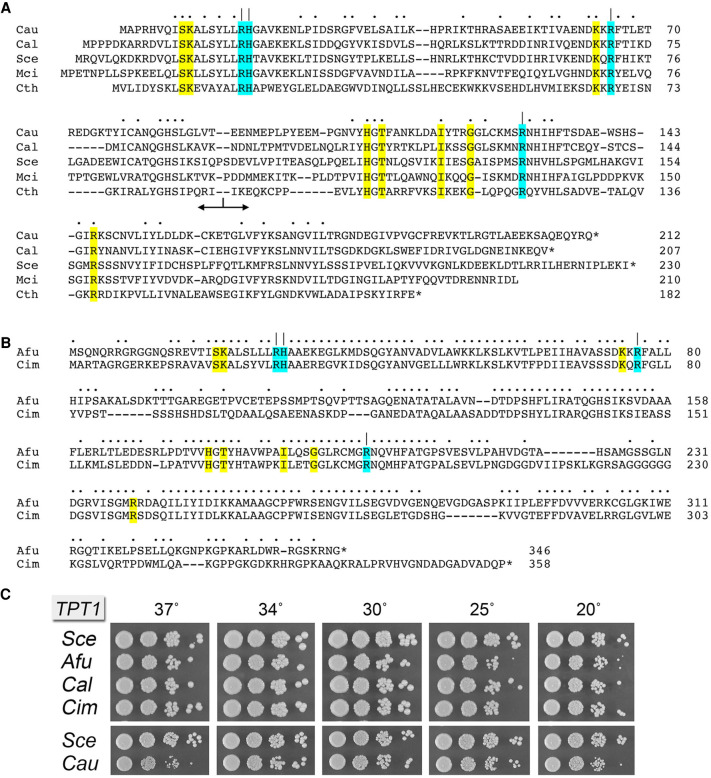
FIGURE 1.
Tpt1 orthologs from pathogenic fungi. (A) Alignment of the amino acid sequences of Tpt1 proteins from Candida auris (Cau, Genbank PIS54536.1), Candida albicans (Cal, Genbank AOW28085.1), Saccharomyces cerevisiae (Sce, Genbank NP_014539.1), Mucor circinelloides (Mci, Genbank EPB89638.1), and Clostridium thermocellum (Cth, ABN54255.1). Positions of amino acid side chain identity/similarity are indicated by dots above the alignment. Gaps in the alignment are indicated by dashes. The border between the amino-terminal RNA lobe and the carboxy-terminal NAD+ lobe is indicated by the bidirectional arrow below the alignment. The conserved Arg–His–Arg–Arg catalytic tetrad is highlighted in cyan shading and denoted by | above the alignment. Other conserved amino acids that contact the RNA 2′-PO4 nucleotide or the ADP-ribose moiety of NAD+ are highlighted in yellow shading. (B) Alignment of the amino acid sequences of Tpt1 proteins from Aspergillus fumigatus (Afu, Genbank EDP53951.1) and Coccidioides immitis (Cim, Genbank KJF61580.1), annotated as in A. (C) Complementation of S. cerevisiae tpt1Δ by expression of Tpt1 orthologs from pathogenic fungi. Complementation was assayed by plasmid shuffle. Viable FOA-resistant tpt1Δ p413–TPT1 strains as specified were grown in YPD-Ad liquid medium at 30°C to mid-log phase, then diluted to attain A600 of 0.1, and aliquots (3 µL) of serial 10-fold dilutions were spotted on YPD agar plates and incubated at 20°C, 25°C, 30°C, 34°C, 30°C, and 37°C. Photographs of the plates are shown.