Abstract
tRNAs undergo an extensive maturation process including posttranscriptional modifications that influence secondary and tertiary interactions. Precursor and mature tRNAs lacking key modifications are often recognized as aberrant and subsequently targeted for decay, illustrating the importance of modifications in promoting structural integrity. tRNAs also rely on tRNA chaperones to promote the folding of misfolded substrates into functional conformations. The best characterized tRNA chaperone is the La protein, which interacts with nascent RNA polymerase III transcripts to promote folding and offers protection from exonucleases. More recently, certain tRNA modification enzymes have also been demonstrated to possess tRNA folding activity distinct from their catalytic activity, suggesting that they may act as tRNA chaperones. In this review, we will discuss pioneering studies relating posttranscriptional modification to tRNA stability and decay pathways, present recent advances into the mechanism by which the RNA chaperone La assists pre-tRNA maturation, and summarize emerging research directions aimed at characterizing modification enzymes as tRNA chaperones. Together, these findings shed light on the importance of tRNA folding and how tRNA chaperones, in particular, increase the fraction of nascent pre-tRNAs that adopt a folded, functional conformation.
Keywords: tRNA, posttranscriptional modifications, RNA chaperones, La protein, tRNA quality control, RNA folding
tRNA PROCESSING AND MATURATION IS COMPLEX AND MULTISTEPPED
Before assuming a role in translation, tRNAs undergo an extensive maturation process to produce a folded, functional tRNA. Precursor tRNAs (pre-tRNAs) are subject to numerous processing activities including endo- and exonucleolytic digestion to yield the mature 5′ and 3′ ends, post-transcriptional modification, and CCA addition (for reviews, see Hopper and Phizicky 2003; Phizicky and Hopper 2010). While tRNA processing is largely coordinated, a major challenge involves folding into the correct structure. The RNA folding problem consists of a kinetic problem, where RNAs exhibit a tendency to become kinetically trapped in alternate, aberrant conformations, and a thermodynamic problem, where the correct tertiary structure may not be energetically favored over competing structures (Herschlag 1995). Structured noncoding RNAs (ncRNAs), including tRNA, have difficulty assuming the correct tertiary structure, as tertiary interactions are less enthalpically stable than Watson–Crick interactions and occur at a slower rate (Zarrinkar et al. 1996; Sclavi et al. 1998; Woodson 2010). Moreover, biophysical investigations of tRNA folding demonstrated the occurrence of kinetically trapped folding intermediates similar to the folding of larger RNAs (Serebrov et al. 2001). Early studies in tRNA structure illustrated the importance of correct structure for functionality, showing that although two alternate conformations of tryptophanyl tRNA were isolated from E. coli, only one conformation was efficiently aminoacylated (Gartland and Sueoka 1966). Understanding how tRNAs assume the correct, “active” conformation has been the subject of ongoing research for more than 50 yr and is often addressed either by studies on tRNA modification enzymes or by investigations of tRNA chaperones.
POSTTRANSCRIPTIONAL MODIFICATIONS INFLUENCE tRNA STABILITY BY PROMOTING BASE-PAIRING AND TERTIARY INTERACTIONS
tRNAs are among the most highly modified RNA species, with an average of 13 posttranscriptional modifications on nuclear-encoded tRNAs (Fig. 1; Pan 2018). A number of studies have focused on modifications in the anticodon stem–loop (ASL), which primarily affect translational fidelity by maintaining the correct open reading frame and influencing codon–anticodon base-pairing (for review, see Tuorto and Lyko 2016), although modifications outside the ASL (henceforth referred to as body modifications) have similarly important functions in influencing tRNA structure. Early studies comparing the structures of modified and unmodified tRNAs provided the framework for a model stating that modifications promote native structure by influencing base-pairing and tertiary interactions, leading to the formation of the canonical cloverleaf secondary and L-shaped tertiary structures. Enzymatic and chemical probing of cellular and in vitro-transcribed human mitochondrial tRNA LysUUU revealed that the native tRNA folds into the expected cloverleaf structure, while unmodified tRNA assumes an alternate structure characterized by an elongated, bulged hairpin in place of the anticodon stem. Aberrant hairpin formation was traced to the absence of m1A9 modification, the presence of which inhibits base-pairing with U64 to promote structural rearrangement and formation of the canonical anticodon stem (Helm et al. 1998). Synthesis of tRNA LysUUU containing m1A9 confirmed that the single modification alone was sufficient for cloverleaf folding, prompting the idea that tRNA modifications have a role in influencing tRNA structure and folding (Helm et al. 1999). It is worth noting that mitochondrial tRNAs, including tRNA LysUUU, often differ in structure from canonical cytoplasmic tRNAs and as such, conclusions drawn from these studies should be interpreted with this caveat in mind. The importance of body modifications can also be inferred from the numerous human diseases characterized by mutations to tRNA modification enzymes (for review, see Pereira et al. 2018). Well-studied examples include mutations to the tRNA methyltransferase Trmt1 or the pseudouridine synthase Pus3 that result in intellectual disabilities, as well as the decreased expression of the mitochondrial tRNA methyltransferase Trmt61b associated with Alzheimer's disease (Sekar et al. 2015; Shaheen et al. 2016; Dewe et al. 2017; Zhang et al. 2020).
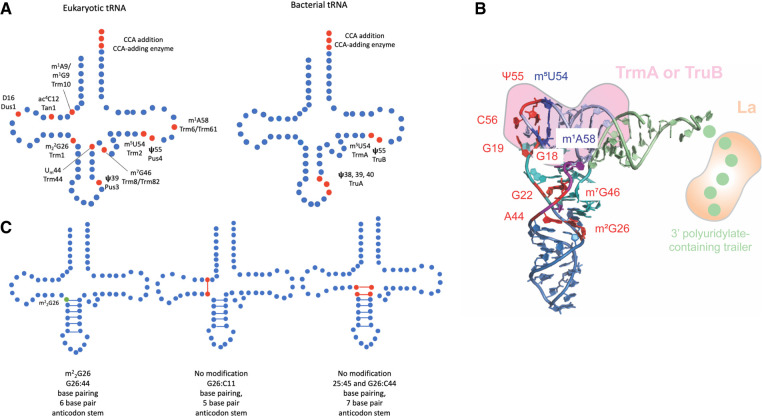
FIGURE 1.
(A) Schematic of eukaryotic and bacterial tRNA modifications discussed and the respective enzymes responsible for each modification. Modified nucleotides are indicated in red. (B) Three-dimensional tRNA structure highlighting tertiary base pairs and interaction sites of tRNA chaperones. The different tRNA arms are color-coded with the acceptor arm in light green, the D arm in cyan, the acceptor arm in blue, the variable arm in purple, and the T arm in light blue (structure 4TRA from Westhof et al. 1988), visualized in PyMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC). Critical tertiary interactions involving modified nucleotides connecting different tRNA arms are highlighted in red: m2G26—A44 (hinge region), G22—m7G46, G18—Ψ55, and G19—C56. The tertiary interaction of m5U54 and m1A58 in the T-loop is depicted in blue. La (orange) binds to the 3′ polyuridylate-containing trailer. TrmA and TruB (pink) both interact with the T arm causing the disruption of tertiary base pairs to the D arm. (C) The influence of m22G26 modification on base-pairing and anticodon length (Steinberg and Cedergren 1995). m22G26 modification is indicated in green and noncanonical base pairs resulting from unmodified G26 are indicated in red.
Continued advances in structural and computational biology have provided further insight into the role of additional tRNA modifications in structural stability. D-loop structure is influenced by the presence of dihydrouridine, which promotes the formation of a hairpin with a stable stem and flexible loop (Dyubankova et al. 2015). The loss of the C5–C6 double bond in dihydrouridine decreases stacking interactions, resulting in increased flexibility that promotes conformational dynamics (Sundaralingam et al. 1971; Suck and Saenger 1973). Nuclear magnetic resonance (NMR) studies revealed that dihydrouridine also destabilizes the C3′-endo conformation, compared to the rapid equilibrium between C2′- and C3′-endo conformations observed for uridine, thus facilitating tertiary interactions that contribute to three-dimensional tRNA folding (Dalluge et al. 1996). Studies conducted in budding yeast identified Dus1 and Dus2 as the enzymes responsible for modifying uridine to dihydrouridine on pre-tRNAs in vivo and in vitro (Xing et al. 2002). Importantly, mutations to Dus2, which modifies U20, result in reduced levels of a mutant allele of tRNA SerCGA, suggesting a role for dihydrouridine in modulating tRNA stability (Xu et al. 2018). N4-acetylcytidine at C12 of leucine and serine tRNAs is another highly conserved modification found in the D-loop (Sprinzl et al. 1998). Similar to dihydrouridine, D-loop acetylation promotes tRNA stability, with deletion of the budding yeast tRNA acetyltransferase Tan1 also leading to a decrease in the steady state levels of mature tRNA SerCGA (Johansson and Byström 2004).
Analogous to D-loop modifications, modification of T-loop nucleotides has also been demonstrated to contribute to stability. The formation of pseudouridine (Ψ) at position 55 favors the C3′-endo pucker conformation, resulting in increased base-stacking ability, thereby promoting tRNA stabilization (Davis 1995; Durant and Davis 1999; Newby and Greenbaum 2001, 2002; Xu et al. 2016). Adjacent to Ψ55, there is a methyluridine, m5U54, in all tRNAs which increases the thermal stability of tRNA (Sengupta et al. 2000). Another well-characterized T-loop modification is m1A58, which is catalyzed by the Trm6–Trm61 complex in budding yeast and is required for stability of tRNAi MetCAU. Importantly, the growth defect observed for Trm6 mutants grown at elevated temperatures can be suppressed by overexpression of tRNAi MetCAU or the RNA chaperone La, suggesting the lack of modification may result in tRNA misfolding and degradation (discussed in more detail below) (Anderson et al. 1998). 2′-O-methylation at U44 by Trm44, which is conserved among metazoans and fungi, is also required for growth at elevated temperatures in budding yeast strains lacking Tan1, due to reduced levels of tRNA SerCGA/UGA (Kotelawala et al. 2008). The finding that modifications influence base-pairing has been further characterized with computational studies revealing that modified base pairs within the T-loop, including the 54:58 pair, stabilize the T-loop by increasing the interaction energy of the base pair. The modified 54:58 base pair also serves to promote tertiary interactions between the D- and T-loops by enhancing the interaction involving the 18:55 and 19:56 base pairs stacking on top of the 54:58 base pair (Fig. 1B; Seelam et al. 2017).
Another important tRNA structural determinant relies on the clustering of stabilizing modifications at the junctures between stems, including positions 9 and 10 at the interface of the D-stem and acceptor stem and position 26 at the junction of the D-stem and anticodon stem. In particular, position 26 as well as positions 44 and 45 at the junction of the anticodon and variable arms are known as the important hinge region in tRNA, enabling distortion of the tRNA during interactions with the ribosome (Fig. 1B; Valle et al. 2003). Methylated guanosines at positions 10 and 26 are conserved in eukaryotes and archaea and the presence of methyl groups on the Watson–Crick face prevent canonical base-pairing, leading to increased flexibility and conformational dynamics (Seelam et al. 2017). Consistent with the need for flexibility at junctions, analysis of the tRNA sequence database (Jühling et al. 2009) identified a frequency of methylated base pairs at positions 10:25, 26:44, and 49:65 (Seelam et al. 2017). Further, although 5-methylcytosine at positions 48 and 49 at the junction of the variable loop and T-stem is not located on the Watson–Crick face and therefore does not influence base-pairing, the modification nevertheless promotes tRNA stability, further corroborating the importance of junction modifications (Tuorto et al. 2012).
A number of studies have shown that stability of the hinge region is influenced by the presence of m22G26. Dimethylation of G26 is critical in maintaining the correct balance between hinge flexibility and rigidity by adjusting the angle of the D-stem (Edqvist et al. 1995). The importance of G26 dimethylation in preventing base-pairing with cytidine at position 11 or 44 that would result in alternative tertiary structures is widely conserved. Sequence analysis of nuclear-encoded tRNAs revealed a correlation between the ability to fold into alternate conformers—including noncanonical 5 bp or 7 bp anticodon stems—and the presence of a dimethylguanosine, suggesting that the modification has a role in preventing alternate folds (Fig. 1C; Steinberg and Cedergren 1995; Vakiloroayaei et al. 2017). tRNAs lacking G26 dimethylation also show structural rearrangement of the tRNA core due to a lack of base-stacking and the resulting loss of tertiary interactions (Sonawane et al. 2016). Further support linking G26 dimethylation to hinge region stability comes from chemical probing experiments demonstrating differences in sensitivity between G26-modified and unmodified tRNA SerUGA, particularly in the A-U rich anticodon stem and nucleotides in the hinge region. The increased accessibility of the hinge region to chemical probing in the absence of G26 dimethylation suggests that in addition to its previously described function in preventing base-pairing with C44, the modification also has a role in increasing the stability of the hinge region (Vakiloroayaei et al. 2017).
Evidence that m22G is important for structure and function in vivo comes from studies using the fission yeast Schizosaccharomyces pombe. Deletion of Trm1, the tRNA methyltransferase that catalyzes m22G modification, results in impaired function of a suppressor tRNA mutant derived from tRNA SerUGA. Since suppressor tRNA constructs contain destabilizing mutations that lead to tRNA misfolding, it was suggested that Trm1-catalyzed modification promotes tRNA function by reinforcing the native fold (Arimbasseri et al. 2015; Vakiloroayaei et al. 2017). It is worth noting that Trm1 is not required for functionality of the ochre suppressor tRNA SUP4 in budding yeast, although Trm1 deletion in combination with the deletion of anticodon modification enzymes impairs suppressor tRNA activity (Klassen and Schaffrath 2018). The role of m22G26 in promoting tRNA functionality likely arises from the additional structural stability imparted by the modification. As it can be challenging to measure tRNA folding in vivo, aminoacylation is often used as an indirect readout of proper tRNA folding. Fission yeast lacking Trm1 and Sla1, the yeast homolog of the tRNA chaperone La, show decreased aminoacylation efficiency of tRNA SerCGA/UGA, indicative of tRNA misfolding (Vakiloroayaei et al. 2017). Collectively, the data suggest that modifications that influence structural stability of the hinge region also modulate tRNA function by reinforcing native structure and promoting aminoacylation.
FUNCTIONAL LINKS BETWEEN tRNA MODIFICATION AND QUALITY CONTROL PATHWAYS
Another indication that posttranscriptional modifications are important structural determinants for tRNA function comes from extensive findings that hypomodified tRNAs are targeted for decay. Several tRNA quality control pathways that monitor tRNA structure and degrade aberrant tRNAs have been discovered, including the nuclear surveillance pathway. The nuclear surveillance pathway, best characterized in budding yeast, involves the targeting and degradation of aberrant precursor transcripts, including pre-tRNA (Mitchell et al. 1997; LaCava et al. 2005). Nuclear surveillance is largely mediated by the TRAMP complex (Trf4/Air2/Mtr4), which consists of the poly(A) polymerase Trf4 or the closely related Trf5, the zinc knuckle protein Air2 or its homolog Air1, and the RNA helicase Mtr4 (LaCava et al. 2005). TRAMP facilitates recognition and polyadenylation of aberrant tRNAs, followed by 3′ → 5′ exonucleolytic degradation by the nuclear exosome (LaCava et al. 2005; Wyers et al. 2005; Hamill et al. 2010).
Initial studies on the structure of tRNAi MetCAU demonstrated that the loss of m1A58 modification results in increased susceptibility to nuclear surveillance due to weakened tertiary structure relative to its fully modified counterpart (Anderson et al. 1998). m1A58 modification, catalyzed by the Trm6–Trm61 heterodimer, is also critical for maintaining steady-state tRNA levels. A Trm6 mutant exhibited impaired growth at high temperature that can be linked to degradation of the hypomodified pre-tRNAi MetCAU, although this growth defect can be suppressed by overexpression of the RNA chaperone La (Anderson et al. 1998; Kadaba et al. 2004). Hypomodified pre-tRNAi MetCAU was also found to be polyadenylated by the noncanonical poly(A) polymerase Trf4 prior to decay (Kadaba et al. 2004). Certain tRNAs are less sensitive to a loss of m1A58 modification, with steady-state levels of tRNA LeuCAA remaining unchanged in the absence of a functional Trm6–Trm61 dimer. It has been proposed that differences in susceptibility to nuclear surveillance arise from structural differences among individual tRNAs, as well as different requirements for stabilizing modifications (Kadaba et al. 2004). Exosome-mediated decay occurs at an early point in tRNA biogenesis, as polyadenylated substrates still possess a 5′ leader and 3′ trailer. While the mechanism by which hypomodified tRNAs are recognized by TRAMP still remains unclear, the disrupted interaction between nucleotides 54 and 58 resulting from a lack of m1A modification may lead to an alternate structure recognized as aberrant by the nuclear surveillance machinery (Kadaba et al. 2006). Building on the idea that structurally defective tRNAs are targeted for decay by the nuclear surveillance machinery, additional studies in fission yeast revealed that the RNA chaperone La binds to the 3′ end of pre-tRNAs to protect against degradation by the exosome exonuclease Rrp6 (Huang et al. 2006). Similar polyadenylation-dependent tRNA decay pathways exist in bacteria, with thermodynamically unstable mutant tRNAs getting polyadenylated by the poly(A) polymerase PAP and degraded by PNPase (Li et al. 2002).
The budding yeast rapid tRNA decay (RTD) pathway relies on a different set of exonucleases to target structurally unstable mature tRNAs in the cytoplasm. While RTD also targets hypomodified tRNAs, it occurs on a much different time scale that resembles mRNA degradation kinetics (Grigull et al. 2004; Alexandrov et al. 2006). Deletion of Trm8 or Trm82, which form a complex that catalyzes m7G46 modification, in combination with the deletion of any one of seven nonessential body modification enzymes results in a temperature-sensitive growth defect accompanied by the rapid deacylation and decay of tRNA ValAAC (Alexandrov et al. 2006). Similar rapid degradation of tRNA SerCGA/UGA was observed in strains lacking Trm44 and Tan1, which are responsible for Um44 and ac4C12, respectively. RTD involves the exonucleases Rat1 and Xrn1, which degrade mature tRNA from the 5′ end, enabling degradation of aminoacylated and deacylated tRNAs (Chernyakov et al. 2008). The phosphatase Met22 is also indirectly involved in modulating RTD, since Met22 catalyzes the removal of the 3′ phosphate of the Rat1 and Xrn1 inhibitor pAP (Dichtl 1997; Chernyakov et al. 2008). The RTD pathway primarily acts on substrates with weakened acceptor and T-stems, which increases accessibility to the 5′ end. It is therefore likely that hypomodified tRNAs are targeted by the RTD pathway because the lack of modifications including m7G46, Um44, and ac4C12 weaken tertiary interactions, thereby increasing accessibility of the acceptor stem for the degradation machinery (Chernyakov et al. 2008; Whipple et al. 2011). RTD also plays a role in temperature-dependent tRNA quality control, with higher temperatures resulting in weaker tRNA stability and increased accessibility of the 5′ end to Rat1 and Xrn1 (Payea et al. 2018). An additional Met22-dependent decay pathway has recently been revealed to act on intron-containing pre-tRNAs containing disrupted exon–intron structures. While the mechanism by which this decay pathway occurs remains ambiguous, it has been proposed that it may involve tRNA splicing machinery (Payea et al. 2020).
Another mechanism budding yeast use to ensure that cytoplasmic tRNAs are structurally stable and correctly end-processed is the retrograde tRNA transport pathway. Reimport of mature cytoplasmic tRNAs into the nucleus is Ran-dependent and utilizes the importin-beta family member Mtr10 (Shaheen and Hopper 2005; Takano et al. 2005). These initial studies prompted the hypotheses that the nucleus could act as a reservoir for mature tRNAs under certain cellular stresses or that nuclear import serves as a proof-reading mechanism following cytoplasmic tRNA maturation steps including splicing (Shaheen and Hopper 2005). These two possibilities are likely not mutually exclusive, as the findings that glucose and amino acid starvation trigger nuclear accumulation provided evidence supporting the first hypothesis (Whitney et al. 2007), while more recent studies demonstrated a role for nuclear reimport in tRNA quality control (Kramer and Hopper 2013). Leader- and trailer-containing tRNA IleUAU that has already undergone cytoplasmic splicing and m22G26-hypomodified tRNA LysUUU and TyrGUA accumulated in budding yeast cells with impaired retrograde tRNA import, suggesting that incorrectly end-processed and hypomodified tRNAs are normally reimported to the nucleus although it remains unclear whether reimported tRNAs are subject to repair or degradation (Kramer and Hopper 2013). The retrograde tRNA transport pathway is likely evolutionarily conserved, with similar pathways described in various mammalian systems (Shaheen et al. 2007; Barhoom et al. 2011; Miyagawa et al. 2012; Watanabe et al. 2013). In particular, oxidative stress triggers nuclear import of select cytoplasmic tRNAs in several human cell lines, including tRNAs with truncated 3′ ends (Schwenzer et al. 2019). Nuclear reimport during nutrient starvation in budding yeast also involves the Hsp70 chaperone Ssa2p (Takano et al. 2015; Nostramo and Hopper 2020). Ssa2p was demonstrated to act as a tRNA-binding protein in vivo and in vitro and further, exhibits increased affinity for tRNAs with destabilized acceptor stems, prompting the idea that it may function in quality control pathways through the recognition of aberrant cytoplasmic tRNAs (Takano et al. 2015).
Nuclear export has similarly been proposed to act as an additional tRNA quality control step to monitor tRNA ends (Lund and Dahlberg 1998; Cook et al. 2009; Kramer and Hopper 2013). Consistent with the idea that end processing, including aminoacylation, affects tRNA export, budding yeast strains with mutated tRNA aminoacyl synthases showed defects in tRNA export (Sarkar et al. 1999). Nuclear export proteins also discriminate natively folded from misfolded tRNAs, with the vertebrate homolog exportin-t recognizing tertiary interactions including interactions between the TΨC-loop and D-loop (Arts et al. 1998). The fission yeast nuclear export protein Los1 binds to the 5′ and 3′ ends of tRNAs during nuclear export, which allows for discrimination between correctly and incorrectly end-processed tRNAs, the former of which is characterized by a single-stranded 3′ CCA tail accommodated by conserved positive residues (Lys177, Arg181, and Lys259 in the fission yeast Los1) (Cook et al. 2009).
The wealth of studies characterizing tRNA quality control pathways suggest a tendency for hypomodified tRNAs to be highly susceptible to decay although it is not necessarily the loss of modification that results in targeting for decay, but rather the structural instability occurring in the absence of modification. The exact mechanism by which aberrant pre-tRNAs are targeted by the nuclear surveillance pathway remains under debate, but several groups have posited that the alternate structures assumed by hypomodified pre-tRNAs lead to slower maturation, aminoacylation, and assembly into ribonucleoprotein (RNP) complexes, resulting in exposure of the 3′ end to polyadenylation and decay (Li et al. 2002; Houseley et al. 2006; Reinisch and Wolin 2007). There is a more straightforward explanation for RTD targeting, where a lack of key modifications destabilizes the acceptor stem to make it more accessible to 5′ → 3′ exonucleases (Alexandrov et al. 2006; Chernyakov et al. 2008; Whipple et al. 2011). The RTD pathway may also function redundantly with retrograde tRNA transport, as budding yeast require at least one of the two pathways for viability (Kramer and Hopper 2013). Recognition and elimination of hypomodified tRNAs by quality control pathways is of great import, since hypomodified tRNAs—but not truncated or improperly end-processed tRNAs—may still be accommodated by the ribosome and therefore able to participate in translation, where their compromised stability can lead to deleterious effects such as impaired translation of certain codons (Urbonavičius et al. 2002).
tRNA FOLDING IS PROMOTED BY RNA CHAPERONES
RNA chaperones are a class of RNA binding proteins that prevent or resolve misfolds and nonfunctional, inhibitory structures without external energy input (Herschlag 1995; Rajkowitsch et al. 2007). A kinetic partitioning model posits that only a small fraction of nascent RNA directly folds into a native conformation, and RNA chaperones can assist the remaining RNAs that are kinetically trapped in intermediate or misfolded conformations (Pan et al. 2000; Rajkowitsch et al. 2007; Woodson 2010). RNA chaperones have been reported to destabilize base pairs to unwind misfolded structures, thereby giving RNAs additional chances to fold into native structures, although the mechanisms used by RNA chaperones remain poorly understood. In the absence of ATP hydrolysis to provide the energy necessary to disrupt base pairs, RNA chaperones have been hypothesized to make use of the thermodynamic differences between native and intermediate folding states to promote RNA folding (Woodson 2010). The effectiveness of RNA chaperone activity is largely dependent on transient substrate binding and rapid dissociation, allowing unfolded RNA structures the chance to refold into native conformations. Strong binding has been hypothesized to result in the stabilization of alternate structures, as has been demonstrated by the bacterial RNA-binding protein StpA, which exhibits increased chaperone activity when binding is partially impaired (Mayer et al. 2007; Rajkowitsch et al. 2007; Woodson 2010). Basic patches and disordered regions are common features of proteins that display RNA chaperone activity. Basic regions buffer the electrostatic interactions between RNA and the surrounding divalent cations, allowing for the disruption of aberrant tertiary structures (Woodson 2010). The role of disordered regions in RNA chaperone activity is less transparent, but it has been suggested that disordered regions may become structured upon substrate binding, with the binding event acting as an entropy transfer to facilitate the local unwinding of misfolded RNA structures (Tompa and Csermely 2004).
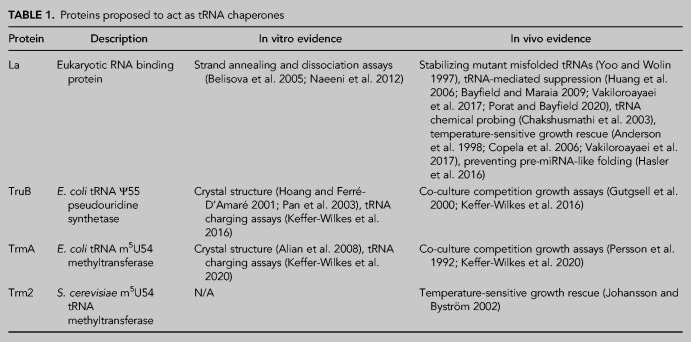
While RNA chaperones have been extensively studied in the context of bacterial small RNAs (for review, see Santiago-Frangos and Woodson 2018), virus-encoded nucleocapsid proteins, and ribosomal proteins (for review, see Semrad 2011), few proteins have been described to possess tRNA-specific chaperone activity. Here, we will describe the best-characterized examples of tRNA chaperones, namely the RNA-binding protein La as well as the bacterial tRNA modification enzymes TrmA and TruB which are responsible for formation of m5U54 and Ψ55 in the T-loop, respectively (Table 1; Fig. 1B).
TABLE 1.
Proteins proposed to act as tRNA chaperones
The most extensively characterized tRNA chaperone is the La protein, an RNA binding protein which associates with nascent RNAPIII transcripts, including pre-tRNA, through the ubiquitous 3′ polyuridylate-containing trailer that corresponds to the RNAPIII termination sequence (Rinke and Steitz 1982; Reddy et al. 1983; Mathews and Francoeur 1984; Stefano 1984). While La is essential in higher eukaryotes, its dispensability in budding and fission yeast enabled mechanistic studies into the function of La in pre-tRNA stability and maturation (Yoo and Wolin 1994; Van Horn et al. 1997). As 3′ end binders, the budding and fission yeast La proteins (Lhp1 and Sla1, respectively) influence the order of tRNA end processing. In the La-dependent pathway, La binding to the 3′ trailer blocks 3′ → 5′ exonucleases, resulting in 5′ endonucleolytic cleavage of the leader by RNase P followed by endonucleolytic cleavage of the 3′ trailer by RNase Z and the subsequent dissociation of La. In contrast, 3′ exonucleolytic nibbling of the 3′ trailer to yield the mature 3′ end occurs before 5′ cleavage when La is absent (Van Horn et al. 1997; Yoo and Wolin 1997; Copela et al. 2008). The role of La in 3′ end protection also functions to protect pre-tRNAs from the nuclear surveillance pathway. Polyuridylate trailer binding by La shields the 3′ end from TRAMP-mediated polyadenylation and subsequent exosome-mediated decay, thereby preventing both native, and to some extent misfolded, tRNAs from decay (Huang et al. 2006). 3′ end processing in budding yeast involves the interplay between the La protein and exo- and endonucleases (Skowronek et al. 2014). tRNAs with shorter 3′ trailers are preferentially processed by exonucleases like Rex1 to generate the mature 3′ end, while the endonuclease Trz1 acts on pre-tRNAs with longer trailers (Skowronek et al. 2014). The balance between end maturation and decay is also dependent on the competition between Rex1 and La for 3′ end binding, with Rex1 binding leading to the generation of TRAMP substrates by 3′ trimming and La binding resulting in pre-tRNA stabilization and protection from TRAMP-mediated degradation (Copela et al. 2008).
Numerous structural studies led to insights into the polyuridylate binding mode of La and how it contributes to pre-tRNA binding and maturation (for review, see Bayfield et al. 2019). The crystal structure of the amino-terminal domain of the human La protein, consisting of the La motif and an RNA recognition motif (RRM1), revealed that three terminal uridylates are sequestered in the basic- and aromatic-rich cleft between the La motif and RRM1 (Fig. 2A,C; Teplova et al. 2006; Kotik-Kogan et al. 2008). While earlier structural studies proposed that the four-stranded β-sheet in RRM1 participates in polyuridylate binding (Alfano et al. 2004), the crystal structure demonstrated that using the edge of RRM1 for binding the penultimate uridylate leaves an exposed face of the β-sheet available for additional RNA contacts (Teplova et al. 2006). In agreement with this additional proposed binding site on the face of the RRM1 β-sheet, it was found that La has a higher affinity for pre-tRNA than end-processed tRNA and a dissociated polyuridylate-containing trailer, which can be attributed to the additive effects upon pre-tRNA engagement with the polyuridylate binding site and the RRM1 β-sheet binding site. Additional support for an alternate RRM1 binding site comes from the findings that intact pre-tRNAs bind La through contacts to the La motif and RRM1, with both binding sites together having greater affinity than either site alone. Upon trailer cleavage, the end-processed tRNA and trailer each occupy only one binding site and the resulting decreased affinity causes dissociation of both products (Bayfield and Maraia 2009). The importance of the RRM1 in pre-tRNA binding in vivo and in vitro has led to a model suggesting that polyuridylate binding by the La motif contributes to binding specificity by directing La to RNAPIII transcripts including pre-tRNA, while the RRM1 further increases pre-tRNA binding affinity through contacts to the pre-tRNA body (Horke et al. 2004a,b).
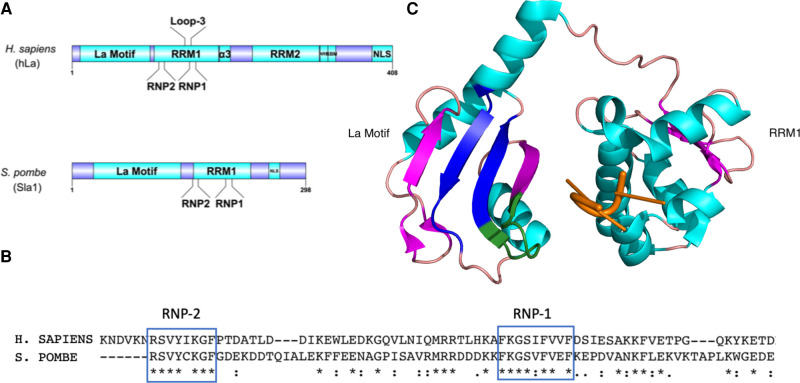
FIGURE 2.
Regions in the RNA recognition motif (RRM1) of the La protein play a role in La's RNA chaperone activity. (A) Schematic of the human and fission yeast La proteins. (NRE) Nuclear retention element. (SBM) Short basic motif. (NLS) Nuclear localization sequence. Schematics were generated with Ren et al. (2009). (B) Multiple sequence alignment (Madeira et al. 2019) of the conserved RNP1 and RNP2 sequences in the RRM1 of the human and fission yeast La proteins (Bayfield and Maraia 2009). (C) Ribbon representation of the high-resolution structure of the La motif and RRM1 of the human La protein in complex with a polyuridylate RNA oligomer (orange) (structure 2VOO from Kotik-Kogan et al. 2008), visualized in PyMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC). α-helices are pictured in cyan, β-sheets in pink. Regions important for RNA chaperone activity are highlighted: RNP1 and RNP2 (blue) and loop-3 (green).
Consistent with the engagement of different pre-tRNA elements—the polyuridylate trailer and pre-tRNA body—the two separate RNA binding surfaces on the La protein mediate distinct activities in pre-tRNA processing. PAR-CLIP analysis mapped La binding to the pre-tRNA trailer as well as the 5′ end of the D-arm, although the precise binding location on La or the full extent of La contacts to the tRNA body remains unknown (Gogakos et al. 2017). Having established La's role in 3′ end protection through trailer binding, several studies were undertaken to understand how the RRM1 β-sheet surface participates in tRNA maturation. La proteins from various species have conserved basic and aromatic patches in the RRM1, referred to as RNP1 and RNP2 (Fig. 2A,B). Mutation of conserved aromatic residues from these patches—Y114 and F155 in human La, which project from the β-sheet of RRM1—had no effect on uridylate binding or 3′ end protection but were required for tRNA functionality in a tRNA-mediated suppression assay that relies on correctly folded tRNAs for activity, indicating that the β-sheet binding surface plays a role in tRNA folding (Huang et al. 2006). This activity was further mapped to include basic residues in loop-3 of the RRM1 (Fig. 2), with mutations to loop-3 impairing tRNA-mediated suppression activity as well as RNA folding in an in vitro cis-splicing assay (Bayfield and Maraia 2009). Similar to other tested RNA chaperones, La is capable of annealing and dissociating RNA duplexes (Belisova et al. 2005; Naeeni et al. 2012), and mutants incapable of strand annealing and dissociation were also inactive in tRNA-mediated suppression, suggesting that La promotes tRNA functionality, at least in part, by using its RNA chaperone activity to assist pre-tRNA folding (Naeeni et al. 2012). Other regions of La, including its unstructured carboxy-terminal domain, have also been implicated in tRNA chaperone activity (Jacks et al. 2003; Kucera et al. 2011; Naeeni et al. 2012; Brown et al. 2016). In budding yeast, binding of the carboxy-terminal domain of La to the tRNA anticodon stem–loop is associated with promoting anticodon stem–loop structure (Kucera et al. 2011). Importantly, La's function as an RNA chaperone in promoting pre-tRNA folding and maturation is conserved among La homologs from various species (Bayfield and Maraia 2009; Naeeni et al. 2012; Porat and Bayfield 2020).
Further links can be drawn between La and tRNA modification enzymes with respect to pre-tRNA maturation and stability. Studies in budding yeast revealed that La functions redundantly with tRNA-stabilizing modification enzymes and proteins that bind tRNA during tRNA biogenesis and maturation. Genetic screens identified a temperature-dependent requirement for La in budding yeast strains with a mutation in tRNA ArgCCG that weakens the anticodon stem (Copela et al. 2006). It is interesting to note that tRNA ArgCCG mutations that lead to a requirement for La have greater effects at low temperatures, which may reflect the propensity for pre-tRNAs to become kinetically trapped in alternate conformations (Chakshusmathi et al. 2003). This, coupled with previous work demonstrating that tRNAs with weak anticodon stems have a propensity for misfolding, suggests that La binding to tRNA results in stabilization of the anticodon stem to promote native fold (Chakshusmathi et al. 2003; Copela et al. 2006). Another indication that La has a role in promoting pre-tRNA stability is the requirement for the stabilizing modification enzymes Pus3 and Pus4 in La knockout strains with a mutant allele of tRNA ArgCCG (Copela et al. 2006). Similarly, deletion of La and the m22G26 methyltransferase Trm1 is synthetic lethal in budding and fission yeast grown at high temperatures where tRNAs are more likely to misfold, suggesting that both proteins function redundantly to prevent pre-tRNA from adopting alternate, aberrant conformations that may lead to decay (Copela et al. 2006; Vakiloroayaei et al. 2017). The synthetic lethal phenotype can be rescued with reexpression of either human or fission yeast La, although rescue is dependent on an intact RRM1 β-sheet surface, illustrating the importance of La's chaperone activity. Based on the additional findings that La and Trm1 cooperate for the accumulation of a select set of G26-containing tRNAs in fission yeast, it is curious that deletion of the nuclear exosome exonuclease Rrp6 does not rescue the synthetic lethality phenotype by preventing pre-tRNA degradation, but rather exacerbates the synthetic lethal phenotype. Such a result prompts the idea that Rrp6 deletion could result in a dominant-negative phenotype, where aberrant pre-tRNAs that would normally be stabilized by La and Trm1 or degraded by the nuclear surveillance pathway persist and interfere with cellular functions (Fig. 3; Vakiloroayaei et al. 2017). It remains to be seen how aberrant pre-tRNAs affect cellular function, as well as how other tRNA modification enzymes cooperate with La to promote pre-tRNA stability. The data converge on a model where La, tRNA modification enzymes, and tRNA binding proteins contribute to pre-tRNA structural stability, resulting in efficient tRNA biogenesis (Copela et al. 2006; Vakiloroayaei et al. 2017).
FIGURE 3.
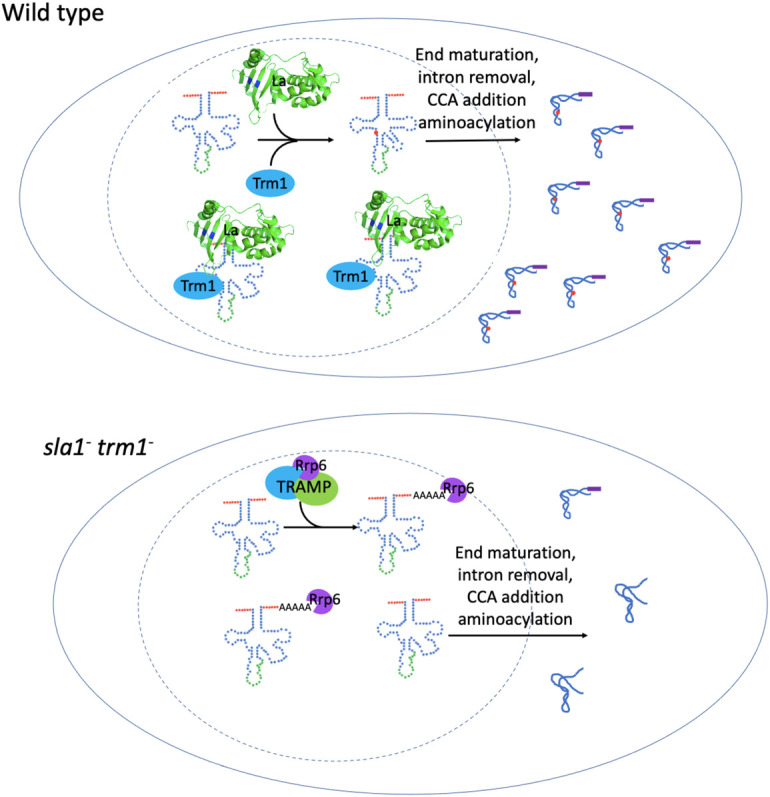
La (Sla1) and Trm1 cooperate to promote tRNA folding, accumulation, and functionality in fission yeast. Sla1 is represented by the amino-terminal domain of the human La protein (structure 2VOO from Kotik-Kogan et al. 2008). A combination of La-mediated tRNA chaperone activity and Trm1 modification ensures proper tRNA folding such that pre-tRNAs accumulate and undergo aminoacylation (top), while misfolded pre-tRNAs are often degraded by the nuclear surveillance machinery in the absence of La and Trm1 (bottom). Some misfolded or unmodified pre-tRNAs may escape nuclear surveillance and are exported to the cytoplasm, where they may get aminoacylated and function in translation (bottom).
Beyond the well-established role for La in promoting pre-tRNA folding, maturation, and functionality, an intriguing study demonstrated that La-mediated tRNA chaperone activity also influences miRNA biogenesis. La depletion in human cell lines results in tRNA fragments being loaded into Ago2 (Hasler et al. 2016). La knockdown also leads to an increase in miR-1983, which is derived from the 3′ end of pre-tRNA IleUAU. In the absence of La binding, the complementary 5′ and 3′ ends of pre-tRNA IleUAU form an alternate hairpin structure that is recognized as a miRNA precursor by the nuclear export protein Xpo5. In the event of La binding to pre-tRNA IleUAU, La's tRNA chaperone activity promotes the native tRNA fold over the alternate hairpin conformation. In this sense, La's function as a tRNA chaperone enables it to decrease tRNA fragment entry into the miRNA biogenesis pathway, preventing RISC loading with aberrant small RNAs that may exert negative effects downstream (Hasler et al. 2016).
A long-outstanding question concerning the La protein and other tRNA chaperones revolves around the mechanism of substrate selection, and whether tRNA chaperones preferentially bind misfolded over native substrates to assist their folding. La and the bacterial pseudouridine synthase TruB, which also functions as a tRNA chaperone, indiscriminately bind folded and misfolded tRNAs, suggesting a general mechanism regarding substrate selection by tRNA chaperones (Keffer-Wilkes et al. 2016; Vakiloroayaei et al. 2017). Both studies proposed that chaperones are recruited to tRNA by features relating to their processing state rather than fold. For La, recognition of the polyuridylate-containing trailer on nascent pre-tRNAs enables binding and subsequent chaperone activity. The time between La binding to the pre-tRNA trailer and dissociation following trailer cleavage allows tRNAs to acquire the proper fold through a combination of La-mediated RNA chaperone activity and post-transcriptional modifications that stabilize structure (Fig. 3; Vakiloroayaei et al. 2017). Similarly, the bacterial pseudouridine synthase TruB preferentially binds and modifies unmodified tRNA, which corresponds to a relatively early step in tRNA biogenesis and exhibits similar binding affinity for folded and misfolded substrates (Keffer-Wilkes et al. 2016; Schultz and Kothe 2020). The bacterial tRNA methyltransferase TrmA also exhibits preferences relating to processing state, binding TruB-modified tRNAs with higher affinity than unmodified tRNAs (Schultz and Kothe 2020). Together, data from these three tRNA chaperones suggest a general model wherein tRNA chaperones can sample all tRNAs at a certain point in the maturation pathway. Under conditions where RNA chaperone levels are limiting relative to the level of pre-tRNA substrates (as has been hypothesized for La [Huang et al. 2005]), this suggests that binding of all tRNAs (misfolded or not) by chaperones may limit chaperone folding efficiency, in which a pool of misfolded tRNAs might not have access to chaperone activity as a result of competition with folded substrates (Vakiloroayaei et al. 2017). It is tempting to speculate that this indiscriminatory mechanism of substrate recognition may have evolved as a result of the difficulty or ambiguity of recognizing misfolds due to the relative homogeneity of nucleotides compared to amino acids (Herschlag 1995; Rajkowitsch et al. 2007; Woodson 2010; Vakiloroayaei et al. 2017). It is not yet known whether this is a common mechanism of all tRNA chaperones, and it is therefore anticipated that additional structural studies on the binding mechanisms of other known and speculated tRNA chaperones will provide a more complete picture.
EARLY WORK POSTULATING tRNA AND rRNA MODIFICATION ENZYMES AS RNA CHAPERONES
Unlike the La protein, tRNA modification enzymes often are not end-binding proteins and as such, would not be expected to offer protection from exonucleolytic degradation. Nevertheless, the increase in tRNA degradation observed in yeast and bacteria lacking particular modification enzymes suggests that these enzymes impart structural stability on their substrates, either through the modification itself or an alternate, catalysis-independent function. The finding that catalytically inactive TruB and TrmA variants rescue growth defects in the respective knockout bacterial strains were among the first indications that tRNA modification enzymes may have additional functions as tRNA chaperones (Persson et al. 1992; Gutgsell et al. 2000). Similar to its prokaryotic homolog TrmA, the eukaryotic homolog Trm2 has also been suggested to promote tRNA folding and thus act as a bona fide tRNA chaperone (Johansson and Byström 2002). Although TRM2 is a nonessential gene in budding yeast, Trm2 knockout combined with a mutant allele of tRNA SerCGA results in a temperature-sensitive growth defect that correlates with reduced tRNA SerCGA steady state levels. The stability of certain mutant tRNA SerCGA alleles, including the T51C mutation in the TΨC-loop, can be restored with a catalytically inactive Trm2 mutant, although the partial rescue of the temperature-sensitive growth defect suggests that while Trm2 may have a role in tRNA maturation independent of catalysis, the modification itself is nevertheless important for tRNA functionality (Johansson and Byström 2002). Further support for this hypothesis came from numerous structural studies revealing that the catalytic mechanisms of tRNA modification enzymes often involve enzyme-mediated tRNA unfolding and structural rearrangement (Hoang and Ferré-D'Amaré 2001; Hoang et al. 2006; Alian et al. 2008; Finer-Moore et al. 2015). Together, these ideas lead to a relatively new model linking modification and folding in the tRNA processing pathway.
Similar models for RNA modification enzymes acting as RNA chaperones have come from the study of rRNA modification enzymes (Grosjean 2005; Lafontaine and Tollervey 2014). The budding yeast rRNA methyltransferase Dim1 was among the first rRNA modification enzymes suggested to have alternate functions unrelated to modification. Dim1 depletion inhibits pre-rRNA cleavage to generate 20S pre-rRNA, although replacement of the modifiable adenosines (A1779 and A1780) with unmodifiable guanosines results in normal pre-rRNA processing (Lafontaine et al. 1995). Further support for the idea that Dim1 possesses an alternate function comes from the finding that yeast strains bearing a catalytically inactive Dim1 mutant grow normally, indicating that Dim1, rather than methylation, is required for pre-rRNA processing (Lafontaine et al. 1998). Similar findings have been reported for RluD, a bacterial pseudouridine synthase responsible for modifying 3 nt in helix 69 in 23S rRNA, which makes contacts to the A and P site tRNA, and which are required for effective translation termination (Ejby et al. 2007). In an E. coli K-12 strain, disruption of the RluD gene results in a growth defect which is caused by the interplay with a defective release factor 2 (RF2) carrying an A246T substitution found in all K-12 strains; notably, deletion of RluD has no significant phenotype in strains carrying a wild-type RF2 gene. Interestingly however, the growth defect in E. coli K-12 strains lacking RluD can be rescued by expression of a catalytically inactive RluD mutant suggesting that RluD may fold helix 69 in 23S rRNA independent of modifying it, thereby enabling a normal interaction with RF2 A246T. Thus, RluD may act as an RNA chaperone similarly as had been previously reported for the related tRNA pseudouridine synthase TruB (Gutgsell et al. 2001; O'Connor and Gregory 2011).
Despite the wealth of speculation into alternative functions for rRNA modification enzymes, it was only recently that certain modification enzymes were demonstrated to possess genuine RNA chaperone activity (Keshav et al. 2020). The bacterial methyltransferase RsmC, which catalyzes the formation of m2G at nucleotide 1207 in 16S rRNA, was found to have a role in rRNA folding, particularly relating to formation of helix 34. In accordance with the need to anneal both strands of helix 34 (the 5h34 strand, comprising nucleotides 1047–1065, and the 3h34 strand, which includes nucleotides 1191–1210), RsmC was shown to slightly enhances duplex formation in vitro. Additional experiments using single-molecule FRET showed that RsmC is also capable of denaturing a hairpin in the 3h34 strand, leading to the model that RsmC may use its RNA chaperone activity to decrease the transition state energy between the hairpin and denatured states of the 3h34 strand, thereby smoothing the RNA folding landscape of rRNA maturation and ribosome assembly (Keshav et al. 2020).
tRNA MODIFICATION ENZYMES CAN ACT AS tRNA CHAPERONES
Evidence that tRNA modification enzymes also function as genuine RNA chaperones has very recently come to the forefront. Pseudouridine synthases typically use a base-flipping mechanism to access their target nucleotides for modification. Since these enzymes rely on local structural rearrangement, it is tempting to speculate that these enzymes might also act as tRNA chaperones to assist tRNA folding. TruB, the bacterial pseudouridine synthase responsible for Ψ55 modification, recognizes the preorganized and folded T stem–loop, with nucleotides 55–57 projecting into the active site cleft (Becker et al. 1997; Gu et al. 1998; Hoang and Ferré-D'Amaré 2001; Pan et al. 2003). Insertion of the histidine imidazole group (H43) into the T-loop forces U55 to flip out to avoid steric clashes with side chains carboxy-terminal to the inserted histidine. Due to the resulting disrupted tertiary interactions between the T- and D-loops (Fig. 1B), the bases of nucleotides 56 and 57 also flip out together with U55, positioning all three bases in the active site of the enzyme (Hoang and Ferré-D'Amaré 2001). The lost tertiary interactions of U55 and C56 with G18 and G19 in the D-loop are compensated by protein–RNA contacts between TruB and the bases of nucleotides 56 and 57 (Pan et al. 2003). It has thus been hypothesized that TruB's ability to disrupt tertiary structure may allow it to act as an RNA chaperone by giving misfolded substrates another chance to form correct tertiary interactions (Hoang and Ferré-D'Amaré 2001). A similar base-flipping mechanism has been described for the pseudouridine synthase TruA, which modifies U38, U39, and U40 in the anticodon stem. Instead of side chain insertion like TruB, a conserved arginine residue in TruA (R62) stabilizes the flipped out nucleotide through stacking interactions (Hur and Stroud 2007). An equivalent arginine is involved in the base-flipping mechanism observed for the pseudouridine synthases RluA, which modifies U746 in 23S rRNA and U32 in tRNA, and RsuA, which modifies U516 in 16S rRNA (Hoang et al. 2006).
While the catalytic mechanisms differ between pseudouridine synthases and methyltransferases, bacterial and eukaryotic tRNA methyltransferases also use base-flipping to access target nucleotides. tRNA methylation by the bacterial methyltransferase TrmA, which is responsible for m5U54 modification, requires rearrangement and refolding of the T-arm to position U54 in the active site of the enzyme. Structural rearrangement also entails breaking canonical base-stacking interactions involving A58 such that A58 occupies the newly vacated space left by U54 and becomes part of alternate base-stacking interactions. Biochemical data suggest that additional interactions of phenylalanine 106 and histidine 125 with the D-loop further contribute to the disruption of tertiary base pairs in tRNA upon binding to TrmA (Keffer-Wilkes et al. 2020). Catalysis of m1A58 formation in tRNA LysUUU by the human Trm6–Trm61 heterodimer also involves disruption of hinge region interactions to access A58. Rearrangement of the D-loop by breaking base-pairing interactions between G18 and G19 with U55 and C56, coupled with rearrangement of nucleotides 55–60 in the T-loop, position A58 in the active site of the catalytic subunit Trm61 (Fig. 1B). Much like TruA, hydrogen bond interactions between rearranged nucleotides and the protein compensate for disrupted base-pairing between the D- and T-loops (Finer-Moore et al. 2015).
The extensive findings that tRNA modification enzymes often refold their substrates, as well as studies suggesting a fitness advantage conferred by catalytically inactive mutants converge upon a model wherein tRNA modification enzymes have dual functions in catalysis and tRNA folding. Several elegant studies have expanded upon so-called classical RNA chaperones, characterizing prokaryotic and eukaryotic tRNA modification enzymes as a novel class of tRNA-specific chaperones. Alternate functions of TruB were explored, revealing that a catalytically inactive TruB variant is capable of folding tRNA in vitro (Keffer-Wilkes et al. 2016). In vitro folding activity correlated with the degree of growth rescue in co-culture competition assays; consequently, amino acid residue substitutions abolishing tRNA binding led to decreased folding activity and bacterial fitness, supporting the idea that binding and chaperone activity, rather than modification, is crucial for fitness (Keffer-Wilkes et al. 2016). Rapid-kinetic stopped-flow experiments using tRNA fluorescently labeled at nucleotide 57 revealed that following TruB binding, the tRNA elbow region is quickly and repeatedly unfolded and refolded before pseudouridylation, thereby revealing the molecular mechanism of TruB's tRNA chaperone activity (Hoang and Ferré-D'Amaré 2001; Keffer-Wilkes et al. 2016). The authors also demonstrated that TruB exhibits specific and high affinity for tRNAs, rather than single-stranded or structured RNA substrates, prompting its classification as a tRNA chaperone. It has been suggested that TruB's slow catalytic rate may have evolved as a mechanism to allow for multiple rounds of tRNA unfolding and refolding by TruB to assist tRNA folding into native conformations (Wright et al. 2011; Keffer-Wilkes et al. 2016). Intriguingly, a catalytically inactive mutant of Pus4, the budding yeast homolog of TruB, has a role in inhibiting RNA viruses, and while the mechanism remains unknown, the finding is suggestive of an evolutionarily conserved dual function for TruB (Zhu et al. 2007). Consistent with this, many of the residues identified by structural and biochemical studies as required for tRNA binding (Hoang et al. 2006; Keffer-Wilkes et al. 2016) are conserved among TruB homologs (Fig. 4A). More recently, a catalytic mutant of the human pseudouridine synthase PUS10, which modifies tRNA at positions 54 and 55, was found to have a role in promoting miRNA biogenesis that is thought to be related to miRNA folding, further supporting the idea of catalytic-independent functions of tRNA modification enzymes (Song et al. 2020).
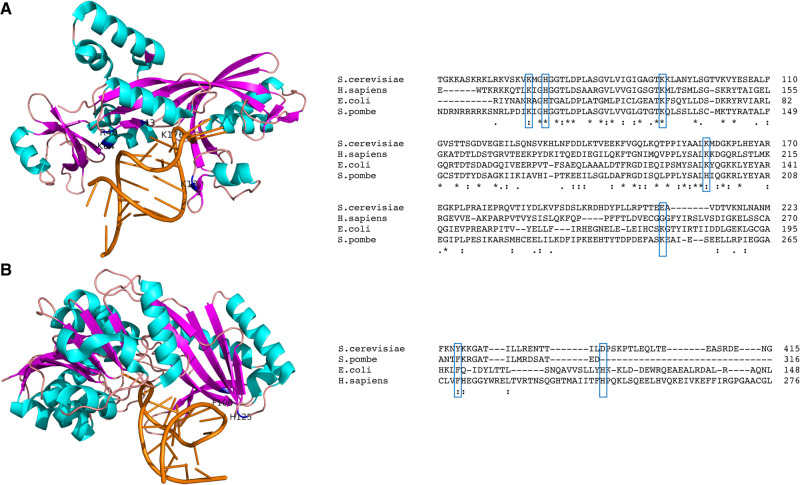
FIGURE 4.
Select tRNA-binding residues are conserved among TruB and TrmA homologs. (A) Sequence alignment (Madeira et al. 2019) and ribbon representation of the high-resolution structure of the bacterial pseudouridine synthase TruB in complex with a T stem–loop RNA (orange) (structure 1K8W from Hoang and Ferré-D'Amaré 2001) and visualized in PyMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC). α-helices are pictured in cyan, β-sheets in pink. Experimentally identified tRNA binding regions (Hoang and Ferré-D'Amaré 2001; Keffer-Wilkes et al. 2016) are indicated in blue. (B) Sequence alignment (Madeira et al. 2019) and ribbon representation of the high-resolution structure of the bacterial methyltransferase TrmA in complex with a T stem–loop RNA (orange) (structure 3BT7 from Alian et al. 2008 and visualized in PyMOL [The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC]). α-helices are pictured in cyan, β-sheets in pink. Experimentally identified tRNA binding regions (Keffer-Wilkes et al. 2020) are indicated in blue.
Additional studies on tRNA methyltransferases raised the possibility that tRNA modification enzymes functioning as tRNA chaperones may be the rule, rather than the exception. Expanding on initial studies demonstrating that the bacterial methyltransferase TrmA is essential for viability but its catalytic activity is not (Persson et al. 1992), phenylalanine and histidine residues (F106 and H125) were identified as critical for tRNA binding (Keffer-Wilkes et al. 2020). Much like TruB, wild-type and catalytically inactive TrmA possess in vitro tRNA folding activity, with substitutions of H125 and F106 showing reduced to no folding activity, respectively. Confirming the importance of tRNA binding in vivo, TrmA knockout strains and knockout strains expressing tRNA binding or catalytic mutants were equally outcompeted by wild-type strains in co-culture competition assays. The implication that an aromatic residue, in particular, is required for tRNA binding and by extension, tRNA chaperone activity, is supported by the high degree of conservation of F106 as an aromatic residue among bacterial and eukaryotic TrmA homologs (Fig. 4B). Integrating previous structural data on TrmA, the authors proposed a model in which F106 and H125 act as wedges to disrupt tertiary interactions in the elbow region through steric clashes with G18 at the interface of the D and T arms, thereby providing tRNA with a second chance at forming correct tertiary interactions (Fig. 1B; Alian et al. 2008; Keffer-Wilkes et al. 2020).
FUTURE PERSPECTIVES AND OUTLOOK
Together, these findings call into question whether RNA chaperone activity could be a more general feature of RNA modification enzymes. This dual activity may be especially critical for structured noncoding RNA like tRNAs, where aminoacylation and subsequent function in translation is fundamentally linked to the ability to assume native conformations (Bhaskaran et al. 2012). While certain tRNA modification enzymes have been demonstrated to promote tRNA folding through a combination of modification and RNA chaperone activity (Fig. 1B), questions have been raised about the relative importance of modifications themselves. It has been proposed that certain modifications may not be very beneficial, but instead serve as a signal for the enzyme to dissociate (Gutgsell et al. 2001; Keffer-Wilkes et al. 2016). In these cases, tRNA would benefit more from RNA chaperone activity than modification. It is also possible that different tRNA species use these dual functions to different extents. Some tRNAs may require one activity over the other, resulting in distinct pools of substrates: a set of modified substrates, and unmodified tRNAs that still bind to the tRNA modifying enzyme, but for the purpose of chaperone activity. Continuing to explore the molecular mechanisms of tRNA chaperone activity and testing additional tRNA modification enzymes for chaperone activity are therefore areas of immediate interest. Structural analysis of tRNA modification enzymes will be instrumental in identifying catalytic residues, residues that contact tRNA, and possible conformational changes induced in the tRNA by the modification enzyme, enabling genetic and biochemical studies to interrogate potential chaperone activity. Moreover, future research should be aimed at assessing the relative contributions of modifications and RNA chaperone activity on tRNA folding. While several recent studies have addressed this question, it remains difficult to uncouple the two activities, as modification is often critical for fitness (Keffer-Wilkes et al. 2020). The development of reporter assays to directly assess tRNA chaperone activity (Porat and Bayfield 2020) (also see Heise 2020) will lead to significant contributions in understanding tRNA chaperones.
Another question of recent interest involves tRNA modification circuits, i.e., the preferential introduction of modifications in a certain order, and how modification circuits impact tRNA structural stability (anticodon modification circuits reviewed in Han and Phizicky 2018). Yeast and bacterial tRNA body modifications, particularly in the elbow region, are installed in a preferential order. Pseudouridylation of nucleotide 55 typically occurs on nascent, unmodified tRNA and once tRNA is modified, Ψ55 promotes subsequent modifications, including methylation at nucleotide 54 (Barraud et al. 2019; Schultz and Kothe 2020). Modification circuits have also been demonstrated for vertebrate tRNAs, with TΨC-loop modifications installed early in tRNA biogenesis, followed by leader and trailer processing, additional body and anticodon modifications, and intron removal (Melton et al. 1980; Nishikura and De Robertis 1981). Such findings may reflect the structural changes arising from modification or potential tRNA chaperone activity, with certain later-acting modification enzymes requiring specific conformations that may be facilitated by earlier-acting modification enzymes at different points in the tRNA maturation pathway. Sure enough, Ψ55 modification by TruB on the bacterial tRNA PheGAA was found to stabilize base-stacking interactions in the elbow region, thereby increasing the affinity of tRNA for the methyltransferase TrmA, which modifies nucleotide 54 (Alian et al. 2008; Schultz and Kothe 2020). Interestingly, the strength and dependence on modification circuits varies depending on the tRNA, with the yeast tRNA PheGAA showing greater perturbations in modification in Pus4 and Trm2 knockout strains than total cellular tRNA, suggesting its increased sensitivity to modification circuits (Barraud et al. 2019). The order of installation of tRNA modifications also likely relies on the subcellular localization of tRNA modification enzymes. Understanding how tRNA modification enzymes act as tRNA chaperones may offer further insight into the mechanisms by which specific tRNA conformations shape body modification circuits.
The role of RNA chaperones in promoting native tRNA folding is an important aspect of tRNA structure and functionality and as such, often interfaces with tRNA decay pathways. As the tRNA chaperone La has been demonstrated to rescue misfolded pre-tRNAs from nuclear surveillance (Calvo et al. 1999; Huang et al. 2006; Vakiloroayaei et al. 2017), it is anticipated that tRNA modification enzymes that also function as tRNA chaperones may have similar roles in preventing tRNA decay. Consistent with this idea, current models state that binding of the La protein and the modification enzymes Trm1, Trm2, Trm3, and Pus4 to mutant or misfolded tRNAs promotes native fold over alternate conformations, with modification enzymes proposed to assume a chaperone-like function distinct from their catalytic activity (Johansson and Byström 2002; Copela et al. 2006). It is interesting to note that tRNA modification enzymes homologous to those that are genetically linked to the RNA chaperone La have been demonstrated or are anticipated to display RNA chaperone activity (Johansson and Byström 2002; Copela et al. 2006; Zhu et al. 2007). One might therefore draw the conclusion that both tRNA stabilization and tRNA chaperone functions are redundant between La and select tRNA modification enzymes. Although the study of RNA chaperones is not new, the finding that tRNA modification enzymes can also function as tRNA chaperones signals a paradigm shift regarding the role of these enzymes in tRNA maturation. Still, chaperone activity is but one facet of tRNA structural stability, with modifications themselves also playing an important role in influencing secondary and tertiary structure. The newfound link between tRNA folding and modification, and how they interface with tRNA quality control pathways, underscores the importance of optimizing structure and stability of tRNAs to ensure their functionality.
ACKNOWLEDGMENTS
The authors thank Rich Maraia for helpful comments on the manuscript. J.P. is supported by a Canada Graduate Scholarship (Doctoral) from the Natural Sciences and Engineering Research Council of Canada. U.K. and M.A.B. are supported by Discovery Grants from NSERC (U.K.: “Mechanism and function of tRNA modification and folding; M.A.B.: “Impact of chemical modification of non-coding RNAs on gene expression in S. pombe”).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078428.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96. 10.1016/j.molcel.2005.10.036 [DOI] [PubMed] [Google Scholar]
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR. 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329. 10.1038/nsmb747 [DOI] [PubMed] [Google Scholar]
- Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. 2008. Structure of a TrmA-RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci 105: 6876–6881. 10.1073/pnas.0802247105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Björk GR, Tamame M, Hinnebusch AG. 1998. The essential Gcd10p-Gcd14p nuclear complex 15 required for 1- methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662. 10.1101/gad.12.23.3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Blewett NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, Maraia RJ. 2015. RNA polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet 11: e1005671. 10.1371/journal.pgen.1005671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW. 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17: 7430–7441. 10.1093/emboj/17.24.7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barhoom S, Kaur J, Cooperman BS, Smorodinsky NI, Smilansky Z, Ehrlich M, Elroy-Stein O. 2011. Quantitative single cell monitoring of protein synthesis at subcellular resolution using fluorescently labeled tRNA. Nucleic Acids Res 39: e129. 10.1093/nar/gkr601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Gato A, Heiss M, Catala M, Kellner S, Tisné C. 2019. Time-resolved NMR monitoring of tRNA maturation. Nat Commun 10: 3373. 10.1038/s41467-019-11356-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield MA, Maraia RJ. 2009. Precursor-product discrimination by La protein during tRNA metabolism. Nat Struct Mol Biol 16: 430–437. 10.1038/nsmb.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayfield MA, Vinayak J, Kerkhofs K, Mansouri-Noori F. 2019. La proteins couple use of sequence-specific and non-specific binding modes to engage RNA substrates. RNA Biol 18: 168–177. 10.1080/15476286.2019.1582955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Motorin Y, Sissler M, Florentz C, Grosjean H. 1997. Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the TΨ-loop of yeast tRNAs. J Mol Biol 274: 505–518. 10.1006/jmbi.1997.1417 [DOI] [PubMed] [Google Scholar]
- Belisova A, Semrad K, Mayer O, Kocian G, Waigmann E, Schroeder R, Steiner G. 2005. RNA chaperone activity of protein components of human Ro RNPs. RNA 11: 1084–1094. 10.1261/rna.7263905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran H, Rodriguez-Hernandez A, Perona JJ. 2012. Kinetics of tRNA folding monitored by aminoacylation. RNA 18: 569–580. 10.1261/rna.030080.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KA, Sharifi S, Hussain R, Donaldson L, Bayfield MA, Wilson DJ. 2016. Distinct dynamic modes enable the engagement of dissimilar ligands in a promiscuous atypical RNA recognition motif. Biochemistry 55: 7141–7150. 10.1021/acs.biochem.6b00995 [DOI] [PubMed] [Google Scholar]
- Calvo O, Cuesta R, Anderson J, Gutiérrez N, García-Barrio MT, Hinnebusch AG, Tamame M. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol 19: 4167–4181. 10.1128/MCB.19.6.4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakshusmathi G, Do Kim S, Rubinson DA, Wolin SL. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J 22: 6562–6572. 10.1093/emboj/cdg625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380. 10.1101/gad.1654308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E. 2009. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461: 60–65. 10.1038/nature08394 [DOI] [PubMed] [Google Scholar]
- Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL. 2006. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA 12: 644–654. 10.1261/rna.2307206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL. 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227. 10.1261/rna.1050408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalluge JJ, Hashizume T, Sopchik AE, Mccloskey JA, Davis DR. 1996. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res 24: 1073–1079. 10.1093/nar/24.6.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026. 10.1093/nar/23.24.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D. 2017. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol Cell Biol 37: MCB.00214-17. 10.1128/MCB.00214-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B. 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J 16: 7184–7195. 10.1093/emboj/16.23.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Davis DR. 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol 285: 115–131. 10.1006/jmbi.1998.2297 [DOI] [PubMed] [Google Scholar]
- Dyubankova N, Sochacka E, Kraszewska K, Nawrot B, Herdewijn P, Lescrinier E. 2015. Contribution of dihydrouridine in folding of the D-arm in tRNA. Org Biomol Chem 13: 4960–4966. 10.1039/C5OB00164A [DOI] [PubMed] [Google Scholar]
- Edqvist J, Ståby KB, Grosjean H. 1995. Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture. Biochimie 77: 54–61. 10.1016/0300-9084(96)88104-1 [DOI] [PubMed] [Google Scholar]
- Ejby M, Sørensen MA, Pedersen S. 2007. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc Natl Acad Sci 104: 19410–19415. 10.1073/pnas.0706558104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer-Moore J, Czudnochowski N, O'Connell JD, Wang AL, Stroud RM. 2015. Crystal structure of the human tRNA m1A58 methyltransferase-tRNA3Lys complex: refolding of substrate tRNA allows access to the methylation target. J Mol Biol 427: 3862–3876. 10.1016/j.jmb.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartland WJ, Sueoka N. 1966. Two interconvertible forms of tryptophanyl sRNA in E. coli. Proc Natl Acad Sci 55: 948–956. 10.1073/pnas.55.4.948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogakos T, Brown M, Garzia A, Meyer C, Hafner M, Tuschl T. 2017. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep 20: 1463–1475. 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547. 10.1128/MCB.24.12.5534-5547.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. 2005. Fine-tuning of RNA functions by modification and editing. Top Curr Genet 12: 442. 10.1007/b95147 [DOI] [Google Scholar]
- Gu X, Yu M, Ivanetich KM, Santi D V. 1998. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry 37: 339–343. 10.1021/bi971590p [DOI] [PubMed] [Google Scholar]
- Gutgsell N, Englund N, Niu L, Kaya Y, Lane BG, Ofengand J. 2000. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 6: 1870–1881. 10.1017/S1355838200001588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutgsell NS, Del Campo M, Raychaudhuri S, Ofengand J. 2001. A second function for pseudouridine synthases: a point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 7: 990–998. 10.1017/S1355838201000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S, Wolin SL, Reinisch KM. 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci 107: 15045–15050. 10.1073/pnas.1003505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Phizicky EM. 2018. A rationale for tRNA modification circuits in the anticodon loop. RNA 24: 1277–1284. 10.1261/rna.067736.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler D, Lehmann G, Murakawa Y, Klironomos F, Jakob L, Grässer FA, Rajewsky N, Landthaler M, Meister G. 2016. The lupus autoantigen La prevents mis-channeling of tRNA fragments into the human microRNA pathway. Mol Cell 63: 110–124. 10.1016/j.molcel.2016.05.026 [DOI] [PubMed] [Google Scholar]
- Heise T, ed. 2020. RNA chaperones: methods and protocols. Springer US, NY. [Google Scholar]
- Helm M, Brulé H, Degoul F, Cepanec C, Leroux JP, Giegé R, Florentz C. 1998. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res 26: 1636–1643. 10.1093/nar/26.7.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Giegé R, Florentz C. 1999. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 38: 13338–13346. 10.1021/bi991061g [DOI] [PubMed] [Google Scholar]
- Herschlag D. 1995. RNA chaperones and the RNA folding problem. J Biol Chem 270: 20871–20874. 10.1074/jbc.270.36.20871 [DOI] [PubMed] [Google Scholar]
- Hoang C, Ferré-D'Amaré AR. 2001. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107: 929–939. 10.1016/S0092-8674(01)00618-3 [DOI] [PubMed] [Google Scholar]
- Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferré-D'Amaré AR. 2006. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol Cell 24: 535–545. 10.1016/j.molcel.2006.09.017 [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. 2003. tRNA transfers to the limelight. Genes Dev 17: 162–180. 10.1101/gad.1049103 [DOI] [PubMed] [Google Scholar]
- Horke S, Reumann K, Schulze C, Grosse F, Heise T. 2004a. The La motif and the RNA recognition motifs of human La autoantigen contribute individually to RNA recognition and subcellular localization. J Biol Chem 279: 50302–50309. 10.1074/jbc.M407504200 [DOI] [PubMed] [Google Scholar]
- Horke S, Reumann K, Schweizer M, Will H, Heise T. 2004b. Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J Biol Chem 279: 26563–26570. 10.1074/jbc.M401017200 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. 2006. RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539. 10.1038/nrm1964 [DOI] [PubMed] [Google Scholar]
- Huang Y, Intine R V, Mozlin A, Hasson S, Maraia RJ. 2005. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol Cell Biol 25: 621–636. 10.1128/MCB.25.2.621-636.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Bayfield MA, Intine R V, Maraia RJ. 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat Struct Mol Biol 13: 611–618. 10.1038/nsmb1110 [DOI] [PubMed] [Google Scholar]
- Hur S, Stroud RM. 2007. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell 26: 189–203. 10.1016/j.molcel.2007.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks A, Babon J, Kelly G, Manolaridis I, Cary PD, Curry S, Conte MR. 2003. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure 11: 833–843. 10.1016/S0969-2126(03)00121-7 [DOI] [PubMed] [Google Scholar]
- Johansson MJO, Byström AS. 2002. Dual function of the tRNA(m5U54) methyltransferase in tRNA maturation. RNA 8: 324–335. 10.1017/S1355838202027851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJO, Byström AS. 2004. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA 10: 712–719. 10.1261/rna.5198204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162. 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240. 10.1101/gad.1183804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521. 10.1261/rna.2305406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keffer-Wilkes LC, Veerareddygari GR, Kothe U. 2016. RNA modification enzyme TruB is a tRNA chaperone. Proc Natl Acad Sci 113: 14306–14311. 10.1073/pnas.1607512113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keffer-Wilkes LC, Soon EF, Kothe U. 2020. The methyltransferase TrmA facilitates tRNA folding through interaction with its RNA-binding domain. Nucleic Acids Res 48: 7981–7990. 10.1093/nar/gkaa548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshav GC, Gyawali P, Balci H, Abeysirigunawardena S. 2020. Ribosomal RNA methyltransferase RsmC moonlights as an RNA chaperone. ChemBioChem 21: 1885–1892. 10.1002/cbic.201900708 [DOI] [PubMed] [Google Scholar]
- Klassen R, Schaffrath R. 2018. Collaboration of tRNA modifications and elongation factor eEF1A in decoding and nonsense suppression. Sci Rep 8: 12749. 10.1038/s41598-018-31158-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelawala L, Grayhack EJ, Phizicky EM. 2008. Identification of yeast tRNA Um44 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA 14: 158–169. 10.1261/rna.811008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S. 2008. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure 16: 852–862. 10.1016/j.str.2008.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EB, Hopper AK. 2013. Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc Natl Acad Sci 110: 21042–21047. 10.1073/pnas.1316579110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera NJ, Hodsdon ME, Wolin SL. 2011. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc Natl Acad Sci 108: 1308–1313. 10.1073/pnas.1017085108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Tollervey D. 2014. Regulatory aspects of rRNA modification and pre-rRNA processing. In Modification and editing of RNA (ed. Grosjean H, Benne R), pp. 281–288. American Society for Microbiology/Wiley, NJ. 10.1128/9781555818296.ch15 [DOI] [Google Scholar]
- Lafontaine D, Vandenhaute J, Tollervey D. 1995. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev 9: 2470–2481. 10.1101/gad.9.20.2470 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Preiss T, Tollervey D. 1998. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol 18: 2360–2370. 10.1128/MCB.18.4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Reimers S, Pandit S, Deutscher MP. 2002. RNA quality control: degradation of defective transfer RNA. EMBO J 21: 1132–1138. 10.1093/emboj/21.5.1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282: 2082–2085. 10.1126/science.282.5396.2082 [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47: W636–W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Francoeur AM. 1984. La antigen recognizes and binds to the 3′-oligouridylate tail of a small RNA. Mol Cell Biol 4: 1134–1140. 10.1128/MCB.4.6.1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. 2007. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res 35: 1257–1269. 10.1093/nar/gkl1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton DA, De Robertis EM, Cortese R. 1980. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature 284: 143–148. 10.1038/284143a0 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466. 10.1016/S0092-8674(00)80432-8 [DOI] [PubMed] [Google Scholar]
- Miyagawa R, Mizuno R, Watanabe K, Ijiri K. 2012. Formation of tRNA granules in the nucleus of heat-induced human cells. Biochem Biophys Res Commun 418: 149–155. 10.1016/j.bbrc.2011.12.150 [DOI] [PubMed] [Google Scholar]
- Naeeni AR, Conte MR, Bayfield MA. 2012. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J Biol Chem 287: 5472–5482. 10.1074/jbc.M111.276071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845. 10.1017/S1355838201002308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. 2002. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci 99: 12697–12702. 10.1073/pnas.202477199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K, De Robertis EM. 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in a spliced transfer RNA. J Mol Biol 145: 405–420. 10.1016/0022-2836(81)90212-6 [DOI] [PubMed] [Google Scholar]
- Nostramo RT, Hopper AK. 2020. A novel assay provides insight into tRNAPhe retrograde nuclear import and re-export in S. cerevisiae. Nucleic Acids Res 48: 11577–11588. 10.1093/nar/gkaa879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M, Gregory ST. 2011. Inactivation of the RluD pseudouridine synthase has minimal effects on growth and ribosome function in wild-type Escherichia coli and Salmonella enterica. J Bacteriol 193: 154–162. 10.1128/JB.00970-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. 2018. Modifications and functional genomics of human transfer RNA. Cell Res 28: 395–404. 10.1038/s41422-018-0013-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Deras ML, Woodson SA. 2000. Fast folding of a ribozyme by stabilizing core interactions: evidence for multiple folding pathways in RNA. J Mol Biol 296: 133–144. 10.1006/jmbi.1999.3439 [DOI] [PubMed] [Google Scholar]
- Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. 2003. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Natl Acad Sci 100: 12648–12653. 10.1073/pnas.2135585100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payea MJ, Sloma MF, Kon Y, Young DL, Guy MP, Zhang X, Zoysa TDE, Fields S, Mathews DH, Phizicky EM. 2018. Widespread temperature sensitivity and tRNA decay due to mutations in a yeast tRNA. RNA 24: 410–422. 10.1261/rna.064642.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payea MJ, Hauke AC, De Zoysa T, Phizicky EM. 2020. Mutations in the anticodon stem of tRNA cause accumulation and Met22-dependent decay of pre-tRNA in yeast. RNA 26: 29–43. 10.1261/rna.073155.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M, Francisco S, Varanda AS, Santos M, Santos MAS, Soares AR. 2018. Impact of tRNA modifications and tRNA-modifying enzymes on proteostasis and human disease. Int J Mol Sci 19: 3738. 10.3390/ijms19123738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson BC, Gustafsson C, Berg DE, Björk GR. 1992. The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc Natl Acad Sci 89: 3995–3998. 10.1073/pnas.89.9.3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat J, Bayfield MA. 2020. Use of tRNA-mediated suppression to assess RNA chaperone function. In RNA chaperones: methods and protocols (ed. Heise T), pp. 107–120. Springer US, New York. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Bläsi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4: 118–130. 10.4161/rna.4.3.5445 [DOI] [PubMed] [Google Scholar]
- Reddy R, Henning D, Tan E, Busch H. 1983. Identification of a La protein binding site in a RNA polymerase III transcript (4.5 I RNA). J Biol Chem 258: 8352–8356. 10.1016/S0021-9258(20)82072-9 [DOI] [PubMed] [Google Scholar]
- Reinisch KM, Wolin SL. 2007. Emerging themes in non-coding RNA quality control. Curr Opin Struct Biol 17: 209–214. 10.1016/j.sbi.2007.03.012 [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. 2009. DOG 1.0: illustrator of protein domain structures. Cell Res 19: 271–273. 10.1038/cr.2009.6 [DOI] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. 1982. Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La Lupus antibodies. Cell 29: 149–159. 10.1016/0092-8674(82)90099-X [DOI] [PubMed] [Google Scholar]
- Santiago-Frangos A, Woodson SA. 2018. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA 9: e1475. 10.1002/wrna.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Azad AK, Hopper AK. 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci 96: 14366–14371. 10.1073/pnas.96.25.14366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz SK-L, Kothe U. 2020. tRNA elbow modifications affect the tRNA pseudouridine synthase TruB and the methyltransferase TrmA. RNA 26: 1131–1142. 10.1261/rna.075473.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenzer H, Jühling F, Chu A, Pallett LJ, Baumert TF, Maini M, Fassati A. 2019. Oxidative stress triggers selective tRNA retrograde transport in human cells during the integrated stress response. Cell Rep 26: 3416–3428.e5. 10.1016/j.celrep.2019.02.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. 1998. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279: 1940–1943. 10.1126/science.279.5358.1940 [DOI] [PubMed] [Google Scholar]
- Seelam PP, Sharma P, Mitra A. 2017. Structural landscape of base pairs containing post-transcriptional modifications in RNA. RNA 23: 847–859. 10.1261/rna.060749.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar S, McDonald J, Cuyugan L, Aldrich J, Kurdoglu A, Adkins J, Serrano G, Beach TG, Craig DW, Valla J, et al. 2015. Alzheimer's disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging 36: 583–591. 10.1016/j.neurobiolaging.2014.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrad K. 2011. Proteins with RNA chaperone activity: a world of diverse proteins with a common task-impediment of RNA misfolding. Biochem Res Int 2011: 532908. 10.1155/2011/532908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta R, Vainauskas S, Yarian C, Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM, Agris PF. 2000. Modified constructs of the tRNA TΨC domain to probe substrate conformational requirements of m1A58 and m5U54 tRNA methyltransferases. Nucleic Acids Res 28: 1374–1380. 10.1093/nar/28.6.1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebrov V, Clarke RJ, Gross HJ, Kisselev L. 2001. Mg2+-induced tRNA folding. Biochemistry 40: 6688–6698. 10.1021/bi002241p [DOI] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK. 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295. 10.1073/pnas.0503836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. 2007. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci 104: 8845–8850. 10.1073/pnas.0700765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, Alkuraya FS. 2016. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet 135: 707–713. 10.1007/s00439-016-1665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek E, Grzechnik P, Späth B, Marchfelder A, Kufel J. 2014. TRNA 3′ processing in yeast involves tRNase Z, Rex1, and Rrp6. RNA 20: 115–130. 10.1261/rna.041467.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane KD, Bavi RS, Sambhare SB, Fandilolu PM. 2016. Comparative structural dynamics of tRNAPhe with respect to hinge region methylated guanosine: a computational approach. Cell Biochem Biophys 74: 157–173. 10.1007/s12013-016-0731-z [DOI] [PubMed] [Google Scholar]
- Song J, Zhuang Y, Zhu C, Meng H, Lu B, Xie B, Peng J, Li M, Yi C. 2020. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol 16: 160–169. 10.1038/s41589-019-0420-5 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 15: r53–r188. 10.1093/nar/15.suppl.r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154. 10.1016/0092-8674(84)90083-7 [DOI] [PubMed] [Google Scholar]
- Steinberg S, Cedergren R. 1995. A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. RNA 1: 886–891. [PMC free article] [PubMed] [Google Scholar]
- Suck D, Saenger W. 1973. The crystal and molecular structure of O2,2′-cyclouridine. Influence of O(2)–C(2′) cyclization on the sugar conformation of pyrimidine nucleosides. Acta Crystallogr B Struct Crystallogr Cryst Chem 29: 1323–1330. 10.1107/S0567740873004449 [DOI] [Google Scholar]
- Sundaralingam M, Rao ST, Abola J. 1971. Molecular conformation of dihydrouridine: puckered base nucleoside of transfer RNA. Science 172: 725–727. 10.1126/science.172.3984.725 [DOI] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T. 2005. Molecular biology: tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142. 10.1126/science.1113346 [DOI] [PubMed] [Google Scholar]
- Takano A, Kajita T, Mochizuki M, Endo T, Yoshihisa T. 2015. Cytosolic Hsp70 and co-chaperones constitute a novel system for tRNA import into the nucleus. Elife 4: e04659. 10.7554/eLife.04659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ. 2006. Structural basis for recognition and sequestration of UUUOH 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell 21: 75–85. 10.1016/j.molcel.2005.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Csermely P. 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 18: 1169–1175. 10.1096/fj.04-1584rev [DOI] [PubMed] [Google Scholar]
- Tuorto F, Lyko F. 2016. Genome recoding by tRNA modifications. Open Biol 6: 160287. 10.1098/rsob.160287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- Urbonavičius J, Durand JMB, Björk GR. 2002. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J Bacteriol 184: 5348–5357. 10.1128/JB.184.19.5348-5357.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakiloroayaei A, Shah NS, Oeffinger M, Bayfield MA. 2017. The RNA chaperone La promotes pre-tRNA maturation via indiscriminate binding of both native and misfolded targets. Nucleic Acids Res 45: 11341–11355. 10.1093/nar/gkx764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. 2003. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906. 10.1038/nsb1003 [DOI] [PubMed] [Google Scholar]
- Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL. 1997. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3: 1434–1443. [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Miyagawa R, Tomikawa C, Mizuno R, Takahashi A, Hori H, Ijiri K. 2013. Degradation of initiator tRNAMet by Xrn1/2 via its accumulation in the nucleus of heat-treated HeLa cells. Nucleic Acids Res 41: 4671–4685. 10.1093/nar/gkt153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Dumas P, Moras D. 1988. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals. Acta Crystallogr A 44: 112–123. 10.1107/S010876738700446X [DOI] [PubMed] [Google Scholar]
- Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev 25: 1173–1184. 10.1101/gad.2050711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK. 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686. 10.1091/mbc.e07-01-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA. 2010. Taming free energy landscapes with RNA chaperones. RNA Biol 7: 677–686. 10.4161/rna.7.6.13615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JR, Keffer-Wilkes LC, Dobing SR, Kothe U. 2011. Pre-steady-state kinetic analysis of the three Escherichia coli pseudouridine synthases TruB, TruA, and RluA reveals uniformly slow catalysis. RNA 17: 2074–2084. 10.1261/rna.2905811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. 10.1016/j.cell.2005.04.030 [DOI] [PubMed] [Google Scholar]
- Xing F, Martzen MR, Phizicky EM. 2002. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA 8: 370–381. 10.1017/S1355838202029825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Vanommeslaeghe K, Aleksandrov A, MacKerell AD, Nilsson L. 2016. Additive CHARMM force field for naturally occurring modified ribonucleotides. J Comput Chem 37: 896–912. 10.1002/jcc.24307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhou Y, Byström AS, Johansson MJO. 2018. Identification of factors that promote biogenesis of tRNACGASer. RNA Biol 15: 1286–1294. 10.1080/15476286.2018.1526539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. 1994. La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol Cell Biol 14: 5412–5424. 10.1128/MCB.14.8.5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402. 10.1016/S0092-8674(00)80220-2 [DOI] [PubMed] [Google Scholar]
- Zarrinkar PP, Wang J, Williamson JR. 1996. Slow folding kinetics of RNase P RNA. RNA 2: 564–573. [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Lentini JM, Prevost CT, Hashem MO, Alkuraya FS, Fu D. 2020. An intellectual disability-associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity. Hum Mutat 41: 600–607. 10.1002/humu.23976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Gopinath K, Murali A, Yi G, Hayward SD, Zhu H, Kao C. 2007. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci 104: 3129–3134. 10.1073/pnas.0611617104 [DOI] [PMC free article] [PubMed] [Google Scholar]