Abstract
The enzyme Tpt1 is an essential agent of fungal tRNA splicing that removes an internal RNA 2′-PO4 generated by fungal tRNA ligase. Tpt1 performs a two-step reaction in which: (i) the 2′-PO4 attacks NAD+ to form an RNA-2′-phospho-(ADP-ribose) intermediate; and (ii) transesterification of the ADP-ribose O2″ to the RNA 2′-phosphodiester yields 2′-OH RNA and ADP-ribose-1″,2″-cyclic phosphate. Because Tpt1 does not participate in metazoan tRNA splicing, and Tpt1 knockout has no apparent impact on mammalian physiology, Tpt1 is considered a potential antifungal drug target. Here we characterize Tpt1 enzymes from four human fungal pathogens: Coccidioides immitis, the agent of Valley Fever; Aspergillus fumigatus and Candida albicans, which cause invasive, often fatal, infections in immunocompromised hosts; and Candida auris, an emerging pathogen that is resistant to current therapies. All four pathogen Tpt1s were active in vivo in complementing a lethal Saccharomyces cerevisiae tpt1Δ mutation and in vitro in NAD+-dependent conversion of a 2′-PO4 to a 2′-OH. The fungal Tpt1s utilized nicotinamide hypoxanthine dinucleotide as a substrate in lieu of NAD+, albeit with much lower affinity, whereas nicotinic acid adenine dinucleotide was ineffective. Fungal Tpt1s efficiently removed an internal ribonucleotide 2′-phosphate from an otherwise all-DNA substrate. Replacement of an RNA ribose-2′-PO4 nucleotide with arabinose-2′-PO4 diminished enzyme specific activity by ≥2000-fold and selectively slowed step 2 of the reaction pathway, resulting in transient accumulation of an ara-2′-phospho-ADP-ribosylated intermediate. Our results implicate the 2′-PO4 ribonucleotide as the principal determinant of fungal Tpt1 nucleic acid substrate specificity.
Keywords: 2′-phosphotransferase, antifungal target, nicotinamide adenine dinucleotide, tRNA splicing
INTRODUCTION
tRNA 2′-phosphotransferase (Tpt1) is an essential enzyme in the fungal and plant tRNA splicing pathways that removes the 2′-PO4 at the splice junction generated by fungal and plant tRNA ligases (Culver et al. 1997). Tpt1 catalyzes a two-step reaction in which: (i) the internal RNA 2′-PO4 attacks NAD+ to form an RNA-2′-phospho-(ADP-ribose) intermediate; and (ii) transesterification of the ribose O2″ to the 2′-phosphodiester yields 2′-OH RNA and ADP-ribose-1″,2″-cyclic phosphate products (McCraith and Phizicky 1991; Culver et al. 1993; Spinelli et al. 1999; Steiger et al. 2005; Munir et al. 2018a). Tpt1 enzymes are present in bacteria, metazoa, and archaea (Spinelli et al. 1998; Sawaya et al. 2005; Munir et al. 2018a,b), but their physiological roles and endogenous substrates in these taxa are unknown.
We view Tpt1 as a promising target for discovery of antifungals, based on the fact that the enzymatic mechanism of tRNA splicing in metazoa—mediated by the RNA ligase RtcB—is entirely different from that of fungi and does not result in a junction 2′-PO4 (Chakravarty et al. 2012; Popow et al. 2012). Whereas mammals do have Tpt1, it plays no essential role in mammalian physiology, insofar as a tpt1-KO mouse develops normally and has no defects in protein synthesis (Harding et al. 2008). This contrasts with fungi where Tpt1 is essential for viability, for example, in the model fungi Saccharomyces cerevisiae and Schizosaccharomyces pombe and in the pathogenic fungus Candida albicans (Culver et al. 1997; Segal et al. 2018).
To fortify the case for fungal Tpt1 as a drug target, it is imperative to understand the properties of Tpt1 enzymes produced by fungi that cause human disease. It would be naïve to assume that all fungal pathogen Tpt1s have the same characteristics as Tpt1 from the nonpathogenic fungi S. cerevisiae and Chaetomium thermophilum, or the bacterial Tpt1s from Escherichia coli, Runella slithyformis, and Clostridium thermocellum, or the archaeal Tpt1s from Aeropyrum pernix, Pyrococcus horikoshii, and Archaeoglobus fulgidus that have been studied biochemically and/or structurally (Spinelli et al. 1999; Kato-Murayama et al. 2005; Steiger et al. 2005; Munir et al. 2018a,b; Banerjee et al. 2019). To that end, we have conducted here an analysis of Tpt1 from four human fungal pathogens.
Coccidioidomycosis (Valley Fever) is a systemic infection caused by the dimorphic fungus Coccidioides immitis. Infection is by inhalation and can elicit disease even in healthy individuals (Kirkland and Fierer 2018; McCotter et al. 2019). Clinical severity ranges from a self-limited outpatient pneumonia to severe disseminated infection that can require life-long therapy. Extrapulmonary dissemination of C. immitis infection, particularly meningitis, can be deadly. The incidence of reported cases of coccidioidomycosis in the US has increased greatly over the past two decades, especially in California and Arizona (McCotter et al. 2019; www.cdc.gov/fungal/diseases/coccidioidomycosis/statistics.html). Recently, the endemic area has expanded to include Washington State. Estimates of the annual burden of symptomatic coccidioidomycosis suggest that the true number of cases is 6 to 14 times greater than that reported to public health agencies. Coccidioidomycosis causes significant morbidity, especially in elderly and immune-compromised individuals and for the concentrated inmate population of the prisons located in the endemic California area.
Aspergillus fumigatus is a saprophytic soil fungus that causes lung disease when airborne conidia are inhaled. Immunocompetent individuals may develop aspergilloma or allergic bronchopulmonary aspergillosis. Immunosuppressed individuals, especially those with hematological malignancies or who have undergone bone marrow or solid organ transplantation, may develop invasive aspergillosis, a severe and often fatal infection (Dagenais and Keller 2009).
Candida albicans, a commensal yeast colonizer of humans, causes a spectrum of illnesses ranging from local infections of oral, esophageal, and genital mucosa to life-threatening invasive systemic infections in immunocompromised hosts (Pappas et al. 2018). Candida auris, first isolated in 2009 in Japan, has since emerged globally as a multidrug-resistant invasive fungal pathogen that poses a dire health threat in hospitalized patient populations (ElBaradei 2020; Kean et al. 2020). Clinical cases of C. auris infections in the U.S. have clustered primarily in New York, Illinois, and New Jersey (www.cdc.gov/fungal/candida-auris/tracking-c-auris.html#states). There is a clear need for new therapeutic options against this organism.
Here we interrogate the activities and substrate specificities of Tpt1 enzymes from the four aforementioned fungi that cause human disease. By studying analogs of NAD+ and 2′-PO4 RNA, we identify chemical modifications that affect substrate reactivity (and affinity, in the case of nicotinamide hypoxanthine dinucleotide) or result in the accumulation of an ADP ribosylated intermediate (upon replacement of the ribose 2′-PO4 with arabinose 2′-PO4). We find that fungal Tpt1 enzymes do not discriminate RNA versus DNA flanking the 2′-PO4 ribonucleotide.
RESULTS
Genetic complementation in yeast confirms biological activity of Tpt1 homologs from pathogenic fungi
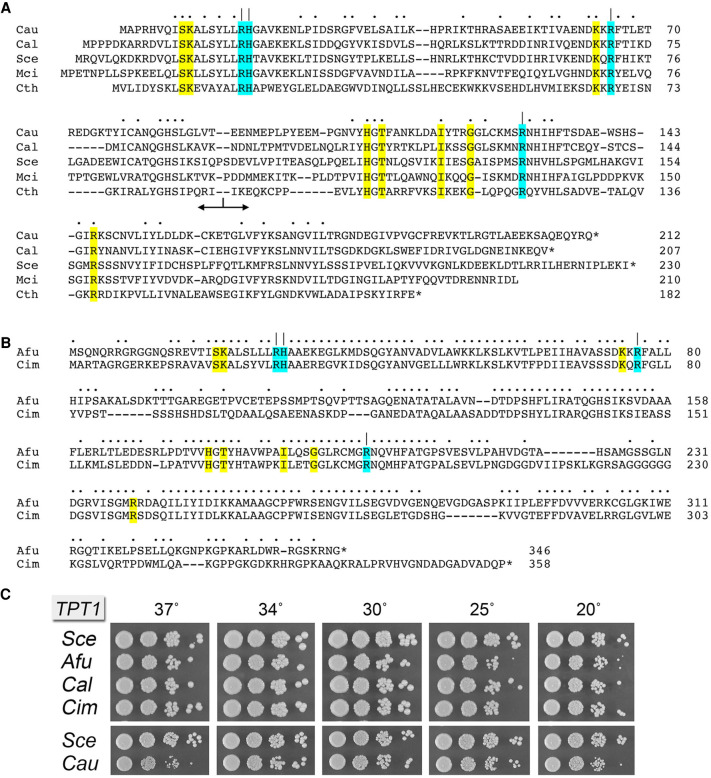
The 212-aa Candida auris Tpt1 (CauTpt1) protein belongs to the “smaller-size” clade of Tpt1 enzymes found in bacteria (e.g., Clostridium thermocellum 182-aa), budding yeasts Saccharomyces cerevisiae (230-aa) and Candida albicans (207-aa), and the soil fungus Mucor circinelloides (210-aa). A primary structure alignment of these five Tpt1s highlights 51 positions of side chain identity/similarity (Fig. 1A). Within the Candida genus, the C. auris and C. albicans Tpt1 enzymes share 127 positions of identity/similarity. The Tpt1 proteins of several other fungal pathogens comprise a “larger-size” clade of Tpt1s, for example, from Histoplasma capsulatum (395-aa), Blastomyces dermatididis (384-aa), Coccidioides immitis (358-aa), Cryptococcus neoformans (356-aa), and Aspergillus fumigatus (346-aa). A primary structure alignment of the C. immitis and A. fumigatus Tpt1s reveals 223 positions of side chain identity/similarity (Fig. 1B). The larger size of the C. immitis Tpt1 vis-à-vis the “minimized” 182-aa bacterial enzyme reflects the presence of a 10-aa amino-terminal extension, a 78-aa carboxy-terminal extension, and three internal insertions (aa 80–131, 147–155, and 201–230) that have no counterpart in the bacterial protein.
FIGURE 1.
Tpt1 orthologs from pathogenic fungi. (A) Alignment of the amino acid sequences of Tpt1 proteins from Candida auris (Cau, Genbank PIS54536.1), Candida albicans (Cal, Genbank AOW28085.1), Saccharomyces cerevisiae (Sce, Genbank NP_014539.1), Mucor circinelloides (Mci, Genbank EPB89638.1), and Clostridium thermocellum (Cth, ABN54255.1). Positions of amino acid side chain identity/similarity are indicated by dots above the alignment. Gaps in the alignment are indicated by dashes. The border between the amino-terminal RNA lobe and the carboxy-terminal NAD+ lobe is indicated by the bidirectional arrow below the alignment. The conserved Arg–His–Arg–Arg catalytic tetrad is highlighted in cyan shading and denoted by | above the alignment. Other conserved amino acids that contact the RNA 2′-PO4 nucleotide or the ADP-ribose moiety of NAD+ are highlighted in yellow shading. (B) Alignment of the amino acid sequences of Tpt1 proteins from Aspergillus fumigatus (Afu, Genbank EDP53951.1) and Coccidioides immitis (Cim, Genbank KJF61580.1), annotated as in A. (C) Complementation of S. cerevisiae tpt1Δ by expression of Tpt1 orthologs from pathogenic fungi. Complementation was assayed by plasmid shuffle. Viable FOA-resistant tpt1Δ p413–TPT1 strains as specified were grown in YPD-Ad liquid medium at 30°C to mid-log phase, then diluted to attain A600 of 0.1, and aliquots (3 µL) of serial 10-fold dilutions were spotted on YPD agar plates and incubated at 20°C, 25°C, 30°C, 34°C, 30°C, and 37°C. Photographs of the plates are shown.
Initial insights into substrate recognition and the mechanism of the transesterification step emerged from a crystal structure of Clostridium thermocellum Tpt1 (CthTpt1) in a product-mimetic complex with ADP-ribose-1″-PO4 in the NAD+ site and pAp in the RNA site (Banerjee et al. 2019) and from kinetic and mutational analyses of Runella slithyformis Tpt1 that identified a Arg–His–Arg–Arg catalytic tetrad in the active site (Munir et al. 2019). This catalytic tetrad, shaded cyan in Figure 1A,B, is conserved in all Tpt1 homologs. Other conserved amino acids that contact the pAp in the RNA site or the ADP-ribose moiety of NAD+ are highlighted in yellow in Figure 1A,B.
We cloned ORFs encoding Tpt1 enzymes from four fungal pathogens, Aspergillus fumigatus (Afu), Coccidioides immitis (Cim), Candida albicans (Cal), and Candida auris (Cau) into yeast CEN plasmids wherein their expression is driven by the S. cerevisiae TPI1 promoter. We tested Tpt1 complementation by plasmid shuffle in S. cerevisiae tpt1Δ (Sawaya et al. 2005). All four pathogen Tpt1 orthologs complemented tpt1Δ. The AfuTPT1, CimTPT1, CalTPT1, and CauTPT1 strains thrived when spot-tested for growth on YPD agar medium at 20°C–37°C, though the CauTPT1 strain was slower growing at 37°C as gauged by colony size (Fig. 1C).
Fungal Tpt1 proteins have RNA 2′-phosphotransferase activity in vitro
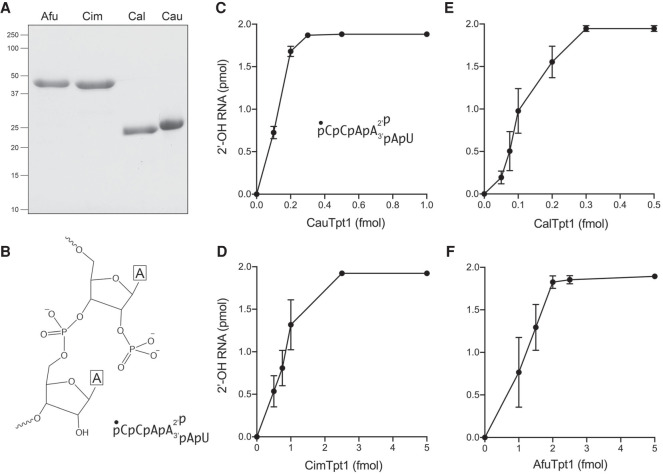
Having shown that the four fungal Tpt1s are biologically active, we proceeded to make recombinant fungal Tpt1 proteins and elucidate their properties. We produced the full-length Tpt1 proteins in E. coli as His10Smt3 fusions and purified them from soluble extracts by sequential Ni-affinity chromatography/imidazole elution, removal of the His10Smt3 tag by treatment with Ulp1 protease, recovery of the tag-free Tpt1 protein in the flow-through of a second Ni-affinity column, and a final Superose-200 gel filtration step. SDS-PAGE affirmed the purity of the enzyme preparations (Fig. 2A). We assayed the four fungal Tpt1 enzymes for RNA 2′-phosphotransferase activity in the presence of 0.2 µM 5′ 32P-labeled 6-mer 2′-PO4 RNA oligonucleotide (shown in Fig. 2B) and 1 mM NAD+ as described (Munir et al. 2018a). After a 30 min incubation at 37°C, the reaction products were resolved by urea-PAGE, which showed that each of the Tpt1 enzymes converted all of the input 2′-PO4 RNA substrate to a slower-migrating 2′-OH RNA product. We did not detect accumulation of a slower-migrating ADP-ribosylated RNA intermediate at limiting enzyme concentrations. The extents of product formation as a function of input Tpt1 are plotted in Figure 2C–F. Tpt1 concentrations of ≤0.25 nM sufficed to dephosphorylate 200 nM 2′-PO4-branched RNA.
FIGURE 2.
Fungal Tpt1 proteins have RNA 2′-phosphotransferase activity in vitro. (A) Aliquots (5 µg) of purified recombinant AfuTpt1, CimTpt1, CalTpt1, and CauTpt1 as specified were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. (B) Structure of the 2′-PO4 dinucleotide in the 5′ 32P-labeled 6-mer RNA substrate. (C–F) Reaction mixtures (10 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 1 mM NAD+, 0.2 µM (2 pmol) 5′ 32P-labeled 6-mer 2-PO4 RNA, and CauTpt1 (C), CimTpt1 (D), CalTpt1 (E), or AfuTpt1 (F) as specified on the x-axis were incubated at 37°C for 30 min. The reaction products were analyzed by urea-PAGE. The extents of formation of the 2′-OH product are plotted as a function of input Tpt1. Each datum is the average of three independent titration experiments ±SEM.
NAD+ analogs as substrates for fungal Tpt1 enzymes
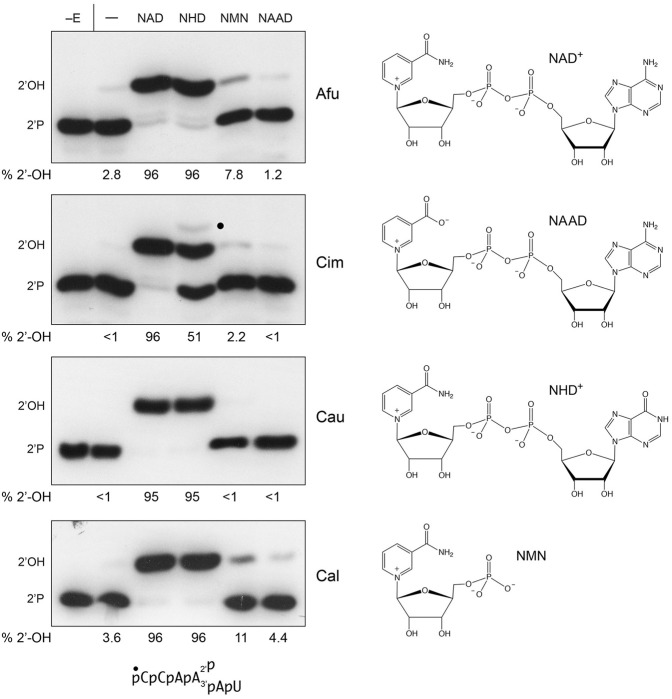
Initial characterization of S. cerevisiae Tpt1 by McCraith and Phizicky (1991) indicated that the requirement for NAD+ could not be fulfilled by NADH, NADP+, NADPH, ADP-ribose, or nicotinamide; the low levels of activity detected at high concentrations of NADH and NADP+ were attributed to trace contamination of the NADH and NADP+ reagents with NAD+. The structure of CthTpt1 in complex with ADP-ribose-1″-PO4 made clear that the adenosine-2′-PO4 moiety of NADP+ would preclude binding to the NAD+ site of the enzyme (Banerjee et al. 2019). The mechanism of Tpt1 step1 catalysis, entailing attack of the 2′-PO4 on the C1″ of NAD+ with displacement of nicotinamide (Spinelli et al. 1999), explains the failure of ADP-ribose or nicotinamide to function in lieu of NAD+. Here we tested NAD+ analogs nicotinamide hypoxanthine dinucleotide (NHD+), nicotinamide mononucleotide (NMN), and nicotinic acid adenine dinucleotide (NAAD) as substrates for the four fungal pathogen Tpt1 enzymes. These compounds can, in principle, occupy the NMN site of the NAD+ pocket and each has a C1 glycosidic linkage to the step 1 leaving group (Fig. 3). Reactions containing 0.2 µM 5′ 32P-labeled 6-mer 2′-PO4 RNA, 1 nM Tpt1, and either no added nucleotide or 1 mM NAD+, NHD+, NMN, or NAAD were incubated for 30 min and the radiolabeled products were resolved by urea-PAGE (Fig. 3). 1 mM NAD+ and NHD+ were effective substrates for AfuTpt1, CauTpt1, and CalTpt1 with respect to formation of the 2′-OH RNA product (comprising 95%–96% of total 32P-RNA; Fig. 3), signifying that adenine is not strictly required as the NAD+ nucleobase. The CimTpt1 reaction with NHD+ was halfway complete after 30 min and generated a residual intermediate species, presumably RNA-2′-phoshospho-(HDP-ribose), denoted by a black dot in Figure 3 (and comprising 2.2% of total 32P-RNA), that was not seen with the other Tpt1 enzymes. NMN at 1 mM concentration supported low levels of activity by AfuTpt1 and CalTpt1, converting 6% of the input 2′-PO4 substrate to 2′-OH product above the level seen in the no nucleotide control reactions. CauTpt1 and CimTpt1 were inactive with 1 mM NMN (Fig. 3). We infer that the AMP moiety of NAD+ is important for the binding and/or proper positioning of the reactive NMN moiety during step1 catalysis. NAAD elicited no 2′-OH RNA product formation versus the no nucleotide controls (Fig. 3), indicating that the carboxylate substitution for the amide of nicotinamide is inimical to Tpt1 activity.
FIGURE 3.
NAD+ analogs as substrates for fungal Tpt1 enzymes. (Left panel) Reaction mixtures (10 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 0.2 µM (2 pmol) 5′ 32P-labeled 6-mer 2-PO4 RNA (shown at bottom), 1 mM nicotinamide adenine dinucleotide (NAD), nicotinamide hypoxanthine dinucleotide (NHD), nicotinamide mononucleotide (NMN), or nicotinic acid adenine dinucleotide (NAAD) as indicated above the lanes, and 10 fmol AfuTpt1, CimTpt1, CauTpt1, or CalTpt1 as indicated on the right were incubated at 37°C for 30 min. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. Tpt1 was omitted from reactions shown in lanes −E. Control reactions containing Tpt1 but no NAD+ are included in lanes −. The species corresponding to the 2′-phosphate substrate and 2′-OH product are indicated on the left. The RNA-2′-phospho-HDPR intermediate detected in the CimTpt1 reaction with NHD is denoted by a dot. The extents of formation of the 2′-OH product (as percent of the total 32P-labeled RNA) are specified below the lanes. (Right panel) The chemical structures of the substrates are shown.
Hypoxanthine substitution for NAD+ adenine affects substrate affinity
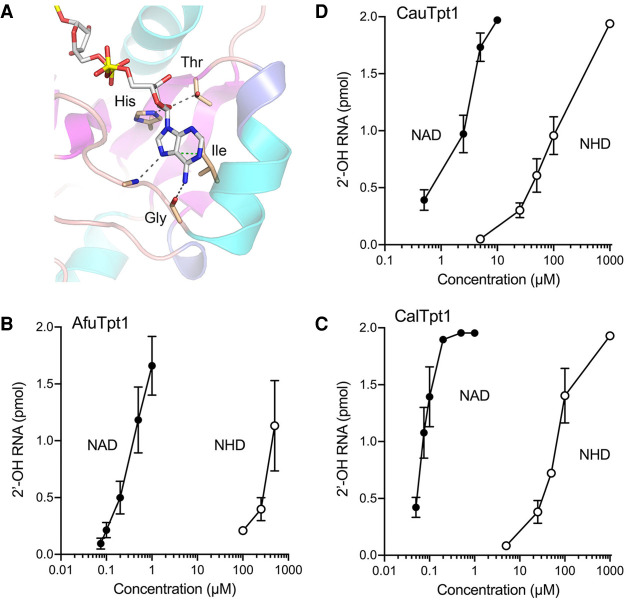
The CthTpt1 crystal structure highlighted atomic contacts to the adenosine nucleoside that contribute to NAD+ binding and, perhaps, adenine specificity (Fig. 4A). The ribose 2′-OH makes hydrogen bonds to histidine and threonine side chains of an HGT motif conserved in all Tpt1 homologs (highlighted in yellow in Fig. 1A,B). A double-alanine mutation of the His and Thr of this motif is lethal in vivo and effaces Tpt1 activity in vitro (Sawaya et al. 2005; Banerjee et al. 2019). A conserved isoleucine situated 8-aa downstream from the HGT motif (shaded yellow in Fig. 1A,B) stacks on and makes van der Waals contacts to the adenine nucleobase (Fig. 4A). The main-chain carbonyl of a conserved glycine located 4-aa downstream from this isoleucine makes a hydrogen bond to adenine-N6 (Fig. 4A). The main chain amide 2-aa downstream from the glycine donates a hydrogen bond to adenine-N7 (Fig. 4A). The glycine main chain contact to adenine-N6 would, in principle, engender a preference for adenine versus hypoxanthine (which has an O6 substituent). Because this contact is remote from the enzymic site where reaction chemistry is occurring, and given that NHD+ is capable of supporting catalysis at 1 mM concentration, we considered the prospect that the hypoxanthine analog might affect Tpt1 affinity for the phosphoacceptor substrate. Thus, we back-titrated the NAD+ and NHD+ concentrations in Tpt1 reactions performed under conditions of RNA excess over enzyme. Plots of the extent of 2′-OH RNA product formation by AfuTpt1, CalTpt1, and CauTpt1 as a function of NAD+ or NHD+ concentration are shown in Figure 4B–D. These fungal Tpt1 enzymes were very efficient at scavenging submicromolar (AfuTpt1 and CalTpt1) or low micromolar (CauTpt1) concentrations of NAD+, akin to what had been reported previously for Runella Tpt1 (Munir et al. 2018a). The NHD+ titration curves were shifted to the right in each case, indicative of ∼1000-fold lower affinity of AfuTpt1 and CalTpt1, and ∼100-fold lower affinity of CauTpt1, for NHD+ versus NAD+ (Fig. 4B–D).
FIGURE 4.
Hypoxanthine substitution for NAD+ adenine affects substrate affinity. (A) View of the interface of CthTpt1 with ADP-ribose in the crystal structure of a product-mimetic complex (from pdb 6E3A). Atomic contacts to the adenosine nucleoside are denoted by dashed lines. The indicated His, Thr, and Gly residues that mediate these contacts are conserved among Tpt1 orthologs. (B–D) Reaction mixtures (10 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 0.2 µM (2 pmol) 5′ 32P-labeled 6-mer 2′-PO4 RNA substrate, 2 mM DTT, 2.5 fmol AfuTpt1 (B), 2.5 fmol CalTpt1 (C), or 2.5 fmol CauTpt1 (D), and NAD+ or NHD+ at the concentrations specified on the x-axes were incubated at 37°C for 30 min. The extents of formation of the 2′-OH product are plotted as a function of NAD+ or NHD+ concentration (log scale on the x-axis). Each datum is the average of three independent titration experiments ±SEM.
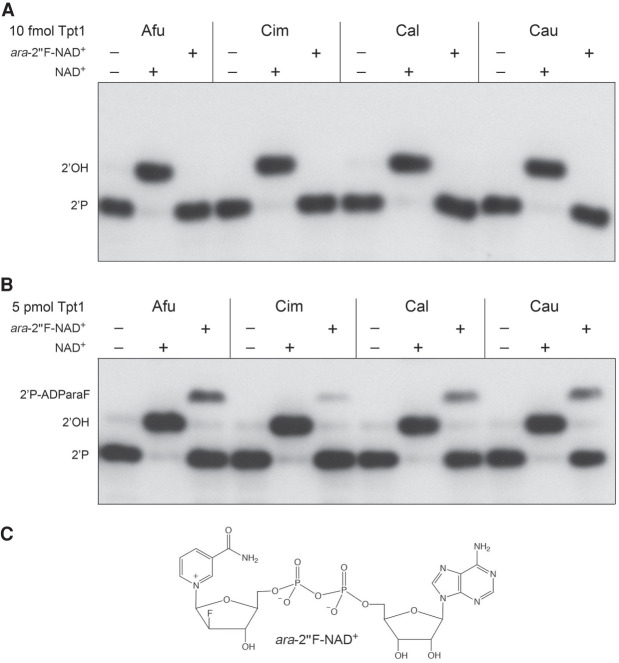
Test of ara-2″-fluoro NAD+ as substrate for fungal Tpt1 enzymes
The ara-2″F analog of NAD+ (shown in Fig. 5C) could, in principle, be able to support the first step in the Tpt1 pathway, but would be unable to undergo transesterification in the second step for lack of an O2″ nucleophile. Previously, we surveyed Tpt1 enzymes from Runella slithyformis, Clostridium thermocellum, Chaetomium thermophilum, and Homo sapiens at 0.5 µM concentration for activity in the presence of 0.2 µM 5′ 32P-labeled 6-mer 2′-PO4-branched RNA oligonucleotide and either 50 µM NAD+ or ara-2″F-NAD+ (Dantuluri et al. 2020). When provided with 50 µM ara-2′′F-NAD+, the Runella and Clostridium enzymes converted nearly all of the substrate into an RNA-2′-phospho-(ADP-fluoroarabinose) dead-end product of step 1 of the Tpt1 pathway. Chaetomium Tpt1 also formed the dead-end product in the presence of 50 µM ara-2″F-NAD+, though the extent of substrate conversion was lower. In contrast, human Tpt1 effected no detectable reaction of the 2′-PO4 RNA substrate in the presence of 50 µM ara-2″F-NAD+. We surmised that Tpt1 enzymes from different sources may vary in their sensitivity to the arabinose sugar modification of NAD+, but the utilization of ara-2″F-NAD+ as a substrate by Tpt1 does indeed result in trapping of the step 1 reaction product (Dantuluri et al. 2020).
FIGURE 5.
Test of ara-2″-fluoro NAD+ as substrate for fungal Tpt1 enzymes. Reaction mixtures (10 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 0.2 µM (2 pmol) 5′ 32P-labeled 6-mer 2-PO4 RNA, 50 µM NAD+ or ara-2″F-NAD+ (shown in panel C) where indicated by +, and either 10 fmol Tpt1 (A) or 5 pmol Tpt1 (B) were incubated at 37°C for 30 min. The reaction products were analyzed by urea-PAGE and visualized by autoradiography. The species corresponding to the 2′-phosphate substrate, 2′-OH product, and 2′-phospho-ADParaF intermediate are indicated on the left.
Here we tested the four fungal pathogen Tpt1 enzymes for their ability to use 50 µM ara-2″F-NAD+ as a substrate to dephosphorylate the 6-mer 2′-PO4 RNA (0.2 µM) under conditions of RNA excess (1 nM Tpt1) or enzyme excess (0.5 µM Tpt1). Control reactions with 50 µM NAD+ were performed in parallel. Whereas 1 nM of each Tpt1 sufficed to dephosphorylate the RNA in the presence of NAD+, no product was formed in the presence of ara-2″F-NAD+ (Fig. 5A). However, when Tpt1 was in excess over the 2′-PO4-branched RNA, the fungal Tpt1 enzymes did use ara-2″F-NAD+ to convert a minority of the RNA substrate into an RNA-2′-phospho-(ADP-fluoroarabinose) dead-end step 1 product (Fig. 5B): the fraction converted being 18% by AfuTpt1, 3% by CimTpt1, 17% by CalTpt1, and 20% by CauTpt1. A scant amount of 2′-OH RNA end-product was detected in reactions lacking exogenous NAD+ (Figs. 3, 5B); we attribute this to residual NAD+ in the Tpt1 preparations that was associated with the recombinant proteins during purification. We conclude that the Tpt1 enzymes from pathogenic fungi are extremely feeble in their capacity to use ara-2″F-NAD+ in lieu of NAD+.
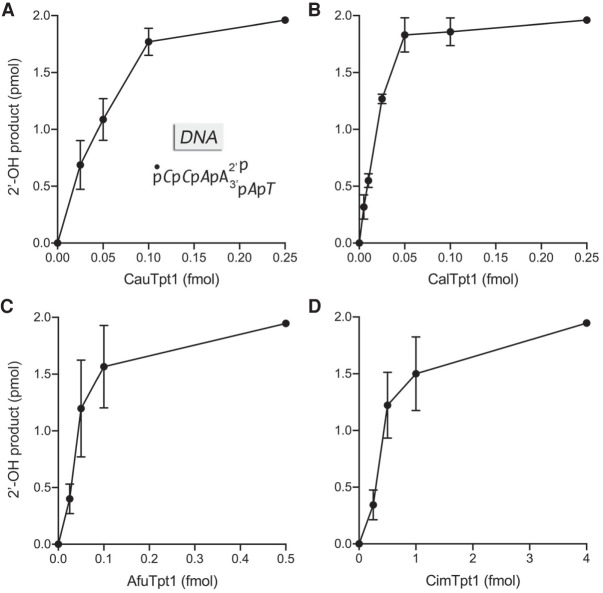
Fungal Tpt1s remove an internal ribonucleoside 2′-phosphate from a DNA substrate
To query the RNA requirement for Tpt1 activity, we exploited a 5′ 32P-labeled analog of the 6-mer 2′-PO4 oligonucleotide substrate (Dantuluri et al. 2020) in which the five nucleotides flanking the 2′-PO4 ribonucleotide were replaced by deoxynucleotides (Fig. 6). The 2′-PO4 DNA substrate (at 0.2 µM concentration) was reacted for 30 min with the four fungal Tpt1s in the presence of 1 mM NAD+. All four Tpt1s were capable of quantitatively converting the 2′-PO4 DNA into the 2′-OH product. The Tpt1 titration profiles highlighted that the specific activities of the Cau, Cal, and Afu enzymes were higher with the DNA scaffold substrate (Fig. 6) than with the all-RNA substrate (Fig. 2) by approximately twofold, sixfold, and 15-fold, respectively, whereas CimTpt1 specific activity was similar for the DNA and RNA scaffolds flanking the 2′-PO4.
FIGURE 6.
Fungal Tpt1s efficiently remove an internal 2′-phosphate from a DNA substrate. Reaction mixtures (10 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 1 mM NAD+, 0.2 µM (2 pmol) 5′ 32P-labeled 6-mer DNA with a single internal ribonucleotide 2-PO4 (shown in A with deoxynucleotides in italics), and CauTpt1 (A), CalTpt1 (B), AfuTpt1 (C), or CimTpt1 (D) as specified on the x-axes were incubated at 37°C for 30 min. The extents of formation of the 2′-OH product are plotted as a function of input Tpt1. Each datum is the average of three independent titration experiments ±SEM.
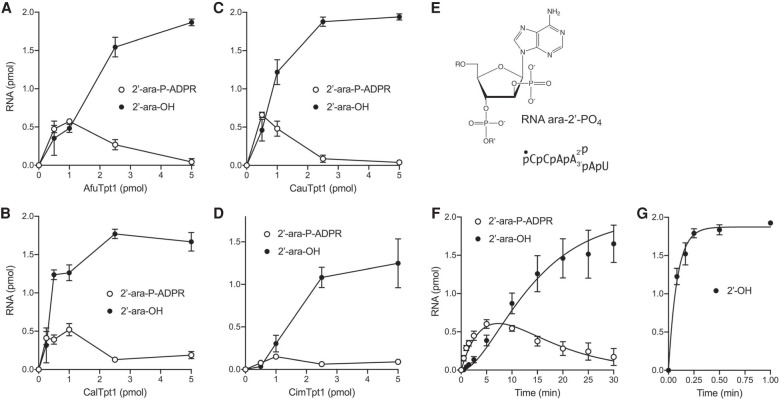
Effect of an arabinose sugar at the 2′-phosphate nucleotide
We previously tested a 6-mer RNA with an internal arabinose-2′-phosphate moiety (shown in Fig. 7E) as substrate for the Runella and Clostridium Tpt1 enzymes. Replacement of the ribose-2′-PO4 nucleotide with arabinose-2′-PO4 selectively slowed step 2 of the reaction pathway and resulted in the transient accumulation of high levels of the RNA-2′-arabino-phospho-(ADP-ribose) reaction intermediate (Dantuluri et al. 2020). Here we tested activity of the four fungal pathogen Tpt1s with the RNA ara-2′-PO4 substrate. A 30 min reaction of 0.2 µM (2 pmol) 5′ 32P-labeled ara-2′-PO4 RNA with increasing concentrations of Afu, Cal, and Cau Tpt1s in the presence of 1 mM NAD+ resulted in the progressive accumulation of ara-2′-phospho-ADP-ribosylated intermediate, to peak levels of 26% to 33% of total RNA at 0.5 pmol (50 nM) to 1 pmol (100 nM) enzyme (Fig. 7A–C). At higher levels of Afu, Cal, and Cau Tpt1 (2.5 to 5 pmol), 89 to 97% of the input RNA was converted to the ara-2′-OH product (Fig. 7A–C). Comparing the titration profiles of these enzyme with the ribo-2′-PO4 and ara-2′-PO4 substrates (Figs. 2, 7), especially the levels of enzyme needed to attain maximal conversion to 2′-OH product, indicated that the ara-2′-PO4 modification reduced the specific activity of 2′-OH product formation by 2000-fold (AfuTpt1), 5000-fold (CalTpt1), and 10,000 fold (CauTpt1). In the case of CimTpt1 and its reaction with the RNA ara-2′-PO4 substrate, the RNA-2′-arabino-phospho-(ADP-ribose) intermediate comprised 8% of total RNA at 1 pmol enzyme and the extent of conversion to ara-2′-OH product was 63% at 5 pmol of enzyme (Fig. 7D), indicative of a ∼2000-fold decrement in 2′-OH product formation compared to the unmodified ribo-2′-PO4.
FIGURE 7.
Effect of an arabinose sugar at the 2′-phosphate branchpoint. (A–D) Tpt1 titrations. Reaction mixtures (10 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 1 mM NAD+, 0.2 µM (2 pmol) 5′ 32P-labeled 6-mer ara-2′-PO4 substrate (shown in E, highlighting the chemical structure of the arabinose-2′-PO4 branchpoint), and AfuTpt1 (A), CalTpt1 (B), CauTpt1 (C), or CimTpt1 (D) as specified on the x-axes were incubated at 37°C for 30 min. The reaction products were analyzed by urea-PAGE and quantified by scanning the gels. The extents of formation of the 2′-OH product and the ADP-ribosylated intermediate are plotted as a function of input Tpt1. Each datum in the graphs is the average of three separate experiments ±SEM. (F,G) Single-turnover kinetics. Reaction mixtures (100 µL) containing 100 mM Tris-HCl, pH 7.5, 2 mM DTT, 1 mM NAD+, 0.2 µM 5′ 32P-labeled 6-mer ara-2′-PO4 substrate (F) or unmodified 6-mer RNA 2′-PO4 substrate (G), and 0.5 µM AfuTpt1 were incubated at 37°C. The reactions were initiated by adding enzyme to a prewarmed reaction mixture. Aliquots (10 µL, containing 2 pmol of RNA) were withdrawn at the times specified on the x-axis and quenched immediately with three volumes of cold 90% formamide, 50 mM EDTA. The extents of formation of the 2′-OH product and the ADP-ribosylated intermediate are plotted as a function of reaction time. Each datum is the average of three independent titration or time-course experiments (±SEM). The data in F were fit by nonlinear regression in Prism to a unidirectional two-step mechanism. The data in G were fit to a single exponential.
A kinetic analysis of the AfuTpt1 reaction under conditions of enzyme excess (0.2 µM ara-2′-PO4 RNA; 0.5 µM AfuTpt1) is shown in Figure 7F and affirmed a precursor-product relationship whereby the ADP-ribosylated RNA intermediate accumulated steadily over 2.5 min and peaked at 5 min, when it comprised 30% of total RNA, before declining thereafter as the ara-2′-OH product was formed after an initial lag. The data fit by nonlinear regression in Prism to a unidirectional two-step mechanism with apparent step 1 and step 2 rate constants of 0.114 ± 0.0112 min−1 and 0.162 ± 0.0174 min−1, respectively (Fig. 7F). Control reactions of 0.5 µM AfuTpt1 with the unmodified ribo-2′-PO4 RNA showed that 61% of input RNA was converted to 2′-OH product in 5 sec (at which time the ADP-ribosylated RNA intermediate comprised 1% of the total RNA) and that the reaction endpoint was achieved in 15 sec (Fig. 7G). The kinetics of 2′-OH RNA formation were fit to a single exponential with an apparent rate constant of 11.7 ± 1.3 min−1. Because it is clear that the rate of step 2 is much faster than the step 1 rate for AfuTpt1′s reaction with the standard 2′-PO4 branched RNA, that is, step 1 is rate-limiting, we can regard the apparent rate constant of 11.7 min−1 as virtually synonymous with the step1 rate constant. Thus, we surmise that the ara-2′-PO4 modification slows AfuTpt1 step 1 by a factor of 100 and exerts an even more profound slowing effect on step 2 of the reaction pathway.
DISCUSSION
The present study illuminates the activities and substrate specificities of Tpt1 enzymes from four different fungi that cause human disease. The fungal enzymes display high affinity and selectivity in utilization of NAD+ as the substrate for phosphoryl transfer. The AMP moiety of NAD+, though not directly involved in reaction chemistry, is essential for Tpt1 activity (i.e., NMN is a feeble substrate), consistent with the extensive enzymic interface with the AMP moiety seen in the crystal structure of CthTpt1. The fungal enzymes can utilize NHD+, albeit with much lower affinity than NAD+, which we presume reflects the antagonistic effect replacing the adenine-N6 amine with a hypoxanthine-O6 carbonyl on the atomic contact to the nearby main-chain carbonyl of a conserved glycine in the NAD+ lobe of the enzyme. The fungal enzymes are incapable of using NAAD as a substrate, suggesting that there are essential enzymic interactions with the amide moiety of nicotinamide that are either lost, or antagonized, when the amide is replaced by a carboxylate. A detailed rationale for this effect is elusive in the absence of a structure of Tpt1 in a binary complex with NAD+. Replacing the NMN ribose of NAD+ with 2′-fluoroarabinose can trap RNA-2′-phospho-(ADP-fluoroarabinose) as a dead-end step 1 product in the case of the Runella and Clostridium Tpt1 enzymes, but human Tpt1 was unreactive with ara-2″F-NAD+ (Dantuluri et al. 2020). Here we found that the four fungal Tpt1 enzymes were feeble in their use of ara-2″F-NAD+ as a substrate for step 1 of the Tpt1 pathway.
Replacement of each of the ribose sugars flanking the internal 2′-PO4 with a deoxynucleotide did not adversely affect the efficiency of 2′-PO4 removal by fungal Tpt1; rather their specific activities were higher with the DNA 6-mer substrates versus the RNA 2′-PO4 6-mer. In contrast, replacing the ribose of the 2′-PO4 nucleotide with arabinose reduced the specific activity of 2′-OH product formation by the fungal Tpt1 enzymes by ≥2000-fold and resulted in the build-up of an ara-2′-phospho-ADP-ribosylated intermediate. Single-turnover kinetic analysis showed that the ara-2′-PO4 slows AfuTpt1 step 1 catalysis by 100-fold and is even more deleterious for step 2. These results pinpoint the 2′-PO4 nucleotide as the principal determinant of Tpt1 nucleic acid substrate specificity.
Tpt1 collaborates with the trifunctional Trl1 enzyme (composed of 2′,3′-cyclic phosphodiesterase, GTP-dependent 5′-OH kinase, and ATP-dependent RNA ligase domains) to perform the end-healing and end-sealing steps of tRNA splicing in model yeast species such as S. cerevisiae, S. pombe, and C. albicans. Genetic analyses established that Trl1 and Tpt1 are essential for growth of these three taxa (Phizicky et al. 1992; Culver et al. 1997; Sawaya et al. 2003, 2005; Kim et al. 2010; Segal et al. 2018). Here and previously (Remus et al. 2016, 2017), we have characterized the Trl1 and Tpt1 enzymes from several human fungal pathogens. In contrast, the tRNA end-joining steps of mammalian tRNA splicing are performed by a single essential enzyme, RtcB, via a completely different set of chemical reactions (Popow et al. 2012). A key question with respect to tRNA splicing as a plausible antifungal drug target is whether the fungi that cause human disease possess an alternative RtcB pathway of RNA ligation that could sustain them if Trl1 and Tpt1 were inhibited pharmacologically. Our survey of the proteomes of C. albicans, C. auris, A. fumigatus, and C. immitis indicates that they do not have homologs of RtcB. Other human fungal pathogens—including Blastomyces, Histoplasma, and Cryptococcus—encode Trl1 and Tpt1 enzymes but have no homologs of RtcB. This augurs well for tRNA splicing as a therapeutic target in a broad spectrum of fungal disease settings.
That said, an even more salient question is whether all fungi are monophyletic with respect to their mechanism of tRNA ligation. To our inspection, they might not be, insofar as a PSI-BLAST search of available fungal proteomes (www.ncbi.nlm.nih.gov/blast) identified more than 90 fungal taxa that encode a convincing homolog of RtcB (Supplemental Table S1). The “RtcB clade” embraces species from six phyla of the fungal kingdom, especially Ascomycota, Basidiomycota, Microsporidia, and Mucoromycota (Supplemental Table S1). The fungi that have RtcB also encode a Trl1 homolog (Supplemental Table S1), with the exception of the Microsporidia which do not have a recognizable Trl1 enzyme in their proteomes. Thus, whereas model yeasts and the various human fungal pathogens elaborate only the Trl1/Tpt1 pathway, there are quite a number of fungal species that have two potential pathways of RNA sealing and tRNA splicing.
MATERIALS AND METHODS
Recombinant Tpt1 proteins of pathogenic fungi
BamHI-XhoI DNA fragments encoding the Aspergillus fumigatus, Coccidioides immitis, Candida albicans, and Candida auris Tpt1 proteins were inserted between the BamHI and XhoI sites of pET28b–His10Smt3 so as to fuse the Tpt1 polypeptides in-frame to an amino-terminal His10Smt3 tag. In the case of CauTpt1, the codons for Ser13, Ser128, and Ser143, which are CUG in the C. auris genome (CUG specifies Leu in the standard genetic code but is translated as Ser in Candida species; Miranda et al. 2006) were changed to UCU to program Ser at these positions when the CauTPT1 ORF is expressed in E. coli or S. cerevisiae. The pET28b–His10Smt3–Tpt1 plasmids were transformed into Escherichia coli BL21(DE3)-CodonPlus cells. Cultures (1 L) derived from single kanamycin and chloramphenicol resistant transformants were grown at 37°C in Luria–Bertani medium containing 50 µg/mL kanamycin and 35 µg/mL chloramphenicol until the A600 reached 0.6, at which time the cultures were cooled to 4°C and adjusted to 0.5 mM isopropyl-β-d-thiogalactoside and 2% (v/v) ethanol and then incubated for 16 h at 17°C with continuous shaking at 200 rpm. Cells were harvested by centrifugation and the pellets were stored at −80°C. All subsequent purification steps were carried out at 4°C. Thawed cell pellets were suspended in 50 mL buffer A (50 mM Tris-HCl, pH 7.5, 1 M NaCl, 20 mM imidazole, 4 mM DTT, 20% glycerol, 0.05% Triton X-100) and lysozyme was added to a final concentration of 1 mg/mL along with one protease inhibitor tablet (Roche). After mixing for 30 min, the resulting lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation for 45 min at 14,000 rpm. The soluble extracts were mixed for 1 h with 5 mL of His60 Ni Superflow resin (Clontech) that had been equilibrated in buffer A. The resin was washed twice with 50 mL of buffer A, then serially with 30 mL of buffer B (50 mM Tris-HCl, pH 7.5, 3 M KCl, 4 mM DTT, 0.05% Triton X-100), and 50 mL of 50 mM imidazole in buffer C (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 4 mM DTT, 10% glycerol, 0.05% Triton X-100). The resin was then poured into a column and the bound material was eluted with 500 mM imidazole in buffer C. The elution profiles of the fungal Tpt1s were monitored by SDS-PAGE. Peak fractions containing His10Smt3–Tpt1 proteins were pooled, supplemented with Smt3-specific protease Ulp1 (to attain a His10Smt3–Tpt1:Ulp1 ratio of 500:1) and then dialyzed overnight (in a Thermo Scientific dialysis cassette, 10 MWCO) against 20 mM imidazole in buffer C. The tag-less Tpt1 proteins were separated from the His10Smt3 tag by applying the dialysates to 5 mL of His60 Ni Superflow resin that had been equilibrated in buffer C containing 20 mM imidazole. After pouring the resin into a column, the Tpt1 proteins were recovered in the flow-through and 50 mM imidazole eluate fractions. The His10Smt3 tag was bound to the resin and recovered in the 250 mM and 500 mM imidazole eluate fractions. The fungal Tpt1 proteins were concentrated by centrifugal ultrafiltration (Amicon,10,000 MWCO) to a final volume of 5 mL and then gel-filtered through a 120 mL 16/60 HiLoad Superdex-200 column (GE Healthcare) equilibrated with buffer D (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 4 mM TCEP, 5% glycerol; 0.05% Triton X-100) at a flow rate of 0.8 mL/min, while collecting 1.5 mL fractions. The peak Tpt1 fractions were pooled and concentrated by centrifugal ultrafiltration. Protein concentrations were determined by using the Biorad dye reagent with bovine serum albumin as the standard. The yields of purified Tpt1 from 1-liter bacterial cultures were as follows: 2 mg AfuTpt1; 2 mg CimTpt1; 3.5 mg CalTpt1; 5 mg CauTpt1.
5′ 32P-labeled oligonucleotide substrates
2′-PO4 branchpoint-containing 6-mer oligonucleotides (unmodified or modified versions of 5′-CCAA2′PAU) were synthesized, deprotected, and purified as described previously (Munir et al. 2018a; Dantuluri et al. 2020). The 6-mers were 5′ 32P-labeled by reaction with phosphatase-dead T4 polynucleotide kinase (Pnkp-D167N) in the presence of [γ32P]ATP. The radiolabeled 6-mers were gel-purified by electrophoresis through a 40-cm 20% nondenaturing polyacrylamide gel containing 45 mM Tris-borate, 1 mM EDTA. The radiolabeled oligonucleotides were eluted from excised gel slices during overnight incubation in 10 mM Tris-HCl, pH 6.8, 1 mM EDTA and then stored at −20°C.
Assay of Tpt1 activity
Reaction mixtures containing 100 mM Tris-HCl (pH 7.5), 0.2 µM 5′ 32P-labeled nucleic acid substrates, NAD+ or NAD+ analogs as specified, and Tpt1 as specified in the figure legends were incubated at 37°C. The reactions were quenched at the times specified in the figure legends by addition of three volumes of cold 90% formamide, 50 mM EDTA. The products were analyzed by electrophoresis (at 55 W constant power) through a 40-cm 20% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate, 1 mM EDTA and visualized by autoradiography and/or scanning the gel with a Fujifilm FLA-7000 imaging device. The products were quantified by analysis of the gel scans in ImageQuant. Ara-2″F-NAD+ was purchased from BIOLOG (Bremen, Germany; Cat. No. D148). NMN (nicotinamide mononucleotide), NAAD (nicotinic acid adenine dinucleotide), and NHD (nicotinamide hypoxanthine dinucleotide) were purchased from Sigma.
Complementation of S. cerevisiae tpt1Δ by Tpt1 orthologs from pathogenic fungi
The S. cerevisiae tpt1Δ haploid strain YBS501 (MATa ura3–1 ade2–1 trp1–1 his3–11,15 leu2–3,11–2 can1–100 tpt1::LEU2 p360-TPT1), in which the TPT1 ORF was deleted and replaced by LEU2, is dependent for viability on the p360-TPT1 plasmid (CEN URA3 SceTPT1) (Schwer et al. 2004). YBS501 was transformed with: (i) a p413-SceTPT1 plasmid (CEN HIS3 SceTPT1) plasmid as a positive control; (ii) the empty CEN HIS3 vector as negative control; and (iii) p413 (CEN HIS3) plasmids expressing the AfuTPT1, CimTPT1, CalTPT1, or CauTPT1 open reading frames under the control of a constitutive yeast TPI1 promoter. Transformants were selected at 30°C on His− agar medium. Three individual His+ colonies from each transformation were patched to His− agar medium and cells from each isolate were then streaked on agar medium containing 0.75 mg/mL 5-FOA (5-fluoroorotic acid). The plates were incubated at 30°C. The p413–TPT1 plasmids all supported the formation of FOA-resistant colonies. In contrast, the vector did not allow formation of FOA-resistant colonies after 7 d at any of the temperatures tested. Viable FOA-resistant tpt1Δ p413–TPT1 colonies were grown in YPD-Ad (yeast extract, peptone, 2% dextrose, 0.1 mg/mL adenine) liquid medium at 30°C to mid-log phase (A600 0.5 to 0.7), then diluted to attain A600 of 0.1, and aliquots (3 µL) of serial 10-fold dilutions were spotted on YPD agar plates and incubated at 20°C, 25°C, 30°C, 34°C, and 37°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the U.S. National Institutes of Health (R35-GM126945 to S.S.) and the National Science and Engineering Council of Canada (Discovery grant to M.J.D.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078660.120.
REFERENCES
- Banerjee A, Munir A, Abdullahu L, Damha MJ, Goldgur Y, Shuman S. 2019. Structure of tRNA splicing enzyme Tpt1 illuminates the mechanism of RNA 2′-PO4 recognition and ADP-ribosylation. Nat Commun 10: 218. 10.1038/s41467-018-08211-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Subbotin R, Chait BT, Shuman S. 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci 109: 6072–6077. 10.1073/pnas.1201207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Zillman M, Kierzek R, Michaud N, LaReau RD, Turner DH, Phizicky EM. 1993. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1″-2″ cyclic phosphate. Science 261: 206–208. 10.1126/science.8392224 [DOI] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM. 1997. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem 272: 13203–13210. 10.1074/jbc.272.20.13203 [DOI] [PubMed] [Google Scholar]
- Dagenais TRT, Keller NP. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 22: 447–465. 10.1128/CMR.00055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuluri S, Abdullahu L, Munir A, Katolik A, Damha MJ, Shuman S. 2020. Substrate analogs that trap the 2′-phospho-ADP-ribosylated RNA intermediate of the Tpt1 (tRNA 2′-phosphotransferase) reaction pathway. RNA 26: 373–381. 10.1261/rna.074377.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElBaradei A. 2020. A decade after the emergence of Candida auris: what do we know? Eur J Clin Microbiol Infect Dis 39: 1617–1627. 10.1007/s10096-020-03886-9 [DOI] [PubMed] [Google Scholar]
- Harding HP, Lackey JG, Hsu HC, Zhang Y, Deng J, Xu RM, Damha MJ, Ron D. 2008. An intact unfolded protein response in Trpt1 knockout mice reveals phylogenic divergence in pathways for RNA ligation. RNA 14: 225–232. 10.1261/rna.859908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Murayama M, Bessho Y, Shirouzu M, Yokoyama S. 2005. Crystal structure of the RNA 2′-phosphotransferase from Aeropyrum pernix K1. J Mol Biol 348: 295–305. 10.1016/j.jmb.2005.02.049 [DOI] [PubMed] [Google Scholar]
- Kean R, Brown J, Gulmez D, Ware A, Ramage G. 2020. Candida auris: a decade of understanding of an enigmatic pathogenic yeast. J Fungi 6: 30. 10.3390/jof6010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623. 10.1038/nbt.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland TN, Fierer J. 2018. Coccidioides immitis and posadasii; a review of their biology, genomics, pathogenesis, and host immunity. Virulence 9: 1426–1435. 10.1080/21505594.2018.1509667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCotter OZ, Benedict K, Engelthaler DM, Komatsu K, Lucas KD, Mohle-Boetani JC, Oltean H, Vugia D, Chiller TM, Sondermeyer Cooksey GL, et al. 2019. Update on the epidemiology of coccidioidomycosis in the United States. Med Mycol 57: S30–S40. 10.1093/mmy/myy095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith SM, Phizicky EM. 1991. An enzyme from Saccharomyces cerevisiae uses NAD+ to transfer the splice junction 2′-phosphate from ligated tRNA to an acceptor molecule. J Biol Chem 266: 11986–11992. 10.1016/S0021-9258(18)99054-X [DOI] [PubMed] [Google Scholar]
- Miranda I, Silva R, Santos MA. 2006. Evolution of the genetic code in yeasts. Yeast 23: 203–213. 10.1002/yea.1350 [DOI] [PubMed] [Google Scholar]
- Munir A, Abdullahu L, Damha MJ, Shuman S. 2018a. Two-step mechanism and step-arrest mutants of Runella slithyformis NAD+-dependent tRNA 2′-phosphotransferase Tpt1. RNA 24: 1144–1157. 10.1261/rna.067165.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Banerjee A, Shuman S. 2018b. NAD+-dependent synthesis of a 5′-phospho-ADP-ribosylated RNA/DNA cap by RNA 2′-phosphotransferase Tpt1. Nucleic Acids Res 46: 9617–9624. 10.1093/nar/gky792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir A, Abdullahu L, Banerjee A, Damha MJ, Shuman S. 2019. NAD+-dependent RNA terminal 2′ and 3′ phosphomonoesterase activity of a sub-set of Tpt1 enzymes. RNA 25: 783–792. 10.1261/rna.071142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. 2018. Invasive candidiasis. Nat Rev Dis Primers 4: 18026. 10.1038/nrdp.2018.26 [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Consaul SA, Nehrke KW, Abelson J. 1992. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem 267: 4577–4582. 10.1016/S0021-9258(18)42872-4 [DOI] [PubMed] [Google Scholar]
- Popow J, Schlieffer A, Martinez J. 2012. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci 69: 2657–2670. 10.1007/s00018-012-0944-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus BS, Schwer B, Shuman S. 2016. Characterization of the tRNA ligases of pathogenic fungi Aspergillus fumigatus and Coccidioides immitis. RNA 22: 1500–1509. 10.1261/rna.057455.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus BS, Goldgur Y, Shuman S. 2017. Structural basis for the GTP specificity of the RNA kinase domain of fungal tRNA ligase. Nucleic Acids Res 4522: 12945–12953. 10.1093/nar/gkx1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Schwer B, Shuman S. 2003. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J Biol Chem 278: 43928–43938. 10.1074/jbc.M307839200 [DOI] [PubMed] [Google Scholar]
- Sawaya R, Schwer B, Shuman S. 2005. Structure-function analysis of the yeast NAD+-dependent tRNA 2′-phosphotransferase Tpt1. RNA 11: 107–113. 10.1261/rna.7193705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Sawaya R, Ho CK, Shuman S. 2004. Portability and fidelity of RNA-repair systems. Proc Natl Acad Sci 101: 2788–2793. 10.1073/pnas.0305859101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal ES, Gritsenko V, Levitan A, Yadav B, Dror N, Steenwyk JL, Silberberg Y, Meilich K, Rokas A, Gow NA, et al. 2018. Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans. MBio 9: e02048-18. 10.1128/mBio.02048-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Malik HS, Consaul SA, Phizicky EM. 1998. A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli. Proc Natl Acad Sci 95: 14136–14141. 10.1073/pnas.95.24.14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SL, Kierzek R, Turner DH, Phizicky EM. 1999. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J Biol Chem 274: 2637–2644. 10.1074/jbc.274.5.2637 [DOI] [PubMed] [Google Scholar]
- Steiger MA, Jackman JE, Phizicky EM. 2005. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: evidence for a conserved two-step reaction mechanism. RNA 11: 99–106. 10.1261/rna.7194605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.