxicity predictions. MD simulations validated that these complexes were stable. Further in vitro tests with cell lines and affinity optimization studies are needed to validate this predicted lead compound for the treatment of COVID-19. The newly identified small molecules against SARS-COV-2 protease may also serve as inhibitors for other similar viral proteases.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104362.
[37].
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.McIntosh K., Becker W.B., Chanock R.M. Growth in suckling-mouse brain of "IBV-like" viruses from patients with upper respiratory tract disease. Proc. Natl. Acad. Sci. U. S. A. 1967;58(6):2268. doi: 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Witte K.H., Tajima M., Easterday B.C. Morphologic characteristics and nucleic acid type of transmissible gastroenteritis virus of pigs. Arch. Gesamte Virusforsch. 1968;23(1–2):53–70. doi: 10.1007/BF01242114. [DOI] [PubMed] [Google Scholar]
- 3.Tyrrell D.A.J., Almeida J.D., Cunningham C.H., Dowdle W.R., Hofstad M.S., McIntosh K., Tajima M., Zakstelskaya L.Y., Easterday B.C., Kapikian A., Bingham R.W. Coronaviridae. Intervirology. 1975;5(1–2):76. doi: 10.1159/000149883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual screening of natural products against Type II Transmembrane Serine Protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020;25(10):2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman N., Ali F., Basharat Z., Shehroz M., Khan M.K., Jeandet P., Nepovimova E., Kuca K., Khan H. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-CoV-2: an immunoinformatics approach. Vaccines. 2020;8(3):423. doi: 10.3390/vaccines8030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman N., Muhammad I., Afridi S.G., Khan A., Nayab G.E., Ahmad I., Khan H., Belwal T., Devkota H.P., Milella L. Inhibitory effects of plant extracts and in silico screening of the bioactive compounds against α-glucosidase. South Afr. J. Bot. 2020 [Google Scholar]
- 11.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Y.T., Li W., Zhang Q., Jin Y., Rao W.W., Zeng L.N., Lok G.K., Chow I.H., Cheung T., Hall B.J. Timely research papers about COVID-19 in China. Lancet. 2020;395(10225):684–685. doi: 10.1016/S0140-6736(20)30375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discoveries & Therapeutics. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 17.Cross T.J., Takahashi G.R., Diessner E.M., Crosby M.G., Farahmand V., Zhuang S., Butts C.T., Martin R.W. Sequence characterization and molecular modeling of clinically relevant variants of the SARS-CoV-2 main protease. Biochemistry. 2020;59(39):3741–3756. doi: 10.1021/acs.biochem.0c00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmond Molecular Dynamics System. D. E. Shaw Research, New York, NY . Schrödinger; New York, NY: 2020. Maestro-Desmond Interoperability Tools. 2020. [Google Scholar]
- 19.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q., Akdis C.A., Gao Y.D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Li W., Shi X., Chen Z., Jiang B., Liu J., Wang D., Liu C., Meng Y., Cui L., Yu J. Early antiviral treatment contributes to alleviate the severity and improve the prognosis of patients with novel coronavirus disease (COVID-19) J. Intern. Med. 2020;288(1):128–138. doi: 10.1111/joim.13063. [DOI] [PubMed] [Google Scholar]
- 22.Verma S., Twilley D., Esmear T., Oosthuizen C.B., Reid A.M., Nel M., Lall N. Anti-SARS-CoV natural products with the potential to inhibit SARS-CoV-2 (COVID-19) Front. Pharmacol. 2020;11:1514. doi: 10.3389/fphar.2020.561334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dwarka D., Agoni C., Mellem J.J., Soliman M.E., Baijnath H. Identification of potential SARS-CoV-2 inhibitors from South African medicinal plant extracts using molecular modelling approaches. South Afr. J. Bot. 2020;133:273–284. doi: 10.1016/j.sajb.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Upadhyay S., Tripathi P.K., Singh M., Raghavendhar S., Bhardwaj M., Patel A.K. Evaluation of medicinal herbs as a potential therapeutic option against SARS-CoV-2 targeting its main protease. Phytother Res. 2020;34(12):3411–3419. doi: 10.1002/ptr.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanjanasirirat P., Suksatu A., Manopwisedjaroen S., Munyoo B., Tuchinda P., Jearawuttanakul K., Seemakhan S., Charoensutthivarakul S., Wongtrakoongate P., Rangkasenee N., Pitiporn S. High-content screening of Thai medicinal plants reveals Boesenbergia rotunda extract and its component Panduratin A as anti-SARS-CoV-2 agents. Sci. Rep. 2020;10(1):19963. doi: 10.1038/s41598-020-77003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek Y.S., Ryu Y.B., Curtis-Long M.J., Ha T.J., Rengasamy R., Yang M.S., Park K.H. Tyrosinase inhibitory effects of 1, 3-diphenylpropanes from Broussonetia kazinoki. Bioorg. Med. Chem. 2009;17(1):35–41. doi: 10.1016/j.bmc.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Niu X., Dahse H.M., Menzel K.D., Lozach O., Walther G., Meijer L., Grabley S., Sattler I. Butyrolactone I derivatives from Aspergillus terreus carrying an unusual sulfate moiety. J. Nat. Prod. 2008;71(4):689–692. doi: 10.1021/np070341r. [DOI] [PubMed] [Google Scholar]
- 29.Roumeliotis T.I., Halabalaki M., Alexi X., Ankrett D., Giannopoulou E.G., Skaltsounis A.L., Sayan B.S., Alexis M.N., Townsend P.A., Garbis S.D. Pharmacoproteomic study of the natural product ebenfuran III in DU-145 prostate cancer cells: the quantitative and temporal interrogation of chemically induced cell death at the protein level. J. Proteome Res. 2013;12(4):1591–1603. doi: 10.1021/pr300968q. [DOI] [PubMed] [Google Scholar]
- 30.Hanáková Z., Hošek J., Babula P., Dall'Acqua S., Václavík J., Šmejkal K. C-geranylated flavanones from Paulownia tomentosa fruits as potential anti-inflammatory compounds acting via inhibition of TNF-α production. J. Nat. Prod. 2015;78(4):850–863. doi: 10.1021/acs.jnatprod.5b00005. [DOI] [PubMed] [Google Scholar]
- 31.Kar P., Sharma N.R., Singh B., Sen A., Roy A. Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: an in silico investigation. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1780947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J. Biomol. Struct. Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T.A. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3, 3'-digallate (TF3) Evid. base Compl. Alternative Med. 2005;2(2):209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haberl F., Othersen O., Seidel U., Lanig H., Clark T. High Performance Computing in Science and Engineering, Garching/Munich 2007. Springer; Berlin, Heidelberg: 2009. Investigating protein-protein and protein-ligand interactions by molecular dynamics simulations; pp. 153–164. [Google Scholar]
- 35.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9:923. doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar H., Devaraji V., Joshi R., Jadhao M., Ahirkar P., Prasath R., Bhavana P., Ghosh S.K. Antihypertensive activity of a quinoline appended chalcone derivative and its site-specific binding interaction with a relevant target carrier protein. RSC Adv. 2015;5(80):65496–65513. [Google Scholar]
- 37.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.