CRISPR technology is an established tool for the generation of knockout plants (Zhang et al., 2019), yet limitations remain. First, the manipulation of individual genes may fail to produce phenotypes for groups of genes with redundant or synergistic functions. While this has been partially addressed by multiplexing guide RNAs (gRNAs), there is concern that as the number of targets increases, the chances of obtaining higher‐order knockouts diminish (Zhang et al., 2016). Second, knocking out fundamentally important genes can cause severe pleiotropic phenotypes or lethality. Tissue‐specific knockout of genes in somatic tissues can overcome this limitation (Decaestecker et al., 2019; Liang et al., 2019; Wang et al., 2020). However, the efficiency of simultaneously targeting more than three genes in a tissue‐specific context is unexplored. Here, by multiplexing gRNAs in Arabidopsis thaliana plants expressing Cas9 either ubiquitously (pPcUBI) or root cap specifically (pSMB), we show that six genes can be simultaneously mutated with high efficiency, generating higher‐order mutant phenotypes already in the first transgenic generation (T1). The mutation frequencies for all target genes were positively correlated and unaffected by the order of the gRNAs in the vector, showing that efficient higher‐order mutagenesis in specific plant tissues can be readily achieved.
We selected six efficient gRNAs (Decaestecker et al., 2019 and unpublished results) to target the coding sequences of six genes (GFP, and the Arabidopsis genes SMB, EXI1, GL1, ARF7 and ARF19; Figure 1a) whose knockout lead to easy‐to‐score phenotypes in T1 seedlings (gfp: loss of GFP signal, smb: accumulation of root cap cells (Fendrych et al., 2014), gl1: absence of trichomes (Herman and Marks, 1989)) and do not severely affect plant growth or reproduction. Since position effects within gRNA arrays had been a concern regarding mutation efficiency (Zhang et al., 2016), we generated two vectors (hereafter, pPcUBI(I) and pPcUBI(II)) combining Cas9‐mTagBFP2 driven by the ubiquitous pPcUBI promoter and the six gRNAs in an inverted order (Figure 1b) and transformed these into an Arabidopsis line with ubiquitous expression of a nuclear‐localized GFP (pHTR5:NLS‐GFP‐GUS (Decaestecker et al., 2019) hereafter, NLS‐GFP).
Figure 1.
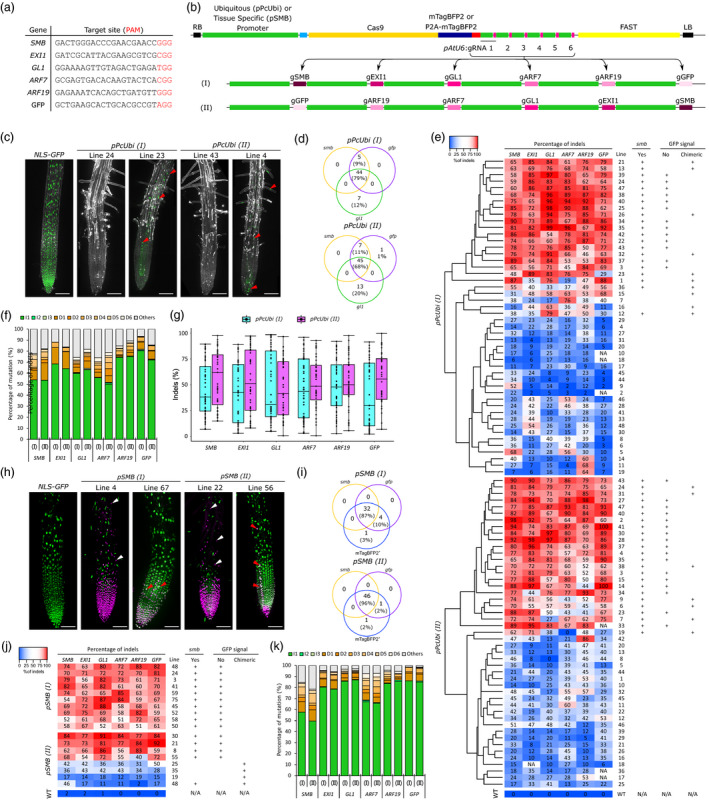
Ubiquitous and root cap‐specific knockout of 6 genes in T1 via CRISPR and CRISPR‐TSKO. (a) gRNA Target sequences. (b) Diagram of the pPcUBI (Petroselinum crispum UBIQUITIN promoter) and pSMB vectors with gRNAs cloned in an inverted order, (I) and (II). (c) Maximum intensity projections of root tips of a representative NLS‐GFP seedling, two pPcUBI(I) and two pPcUBI(II) T1 seedlings showing the complete (left) and chimeric (right) absence of GFP signal and smb phenotype. GFP is in green, propidium iodide (PI) in grey. Red arrowheads indicate root cells still expressing GFP. Scale bars, 100 µm. (d) Venn diagram showing the number of plants displaying smb, gfp and gl1 mutant phenotype in 96 pPcUBI(I) and 95 pPcUBI(II) T1 seedlings. (e) Genotype analysis by amplicon sequencing. Phenotypes are indicated on the right panel. (f) Frequency of the main mutation types in both pPcUBI(I) and pPcUBI(II) plants. I1–I3: 1‐ to 3‐bp insertion, D1–D6: 1‐ to 6‐bp deletion, Others: bigger deletions (>6‐bp), insertions (>3‐bp) or complex repair outcomes containing both insertions and deletions. (g) Percentage of indels observed in pPcUBI(I) and pPcUBI(II) T1 plants. (h) Maximum intensity projections of root tips of a representative NLS‐GFP seedling, two pSMB(I) and two pSMB(II) T1 seedlings grown on 1 µM brassinazole showing the complete (left) and chimeric (right) absence of GFP and presence of mTagBFP2 signal specific to root cap cells. GFP is in green, mTagBFP2 in magenta. White arrowheads indicate live root cap cells with nuclear mTagBFP2 signal covering the elongation zone. Red arrowheads indicate root cells still expressing GFP. Scale bars, 100 µm. (i) Venn diagrams showing the number of plants displaying strong mTagBFP2 signal, smb and gfp phenotype in 86 pSMB(I) and 88 pSMB(II) T1 seedlings. (j) Genotype analysis of BFP+ sorted cells of pSMB(I) and pSMB(II) T2 seedlings by amplicon sequencing. (k) Frequency of the main mutations types in pSMB(I) and pSMB(II) plants.
Forty‐nine out of 96 pPcUBI(I) and 52 out of 95 pPcUBI(II) T1 seedlings displayed both gfp and smb phenotypes in roots, indicating simultaneous mutations (Figure 1c,d). Additionally, 44 out of 96 pPcUBI(I) and 45 out of 95 pPcUBI(II) T1 seedlings also lacked trichomes on the first two true leaves, revealing a high mutation frequency for GL1. Altogether, 79% of the pPcUBI(I) and 68% of the pPcUBI(II) T1 seedlings with at least one detectable knockout phenotype also showed triple gfp smb gl1 mutant phenotypes. When selecting plants based on the loss of GFP, 90% of the pPcUBI(I) and 85% of the pPcUBI(II) T1 seedlings displayed triple mutant phenotypes, indicating a strong correlation of mutagenesis efficiencies.
We quantified indel frequencies in 48 pPcUBI(I), 47 pPcUBI(II) and a control NLS‐GFP plant. The targeted loci were PCR‐amplified from root tips and sequenced using Illumina sequencing. Plants showing total or partial gfp and smb phenotypes had high indel frequencies in GFP (27%–100%) and SMB (38%–98%), as well as in all other target genes. Hierarchical clustering showed that transgenic T1 plants fell in two major classes that had either high or low levels of mutagenesis for all target genes (Figure 1e). In agreement with previous reports (Feng et al., 2014), 1‐bp indels were the predominant repair outcome (50%–80% and 1%–15% respectively), in‐frame indels were rare (2%–8%), and 6%–26% of mutations were bigger deletions (>6‐bp), insertions (>3‐bp) or complex repair outcomes (Figure 1f).
We compared indel frequencies for each target between the two constructs to test the effect of the gRNA position (Figure 1g). The overall indel frequencies were higher for pPcUBI(II), though the difference was only significant for GFP. As all other gRNAs had no substantial changes in indel frequencies, our data do not support a position effect in gRNA arrays, thus reducing the complexity of future experimental design.
We then tested whether six genes can be efficiently mutated in a tissue‐specific context by making two vectors expressing Cas9‐P2A‐mTagBFP2 under the root cap‐specific pSMB promoter with the same arrangement of gRNAs (hereafter, pSMB(I) and pSMB(II)). Plants were grown in the presence of 1 µM brassinazole (BRZ) to facilitate smb phenotyping. This treatment leads to a root covered by living root cap cells in smb mutants (Fendrych et al., 2014) and was easily recognizable due to the presence of nuclear mTagBFP2 signal in living root cap cells (Figure 1h).
Thirty‐two out of 86 pSMB(I) and 46 out of 88 pSMB(II) T1 seedlings showed both gfp and smb phenotypes, as well as a strong mTagBFP2 signal specifically in root cap nuclei as determined by confocal microscopy (Figure 1i). In agreement with our previous report (Decaestecker et al., 2019), mTagBFP2 signal intensity could be used as a proxy for the penetrance of gfp and smb knockout phenotypes. To determine mutagenesis efficiency in all target genes specifically in Cas9‐expressing root cap cells, we collected root‐tip protoplasts expressing mTagBFP2 (BFP+, Cas9 expressing cells) using fluorescence‐activated cell sorting from T2 seedlings of ten pSMB(I) and eight pSMB(II) independent lines. We chose four pSMB(II) lines (19, 25, 35 and 48) with weak or chimeric gfp and smb T1 mutant phenotypes and four pSMB(I) and(II) lines with highly penetrant smb and gfp T1 mutant phenotypes.
The targeted loci were PCR‐amplified directly from sorted protoplast populations and sequenced by NGS. In pSMB(I) and (II), T2 seedlings coming from a T1 parent with strong smb and gfp phenotypes, the Cas9‐expressing BFP+ populations had indel frequencies between 51% and 92% for all six target loci (Figure 1j). As expected, the BFP+ populations of the pSMB(II) lines that with weak or chimeric gfp and smb phenotypes in T1 had lower indel frequencies (2%–50%). These results confirmed that in lines with high GFP and SMB mutagenesis activity, all genes were simultaneously mutated with high efficiency. Similarly to the ubiquitous lines, the alleles generated were largely consistent across events, with 1‐bp indels being the predominant repair outcome (50%–87% and 2%–10%), in‐frame insertion or deletions were rare (0%–5%), and 3%–21% of mutations were bigger indels (>3‐ and >6‐bp) or combination of indels (Figure 1k).
In conclusion, we show that ubiquitous CRISPR and CRISPR‐TSKO approaches allow fast and simultaneous disruption of six genes in the first transgenic generation with high efficiency. As mutation efficiencies over all loci are correlated, we suggest the use of a target gene with an easy‐to‐score, non‐detrimental loss‐of‐function phenotype as a proxy for highly mutagenized lines. As an alternative to endogenous genes (Li et al., 2020), loss of GFP in a reporter line can also be used as a proxy. We foresee this approach to be a powerful tool to dissect genetic networks in model and crops species alike.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
N.B., R.A.B., M.K.N. and T.B.J. designed the experiments and wrote the manuscript. N.B. and R.A.B. performed the experiments and analysed the data.
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 639234 and 864952) and from the Research Foundation – Flanders (FWO, project No. G041118N). We thank Dr. Jonas Blomme for providing the pPcUBI and pGG‐D‐mTagBFP2‐E vectors. We thank the VIB Flow Core for support and access to the instrument.
Bollier, N. , Andrade Buono, R. , Jacobs, T. B. and Nowack, M. K. (2021) Efficient simultaneous mutagenesis of multiple genes in specific plant tissues by multiplex CRISPR. Plant Biotechnol J, 10.1111/pbi.13525
Contributor Information
Thomas B. Jacobs, Email: thomas.jacobs@vib.be.
Moritz K. Nowack, Email: moritz.nowack@vib.be.
References
- Decaestecker, W. , Buono, R.A. , Pfeiffer, M.L. , Vangheluwe, N. , Jourquin, J. , Karimi, M. , Van Isterdael, G. et al. (2019) CRISPR‐TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell, 31, 2868–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych, M. , Van Hautegem, T. , Van Durme, M. , Olvera‐Carrillo, Y. , Huysmans, M. , Karimi, M. , Lippens, S. et al. (2014) Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 24, 931–940. [DOI] [PubMed] [Google Scholar]
- Feng, Z. , Mao, Y. , Xu, N. , Zhang, B. , Wei, P. , Yang, D.L. , Wang, Z. et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas‐induced gene modifications in Arabidopsis. Proc. Natl Acad. Sci. 111, 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P.L. and Marks, M.D. (1989) Trichome development in Arabidopsis thaliana. II. Isolation and complementation of the GLABROUS1 gene. Plant Cell, 1, 1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. , Vavrik, C. and Danna, C.H. (2020) Proxies of CRISPR/Cas9 activity to aid in the identification of mutagenized Arabidopsis plants. G3 (Bethesda), 10, 2033–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Eudes, A. , Yogiswara, S. , Jing, B. , Benites, V.T. , Yamanaka, R. , Cheng‐Yue, C. et al. (2019) A screening method to identify efficient sgRNAs in Arabidopsis, used in conjunction with cell‐specific lignin reduction. Biotechnol. Biofuels, 12, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Ye, L. , Lyu, M. , Ursache, R. , Löytynoja, A. and Mähönen, A.P. (2020) An inducible genome editing system for plants. Nat. Plants, 6, 766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Malzahn, A.A. , Sretenovic, S. and Qi, Y. (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants, 5, 778–794. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Mao, Y. , Ha, S. , Liu, W. , Botella, J.R. and Zhu, J.K. (2016) A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 35, 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]


