Abstract
Purpose
Compared with their European American (EA) counterparts, African American (AA) women are more likely to die from breast cancer in the United States. This disparity is greatest in hormone receptor–positive subtypes. Here we uncover biological factors underlying this disparity by comparing functional expression and prognostic significance of master transcriptional regulators of luminal differentiation.
Experimental Design
Data and biospecimens from 262 AA and 293 EA patients diagnosed with breast cancer from 2001 to 2010 at a major medical center were analyzed by IHC for functional biomarkers of luminal differentiation, including estrogen receptor (ESR1) and its pioneer factors, FOXA1 and GATA3. Integrated comparison of protein levels with network-level gene expression analysis uncovered predictive correlations with race and survival.
Results
Univariate or multivariate HRs for overall survival, estimated from digital IHC scoring of nuclear antigen, show distinct differences in the magnitude and significance of these biomarkers to predict survival based on race: ESR1 [EA HR = 0.47; 95% confidence interval (CI), 0.31–0.72 and AA HR = 0.77; 95% CI, 0.48–1.18]; FOXA1 (EA HR = 0.38; 95% CI, 0.23–0.63 and AA HR = 0.53; 95% CI, 0.31–0.88), and GATA3 (EA HR = 0.36; 95% CI, 0.23–0.56; AA HR = 0.57; CI, 0.56–1.4). In addition, we identify genes in the downstream regulons of these biomarkers highly correlated with race and survival.
Conclusions
Even within clinically homogeneous tumor groups, regulatory networks that drive mammary luminal differentiation reveal race-specific differences in their association with clinical outcome. Understanding these biomarkers and their downstream regulons will elucidate the intrinsic mechanisms that drive racial disparities in breast cancer survival.
Introduction
The incidence of invasive breast cancer in the United States will approach 260,000 this year with over 40,000 annual deaths. Although overall breast cancer mortality has declined, the survival gap between African American (AA) and European American (EA) women continues to widen (1–9). Women of African heritage suffer higher frequencies of triple-negative breast cancer (TNBC), a more aggressive form of breast cancer characterized by the absence of the estrogen receptor (ER), the progesterone receptor (PR), and nonamplified expression of the HER2 (10–12). Though recent studies have identified genetic components associated with African heritage that is linked to the higher frequency of TNBC (13), other studies have also shown significant race-based disparities in patients with hormone receptor–positive breast cancer (2, 3, 14). These differences persist even after controlling for socioeconomic status (2, 3, 15–17), thus implicating roles for intrinsic biological factors.
The transcriptional program driven by ER plays a major role in mammary biology. Throughout the menstrual and reproductive cycles, its activity and levels regulate dynamic shifts in glandular proliferation and differentiation and play definitive roles during lactation and mammary gland involution (18, 19). Once bound to ligand, ER orchestrates major changes in chromatin structure that facilitate entry and assembly of large multicomponent transcriptional complexes charged with executing cell-specific gene expression programs that influence tumor growth and initiation (18, 19). This action provides the theoretical foundation for many endocrine-based therapeutic strategies (20, 21).
FOXA1 and GATA3 are sequence-specific DNA-binding transcription factors that function as chromatin pioneer factors essential for ER function (22–26). As pioneer transcription factors, they interact directly with histones to facilitate nucleosome displacement, chromatin remodeling, and the subsequent entry or binding of ER (22, 24, 27). Both factors play a significant role in sustaining the estrogen response because they are both induced and reciprocally activated by ER (26, 28, 29). FOXA1 and GATA3 play unique and overlapping roles in maintaining epithelial differentiation by activating genes responsible for luminal features while repressing genes associated with basal or mesenchymal phenotypes (26, 30–32). Unlike FOXA1, GATA3 is frequently altered (∼10%) in breast cancer often with mutations limited to one allele suggesting a gain of function (22). However, many known breast cancer-associated gene variants occur at genetic loci containing FOXA1 binding sites (33). Interestingly, AA women show parity-associated reductions in FOXA1 expression because of promoter methylation (34), although, in contrast, FOXA1 promoter methylation is reduced by BRCA1, whose transcription is controlled by ER (ESR1; ref. 35). These diverse interdependent modes of regulatory function and control exemplify how ESR1, FOXA1, and GATA3 act as master regulators to exert profound influence on breast cancer differentiation, prognosis, and response to therapy.
In this study, we explore the racial differences in the relationship between the protein expression of the ER, FOXA1, and GATA3 master regulators and overall breast cancer survival. Moreover, we identify intrinsic differences in the downstream transcriptional regulatory activity they govern to reveal new and novel gene classes that are predictive of race and 3-year survival.
Materials and Methods
Study population, tissue microarray construction, and analysis
Following IRB approval from East Carolina University and the NIH intramural research program, de-identified formalin-fixed and paraffin-embedded tissue samples and de-identified clinical information abstracted from the medical records were requisitioned and initially procured for 733 patients with breast cancer who underwent surgery for stage 0 to stage IV breast cancer between 2001 and 2010 at Pitt County Memorial Hospital (now Vidant Medical Center), Greenville, NC. All patient samples and data obtained were de-identified and approved by the East Carolina University Institutional Review Board as a human subject exempt project, for which no informed consent is needed. The study was conducted in accordance with the Declaration of Helsinki. Race and/or ethnicity were self-reported at the initial visit and captured in the medical record. Survival was recorded retrospectively from the medical records and the cancer registry. Median follow-up is 8.5 years. A total of 588 patient blocks from this cohort were found suitable for use in the construction of a tissue microarray. Replicate tissue microarrays were constructed using 1 mm cores in accordance with previously described methods (36, 37), with a complete representation of 555 patients. Detailed methods for IHC, scoring, and the assignment of clinical variables are provided in the supplemental data.
Gene expression profiling
Analysis of a portion of the breast cancer samples (Total N = 126; EA N = 61; AA N = 65) was carried out by RNA-seq. Following a review of H&E-stained slides, areas for tumor with >80% nuclei were circled, and 2.5 × 2 to 3 mm tissue cores were extracted from the corresponding regions of FFPE tissue blocks. Cores were shipped to the Beijing Genomics Institute (BGI; Beijing, China), where RNA was extracted and sequenced (60M paired-end reads per sample) as described previously (38, 39). Detailed methods for sequencing and description of the analytical pipeline is provided in the Supplementary Data.
Statistical analysis
A linear model estimating outcomes for overall survival, 3-year survival, 5-year survival, and race was applied to measure differences in the association of the digital score of nuclear proteins (OR, confidence interval, and P value) while controlling for clinical factors including age, stage, grade, subtype, and lymph node status (40). A comparison of IHC scoring was performed by the two-sided t test and plotted as described previously (41). Multivariate Cox proportional-hazards model was used to test the independent and combined prognostic values of proteins of interest with/without the presence of selected clinical variables. A Spearman rank correlation was performed to test the relationship between protein H-score and gene expression (RPKM) values (42). The significance of individual hazard ratios was estimated by Wald test. Unsupervised hierarchical clustering of digital IHC protein data from all breast samples was performed using complete linkage and distance correlations with bootstrap resampling and estimated stability of clustering using the “pvclust” R package (43). Optimal cutoff points for H-score were determined as described previously (44). Prediction ability for race and 3-year survival by the regulon genes downstream of master regulators was determined univariately by AUC ROC (45). To define genes that optimized prediction (AUC), genes were added one by one, according to their ranking (univariate, high to low), to the logistic model in Monte Carlo simulations. Protein interaction networks were generated with STRING using the minimum required interaction score of 0.15 (46). Detailed statistical methods are provided in the Supplementary Data. R/Bioconductor version 3.5.1 was used for the entire analysis.
Results
Racial differences in survival outcome of ER-positive versus ER-negative breast cancer
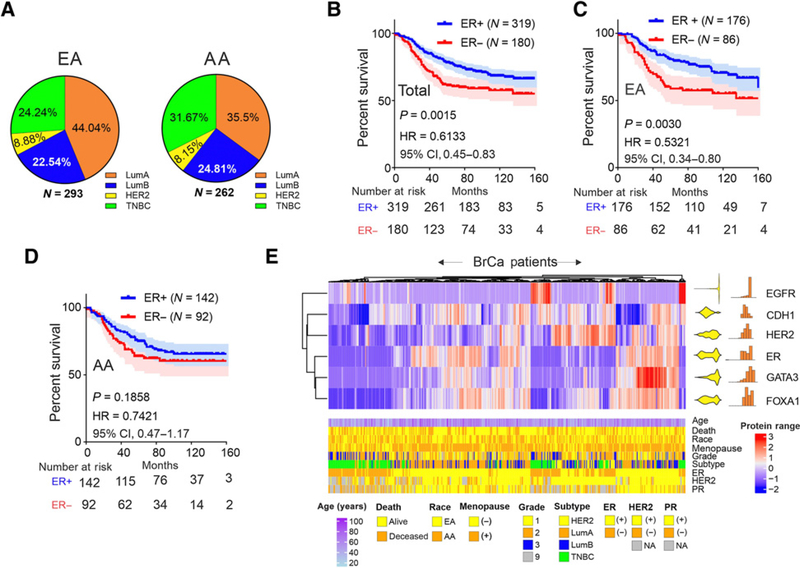
The breast cancer cohort profiled in this study is racially diverse (53% European, N = 293; 47% African, N = 262; Fig. 1A). Correlation between race, clinical, and pathologic characteristics are provided in Table 1. As reported in prior studies, Luminal A subtype frequency is lower in AA compared with EA women, whereas the frequency of TNBC is higher in women of African heritage (Fig. 1A; Table 1). This trend is consistent with those reported by other larger studies in the United States (10, 47, 48) and is representative of the subtype distribution in the parent population in the East Carolina cancer registry (Supplementary Fig. S1). Kaplan–Meier analysis of overall survival associated with ER status confirm the know survival advantage for ER positive (ER+) compared with ER negative (ER−) patients with breast cancer (Fig. 1B). However, this receptor positive survival advantage differs significantly by race, that is, ER+ EA women show much more favorable survival than their AA counterparts (Fig. 1C and D).
Figure 1.
Racial differences in the association of ER expression and survival in a diverse breast cancer cohort. A, Subtype distribution of EA (N = 292) and AA (N = 260) patients with breast cancer. B, Kaplan–Meier analysis of overall survival comparing ER+ to ER− patients with breast cancer. C, Survival profiling of ER+ versus ER− breast cancers in EA patients. D, Survival profiling of ER+ versus ER− AA patients with breast cancer. E, Hierarchical clustering of quantitative IHC expression (H-score) of EGFR, E-cadherin (Ecad), HER2, ER, GATA3, and FOXA1 with data distribution (right, yellow) and histogram (right, red). The scale bar represents the color distribution of a range of protein values. (underneath) Patient demographics and tumor characteristics. Color coding is indicated.
Table 1.
Univariate comparison of patient’s clinical characteristics with respect to race by fitting to linear model (continuous variable) or logistic model (categorical variable).
| Variable | Total sample | EA (N = 293) | AA (N = 262) | HR (95% CI) | P value |
|---|---|---|---|---|---|
| Age (median) | 555 | 60.34 | 56.4 | 0.9947 (0.9917–0.9977) | 0.00065 |
| Menopause status | |||||
| Premenopause (age <50) | 150 | 66 (12%) | 84 (15%) | 1 | |
| Postmenopause (age ≥50) | 405 | 227 (41%) | 178 (32%) | 1.1281 (1.0275–1.2384) | 0.0115 |
| Grade | |||||
| Low | 147 | 85 (15%) | 62 (11%) | 1 | |
| Moderate | 258 | 135 (25%) | 123 (22%) | 1.0565 (0.9547–1.1692) | 0.2869 |
| High | 88 | 40 (7%) | 48 (9%) | 1.1317 (0.9916–1.2916) | 0.0665 |
| NA | 62 | 33 (6%) | 29 (5%) | ||
| Stage | |||||
| 0 | 56 | 31 (6%) | 25 (5%) | 1 | |
| 1 | 184 | 116 (21%) | 68 (12%) | 0.9260 (0.7985–1.0739) | 0.3085 |
| 2 | 185 | 91 (16%) | 94 (17%) | 1.0636 (0.9173–1.2333) | 0.4134 |
| 3 | 74 | 28 (5%) | 46 (8%) | 1.1915 (1.0033–1.4149) | 0.0458 |
| 4 | 38 | 19 (3%) | 19 (3%) | 1.055 (0.8604–1.2938) | 0.6061 |
| NA | 18 | 8 (2%) | 10 (2%) | ||
| Node | |||||
| LN− | 308 | 178 (32%) | 130 (23%) | 1 | |
| LN+ | 201 | 92 (17%) | 109 (20%) | 1.1277 (1.0322–1.2320) | 0.0078 |
| NA | 46 | 23 (4%) | 23 (4%) |
Note: Percentages (%) provided indicate percent of total sample (N = 555) for each variable. HRs are presented with EA patients (presumed from self-reporting) as the referent. Continuous variable = age; unit = years. All other variables are categorical. NA, not available; LN, lymph node. HRs for clinical variables are calculated on the basis of racial differentiation (i.e., EA vs. AA) for each corresponding variable.
Coexpression analysis of ER and other biomarkers that distinguish luminal versus mesenchymal differentiation (FOXA1, GATA3, E-cadherin, HER2, vs. EGFR) reveals significant biphasic correlations between ER expression and its pioneer factors (FOXA1 and GATA3; Fig. 1E).The biphasic nature of the distribution of ER, FOXA1, and, to a lesser extent GATA3, is consistent with the clustering by receptor status abstracted from the medical records, older age, menopausal status, and intrinsic subtype (also see, Table 2). Within the multivariate setting, overall survival is independently associated with age and subtype (Table 2). As has been described for the ER+ classification, LumA subtype when compared with TNBC is associated with a favorable survival (Table 2; Supplementary Fig. S2). However, consistent with the differential racial association of ER status with overall survival, the relative hazard of LumA subtype decreases for EA women whereas it increases for AA women (Supplementary Fig. S2). Comparison of relative Luminal A breast cancer survival between AA and EA women shows a nonsignificant trend toward lower survival in women of African heritage with negligible difference in survival for TNBC (Supplementary Fig. S3).
Table 2.
Univariate and multivariate correlation of patient clinicopathologic characteristics with overall survival by Cox regression analysis.
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | 1.013 | (1.001–1.03) | 0.03 | 1.02 | (1.01–1.04) | 0.002 |
| Race | ||||||
| European | 1 | |||||
| African | 1.06 | (0.77–1.4) | 0.73 | 1.17 | (0.88–1.57) | 0.29 |
| Menopause status | ||||||
| Postmenopause | 1 | |||||
| Premenopause | 0.8 | (0.56–1.2) | 0.24 | 1.18 | (0.72–1.96) | 0.51 |
| Subtype | ||||||
| Lum A | 1 | |||||
| Lum B | 1.775 | (1.174–2.684) | 0.00653 | 1.743 | (1.1518–2.636) | 0.0009 |
| HER2+ | 2.06 | (1.15–3.691) | 0.01518 | 2.145 | (1.1955–3.847) | 0.011 |
| TNBC | 3.253 | (2.264–4.676) | 1.81E–10 | 3.552 | (2.4596–5.128) | 1.37E–11 |
Note: CI is given for overall survival. Multivariate analysis controlled for age, race, menopause status, and subtype, respectively. Criteria for subtype assignments are provided in the Supplementary Materials and Methods. Race referent, EA (presumed from self-reporting); menopause referent, postmenopause; subtype referent, Lum A.
The association between master regulators of luminal differentiation and overall survival in patients with breast cancer differs by race
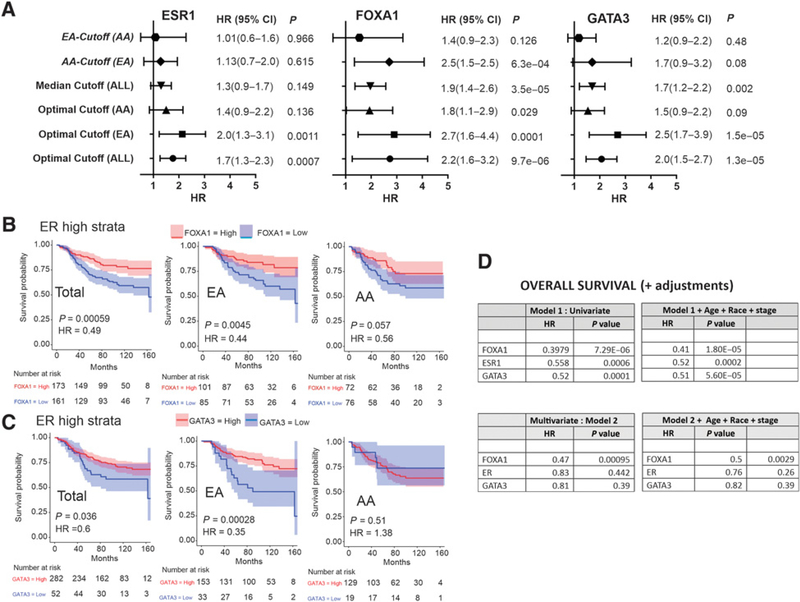
To evaluate the independent predictive value of ER, and the pioneer proteins FOXA1 and GATA3, IHC scores and overall survival outcomes were compared across the cohort before and after stratification by race (Supplementary Fig. S2). Optimum cutoffs for ESR1, FOXA1, and GATA3 histologic scores were defined by exact distribution of maximally selected rank statistic. Using the population cutoff score for each antigen, Kaplan–Meier analysis of the total cohort before and after stratification by race is shown in Fig. 2A–C. For all biomarkers, including ESR1, FOXA1, and GATA3, application of the optimized cutoff is predictive of favorable survival in the total cohort population. However, these predictive values show significantly less favorable or nonsignificant HRs in AA compared with EA women (Fig. 2A–C). Notably, this difference in survival exists despite the absence of any significant racial difference in the levels of either ESR1, FOXA1, GATA3, or the other biomarkers associated with luminal differentiation (CDH1, EGF, HER2; Supplementary Fig. S4). Such observations strongly implicate influences downstream of ESR1, FOXA1, and GATA3 as possible contributors to the racial difference in survival outcome.
Figure 2.
Racial differences in the association of expression of master regulators of luminal differentiation with survival. Kaplan–Meier analysis of the association of ER expression (A), FOXA1 expression (B) confirms GATA3 expression (C) with survival in the total population (left), EA patients (center) and AA patients (right). D, Determination of maximally selected rank statistic to define optimal H-score cutoff (black dashed line) for ESR1, FOXA1, and GATA3, analyzed for the total and race-stratified cohort. Blue points represent protein expression values below optimum cutoff, and red points represent protein values above the optimum cutoff.
To examine whether or not race-specific cutoff for these biomarkers might influence their predictive value, the optimal cutoff for ESR1, FOXA1, and GATA3 were again defined by determining the exact distribution of the maximally selected rank statistic for these antigens separately for EA and AA patients (Fig. 2D). For both ESR1 and FOXA1, the maximally selected cutoff for AA patients is higher than either those of EA or the total population (Fig. 2D, top). In contrast, GATA3, one of the most highly mutated genes in breast cancer with higher frequencies in American women (49), showed an optimal cutoff, in AA patients that is significantly lower than EA women or the total population (Fig. 2D, bottom).
A comparison of race-based biomarker cutoffs
Comparative analysis of the predictive value of race-based cutoffs for ESR1, FOXA1, or GATA3 expression, across the total breast cancer cohort, reveals that the cutoff for AA patients is considerably less predictive or nonsignificant in determining favorable overall survival (Fig. 3A). In each instance, either the total population optimized cutoff, or the cutoff optimized in the European population has the highest predictive discrimination within the entire breast cancer cohort. This relationship persists even when the race-optimized cutoffs are applied across races [e.g., EA-Cutoff (AA), Fig. 3A]. Although the influence of other nonbiological factors that operate differently by race cannot be excluded (e.g., access to care, time of treatment, and type of treatment); such findings suggest that these master regulators of luminal differentiation, may either be functionally less efficient or have reduced transcriptional activity in the downstream regulatory pathways in AA patients.
Figure 3.
ER, FOXA1, and GATA3 have different predictive values for overall survival based on race. A, Forest plot of HR of overall breast cancer survival using median and population optimized cutoff H-scores (ALL, EA, and AA) for ESR1, FOXA1, and GATA3 expression. B, Association between FOXA1 expression and survival in low-risk, high-ER-expressing patient with breast cancer, comparing the total population (left) with EA patients (center) and AA patients (right). C, Association between GATA3 expression and survival in low-risk, high-ER-expressing patients with breast cancer, comparing the total population (left) with EA patients (center) and AA patients (right). D, Univariate and multivariate logistic regression models of overall breast cancer survival based on FOXA1, ESR1, and GATA3 expression adjusted for age, race, and stage. 95% CIs are shown in parentheses.
To determine the relative contribution of the pioneer proteins FOXA1 and GATA3 as established modulators of ER function in predicting overall survival, we compared how expression of FOXA1 or GATA3 stratified the relative hazard of low-risk patients defined by high ER expression. Patients with high ESR1 expression, based on the population optimized cutoff (Fig. 2A), were analyzed for overall survival using each of the optimized cutoff expression values derived from the total cohort, the EA, or the AA patients, respectively (Fig. 3B and C). Within both the total patient cohort and EA patients, expression of either FOXA1 or GATA3 stratifies poor from favorable survival in patients with high ER levels (Fig. 3B and C; left and middle). In contrast, neither FOXA1 nor GATA3 expression provides significant prediction of survival in AA patients (Fig. 3B and C; right).
Univariate modeling demonstrates that FOXA1 measurements significantly outperform both ER and GATA3 as predictors of favorable overall breast cancer survival (Fig. 3D, top left). This relationship persists even after adjusting for age, race, and stage in multivariate analysis (Fig. 3D, top right). Notably, multivariate models adjusting for expression of the other two master regulators, reveal that only FOXA1 is an independent predictor of overall breast cancer survival controlling for either age, race, stage, or the expression of either GATA3 or ESR1 (Fig. 3D, bottom right).
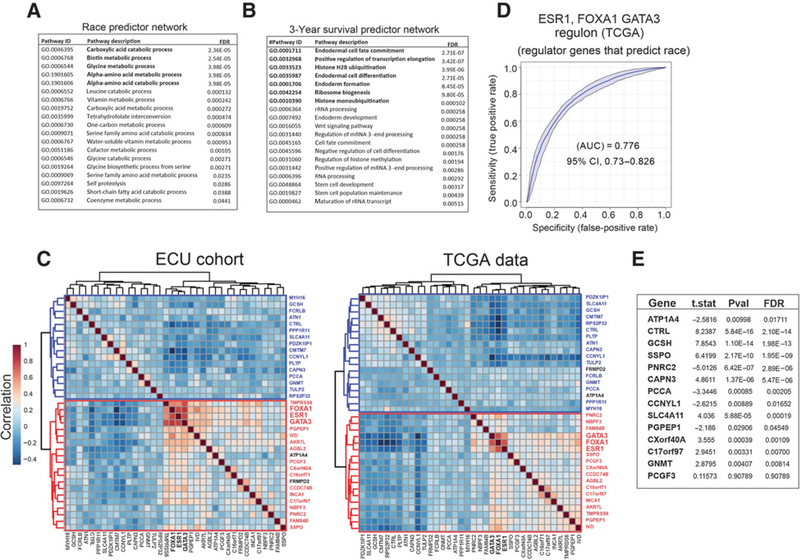
The racial disparity in the association of luminal master regulator expression with breast cancer survival implicates altered activity of downstream transcriptional networks as a source of differences in tumor biology. Recent advances in systems level understanding of transcriptional regulation have developed powerful approaches to define and measure the total transcriptional function and/or “activity” of specific transcription factors by collectively assessing expression of the network of their downstream regulatory targets or “regulons” (50). Computational recognition and construction of these gene networks are available from the collective analysis of publicly available gene expression data sets (50, 51). Using the Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNe; ref. 52) and publicly available human breast cancer gene expression data sets provided through TCGA, Walsh and colleagues defined the regulons controlled by ESR1, FOXA1, and GATA3 (see additional data, ref. 50). The RNA-seq gene expression data for 22% of this cohort (deceased patients, N = 126) was used to uncover genes, controlled by ESR1, FOXA1, or GATA3, which either distinguish race or predict 3-year survival (Fig. 4A and B). Using logistic probability distribution modeling through Monte Carlo simulations, each gene in the regulons of ESR1 (985 genes), FOXA1 (1478 genes), and GATA3 (871 genes; see supplementary material) were combinatorially profiled for their ability to contribute to the prediction of either race or 3-year survival. Optimum predictive value was assessed through AUC determinations based on ROC analysis (Fig. 4A and B). This method identified eleven (11) genes in the ESR1 regulon that contributed to distinguishing race, and eight (8) genes that predicted 3-year survival. Sixteen (16) genes were identified in the FOXA1 regulon that distinguished race and 11 (11) genes that predicted 3-year survival. Finally, in the GATA3 regulon, 12 (12) genes were identified as discriminators of race whereas 12 (12) genes were found to predict 3-year survival (Fig. 4A and B; Supplementary Table S1). Notably, many of these genes are significantly associated with relapse-free survival (RFS) in independent gene expression data sets (Fig. 4C and D; Supplementary Table S2). On the basis of an analysis of known/predicted, direct or functional gene–gene interactions defined within the String database, the linkages of ESR1, FOXA1, and GATA3, the regulon gene groups (race and 3-year survival, respectively) could be assembled into two distinct networks anchored by the ESR1, FOXA1, and GATA3 regulatory triad (Fig 4E and F). The functional cellular processes significantly enriched by inclusion of first-degree interactions of these networks includes multiple metabolic processes involving amino acid, vitamin, and one carbon metabolism (race predictive network; Fig. 5A; Supplementary Table S3); and multiple pathways linked to tissue and cellular differentiation, Wnt signaling, and chromatin modifications (3-year survival predictive network; Fig. 5B; Supplementary Table S3). The gene expression correlation matrix (spearman) of the racial and survival predictors shows strong similarities (discordance in only 2 genes) in clustering of the master regulatory triad expression data in both the ECU patient cohort and the TCGA data set (Fig. 5C). Finally, in validation studies, the ROC analysis of ECU racial predictor genes shows strong agreement with the TCGA data (Fig. 5D and E).
Figure 4.
ROC analysis with a set of optimized genes from ESR1, FOXA1, and GATA3 regulons that, combined, have the highest prediction (AUC) of race (A) or 3-year survival (B) determined by logistic regression. C and D, Volcano plots profiling the association of regulon genes that predict race (C) and 3-year survival (D) with RFS in publicly available breast cancer gene expression data sets (see Supplementary Data). Y-axis, –log P value; X-axis, coefficient of log-scale hazard; green, genes enriched in EA; red, gene enriched in AA. E and F, Regulon genes and gene networks downstream of master regulators of luminal differentiation (ESR1, FOXA1, and GATA3) that optimally predict race and 3-year survival. Lines indicate direct or regulatory interactions. The thickness of the lines indicates the relative strength of the interactions.
Figure 5.
Functional cellular processes that are significantly enriched in first-degree interaction networks assembled from gene predictors of race (A) and 3-year survival (B). Correlation matrix of racial and 3-year survival gene predictors from the ECU (left) and TCGA (right; C). Concordant gene clusters and clustered genes are shown in red and blue. The most highly correlated genes are shown in bold. Discordant genes are shown in black. ROC curve validation of ECU cohort-derived racial predictors using the TCGA expression data set (D) show that many the predictive genes shown (Fig. 4A) have significant activity as a discriminator of race in the TCGA data (E).
Discussion
In this report, we provide an advanced analytical characterization of a retrospective cohort of racially diverse patients with breast cancer collected from a single catchment area in rural East North Carolina. Using this unique cohort, we show that functional predictors of favorable outcome, defined by expression of transcriptional master regulators of mammary luminal differentiation, reveal significant racial differences in their predictive association with favorable outcome. This finding is consistent with other reports, indicating that AA women experience significantly less favorable outcome even when stratified, by biomarker profiling, into forms of breast cancer that typically show favorable outcome in EA women (3, 10, 15, 16). Limitations of this study includes a lack of precise determination of the socioeconomic status of the patients in this cohort, thus the contribution of racial differences in access to care, quality, and adherence to treatment cannot be ruled out (53). Nonetheless, an analysis of the median incomes of the counties in which each patient was diagnosed reveals significant differences for outcome in EA women (HR = 0.6; P = 0.012) compared with a smaller, nonsignificant trend (HR = 0.73; P = 0.13) in AA women (Supplementary Fig S5). In addition, ESR1-positive tumors are less common in AA women, and therefore the sample size for patients with higher expression of FOXA1 and GATA3 is lower (26% and 16%, respectively). Thus, given the samples size, the cutoff determinations may not be totally stable. Other, evidence supporting race-based differences in the intrinsic biology of luminal tumors is provided by two recent reports by Holowatyi and colleagues (54) and Troester and colleagues (55). These studies showed that AA women are more likely to have higher risk assessments in the 21 gene recurrence score (RS) breast cancer assay, and PAM 50 risk of recurrence scoring, even after adjusting for age, clinical stage, tumor grade, and histology (54, 55).
An overarching hypothesis to explain the racial differences in the association of these functional biomarkers with survival outcome, despite similar levels of favorable biomarker expression, is disparate function of the downstream networks governed by these transcriptional master regulators. This could occur through a variety of transcriptionally-linked mechanisms including: (i) polymorphisms in promoter or enhancer transcription factor binding sites; and/or (ii) differences in the coding sequence of the individual constituents of multicomponent transcriptional complexes that disrupt assembly of the complex without influencing the stability of the individual components. Several breast cancer-associated risk loci contain FOXA1 binding sites (33, 56) and current exome sequencing studies have identified multiple variations in the coding sequence of genes in racially diverse populations (57). Many of these variants do not predict protein instability or are of unknown prevalence and consequence in populations of defined genetic ancestry (57). It is conceivable that such “variants of unknown significance” could have substantial roles in determining the downstream transcriptional activity in pathways that play important roles in mammary growth, differentiation and breast cancer outcome. The level, activity and mutational spectrum of the predictive regulon genes, described in this study, provide a cogent starting point for their future investigation as predictive breast cancer biomarkers and functional targets for therapy. Given the role of ESR1, FOXA1, and GATA3 in enhancer function, the role of long-range chromatin interactions, chromosomal domains, and chromatin looping in breast cancer incidence, progression, diagnosis, and treatment, will require extensive future investigation.
Supplementary Material
Translational Relevance.
Quantitative profiling of protein abundance in tumors from a racially diverse breast cancer cohort by digital analysis of IHC-stained tissue reveals gene regulators and gene regulatory networks that are differentially predictive of breast cancer survival based on race. These findings provide a deeper understanding of the association between predictive breast cancer biomarkers and their intrinsic downstream mechanisms and how such associations may differ by race. Such observations offer new insights that will enable the identification of more accurate breast cancer biomarkers with greater population-specific predictive precision.
Acknowledgments
This work was supported by the intramural research programs of the NCI and the National Institute on Minority Health and Health Disparities, Bethesda Maryland, 20892; NIH/NCI Cancer Center Support Grant P30CA013696; the Susan G. Komen (Sponsor ID: SAC160072) Grant in support of the Triple-Negative Breast Cancer in Women with African Ancestry (04/01/2016–07/29/2021); and the Brody School of Medicine Department of Oncology Cancer Research and Education Fund. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations or imply endorsement by the U.S. Government.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Disclosure of Potential Conflicts of Interest
K. Gardner reports receiving commercial research grants from Ultivue Inc., and speakers bureau honoraria from University of California Davis. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.QuickStats: breast cancer death rates* among women aged 50–74 years, by race/ethnicity-national vital statistics system, United States, 2006 and 2016. MMWR Morb Mortal Wkly Rep 2018;67:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Robbins AS, Lin CC, Flanders WD, DeSantis CE, Ward EM, et al. Factors that contributed to black-white disparities in survival among nonelderly women with breast cancer between 2004 and 2013. J Clin Oncol 2017;36:14–24. [DOI] [PubMed] [Google Scholar]
- 3.Warner ET, Tamimi RM, Hughes ME, Ottesen RA, Wong YN, Edge SB, et al. Racial and ethnic differences in breast cancer survival: mediating effect of tumor characteristics and sociodemographic and treatment factors. J Clin Oncol 2015;33:2254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silber JH, Rosenbaum PR, Clark AS, Giantonio BJ, Ross RN, Teng Y, et al. Characteristics associated with differences in survival among black and white women with breast cancer. JAMA 2013;310:389–97. [DOI] [PubMed] [Google Scholar]
- 5.Newman LA, Griffith KA, Jatoi I, Simon MS, Crowe JP, Colditz GA. Meta-analysis of survival in African American and white American patients with breast cancer: ethnicity compared with socioeconomic status. J Clin Oncol 2006;24: 1342–9. [DOI] [PubMed] [Google Scholar]
- 6.DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, et al. Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J Clin 2016;66:290–308. [DOI] [PubMed] [Google Scholar]
- 7.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 2017;67: 439–48. [DOI] [PubMed] [Google Scholar]
- 8.Menashe I, Anderson WF, Jatoi I, Rosenberg PS. Underlying causes of the black-white racial disparity in breast cancer mortality: a population-based analysis. J Natl Cancer Inst 2009;101:993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin 2016;66:31–42. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the carolina breast cancer study. Clin Cancer Res 2010;16:6100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman LA, Kaljee LM. Health disparities and triple-negative breast cancer in African American women: a review. JAMA Surg 2017;152:485–93. [DOI] [PubMed] [Google Scholar]
- 12.Keenan T, Moy B, Mroz EA, Ross K, Niemierko A, Rocco JW, et al. Comparison of the genomic landscape between primary breast cancer in African American versus white women and the association of racial differences with tumor recurrence. J Clin Oncol 2015;33:3621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, et al. Comparison of breast cancer molecular features and survival by African and European ancestry in the cancer genome atlas. JAMA Oncol 2017;3:1654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tichy JR, Deal AM, Anders CK, Reeder-Hayes K, Carey LA. Race, response to chemotherapy, and outcome within clinical breast cancer subtypes. Breast Cancer Res Treat 2015;150:667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sparano JA, Wang M, Zhao F, Stearns V, Martino S, Ligibel JA, et al. Race and hormone receptor-positive breast cancer outcomes in a randomized chemotherapy trial. J Natl Cancer Inst 2012;104:406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershman DL, Unger JM, Barlow WE, Hutchins LF, Martino S, Osborne CK, et al. Treatment quality and outcomes of African American versus white breast cancer patients: retrospective analysis of southwest oncology studies S8814/S8897. J Clin Oncol 2009;27:2157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albain KS, Unger JM, Crowley JJ, Coltman CA Jr, Hershman DL. Racial disparities in cancer survival among randomized clinical trials patients of the southwest oncology group. J Natl Cancer Inst 2009;101:984–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer 2007;7:713–22. [DOI] [PubMed] [Google Scholar]
- 19.Tyson JJ, Baumann WT, Chen C, Verdugo A, Tavassoly I, Wang Y, et al. Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat Rev Cancer 2011;11:523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science 2002;296:1642–4. [DOI] [PubMed] [Google Scholar]
- 21.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009;9:631. [DOI] [PubMed] [Google Scholar]
- 22.Takaku M, Grimm SA, Roberts JD, Chrysovergis K, Bennett BD, Myers P, et al. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat Commun 2018;9:1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaku M, Grimm SA, Shimbo T, Perera L, Menafra R, Stunnenberg HG, et al. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol 2016;17:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 2011;43:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012;481:389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 2013;23: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 2016;37:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perou CM, Borresen-Dale AL. Systems biology and genomics of breast cancer. Cold Spring Harb Perspect Biol 2011;3:pii: a003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol 2004;219:1–7. [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial-mesenchymal transition. J Biol Chem 2010;285:14042–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakshatri H, Badve S. FOXA1 in breast cancer. Expert Rev Mol Med 2009;11:e8. [DOI] [PubMed] [Google Scholar]
- 32.Bernardo GM, Bebek G, Ginther CL, Sizemore ST, Lozada KL, Miedler JD, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene 2013;32:554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 2012;44: 1191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espinal AC, Buas MF, Wang D, Cheng DT, Sucheston-Campbell L, Hu Q, et al. FOXA1 hypermethylation: link between parity and ER-negative breast cancer in African American women? Breast Cancer Res Treat 2017;166:559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong C, Fujino K, Monteiro LJ,Gomes AR,Drost R,Davidson-Smith H, et al. FOXA1 repression is associated with loss of BRCA1 and increased promoter methylation and chromatin silencing in breast cancer. Oncogene 2015;34: 5012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hewitt SM. The application of tissue microarrays in the validation of microarray results. Methods Enzymol 2006;410:400–15. [DOI] [PubMed] [Google Scholar]
- 37.Khoury T, Zirpoli G, Cohen SM, Geradts J, Omilian A, Davis W, et al. Ki-67 expression in breast cancer tissue microarrays: assessing tumor heterogeneity, concordance with full section, and scoring methods. Am J Clin Pathol 2017;148: 108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol 2013;14:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Z, Cheng Y, Tan BC-M, Kang L, Tian Z, Zhu Y, et al. Comprehensive analysis of RNA-seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 2012;30:253. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson GN, Rogers CE. Symbolic description of factorial models for analysis of variance. J R Stat Soc Ser C (Appl Stat) 1973;22:392–9. [Google Scholar]
- 41.Hintze JL, Nelson RD. Violin plots: a box plot-density trace synergism. Am Stat 1998;52:181–4. [Google Scholar]
- 42.Myers JL, Well A, Lorch RF. Research design and statistical analysis. New York: Routledge; 2010. p. 809. [Google Scholar]
- 43.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006;22:1540–2. [DOI] [PubMed] [Google Scholar]
- 44.Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Comput Stat Data Anal 2003;43:121–37. [Google Scholar]
- 45.Hanley JA. Receiver operating characteristic (ROC) methodology: the state of the art. Crit Rev Diagn Imaging 1989;29:307–35. [PubMed] [Google Scholar]
- 46.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45: D362–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allott EH, Geradts J, Cohen SM, Khoury T, Zirpoli GR, Bshara W, et al. Frequency of breast cancer subtypes among African American women in the AMBER consortium. Breast Cancer Res 2018;20:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lund MJ, Trivers KF, Porter PL, Coates RJ, Leyland-Jones B, Brawley OW, et al. Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, GA. Breast Cancer Res Treat 2009;113: 357–70. [DOI] [PubMed] [Google Scholar]
- 49.Pitt JJ, Riester M, Zheng Y, Yoshimatsu TF, Sanni A, Oluwasola O, et al. Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features. Nat Commun 2018;9:4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walsh LA, Alvarez MJ, Sabio EY, Reyngold M, Makarov V, Mukherjee S, et al. An integrated systems biology approach identifies TRIM25 as a key determinant of breast cancer metastasis. Cell Rep 2017;20:1623–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altay G, Mendi O. Inferring genome-wide interaction networks. Methods Mol Biol 2017;1526:99–117. [DOI] [PubMed] [Google Scholar]
- 52.Kushwaha R, Jagadish N, Kustagi M, Tomishima MJ, Mendiratta G, Bansal M, et al. Interrogation of a context-specific transcription factor network identifies novel regulators of pluripotency. Stem Cells 2015;33:367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hershman DL, Tsui J, Wright JD, Coromilas EJ, Tsai WY, Neugut AI. Household net worth, racial disparities, and hormonal therapy adherence among women with early-stage breast cancer. J Clin Oncol 2015;33:1053–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holowatyj AN, Cote ML, Ruterbusch JJ, Ghanem K, Schwartz AG, Vigneau FD, et al. Racial differences in 21-gene recurrence scores among patients with hormone receptor-positive, node-negative breast cancer. J Clin Oncol 2018;36:652–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Troester MA, Sun X, Allott EH, Geradts J, Cohen SM, Tse CK, et al. Racial differences in PAM50 subtypes in the carolina breast cancer study. J Natl Cancer Inst 2018;110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 2008;132:958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghazani AA, Oliver NM, St Pierre JP, Garofalo A, Rainville IR, Hiller E, et al. Assigning clinical meaning to somatic and germ-line whole-exome sequencing data in a prospective cancer precision medicine study. Genet Med 2017;19:787–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.