Plant architecture, especially in important crop plants, has been under various human selection regimes over agricultural history. In plants, shoot form is determined by the complex interplay of hormones which integrate various environmental cues such as light and nutrient availability to influence growth and architecture (Tarancón et al., 2017). Shoot architecture is a highly complex polygenic trait known to play a fundamental role in crop yield. One key example is the shoot architecture in modern maize in comparison to its ancestor teosinte, where a naturally occurring transposon insertion in the TB1 gene results in increased apical dominance causing a single flowering stalk. Selection for this mutation over time has led to the current maize variety from its high‐tillering ancestor (Studer et al., 2011).
One group of hormones that is required for suppressing shoot branching and regulating axillary meristem activity are the Strigolactones (SLs), a class of carotenoid‐derived terpenoid lactones (Umehara et al., 2008). SLs also regulate various developmental processes including internode elongation, leaf shape, secondary stem thickening, as well as root architecture (Waters et al., 2017). SL deficient mutants exhibit increased branching phenotypes caused by a combination of enhanced auxin flux and decreased expression of the TB1‐homolog BRANCHED1 in axillary buds (Bennett et al., 2006; Waters et al., 2017). Given that yield increase in canola (Brassica napus) is a major industry priority, alteration of SL signalling could lead to a highly branched morphotype similar to the dwarfed plants of the green revolution with favourable shoot architecture for addition of more inputs.
In order to examine if suppression of D14 receptor could lead to these desired changes, canola (Westar) were transformed with an RNAi suppression construct driven by the 35SCaMV promoter which targets the SL receptor BnD14. We analysed five independent lines (RiD14; T1 generation) which showed a drastic reduction in BnD14 transcript levels (Figure 1a). Most of T2 generation from these plants exhibited an increased branching phenotype relative to WT plants, (Figure 1b) concomitant with a significant reduction of plant height in all the lines, similar to the classical SL mutants (Figure 1c). Only one line (line 10) exhibited a significant increase in mean total flowers (35%) per plant relative to WT (Figure 1d). The incomplete suppression of D14 in these RNAi lines may have resulted in the observed partial phenotypes but provided valuable evidence that creating D14 knockout lines would be a viable option.
Figure 1.
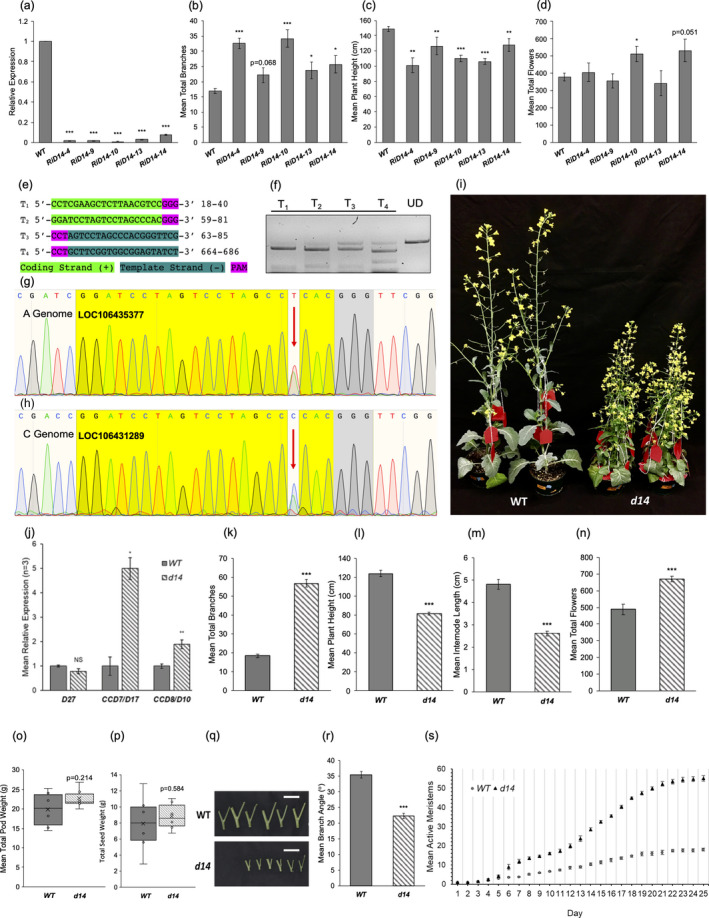
(a) Quantitative RT‐PCR of SL receptor gene BnD14 in leaf tissue from 6‐week old T1 plants. Relative values are normalized to BnACTIN2 (± SEM, n = 4). Comparison of mean number of (b) total branches per plant (c) height (d) total flowers per plant (±SEM, n = 6). (e) Selected CRISPR target sequences highlighted in light green (coding strand) or dark green (template strand), PAM sites in pink. CRISPR targets numbered based on proximity to start codon, position provided in nt distance from start codon. (f) In vitro digestion of BnD14 using recombinant Cas9 endonuclease (UD = undigested control). (g, h) DNA sequencing chromatograms of edited BnD14 A and C genome sequence. Yellow: 20bp Target 2 site, red arrows: insertions in A and C copies, grey: PAM sites. (h) Phenotypic comparison of WT (left) and d14 (right) plants. (j) Quantitative RT‐PCR of SL biosynthetic genes (D27, CCD7/D17 and CCD8/D10) in seedling root tissue from 7‐day old T3 plants. Relative values are normalized to BnACTIN2 (± SEM, n = 3). (k‐n) Mean quantitative analyses of the T2 CRISPR‐Cas9 d14 mutant line (± SEM, n = 6). Comparison of mean, number of total branches per plant (k), plant height (l), internode length (m). total flowers per plant (n). (o) Total pod weight per WT and d14 plant. (p) Total seed weight per plant (± SEM, n = 6). (q) Visual comparison of d14 branch angle relative to WT. Bars = 1cm. (r) Comparison of mean branch angle between d14 and WT plants (±SEM, n = 6). (s) Emergence of active meristems over a 25‐day period beginning with the onset of flowering. (±SEM, n ≥ 5). All statistical significance identified using Student’s T‐test comparing transgenic lines to WT (unpaired test assuming equal variance, *P < 0.05, **P < 0.01, ***P < 0.001).
To achieve this, we generated CRISPR/Cas9‐mediated knockout lines of the genes encoding the SL receptor BnD14. We designed a custom‐made multiplex construct with the coding sequence of S. pyogenes Cas9 under a 35SCaMV constitutive promoter, followed by various single guide RNAs (sgRNAs) (Cong et al., 2013) under the control of different U3 and U6 promoters to target four conserved regions of the BnD14 homeologs (A genome:LOC106435377) (C genome: LOC106431289) (Figure 1e). In vitro digestion of amplified BnD14 sequence with recombinant Cas9 enzyme and in vitro transcribed sgRNA demonstrated the expected activity of Cas9/target sgRNAs (Figure 1f). We transformed Westar canola lines with this construct and isolated two dwarfed lines in T1 primary transformants. Sequencing of the D14 genomic DNA from A and C genomes (tetraploid) of these lines, revealed a single biallelic insertion in all four chromosomal copies of BnD14 (Figure 1g, h).
All T1 CRISPR/Cas9‐edited lines exhibited a similar branched phenotype (Figure 1i) and sequencing showed consistent edits at the specified target sites, and therefore, T2 and T3 seeds from a single line, designated d14, were chosen for further downstream analyses. When SL biosynthetic genes were examined in the roots of d14 plants, characteristic feedback up‐regulation of CCD7 and CCD8 transcripts were observed in the absence of strigolactone perception (Figure 1j).
The d14 plants showed a prolific branching phenotype with an approximately 200% increase in mean total branches per plant relative to the WT as well as a dwarfed phenotype with a 34% reduction in mean plant height and reduced internode length (Figure 1k‐m). Most importantly, the d14 plants exhibited a significant increase of 37% total flowers per plant relative to WT plants (Figure 1n). These observations clearly indicate that lack of BnD14 function leads to promising alterations in yield‐relevant traits.
When we compared yield characteristics from mature d14 and WT plants, a slight, although insignificant, increase of 12.5% (P = 0.214) in total pod weight per plant was found in the d14 line compared to the WT (Figure 1o). Total seed weight per plant showed a slight, however insignificant, increase of 10.4% (P = 0.584) in d14 compared to the WT (Figure 1p). These results show that lesions in SL signalling do not result in detrimental effects on yield in canola.
One key morphology that was altered in the d14 lines is the branch angles or the gravitropic setpoint angle. Typically, lower branch angles are preferred for their tighter architecture and options for greater planting densities. As observed from previous studies (Liang et al., 2016), d14 plants showed a significant reduction in mean gravitropic setpoint angle of 37% relative to WT plants (Figure 1q, r). This trait has been associated with increased planting density in rice crops (Ferrero‐Serrano et al., 2019) and may be of value in canola as well.
When meristem activity was compared in the WT and d14 lines over a 25‐day period beginning with the onset of flowering, the d14 plants exhibited a significant increase in rate of lateral meristem activation over this 25‐day period relative to the WT plants (Figure 1s). This indicates that the increase in total flowers per plant was a result of an elevated axillary meristem activity rather than a prolonged flowering period. Given that canola is grown in many temperate regions with short seasons, augmented meristem activity is always a favoured trait over a prolonged flowering period.
Collectively, we have been able to generate a new morphotype of canola that is quite similar to the dwarfed plants of the green revolution which are best suited for increased resource inputs that could significantly boost yield. The dwarf stature and the lack of increased yield characteristics in SL mutants using model systems such as Arabidopsis and petunia (Simons et al., 2007) may have deterred the exploitation of SL pathway for promoting yield in crop plants. Our observations with the d14 deficient canola lines clearly indicate the potential for tweaking the SL pathway for crop improvement strategies in canola. Recently, it has been shown that specific SL partial loss‐of‐function alleles were also artificially selected for, along with GA mutant alleles, in the generation of elite dwarfed rice varieties during the green revolution (Wang et al., 2020). Incorporation of this trait into elite breeding lines could lead to primary producers having access to a new generation of canola lines with a tighter architecture, increased flowering and a lodging‐tolerant stature amenable for responding to more inputs. As our population grows exponentially, it is imperative that we enhance the resource use efficiency of our existing crop lands to improve yield.
Author contributions
M.S., N.H. and R.D. conducted the experiments. M.S., N.H. and M.A.S. designed the experiments and wrote the manuscript.
Conflict of interests
The authors declare no competing financial interests.
Acknowledgements
This work was supported by the NSERC Strategic Project Grant (10015342) and NSERC Collaborative Research and Development Grant (10023207).
Stanic, M. , Hickerson, N. M. N. , Arunraj, R. , and Samuel, M. A. (2021) Gene‐editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola). Plant Biotechnol. J., 10.1111/pbi.13513
References
- Bennett, T. , Sieberer, T. , Willett, B. , Booker, J. , Luschnig, C. and Leyser, O. (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16, 553–563. [DOI] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. , Hsu, P.D. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero‐Serrano, Á. , Cantos, C. and Assmann, S.M. (2019) The role of dwarfing traits in historical and modern agriculture with a focus on rice. Cold Spring Harb. Perspect. Biol. 11, a034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Ward, S. , Li, P. , Bennett, T. and Leyser, O. (2016) SMAX1‐LIKE7 signals from the nucleus to regulate shoot development in Arabidopsis via partially EAR motif‐independent mechanisms. Plant Cell, 28, 1581–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J.L. , Napoli, C.A. , Janssen, B.J. , Plummer, K.M. and Snowden, K.C. (2007) Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiol. 143, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer, A. , Zhao, Q. , Ross‐Ibarra, J. and Doebley, J. (2011) Identification of a functional transposon insertion in the maize domestication gene tb1 . Nat Genet. 43, 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarancón, C. , González‐Grandío, E. , Oliveros, J.C. , Nicolas, M. and Cubas, P. (2017) A conserved carbon starvation response underlies bud dormancy in woody and Herbaceous species. Front. Plant Sci. 8, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. , Akiyama, K. , Arite, T. , Takeda‐Kamiya, N. , Magome, H. et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Shang, L. , Yu, H. , Zeng, L. , Hu, J. , Ni, S. , Rao, Y. et al. (2020) A strigolactone biosynthesis gene contributed to the green revolution in rice. Mol Plant. 13, 923–932. [DOI] [PubMed] [Google Scholar]
- Waters, M.T. , Gutjahr, C. , Bennett, T. and Nelson, D.C. (2017) Strigolactone signaling and evolution. Annu Rev Plant Biol. 2017, 291–322. [DOI] [PubMed] [Google Scholar]


