Abstract
RtcB enzymes comprise a widely distributed family of manganese- and GTP-dependent RNA repair enzymes that join 2′,3′-cyclic phosphate ends to 5′-OH ends via RtcB-(histidinyl-N)–GMP, RNA 3′-phosphate, and RNA3′pp5′G intermediates. RtcB can ligate either 5′-OH RNA or 5′-OH DNA strands in vitro. The nucleic acid contacts of RtcB are uncharted. Here we report a 2.7 Å crystal structure of Pyrococcus horikoshii RtcB in complex with a 6-mer 5′-OH DNA oligonucleotide HOA1pT2pG3pT4pC5pC6, which reveals enzymic contacts of Asn202 to the terminal 5′-OH nucleophile; Arg238 to the A1pT2 and T2pG3 phosphates; Arg190 and Gln194 to the T2pG3 phosphate; and an Arg190 π-cation interaction with the G3 nucleobase. The structural insights affirm functional studies of E. coli RtcB that implicated the conserved counterpart of Arg238 in engagement of the 5′-OH strand for ligation. The essential active site Cys98 that coordinates two manganese ions is oxidized to cysteine sulfonic acid in our structure, raising the prospect that RtcB activity might be sensitive to modulation during oxidative stress.
Keywords: RNA repair, cysteine sulfonic acid, tRNA splicing
INTRODUCTION
RtcB exemplifies a novel family of RNA ligases implicated in tRNA splicing, RNA repair, nonspliceosomal mRNA splicing, RNA recombination, and biogenesis of circular RNAs (Englert et al. 2011; Popow et al. 2011; Tanaka and Shuman 2011; Tanaka et al. 2011a; Jurkin et al. 2014; Kosmaczewski et al. 2014; Lu et al. 2014; Litke and Jaffrey 2019; Moldovan et al. 2019; Schmidt et al. 2019). Unlike classic RNA and DNA ligases, which join 3′-OH and 5′-phosphate ends, RtcB seals broken RNAs with 5′-OH and either 2′,3′-cyclic phosphate (RNA > p) or 3′-phosphate (RNAp) ends. The chemical mechanism of RtcB end-joining was elucidated via studies of Escherichia coli RtcB, which executes a multistep ligation pathway entailing: (i) reaction of the enzyme with GTP to form a covalent RtcB-(His337-N)-GMP intermediate; (ii) hydrolysis of RNA > p to RNAp; (iii) transfer of guanylate from His337 to the polynucleotide 3′-phosphate to form a polynucleotide-(3′)pp(5′)G intermediate; and (iv) attack of a 5′-OH on the –NppG end to form the splice junction and liberate GMP (Tanaka et al. 2011b; Chakravarty and Shuman 2012; Chakravarty et al. 2012). The RtcB reaction pathway requires manganese as a divalent cation cofactor. The catalytic repertoire of RtcB is not limited to RNA transactions. For example, RtcB is adept at ligating HODNA strands and it efficiently “caps” DNA 3′-phosphate (DNAp) ends to form a stable DNAppG structure (Das et al. 2013; Maughan and Shuman 2015).
Whereas the biochemistry of RtcB is best understood for the E. coli enzyme (EcoRtcB), available structural insights stem from Pyrococcus horikoshii RtcB (PhoRtcB), which has been crystallized in several functional states: as the RtcB apoenzyme; as RtcB complexes with Mn2+ or Mn2+•GTP(αS); and as the covalent RtcB-(His404-Nε)–GMP•Mn2+ intermediate (Okada et al. 2006; Englert et al. 2012; Desai et al. 2013). A key insight from the crystal structures was that RtcB binds two Mn2+ ions, separated by 3.6 Å, one of which (Mn1) coordinates the α phosphate of GTP (and the GMP phosphate in the covalent RtcB–pG complex) while the other (Mn2) contacts the GTP γ phosphate (Desai et al. 2013). Mn1 is coordinated by Cys98, His234, and His329 (conserved as Cys78, His185, and His281 in EcoRtcB); Mn2 is coordinated by Asp95, Cys98, and His203 (conserved as Asp75, Cys78, and His168 in EcoRtcB). The active site cysteine uniquely bridges the two metal ions and its mutation to alanine abolishes all steps of the RtcB nucleic acid ligation pathway (Englert et al. 2012; Maughan and Shuman 2016).
The structural basis for nucleic acid recognition by RtcB is tabula rasa. The presence of sulfate anions flanking the active site in several structures of PhoRtcB has prompted speculation that they mimic phosphodiester phosphates of the nucleic acid strands destined for sealing (Englert et al. 2012; Desai et al. 2013; Maughan and Shuman 2016). However, it is unclear which of these sulfates demarcate the 3′ and 5′ strands and how the respective termini are engaged to the enzyme. In the present study, we begin to close this knowledge gap by reporting the crystal structure of PhoRtcB in a binary complex with a 5′-OH oligonucleotide. The structure clarifies prior functional studies of RtcB mutants (Englert et al. 2012; Maughan and Shuman 2016) and suggests a possible means of regulating RtcB activity via oxidation of the essential active site cysteine.
RESULTS AND DISCUSSION
Structure of PhoRtcB in complex with a 5′-OH oligonucleotide
Crystals grown from a premixture of PhoRtcB with a 6-mer DNA oligonucleotide HOA1pT2pG3pT4pC5pC6p were in space group P43212 with one protomer in the asymmetric unit. The structure was solved by molecular replacement and refined at 2.7 Å resolution to Rwork/Rfree of 0.191/0.216 (Supplemental Table S1). The RtcB protein comprised a single continuous polypeptide from Met1 to Gly481.
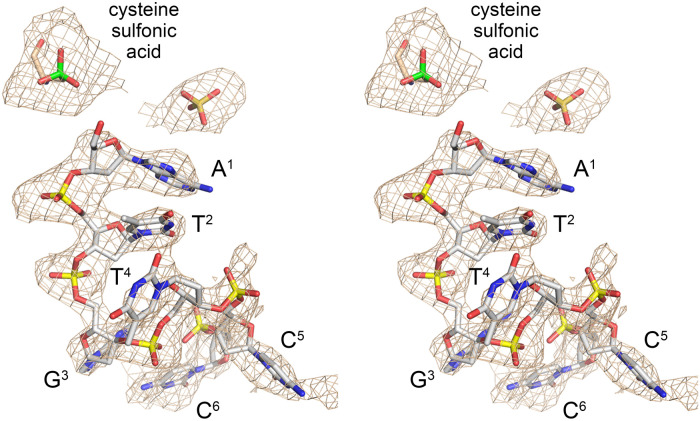
A simulated annealing omit map highlighted two key items. First, there was strong electron density surrounding the sulfur atom of active site metal ligand Cys98 that was inconsistent with an unmodified cysteine side chain. The shape of the omit density was most consistent with oxidation of the cysteine to cysteine sulfonic acid, as modeled in Figure 1. Note that the buffers used for the steps of PhoRtcB purification by Ni-agarose affinity chromatography prior to gel filtration did not contain an added reducing agent and this might account for the observed cysteine oxidation. Second, there was omit density apparent for the 6-mer 5′-OH DNA oligonucleotide, albeit not for the terminal 3′-phosphate group (Fig. 1). There was no evidence of covalent guanylylation of the active site His404 or for the presence of divalent cations in either the Mn1 or Mn2 positions. Omit density indicative of a sulfate or phosphate anion was seen at a site noted in prior RtcB structures to be occupied by sulfate (e.g., in pdb 4ISZ, 4IT0, and 4DWR). We have assigned this ligand as sulfate in the present structure (Fig. 1).
FIGURE 1.
RtcB active site occupancy by 5′-OH oligonucleotide. Stereo view of a simulated annealing difference density omit map of the RtcB active site (beige mesh), contoured at 2 σ with a carve radius of 3.6 Å. Strong electron density surrounding the Cys98 sulfur atom (shown in green) was modeled as cysteine sulfonic acid. A 6-mer DNA oligonucleotide, depicted as a stick model with gray carbons, was placed into density as shown. The nucleobases are labeled and numbered starting from the 5′-OH end. A sulfate anion (stick model) was also placed into density.
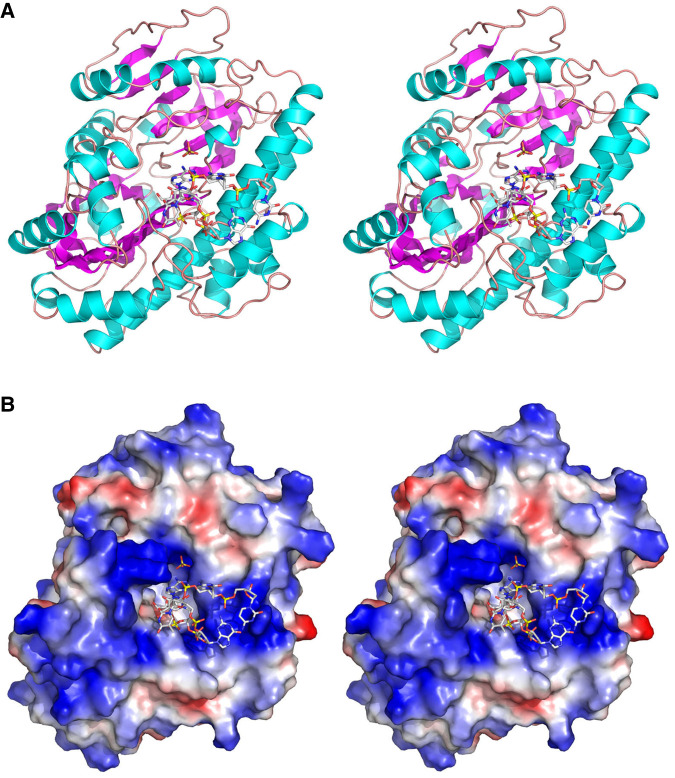
The tertiary structure of the RtcB•oligo complex shown in Figure 2A in stereo view, with the DNA ligand on the anterior surface of the enzyme. The fold consists of a six-strand β sheet and a seven-strand β barrel flanked by 18 helices. A superposition of the 5′-OH DNA complex on the GTP•(Mn)2 complex solved previously revealed minimal changes in protein conformation, with one notable exception involving the loop segment from Gly379 to Thr383 that comprises part of the interface with the guanosine nucleoside of GTP. Gly397-O makes a hydrogen bond to the guanosine ribose 2′-OH and the Gly378, Ser380, and Met381 main chain atoms pack closely against the guanine nucleobase making multiple van der Waals contacts (Desai et al. 2013). This segment undergoes an inward movement in the guanylate-free 5′-OH DNA complex (by 2.1 Å at the Ser380 Cα) that creates a clash with guanosine. We surmise that the guanosine pocket undergoes a modest closed-to-open conformational shift to accommodate the GTP substrate.
FIGURE 2.
Structure of RtcB in complex with 5′-OH oligonucleotide. (A) Stereo view of the RtcB fold, shown as a cartoon model with magenta β strands and cyan helices. The 5′-OH DNA oligonucleotide and sulfate are rendered as stick models. (B) Stereo view of a surface electrostatic model of RtcB (generated in Pymol) in the same orientation as in A, with the 5′-OH DNA and sulfate depicted as stick models.
A surface electrostatic model of the RtcB•oligo complex (Fig. 2B) highlights how the 5′-OH terminal nucleotide penetrates into a deep active site pocket that is surrounded by a positive electrostatic surface (shown in blue) that provides docking sites for several of the backbone phosphates. The sulfate anion is also situated deep inside the active site pocket and is likely to occupy the position of one of the phosphates of the 3′-PO4 strand to which the 5′-OH strand is joined by RtcB.
The conformation of the 6-mer 5′-OH strand is remarkable in several respects. Whereas the A1, T2, and T4 nucleobases form an irregular stack, the G3 nucleoside is splayed out and stacks over the C6 base, leaving the C5 base completely splayed out and pointed away from the enzyme surface (Figs. 1 and 2B). As described in detail below, RtcB recognition of the 5′-OH strand is achieved via direct contact between enzyme and the first three nucleotides of the substrate. The distal nucleotides of the 6-mer that sit on and/or project out from the protein surface are held in place by contacts to a symmetry-related RtcB protomer in the crystal lattice, as shown in Supplemental Figure S1. The terminal 3′-PO4 of the 6-mer strand is not visualized and is presumably disordered; the terminal 3′-OH of the C6 nucleoside is solvent-exposed in the cleft between the symmetry-related protomers. Arg441 of the second RtcB protomer engages the T4pC5 phosphate. The G3, C5, and C6 nucleosides pack against the neighboring RtcB protomer, whose Arg425 makes a bifurcated hydrogen bond to C6-O2. The Gly421 main chain amide makes a hydrogen bond to C5-O2 and the Thr416 and Tyr419 main chain carbonyls receive hydrogen bonds from C5-N4. In contrast, there are no base-specific contacts between the 5′-OH oligonucleotide and the RtcB protomer to which it is bound in the active site, which is sensible given that diverse nucleotide sequences populate the 5′-OH splice junctions of intron-containing tRNAs and other incised RNAs that are substrates for ligation by RtcB. Whereas the physiological significance of cross-protomer nucleic acid contacts is doubtful, they likely contributed to crystal lattice formation in space group P43212, which differed from the space groups P212121 and P21 formed in PhoRtcB crystals reported previously. In this regard, it is worth noting that neither Arg425 nor Arg441 (the side chains in the neighbor protomer that contact the nucleic acid) is conserved in RtcB proteins from other taxa.
Structural consequences of Cys98 oxidation
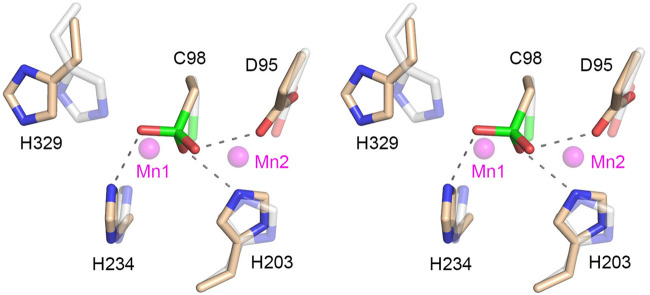
A stereo view of the amino acid manganese ligands in the RtcB active site is shown in Figure 3. The cysteine sulfonate modification engenders several new contacts entailing hydrogen bonds between the sulfonate oxygens and the His234, His203, and Asp95 side chains that normally engage the manganese cofactor. Indeed, superimposing the two manganese ions from the GTP•(Mn)2 structure (depicted as semitransparent magenta spheres) makes clear that the sulfonate would clash with and preclude binding of both metals. The oxidized cysteine also engenders a clash with the normal position of Mn1 ligand His329 that forces a rotamer change. Thus, the cysteine oxidized form of RtcB is incompatible with catalysis.
FIGURE 3.
Structural consequences of Cys98 oxidation. Stereo view of the active site highlighting contacts to the cysteine sulfonic acid moiety (depicted as a stick model with green sulfur atom). Amino acids in the present structure that normally engage the two manganese ions are rendered with beige carbons. Hydrogen bond interactions with the cysteine sulfonic acid are indicated by dashed lines. The equivalent amino acids and the Mn1 and Mn2 manganese ions from PhoRtcB structure 4ISZ are superimposed and depicted as semitransparent stick models with gray carbons and semitransparent magenta spheres, respectively.
Cysteine sulfonate is the end product of sequential cysteine oxidation steps that proceed through cysteine sulfenic acid and cysteine sulfinic acid intermediates. Cysteines in enzyme active sites may be especially sensitive to oxidation, as exemplified by members of the cysteinyl-phosphatase superfamily of enzymes that catalyze metal-independent phosphoryl transfer via a covalent cysteine-S-phosphate intermediate. The crystal structure of mammalian RNA 5′-triphosphatase, which performs the first step of mRNA capping, revealed that the active site cysteine nucleophile had undergone oxidation to cysteine sulfenic acid when crystals were grown in the presence of tungstate (Changela et al. 2001). It was suggested at the time that mRNA processing might be regulated via changes in the redox state of the cells or the microenvironment in which transcription occurs (Changela et al. 2001). It was subsequently reported that intentional treatment of mammalian capping enzyme with hydrogen peroxide effaced its activity and converted the cysteine nucleophile of the RNA triphosphatase enzyme to cysteine sulfonic acid (Mullen and Price 2017). There is a growing appreciation that cysteine oxidation of sensitive protein targets is a key switch underlying a variety of cellular signaling pathways (Paulsen and Carroll 2013).
We expect the pKa of the active site cysteine in RtcB to be lowered, compared to the standard value of 8.3, by virtue of the surrounding positive environment provided by the two manganese ions (in the metal-bound state) and the histidines (in the metal-free state), which would thereby sensitize the cysteine to modification. Whereas cysteine sulfenic acid modification is reversible by treatment with reducing agents, cysteine sulfonic acid is not. It is attractive to think that RtcB ligase activity might be subject to redox regulation via cysteine oxidation. Under mild oxidative stress, conversion of RtcB's cysteine to cysteine sulfenic acid could transiently (and potentially reversibly) dampen ligase activity. Under severe oxidative stress, further irreversible oxidation to cysteine sulfonic acid might occur. To the degree that this might be a situation best avoided, it is conceivable that cellular mechanisms exist to either protect RtcB from this more damaging modification (e.g., via an interacting protein) or to eliminate RtcB that has been irreversibly inactivated. These speculations are food for thought and further experimentation.
RtcB interface with the 5′-OH strand
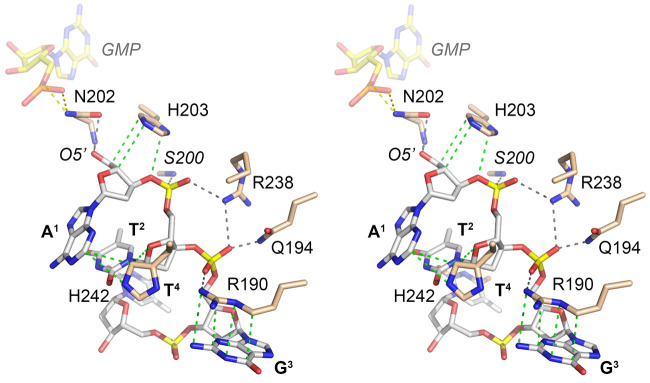
The present structure provides the first look at a network of RtcB interactions with a 5′-OH nucleic acid substrate for sealing. A stereo view of these contacts involving the first three nucleotides of the HOA1pT2pG3pT4pC5pC6 ligand is shown in Figure 4. The 5′-OH nucleophile (that attacks the 3′-phosphate of the RNA3′pp5′G intermediate) is anchored by hydrogen bonds from the Asn202 Oδ and the Asn202 main chain amide. Superimposing the GMP nucleotide from the RtcB–GMP•Mn2+ structure (shown in Fig. 4 as a semitransparent stick model with yellow carbons) indicated that Asn202 Nδ bridges to the GMP phosphate. It was reported that N202A mutation of PhoRtcB did not affect formation of the RtcB–GMP intermediate (Englert et al. 2012). The counterpart of PhoRtcB Asn202 in EcoRtcB is Asn167. Compared to wild-type EcoRtcB, the EcoRtcB N167A mutation slowed the rate of RNA 3′-phosphate guanylylation by 70-fold (Maughan and Shuman 2016). The EcoRtcB N167A mutant was also impaired for DNA 3′-phosphate guanylylation. In reactions with DNAppG and HODNA strands, N167A failed to effect strand ligation and instead catalyzed de-guanylylation of DNAppG (Maughan and Shuman 2016). Thus, the functional data accord with the structure in pinpointing the active site Asn202/Asn167 as an agent of the –NppG/5′-OH strand joining step. A fuller appreciation of the catalytic role of the active site Asn hinges on obtaining a structure of RtcB in complex with DNAppG.
FIGURE 4.
RtcB interface with the 5′-OH strand. Stereo view of the active site highlighting atomic interactions with the 5′-OH DNA oligonucleotide. Hydrogen bonds are indicated by black dashed lines; van der Waals contacts by green dashed lines. The guanylate nucleotide from PhoRtcB structure 4IT0 is superimposed and depicted as a semitransparent stick model with yellow carbons. Putative hydrogen bonds from Asn202 to the guanylate are shown as yellow dashed lines.
PhoRtcB His203, a conserved metal-binding ligand whose position and tautomer are unaffected by the cysteine sulfonate modification (Fig. 3), packs against and makes van der Waals contacts with the 5′-terminal A1 nucleoside sugar (Fig. 4). His242 packs against and makes van der Waals contacts to the A1 nucleobase and the C2 nucleoside sugar (Fig. 4). The first two phosphodiesters receive a rich set of hydrogen bonds from the enzyme. The Ser200 main chain amide coordinates the A1pT2 phosphate and Gln194-Nε contacts the T2pG3 phosphate. Arg190 makes an electrostatic contact with the T2pG3 phosphate and it makes a π-cation stack on the G3 nucleobase (Fig. 4).
The PhoRtcB Arg238 side chain makes bifurcated hydrogen bonds to the A1pT2 and T2pG3 phosphates (Fig. 4). The counterpart of PhoRtcB Arg238 in EcoRtcB is Arg189. Based on single-turnover kinetic data that mutating EcoRtcB Arg189 to alanine slowed the step of RNAppG/HORNA sealing by a factor of 200 compared to wild-type EcoRtcB, while slowing the rate of RNAppG formation by only threefold, it was predicted that Arg189 promotes catalysis of phosphodiester synthesis by coordinating the 5′-terminal phosphodiester of the 5′-OH strand (HON1pN2pN3−) and thereby positioning the 5′-OH for its attack on the 3′-phosphate of RNAppG (Maughan and Shuman 2016). The present structure of the PhoRtcB binary complex with 5′-OH DNA affirms this prediction for Arg238, with the added insight that the arginine engages two vicinal phosphates at the 5′-OH terminus.
MATERIALS AND METHODS
PhoRtcB purification and crystallization
A pET28a-based bacterial expression plasmid with a codon-optimized complete PhoRtcB open reading frame inserted between the NdeI and XhoI restriction sites was purchased from GenScript. His6-tagged PhoRtcB was produced in E. coli BL21(DE3) cells during an overnight induction of a 2-L culture at 18°C with 0.5 mM IPTG. Cells were harvested by centrifugation and the pellets were resuspended in 50 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 1 M NaCl, 20 mM imidazole, 10% glycerol) and stored at −80°C. Subsequent purification was performed at 4°C. The thawed cell suspension in lysis buffer was adjusted to 1 mg/mL lysozyme and supplemented with one EDTA-free protease inhibitor cocktail tablet (Roche; cat. no. 11836170001) and incubated for 30 min. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation in a Fiberlite F20-12X50 rotor for 45 min at 14,000 rpm. The soluble extract was mixed for 1 h with 5 mL of Ni-NTA agarose (Qiagen) that had been equilibrated in lysis buffer. The resin was collected by centrifugation and resuspended in 50 mL of buffer A (50 mM Tris-HCl, pH 8.0, 300 mM NaCl) containing 20 mM imidazole, then recollected by centrifugation. The resin was resuspended in 50 mL of buffer A containing 50 mM imidazole. The resin was then poured into a gravity flow column and the bound material was serially step-eluted with 150, 250, and 500 mM imidazole in buffer A. The elution profiles were monitored by SDS-PAGE. The peak RtcB-containing fractions were pooled, concentrated by centrifugal ultrafiltration, and then subjected to gel filtration through a 120-mL Superdex 200 column that was equilibrated with 20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT, 5% glycerol. Peak fractions were pooled and concentrated to 7 mg/mL by centrifugal ultrafiltration. Protein concentrations were determined by using the BioRad dye reagent with bovine serum albumin as the standard. Crystallization was performed by sitting drop vapor diffusion at room temperature. A 1 µL aliquot of a sample solution containing 125 µM PhoRtcB and 200 µM 6-mer DNA oligonucleotide HOA1pT2pG3pT4pC5pC6p (purchased from IDT) was mixed with 1 µL of Morpheus G6 precipitant/reservoir solution (Gorrec 2009) composed of 0.1 M MOPS/HEPES-Na buffers (pH 7.5), a carboxylic acid additives mixture (20 mM each of sodium formate, ammonium acetate, trisodium citrate, sodium potassium tartrate, sodium oxamate) 20% (v/v) ethylene glycol, 10% (w/v) PEG 8000. Crystals appeared after 2 to 3 d and were harvested after 1 wk. The PhoRtcB crystals were cryoprotected by transfer to a reservoir solution containing 25% glycerol before being flash-frozen in liquid nitrogen.
X-ray diffraction and structure determination
X-ray diffraction data from a single PhoRtcB crystal in space group P43212 were collected at the Advanced Photon Source beamline 24ID–E. The structure was solved by molecular replacement implemented in Phenix (Adams et al. 2010) using pdb 4DWR as the search model. Iterative model building into electron density was performed in O (Jones et al. 1991). Refinements were executed in Phenix. Data collection and refinement statistics are compiled in Supplemental Table S1.
DATA DEPOSITION
Structural coordinates have been deposited in Protein Data Bank under accession code 7LFQ.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health (NIH) grant R35-GM126945 (S.S.) and Deutsche Forschungsgemeinschaft research fellowship 394320208 (A.B.). The MSKCC structural biology core laboratory is supported by National Cancer Institute grant P30-CA008748. X-ray diffraction data were collected at synchrotron facilities supported by grants and contracts from the NIH (P30-GM124165, HEI-S10OD021527) and the Department of Energy (DE-AC02-06CH11357). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078692.121.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystrallogr D Biol Crystallogr 66: 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S. 2012. The sequential 2′,3′ cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res 40: 8558–8567. 10.1093/nar/gks558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty AK, Subbotin R, Chait BT, Shuman S. 2012. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc Natl Acad Sci 109: 6072–6077. 10.1073/pnas.1201207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A HCK, Martins A, Shuman S, Mondragón A. 2001. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J 20: 2575–2586. 10.1093/emboj/20.10.2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Chakravarty AK, Remus BS, Shuman S. 2013. Rewriting the rules for end joining via enzymatic splicing of DNA 3′-PO4 and 5′-OH ends. Proc Natl Acad Sci 110: 20437–20442. 10.1073/pnas.1314289110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KK, Bingman CA, Phillips GN, Raines RT. 2013. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry 52: 2518–2525. 10.1021/bi4002375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR, Söll D. 2011. Archaeal 3′-phosphate RNA splicing ligase characterization identified the missing component in tRNA maturation. Proc Natl Acad Sci 108: 1290–1295. 10.1073/pnas.1018307108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yoa M, Eom SH, Konigsberg WH, Söll D, Wang J. 2012. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining between 3′-terminal phosphate and 5′-OH. Proc Natl Acad Sci 109: 15235–15240. 10.1073/pnas.1213795109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrec F. 2009. The MORPHEUS protein crystallization screen. J Appl Cryst 42: 1035–1042. 10.1107/S0021889809042022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119. 10.1107/S0108767390010224 [DOI] [PubMed] [Google Scholar]
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, Martinez J. 2014. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 33: 2922–2936. 10.15252/embj.201490332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmaczewski SG, Edwards TJ, Han SM, Eckwahl MJ, Meyer BI, Peach S, Hesselberth JR, Wolin SL, Hammarlund M. 2014. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep 15: 1278–1285. 10.15252/embr.201439531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke JL, Jaffrey SR. 2019. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat Biotechnol 37: 667–675. 10.1038/s41587-019-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liang FX, Wang X. 2014. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell 55: 758–770. 10.1016/j.molcel.2014.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan WP, Shuman S. 2015. Characterization of 3′-phosphate RNA ligase paralogs RtcB1, RtcB2, and RtcB3 from Myxococcus xanthus highlights DNA and RNA 5′-phosphate capping activity of RtcB3. J Bacteriol 197: 3616–3624. 10.1128/JB.00631-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan WP, Shuman S. 2016. Distinct contributions of enzymic functional groups to the 2′,3′-cyclic phosphodiesterase, 3′-phosphate guanylylation, and 3′-ppG/5′-OH ligation steps of the Escherichia coli RtcB nucleic acid splicing pathway. J Bacteriol 198: 1294–1304. 10.1128/JB.00913-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan JB, Wang Y, Shuman S, Mills RE, Moran JV. 2019. RNA ligation precedes the retrotransposition of U6/LINE-1 chimeric RNA. Proc Natl Acad Sci 116: 20612–20622. 10.1073/pnas.1805404116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen NJ, Price DH. 2017. Hydrogen peroxide yields mechanistic insights into human mRNA capping enzyme function. PLoS ONE 12: e0186423. 10.1371/journal.pone.0186423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada C, Maegawa Y, Yao M, Tanaka I. 2006. Crystal structure of an RtcB homolog protein (PH1502-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins 63: 1119–1122. 10.1002/prot.20912 [DOI] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. 2013. Cysteine-mediate redox signling: chemistry, biology, and tools for discovery. Chem Rev 113: 4633–4679. 10.1021/cr300163e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lürhmann R, Söll D, et al. 2011. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331: 760–764. 10.1126/science.1197847 [DOI] [PubMed] [Google Scholar]
- Schmidt CA, Giusto JD, Bao A, Hopper AK, Matera AG. 2019. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47: 6452–6465. 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. 2011. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem 286: 7727–7731. 10.1074/jbc.C111.219022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S. 2011a. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem 286: 30253–30257. 10.1074/jbc.C111.274597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Chakravarty AK, Maughan B, Shuman S. 2011b. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286: 43134–43143. 10.1074/jbc.M111.302133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.