Summary
Human population growth has increased the demand for food crops, animal feed, biofuel and biomaterials, all the while climate change is impacting environmental growth conditions. There is an urgent need to develop crop varieties which tolerate adverse growth conditions while requiring fewer inputs. Plant breeding is critical to global food security and, while it has benefited from modern technologies, it remains constrained by a lack of valuable genetic diversity, linkage drag, and an effective way to combine multiple favourable alleles for complex traits. CRISPR/Cas technology has transformed genome editing across biological systems and promises to transform agriculture with its high precision, ease of design, multiplexing ability and low cost. We discuss the integration of CRISPR/Cas‐based gene editing into crop breeding to advance domestication and refine inbred crop varieties for various applications and growth environments. We highlight the use of CRISPR/Cas‐based gene editing to fix desirable allelic variants, generate novel alleles, break deleterious genetic linkages, support pre‐breeding and for introgression of favourable loci into elite lines.
Keywords: gene editing, CRISPR/Cas, domestication, precision breeding, recombination, novel alleles
Introduction
The linking of crop domestication with a century and half of targeted breeding has led to modern cultivars which display a blend of desirable traits. Domestication traits include larger fruit or seeds, loss of natural seed dispersal, altered photoperiod sensitivity and vernalization responses, and improved grain threshability (Doebley et al., 2006). This process involved many complex genetic events and loci shuffling. Numerous domestication genes have been identified and functionally characterized (Olsen and Wendel, 2013). While many genetic variants associated with domestication traits have been fixed within elite germplasm, other improvement traits such as higher yield and nutrition, resistance to biotic and abiotic stress, and improved resource use still vary among crop cultivars and germplasm and are the focus of many breeding programmes (Abberton et al., 2016; Doebley et al., 2006; Meyer et al., 2012; Swinnen et al., 2016). Expanded genetic variation for future improvement of these traits can be found within germplasm collections; however, a much wider range of trait variation is found within landraces and wild relatives (Breseghello and Coelho, 2013; Brozynska et al., 2016). Harnessing beneficial genetic variation and eliminating maladapted genetic material is a major challenge of crop breeding.
Plant breeding is the primary means to reshuffle favourable alleles and develop varieties with superior qualities. Breeding involves inter‐crossing parents with desirable traits to create F1 hybrid lines, and selection of top performing lines from thousands of progenies over multiple, successive generations (F2–F13; Fridman and Zamir, 2012). Depending on the species, growth habit, and starting parental lines, breeding programmes can take between 6 and 15 years to generate a genetically superior cultivar for agricultural production (Acquaah, 2012). Molecular markers associated with major effect quantitative trait loci are used for marker‐assisted selection (MAS). MAS is greatly beneficial in assisting with breeding techniques like backcrossing, for introgression of a locus from a donor line into an elite cultivar, or for combining multiple alleles with gene pyramiding (Chen et al., 2013; Vogel, 2009). These techniques are most often used to deliver simple genetic traits with large effects. However, most economically important traits, such as yield or abiotic stress tolerance, tend to be controlled by many small effect loci (Gilliham et al., 2017). These traits tend to be more genetically complex, and often require utilization of multi‐omics data for selection of favourable traits. MAS enables plant selection based on genomic information, rather than by phenotyping, which expedites the breeding process and mitigates phenotyping limitations (Crossa et al., 2017). Genomic selection uses genome‐wide marker data and integrates genomics estimated breeding values into statistical models or algorithms for genotypic and phenotypic selection.
However, despite the benefit of molecular tools and genomic information, combining multiple desirable agronomic outcomes may still be hindered by genetic correlations between traits. For example, linkage drag occurs when an undesirable locus is located in close genetic proximity to a desirable locus, such that little to no meiotic recombination occurs between the two, and the undesirable locus is co‐inherited with the desirable locus (Choi, 2017, 2017). Unfavourable repulsion linkages, where desirable alleles occur on separate homologous chromosome segments but are unable to recombine (for example, Fhb1 and Sr2 on chromosome 3B in wheat; Anderson et al., 2001; Zhang et al., 2016), pose a challenge for introgression of desirable chromosome segments into a different genetic background. Breaking genetic linkage requires meiotic homologous recombination between the two relevant loci. Depending on the distance between loci, species and population size, meiotic recombination rates can vary widely, and identification of a desirable recombinant individual can be difficult or near impossible (Choi, 2017, 2017; Choi and Henderson, 2017).
Overall, the genetic gain of a breeding programme is a function of heritability and population size, and is largely determined by the capacity to phenotypically evaluate a large number of plants in a high‐throughput manner (Breseghello and Coelho, 2013). The rapid development and integration of CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR associated proteins)‐based gene editing into plant science has created an alternative avenue for crop improvement, and has the potential to increase speed and precision in plant breeding programmes.
CRISPR/Cas‐based gene editing
CRISPR/Cas‐based gene editing enables targeted sequence modification. Since its initial discovery in bacteria and adaptation into a eukaryotic gene editing tool, CRISPR/Cas technology has been used for a variety of genome editing functions. The success of this technology is related to its high precision, ease of design and lower cost compared with other gene editing tools (for example, TALENS or ZFN).
Traditionally, CRISPR/Cas systems comprise of a nuclease with RuvC and HNH domains (SpCas9; Streptococcus pyogenes), and a programmable guide RNA (gRNA) with homology to the target genomic sequence (Doudna and Charpentier, 2014; Jinek et al., 2012). SpCas9 requires a short, specific protospacer adjacent motif (PAM) flanking the 3’‐end of the target site in the genomic DNA. Guided by the programmable spacer region of the gRNA, the Cas nuclease generates a double‐stranded DNA break. These DNA breaks are repaired through one of two endogenous DNA repair pathways: the error‐prone non‐homologous end joining (NHEJ), or the more targeted homology‐directed repair (HDR; Voytas, 2013). Depending on the nature of the DNA break and how it is repaired, several opportunities for genomic editing are created. These include gene knock‐out via generation of null alleles (introduced stop codon or large deletions), and creation of new alleles via specific base‐pair edits or ‘knock‐ins’ (sequence inserts; Murovec et al., 2017).
Advances in CRISPR technology
CRISPR/Cas‐based technology is rapidly expanding, and improvements are being seen in specificity, precision and off‐target effects, editing capabilities, and ease of use in target organisms. Particularly notable is the development of novel SpCas9 variants, Cas9 orthologs and CRISPR‐associated enzymes. For example, the SpCas9 variants known as SpCas9‐nickases create single‐stranded DNA breaks, due to mutations in either the RuvC (Cas9 D10A nickase) or HNH domains (Cas9 H840A nickase). These nickases have been deployed individually or in pairs to create staggered DNA breaks, promoting homology‐directed repair, and also to reduce off‐target effects (Fauser et al., 2014; Jinek et al., 2012; Mikami et al., 2016). A second variant, enhanced SpCas9 or eSpCas9, exhibits modified helicase activity such that mismatches between the guide RNA and target DNA are less energetically favourable, leading to increased specificity (Slaymaker et al., 2016).
Smaller Cas variants not only make lab‐based manipulations significantly simpler, but may also enable delivery of the CRISPR/Cas system to plant cells using viral vectors (Kleinstiver et al., 2015a, 2015b; Murovec et al., 2017; Ran et al., 2015). Cas9 orthologs, such as SaCas9, St1Cas9, and NmCas9, are about a quarter reduced in genomic size as compared to the original SpCas9 (Cas9 ~ 4.2 Kb, SaCas9 ~ 3.2 kb, St1Cas9 ~ 3.4 Kb, NmCas9 ~ 3.2 kb). In addition, these orthologs offer more diverse and longer PAM motifs, thereby increasing specificity and reducing off‐targets effects. The Class 2 Type V effector protein, Cas12a/Cpf1, contains an RuvC‐like domain but lacks the HNH nuclease domain. These Cpf1 proteins (i.e. FnCpf1, AsCpf1, LbCpf1) require a shortened gRNA molecule, a T‐rich PAM motif, and generate a staggered double‐stranded DNA break (Hu et al., 2016; Xu et al., 2017; Zetsche et al., 2015). These alternative properties increase the specificity and versatility of the editing system.
CRISPR/Cas technology has also been adapted into a nucleotide‐editing tool which directs the targeted conversion of a single nucleotide, without requiring double‐stranded DNA breaks, HDR processes or donor DNA templates (Komor et al., 2016). In this system, a cytosine or adenosine deaminase domain is fused to a catalytically inactive CRISPR–Cas9 domain (Cas9 variants dCas9 or Cas9 nickase), and directed to the target DNA via the sequence specific gRNA. The Cas9‐cytosine deaminase fusion converts cytosine (C) to uracil (U) at target sites; in the presence of a uracil glycosylase inhibitor to impede uracil excision, the result is a C → T (G → A) substitution (Komor et al., 2016). Similarly, adenine deaminases, which convert A → G, have been deployed in bacteria and human cells (Gaudelli et al., 2017). This application is especially useful to crops, as a low occurrence of HDR in plants (0.2–5.5%, compared to 5–20% in animals and ~ 100% in bacteria) makes gene targeting by HDR difficult (Bortesi et al., 2016). Moreover, several desirable alleles involve only a single nucleotide polymorphism (SNP), and base editing would be relatively simple in these instances. Already, this approach has been widely deployed in a range of species including rice, wheat, tomato, potato and watermelon (Kang et al., 2018; Li et al., 2018a,b; Lu and Zhu, 2017; Shimatani et al., 2019; Tian et al., 2018; Veillet et al., 2019; Zong et al., 2018).
The recent development of CRISPR prime editing has expanded the gene editing tool kit even further (Anzalone et al., 2019). The prime editing system uses a catalytically inactive Cas9 fused to an engineered reverse transcriptase, target‐programmed with a prime editing guide RNA (pegRNA). Using this system, a wide range of edits have been achieved, including insertions, deletions and point mutations in human cell lines, and also in rice and wheat protoplasts, without double‐stranded DNA breaks or donor DNA templates (Anzalone et al., 2019; Lin et al., 2020). Similar to base editing, CRISPR prime editing does not require HDR, making it highly attractive for use in plants.
Delivery of CRISPR technology into plant cells
The CRISPR/Cas‐based gene editing tool kit has been deployed into many of the staple food crops of the world including maize, wheat and rice (Ansari et al., 2020). In addition, this gene editing tool has also been applied to a variety of fruit crops, such as tomato, watermelon, banana, grapes and cucumber (Wang et al., 2019). In general, agrobacterium‐mediated gene transfer is widely used to transform totipotent cells; however, biolistic gene transfer, protoplasts transformation and microspore transformation are also used (Altpeter et al., 2016; Ferrie et al., 2020). Since Cas9/gRNA activity is not required once edits have been generated, genome editing success has also been found with non‐DNA based delivery systems using in vitro Cas9 ribonucleoprotein complex formulations (delivered by microinjection, particle bombardment etc.), and using viral delivery vectors (Ma et al., 2020; Tsanova et al., 2021). Tissue cell culture and regeneration is usually then required to generate a full plant. Depending on the species, or variety/genotype within a species, regeneration is often considered to be the greatest bottleneck (Altpeter et al., 2016). Even though CRISPR has been realized in a wide variety of crop species, widespread implementation and use is still largely constrained by costly and time‐consuming factors relating to transformation, regeneration and delivery of the CRISPR/Cas‐based technology (Altpeter et al., 2016).
Gene editing and plant breeding for crop improvement
In plant biotechnology, specific genes of interest have been manipulated into loss‐of‐function, gain‐of‐function, altered expression, or truncated proteins, generating novel crop lines with desirable traits in a wide range of species (Weeks, 2017; Zhang et al., 2018b). Gene editing creates an opportunity for fast conversion of undesirable alleles into desirable alleles. This potential is greatly enhanced by the microspore and double haploid technology (Bhowmik et al., 2018; Ferrie et al, 2020), which is already used regularly in many breeding programmes (for example, wheat, B. napus and barley (Hordeum vulgare L.). Importantly, a CRISPR/Cas‐based system can generate a range of DNA edits which are synonymous with those found in natural populations. Moreover, the multiplexing capacity of the CRISPR system means that multiple genetic changes can be achieved in a single generation. Plant breeding plays an essential role in crop development, and established breeding programmes would benefit greatly from the introduction of CRISPR‐based gene editing. However, application of this genetic tool in routine breeding is at its infancy. In this section, we highlight applications of CRISPR/Cas‐based gene editing to achieve a number of goals relevant to inbred crop improvement programmes.
Fixing desirable monogenic traits, and saving near‐miss varieties
Genetic variation in crop germplasm has been molded by domestication and plant selection aimed at developing locally adapted and high‐yielding varieties. As a result, some alleles are consistent or fixed within elite germplasm; these are generally considered domestication genes. For example, domestication type alleles such as the wheat Q allele (Simons et al., 2006; Zhang et al., 2011), maize teosinte branched 1 (Doebley, 2004; Doebley et al., 1995) and tomato fruit size fw2.2 (Alpert et al., 1996; Frary et al., 2000; Nesbitt and Tanksley, 2002) have been fixed through breeder selection. Importantly, there are also several strong effect, favourable alleles which may be missing or not fixed in some locally adapted germplasm. Improvement alleles and/or genes that are segregating in the germplasm require selection through multiple generations. In selecting for these desirable traits, some individuals are culled, and their genetic variation (unrelated to the desirable traits under selection) is lost. Gene editing can potentially fix those strong effect improvement alleles early in the breeding process (Figure 1a,b). In doing so, breeders could avoid the process of plant selection and culling, minimizing the loss of genetic variation and resulting in an expanded population of plants with desirable alleles. This expanded resource can then be exploited in exploring and selecting for other important and more genetically complex traits.
Figure 1.
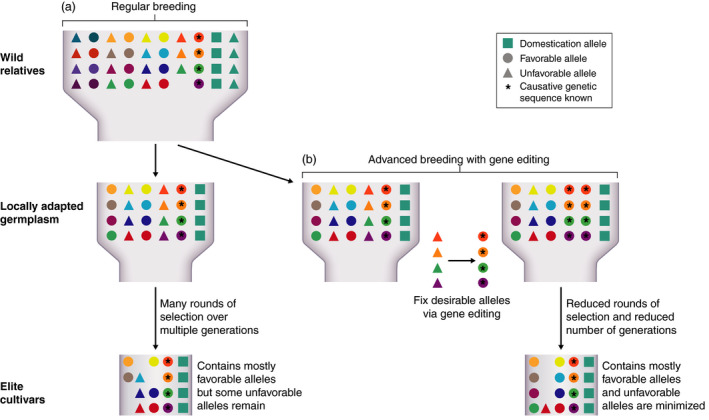
Schematic of advanced domestication using CRISPR/Cas‐based gene editing (a) During domestication and crop breeding wild plant species are developed into locally adapted germplasm. Some genetic variation is lost, while favourable domestication genes/alleles (blue squares) become fixed in the gene pool and unfavourable alleles of domestication genes are lost (blue triangles). Further development of germplasm into elite cultivars requires crosses, multiple rounds of plant selection and multiple generations. Selection of favorable improvement alleles (circles) results in an elite cultivar with mostly favourable alleles but some unfavourable alleles remain (triangles). (b) Using locally adapted germplasm, favorable alleles with known causative genetic sequence (circles with asterisks) are fixed in germplasm by the conversion of unfavourable alleles into favourable alleles through CRISPR/Cas‐based gene editing. Less rounds of selection and less generations are required as many improvement genes are already fixed. Remaining resources can be used to generate cultivars with more favourable alleles (circles) and less unfavourable alleles (triangles). Different coloured circles and triangles represent different genetic loci.
There are many examples of monogenic improvement traits which have functionally characterized corresponding genes/alleles, or for which some genetic sequence information is available (Table 1). For example, the wheat dwarfing alleles Rht‐B1b (formerly Rht1) and Rht‐D1b (Rht2) each contain a single base‐pair mutation which gives rise to a premature STOP codon, resulting in a truncated protein with altered function in gibberellin signal transduction (Table 1). Similarly, semi‐dwarf rice varieties contain an sd1 allele which impacts gibberellin biosynthesis (Hedden, 2003). These alleles confer reduced plant height, resulting in increased harvest index, and contributed to the yield increases exemplified during the green revolution (Hedden, 2003). In these examples, gene editing technology provides a simple and precise approach to generate desirable alleles of dwarfing genes in otherwise agronomically superior cultivars.
Table 1.
Potential gene targets
| Crop | Gene | Allele | Sequence variation | Phenotype | References |
|---|---|---|---|---|---|
| Wheat | Reduced height (Rht)‐B1b and Rht‐D1b | Rht‐B1a, Rht‐B1b, Rht‐D1a, Rht‐D1b | SNP | Reduced plant height | Peng et al. (1999), Ellis et al. (2002) |
| Ppd‐D1 | Ppd‐D1a, Ppd‐D1b | Large deletions/insertions within promoter region Or copy number variation | Photoperiod insensitivity, and flowering under both SD and LD photoperiods. | Beales et al. (2007) | |
| GW2‐A1 | G2373A | SNP | increased TGW | Simmonds et al. (2016) | |
| Pina‐D | Various | Various (mainly SNPs) | Grain hardness | Bhave and Morris (2008), Chen et al. (2012) | |
| NAM‐B1 | Gpc‐B1 | SNP | Grain protein content | Uauy et al. (2006) | |
| Lr34 | Lr34res | SNPs | Disease resistance | Dakouri et al. (2010), Krattinger et al. (2009), Chauhan et al. (2015) | |
| Lr67 | LR67res | SNPs | Disease resistance | Moore et al. (2015) | |
| MLO | mlo | SNPs | Disease resistance | Büschges et al. (1997), Wang et al. (2014) | |
| Rice | GW2 | Loss of function | Increased TGW | Song et al. (2007) | |
| NRT | NRT1.1B | SNP | Higher nitrogen use efficiency | Hu et al. (2015), Li et al. (2018a,b) | |
| Os8N3 | Loss of function | Disease resistance | Kim et al. (2019) | ||
| ALS1 | SNP | Herbicide tolerance | Kuang et al. (2020) | ||
| Oilseeds | FAD2 and FAD3 | SNP | High oleic oil content | Yang et al. (2012), Haun et al. (2014), Okuzaki et al. (2018), Jiang et al. (2017), Morineau et al. (2017) | |
| ALS1 | SNP | Herbicide tolerance | Li et al. (2015) |
The development of CRISPR/Cas‐based gene editing has created an avenue for creation of favourable alleles in germplasm early in the breeding cycle (Figure 1a,b). By fixing these genes, breeders can increase the population size of plants with favourable alleles and less allelic diversity is lost during initial selection rounds. By fixing a collection of improvement traits which are monogenic using CRISPR/Cas‐based gene editing, breeding programmes would go through a ripple of domestication. By fixing a collection of monogenic traits plant breeders could phenotype and perform selections from a population of plants which already have a basic complement of non‐segregating traits (i.e. height; Table 1).
Furthermore, gene editing can be used to save ‘near‐miss’ varieties of inbred crops (Figure 2). During the 8–12 year breeding process, new varieties are developed which exhibit many desirable features such as disease resistance or high yield. However, unanticipated changes in another trait often impact the classification options or market viability of the variety. For example, in wheat, resistance to Fusarium head blight (FHB) is highly desirable; however, susceptibility to FHB is associated with the Rht‐D1b semi‐dwarfing allele, likely through linkage drag (He et al., 2016; Hilton et al., 1999; Srinivasachary, et al., 2008). While FHB resistance is multifaceted, the Rht‐D1b semi‐dwarfing allele is associated with a single base pair polymorphism, making it a simple target for gene editing. Use of gene editing technology with these near‐miss varieties would prevent the loss of those valuable varieties.
Figure 2.
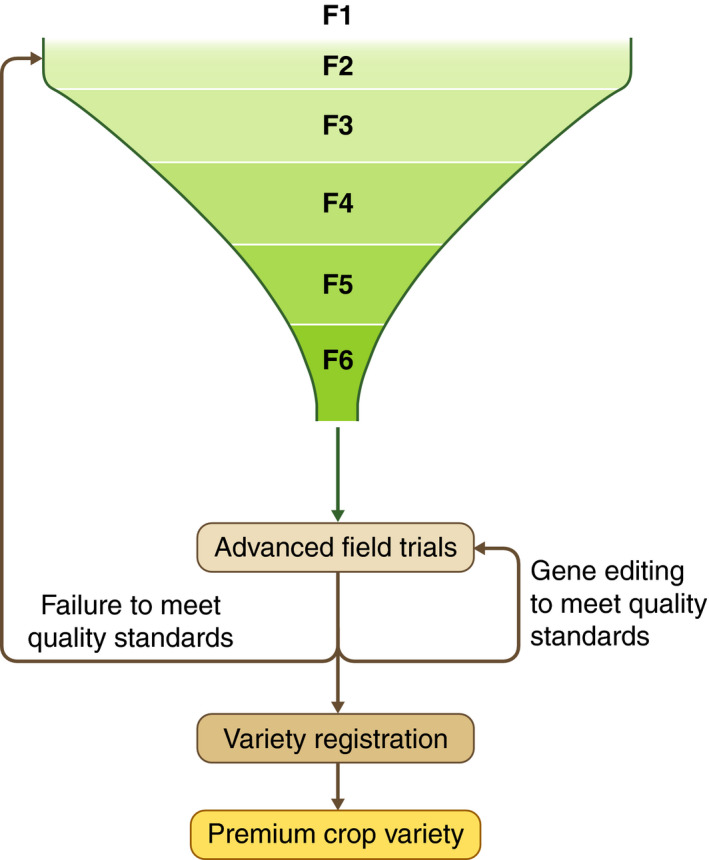
Gene editing to save near‐miss varieties of inbred crops Crop breeding starts at the F1 generation and after multiple rounds of selection, generations and field trials a variety may fail to meet quality standard before final variety registration. Instead of placing these near‐miss varieties back into the breeding cycle, gene editing can be used to rescue these near‐miss varieties saving time and resources.
Potential gene targets
Enhanced yield, yield stability and better seed quality are the most complex yet important objectives of crop breeding. The regulators of yield, adaptation and quality could be the targets for gene editing and modification for crop improvement. A major QTL for rice grain width and weight, grain weight 2 (GW2), was mapped to a gene encoding an ubiquitin E3 ligase. The WY3 allele of GW2 is a null allele and a regulator of cell division, leading to an increase in grain width and weight in rice (Song et al., 2007). Homoeologs of GW2 corresponding to A, B and D genomes have been identified in wheat. Analysis of both hexaploid and tetraploid wheat found that a GW2‐A1 mutant allele significantly increased thousand grain weight (TGW), grain width and grain length (Simmonds et al., 2016). Likewise, null mutations in B and D homoeologs also increase TGW, and combined mutations act additively (Wang et al., 2018; Zhang et al., 2018b). In both rice and wheat, GW2 alleles could be targeted in breeding programmes. As single‐QTL traits, these are attractive targets for genetic fixing using CRISPR/Cas‐based gene editing within a breeding scheme.
Depending on intended end‐use, gene editing could be used to develop a grain quality package consisting of multiple desirable alleles. For example, grain hardness can be addressed by editing the puroindoline‐a and puroindoline‐b (PIN) genes (Matus‐Cadez et al., 2008; Nadolska‐Orczyk et al., 2009). Wheat protein content can be improved by a single base pair edit in the TaNAM‐B1 gene, a NAC transcription factor found in the Gpc‐B1 locus (Uauy et al., 2006). Fixing these alleles through gene editing would reduce or eliminate the need for selection for these desirable alleles. The elite rice indica nitrate transporter allele, NRT1.1B, has recently been edited into japonica rice by altering a single nucleotide, and is another desirable allele which could be fixed in breeding germplasm (Hu et al., 2015; Li et al., 2018a,b). Similarly, in three elite rice varieties, the white pericarp was converted to a desirable wild health‐promoting red pericarp colour without loss of yield. This was achieved through CRISPR‐mediated editing and restoration of the (rc) allele (Zhu et al., 2019).
In oilseeds, the proportions of the unsaturated fatty acids, oleic acid (C18:1), linoleic acid (C18:2) and linolenic acid (C18:3) are impacted by fatty acid desaturase 2 (FAD2) and FAD3. Natural alleles of these genes are targets of allele‐specific markers for high oleic and low linolenic lines in Brassica napus (Yang et al., 2012). Recently, FAD2 and FAD3 have been gene editing targets for successful manipulation of oil content in Camelina sativa, soybean and B. napus (Haun et al., 2014; Jiang et al., 2017; Morineau et al., 2017; Okuzaki et al., 2018). Seed storage proteins in C. sativa have also been targeted to modify seed protein meal properties (Lyzenga et al., 2019).
Robust disease resistance is another characteristic often affected by simple allelic differences. In wheat and barley, natural and edited null mutations in the Mildew resistance locus o (Mlo) gene provides resistance against the pathogen Blumeria graminis f.sp. hordei (Bgh) (Acevedo‐Garcia et al., 2014; Acevedo‐Garcia et al., 2017; Wang et al., 2014). The wheat resistance allele Lr34(res) provides durable resistance against several pathogens, including leaf rust, stripe rust and powdery mildew, and has been widely targeted in wheat breeding programmes (Kolmer et al., 2008). The resistant allele of Lr34(res) differs from the susceptible allele by genetic polymorphisms which change two amino acids in predicted transmembrane helices of an ABC transporter (Krattinger et al., 2009; Risk et al., 2012). Fixing this allele using gene editing would greatly benefit subsequent breeding programmes.
Introduce genetic variation
The processes of domestication followed by intensive breeding have resulted in a genetic bottleneck, and many modern crop germplasms have genomic regions of reduced genetic diversity (Shi and Lai, 2015). Therefore, depending on the species and the traits of interest, breeding programmes may have limited allelic variation from which to select and improve traits. Trait variation from landraces and wild cultivars can be introduced through introgression breeding, but this process can be extremely tedious, time consuming and resource intensive. Incorporating only the beneficial allelic variation into elite lines while leaving behind maladapted genetic material is a major challenge, impacted by recombination rates, homology, species and population size. CRISPR/Cas‐based gene editing has emerged as tool for the generation of novel and superior alleles within crop germplasm or within elite lines (Nogue et al., 2016; Rodríguez‐Leal et al., 2017; Shen et al., 2017). In contrast to random mutagenesis through chemicals (such as ethyl methanesulphonate (EMS)) or gamma irradiation and subsequent genome sequencing (Xu et al., 2017), CRISPR/Cas‐based gene editing can be targeted by multiple gRNAs to genomic regulatory regions of interest such as promoters, developmental regulators and transcription factors to promote constrained mutagenesis within a specific region (Figure 3). Since CRISPR/Cas‐based gene editing can be easily multiplexed, multiple genetic regions can be targeted simultaneously.
Figure 3.
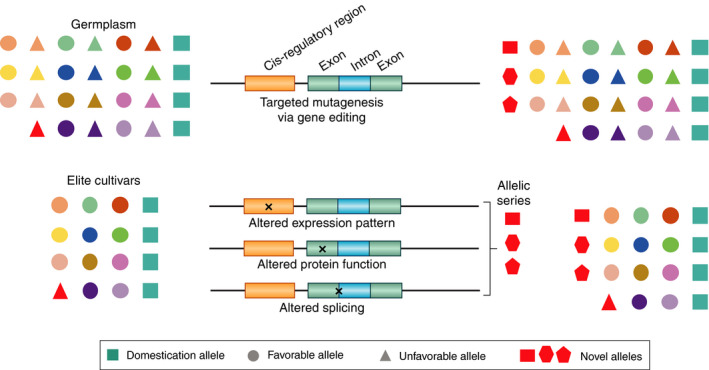
CRISPR/Cas‐based gene editing to introduce genetic variation in specific genomic regions using germplasm or elite cultivars. Semi‐random mutations in cis‐regulatory elements, exons and introns can lead to alteration in expression, altered protein function (loss‐of‐function, gain‐of‐function, and change in activity) and alternative splicing respectively. Mutations can lead to a number of different mutations and generate an allelic series in either germplasm or elite cultivars. Different coloured circles and triangles represent different genetic loci.
Cis‐regulatory elements (CRE), which have likely been a driving force in crop domestication, will be ideal target regions for generating a range of phenotypes (Swinnen et al., 2016). Mutations in CREs generally result in spatial and temporal changes in gene expression, which can result in favourable traits with low pleiotropic effects (Swinnen et al., 2016). CRISPR/Cas‐based gene editing has been deployed to engineer quantitative trait variation by specifically mutagenizing cis‐regulatory regions (Rodríguez‐Leal et al., 2017). This approach, which generates an allelic series for a variety of traits of interest, could be applied to many agriculturally relevant species. In tomato, semi‐random CRISPR‐induced mutations in the promoter region upstream of the CLAVATA (SlCLV3) coding sequence resulted in variable fruit size (Rodríguez‐Leal et al., 2017). A similar approach has been applied to the protein coding region of gene targets to achieve directed evolution for engineering improved or new functions in plants (Butt et al., 2019). For example, a combination of base‐editing‐mediated gene evolution tactics led to the development of novel variants of OsALS1, which confer resistance to the herbicide bispyribac‐sodium (Kuang et al., 2020). Recently, a CRISPR/Cas9 derivative system has been developed in bacteria which likely generates higher levels of mutations and subsequent genomic diversity within a larger genomic region. In the EvolvR system, CRISPR/Cas9 function is merged with an error‐prone DNA polymerase (Halperin et al., 2018). The genomic locus of interest is targeted through the gRNA, and Cas9 generates a single stranded break. The error‐prone polymerase amplifies the strand, introducing errors and thereby generating novel genetic diversity (Halperin et al., 2018). This approach does not require a double‐stranded DNA break or a sophisticated DNA repair pathway, and can produce mutations within a large window (350 bp). The use of this system in eukaryotes and plants remains to be demonstrated.
Recreating adaptive traits for de novo domestication of wild relatives, and evaluating breeding value of exotic germplasm
Wild relatives of modern crops and orphan crops can be regarded as a source of novel genetic variation and desirable traits not found in cultivated crops. However, traits such as small fruit size, low yield and undesirable plant architecture constrain commercial cultivation. Recently, the concept of de novo domestication through gene editing has been explored as a mechanism to domesticate wild and orphan crops quickly, and thus benefit from retained genetic variation as well as from the features of domesticated crops (Zsögön et al., 2017, 2018). This is largely possible, since many traditional domestication genes are ideal candidates for CRISPR/Cas‐based gene editing: they are well characterized, have simple genetic architecture and are monogenetic. In a wild relative of tomato, Solanum pimpinellifolium, six loci important in domestication were simultaneously edited to generate loss‐of‐function alleles. The resulting plants had changes in fruit number (MULT), size (FW2.2, FAS), shape (O gene; OVATE), nutritional content (LYCOPENE BETA CYCLASE) and plant architecture (SP gene; SELF‐PRUNING) (Zsögön et al., 2018). Similarly, domestication genes impacting day‐length insensitivity (SP5G), fruit size (SICLV3, SIWUS), vitamin C levels (SIGGP1) and plant architecture (SP) were stacked in accessions of S. pimpinellifolium with disease and salt tolerance (Li et al., 2018a,b). Another study targeted the orphan crop ‘groundcherry’ (Physalis pruinosa), which has a number of undesirable traits, including sprawling growth pattern, small fruit and strong stem abscission leading to fruit dropping to the ground. CRISPR/Cas‐based gene editing was used to edit the SP5G gene, which resulted in higher concentrations of fruit along each shoot, and the CLV1 gene, which resulted in larger fruits (Lemmon et al., 2018). These studies demonstrate that CRISPR/Cas‐based gene editing can accelerate domestication and increase the value and use of orphan crops or wild relatives (Figure 4).
Figure 4.
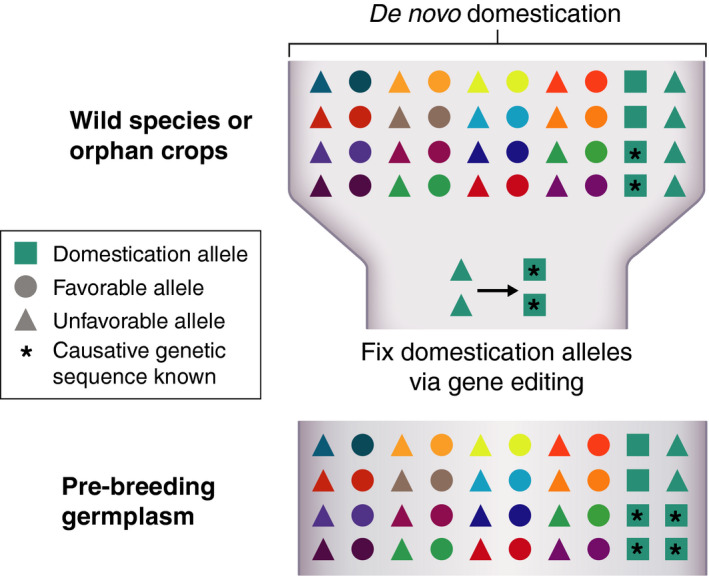
CRISPR/Cas‐based gene editing for de novo domestication. Gene editing of domestication alleles in wild species or orphan crops can generate suitable pre‐breeding germplasm. Different coloured circles and triangles represent different genetic loci.
Determining the average downstream performance, or breeding value, of exotic germplasm is also important in selecting which germplasm to integrate into a breeding programme. While exotic germplasm may have many desirable alleles, the overall breeding value of those alleles can be difficult to assess; variation in phenology, height and flowering time can together mask agronomic potential (Hussain et al., 2018). One approach to assess the breeding value of exotic germplasm is to use hybrid wheat technology. With this strategy, elite lines are crossed with the novel genetic material for which breeding value needs to be determined. The heterozygous hybrids of these crosses express dominant alleles for phenology, thereby minimizing the effects of deleterious alleles. While this approach is feasible, deleterious expression may not be fully masked, especially in cases where desirable alleles governing phenology and adaptation are recessive. An alternative approach is to utilize CRISPR/Cas‐based multiplex gene editing coupled to haploid induction editing technology (HI‐EDIT; Kelliher et al., 2019), to edit domestication genes in the exotic genetic resources. Similar to de novo domestication, this technique could be used to re‐domesticate wheat’s wild progenitors to assess their ‘hidden’ breeding value for multi‐genic traits. For example, the prevalence of undesirable alleles of adaptation traits (height, flowering, photoperiod and grain threshability; Table 2) in the exotic germplasm of wheat masks their agronomic potential. CRISPR/Cas9‐based gene editing can be used to fix the allelic modifications, involving point mutations, deletion or substitution (Table 2), required to recreate adaptive traits in exotic germplasm, thereby unmasking beneficial genetic material and supporting pre‐breeding.
Table 2.
Genome editing targets for de novo domestication of exotic germplasm of wheat
| Trait | Phenotype | Gene | Allele | Sequence variation | References |
|---|---|---|---|---|---|
| Reduced plant height | Short vs. tall | Rht‐B1 | Rht‐B1b (Rht1) | SNP | Peng et al. (1999) |
| Photoperiod response | Insensitive vs. sensitive – Flowering time | Ppd‐A1 | GS100, GS105 | 1.1 kb deletion | Wilhelm et al. (2009) |
| Vernalization response | Insensitive vs. sensitive – Flowering time | VRN2 | Loss of function | SNPs and deletions | Yan et al. (2004) |
| Grain threshability | Naked vs. hulled grains – Free‐threshing | Q (WAP2) | Q | I329V amino acid substitution | Simons et al. (2006) |
CRISPR gene editing to promote recombination at specified genomic regions
Meiotic recombination plays a foundational role in plant breeding, as it allows for allele reshuffling and creates novel allelic combinations. Recombination frequencies can be increased by inducing double‐stranded DNA breaks using chemical agents or physical stress, such as temperature shock or UV exposure (Wijnker and de Jong, 2008). Meiotic recombination is critical in introgresssion of a beneficial locus from a donor line into an elite line through backcrossing (Moose and Mumm, 2008). Ideally, backcrossing would result in a progeny containing just a small locus (introgression) from the donor line. This ideal backcross scenario requires meiotic recombination to occur between the parental chromosomes close to the region of interest. However, meiotic recombination is not evenly distributed along the chromosome, occurring most frequently in regions termed hotspots, and supressed in other regions such as the heterochromatic regions around centromeres (Choi and Henderson, 2017). For example, in wheat, crossover events mainly occur at the distal region of both arms of the chromosomes, while recombination is largely absent in the centromere proximal region (Choulet et al., 2014; Gardiner et al., 2019). As a result of unequal recombination frequency, plants containing a small introgression are rare. In addition, when a desired locus is contained within a non‐recombining chromosomal region, introgression into an elite line is near impossible.
Similarly, meiotic recombination is important to breakup genetic linkages. Desirable allelic composition can be affected generally by linkage drag; when undesirable loci are inherited along with selected desirable loci. Breeders are often faced with the introduction of undesirable phenotypic effects owing to the presence of these unfavourable linked loci, particularly when working with genetically distinct parents (Bai et al., 2013; Brown, 2002; Hospital, 2005). Unintended linkage drag has likely been a routine aspect of breeding programmes (Lin et al., 2014). For example, selection for haplotypes controlling heading date have selected against favourable haplotypes impacting root biomass (Voss‐Fels et al., 2017).
Frequently, the specific DNA sequence that underlies a beneficial locus is unknown, making gene editing of that specific locus not possible. However, the ability to promote homologous recombination at a specific genomic location through CRISPR/Cas‐based gene editing would provide breeders with a precise tool for the introgression of beneficial loci (Figure 5). Because of its ability to target specific genomic regions and ability to generate double‐stranded DNA breaks, CRISPR/Cas gene editing is beginning to be used to promote recombination at specific genomic regions (Filler Hayut et al., 2017; Sarno et al., 2017). In yeast, the left arm of chromosome 7 was targeted with 95 gRNAs to induce mitotic recombination (Sadhu et al., 2016). The resulting homologous recombination events generated a ‘loss of heterozygosity’ panel and allowed for the fine mapping of manganese sensitivity to a single polymorphism. While used in this study for trait mapping, this general approach could also be applied to reshuffle alleles in a low‐recombining regions along a chromosome. In tomato, genomic sections of linked loci represent approximately 25% of the assembled genome (Lin et al., 2014). This is a prime example of where CRISPR/Cas‐based gene editing could be used for generation of recombinant individuals, generating diversity and breaking up these genetic linkages (Figure 5).
Figure 5.
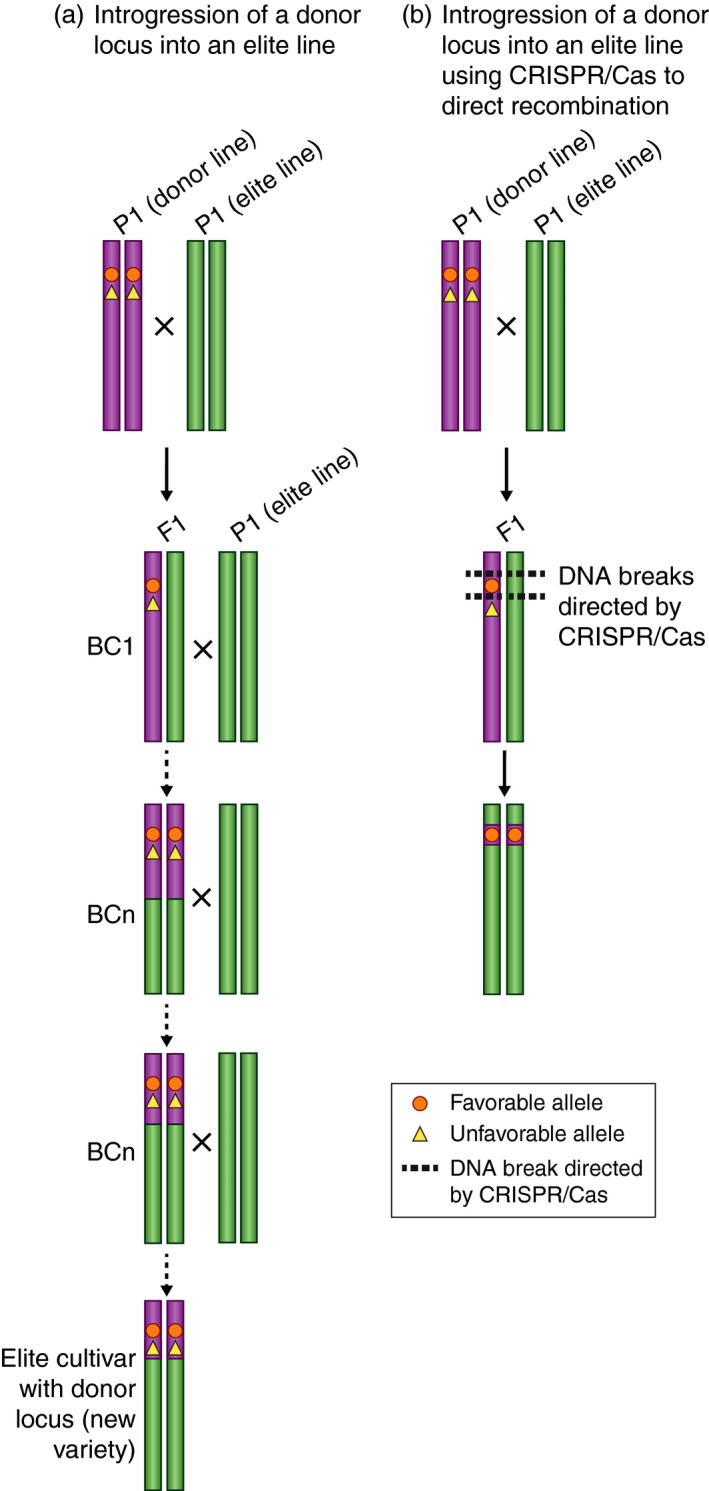
CRISPR/Cas‐based gene editing for introgression and separating linked genetic loci. (a) Schematic showing introgression of a donor locus using backcross breeding. (b) CRISPR/Cas‐based gene editing could be used to promote recombination in an F1 hybrid near a desirable loci resulting in introgression of donor locus and to promote recombination between a desirable and undesirable locus located in close proximately to each other resulting in a break in genetic linkage.
Regulation of genome editing
Only a limited number of countries have developed specific guidelines or regulations regarding gene edited crops. In 2018, the USDA ruled that gene edited crop varieties do not require additional regulatory oversight by the USDA, provided they do not involve plant pests (or contain foreign DNA from plant pests) such as viruses or bacteria (USDA, 2018). However, depending on the traits of gene edited crops they may be subject to regulation through the Environmental Protection Agency (EPA) and /or the Food and Drug Administration (FDA). Recently in 2020, the SECURE (sustainable, ecological, consistent, uniform, responsible, efficient) platform was developed to streamline and update biotechnology approval in the US (Barrangou, 2020).
In contrast to the USA, the European Union (EU) ruled that the regulations for gene edited crop varieties would equivalent to those regulations that exist for genetically modified organism (GMO) products. Other countries, such as Australia, have taken a more nuanced approach and categorize gene edited crops into three groups SDN‐1 (point mutations), SDN‐2 (short insertions or editing of a few base‐pairs by an external DNA‐template sequence) and SDN‐3 (the insertion of longer strands). Each of these groups are subject to different regulations, for example, SDN‐1 type edits are not subject to regulation through the Office of the Gene Technology Regulator (OGTR) while SDN‐2 and SDN‐3 edits are subject to regulation (Menz et al., 2020; Thygesen, 2019).
Canada uses a regulatory system that evaluates the final product, rather than the process used to create the product. As such, gene edited crops fall within the regulation of plants with novel traits (PNTs). Under these regulations, newly developed crops containing novel traits are subject to environmental and safety assessments, regardless of how they were generated (e.g. conventional breeding, mutagenesis, transgenesis or gene editing; Smyth, 2017). China is unique in that, despite heavy investment in genome editing research, it has not provided regulatory guidelines on gene edited crops (Cohen and Desai, 2019). The global landscape of gene editing regulation is unsettled and, in many countries, needs to be updated. Global attitudes towards the fine nuances of gene editing and will have huge impacts on how this technology is implemented and traded across the world.
Conclusions
Domestication and plant breeding have led to high yielding crop varieties which are adapted to local growing conditions. However, the growing human population faces a number of agricultural challenges, including climate change, changes in abiotic and biotic stressors and a loss of arable land, along with a demand for more sustainable and precise agricultural practices. Many crop traits have been fixed through initial waves of domestication and in this review we discussed fixation of another wave of important traits. CRISPR/Cas‐based gene editing provides a means by which we can create naturally occurring allelic variants without the constraints of traditional introgression breeding. In addition, we can create new desirable genetic variants and counteract some of the loss of allelic diversity which has occurred through selective breeding. By directing meiotic recombination through CRISPR/Cas, we may also be able to manipulate genetic allele shuffling and produce plants with more desirable allelic combinations. However, the regulatory framework surrounding gene edited plant lines will impact how and where this technology is realized. Gene editing provides an exciting opportunity to blend functional gene characterization with applied plant breeding.
Conflict of Interest
The authors declare that they have no competing interests.
Author contributions
WJL and SK developed the conceptual structure and outline of the review. WJL prepared the original draft. CJP and SK contributed to specific sections and edited the manuscript.
Acknowledgements
The authors thank Drs. Tim Sharbel and Joanne Ernest for critical reading of the manuscript, and Debbie Maizels (Zoobotanica) for the artwork in figures. The authors acknowledge funding provided by the ‘4D Wheat: Diversity, Domestication, Discovery and Delivery’ project, funded by Genome Canada, Agriculture and Agri‐Food Canada, Western Grains Research Foundation, Saskatchewan Ministry of Agriculture, Saskatchewan Wheat Development Commission, Alberta Wheat Commission, Manitoba Crop Alliance, Ontario Research Fund, Viterra, Canadian Agricultural Partnership, and Illumina. The authors also acknowledge the administrative support of Genome Prairie.
Lyzenga, W. J. , Pozniak, C. J. and Kagale, S. (2021) Advanced domestication: harnessing the precision of gene editing in crop breeding. Plant Biotechnol J, 10.1111/pbi.13576
References
- Abberton, M. , Batley, J. , Bentley, A. , Bryant, J. , Cai, H. , Cockram, J. , de Oliveira, A.C. et al. (2016) Global agricultural intensification during climate change: a role for genomics. Plant Biotechnol. J. 14, 1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo‐Garcia, J. , Kusch, S. and Panstruga, R. (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 204, 273–281. [DOI] [PubMed] [Google Scholar]
- Acevedo‐Garcia, J. , Spencer, D. , Thieron, H. , Reinstädler, A. , Hammond‐Kosack, K. , Phillips, A.L. and Panstruga, R. (2017) mlo‐based powdery mildew resistance in hexaploid bread wheat generated by a non‐transgenic TILLING approach. Plant Biotechnol J. 15, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaah, G. (2012) Principles of plant genetics and breeding. Chichester, UK: Wiley‐Blackwell. [Google Scholar]
- Alpert, K.B. and Tanksley, S.D. (1996) High‐resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato Proc . Natl. Acad. Sci. USA, 93, 15503–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altpeter, F. , Springer, N.M. , Bartley, L.E. , Blechl, A.E. , Brutnell, T.P. , Citovsky, V. , Conrad, L.J. et al. (2016) Advancing Crop Transformation in the Era of Genome Editing. Plant Cell. 28, 1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.A. , Stack, R.W. , Liu, S. , Waldron, B.L. , Fjeld, A.D. , Coyne, C. , Moreno‐Sevilla, B. et al. (2001) DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet. 102, 1164–1168. [Google Scholar]
- Ansari, W.A. , Chandanshive, S.U. , Bhatt, V. , Nadaf, A.B. , Vats, S. , Katara, J.L. , Sonah, H. et al. (2020) Genome Editing in Cereals: Approaches, Applications and Challenges. Int J Mol Sci. 21, 4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone, A.V. , Randolph, P.B. , Davis, J.R. , Sousa, A.A. , Koblan, L.W. and Levy, J.M. (2019) Search‐and‐replace genome editing without double‐strand breaks or donor DNA. Nature, 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, C. , Liang, Y. and Hawkesford, M.J. (2013) Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J Exp Bot. 64, 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou, R. (2020) Finding SECURE ground: USDA edits the biotechnology regulatory framework. CRISPR J. 3, 136–137. [DOI] [PubMed] [Google Scholar]
- Beales, J. , Turner, A. , Griffiths, S. , Snape, J.W. and Laurie, D.A. (2007) A pseudo‐response regulator is misexpressed in the photoperiod insensitive Ppd‐D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115, 721–733. [DOI] [PubMed] [Google Scholar]
- Bhave, M. and Morris, C.F. (2008) Molecular genetics of puroindolines and related genes: allelic diversity in wheat and other grasses. Plant Mol. Biol. 66, 205–219. [DOI] [PubMed] [Google Scholar]
- Bhowmik, P. , Ellison, E. , Polley, B. , Bollina, V. , Kulkarni, M. , Ghanbarnia, K. , Song, H. et al. (2018) Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 8, 6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi, L. , Zhu, C. , Zischewski, J. , Perez, L. , Bassie, L. , Nadi, R. , Forni, G. et al. (2016) Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 14, 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello, F. and Coelho, A.S. (2013) Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J. Agric. Food Chem. 4, 8277–8286. [DOI] [PubMed] [Google Scholar]
- Brown, J.K. (2002) Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 5, 339–344. [DOI] [PubMed] [Google Scholar]
- Brozynska, M. , Furtado, A. and Henry, R.J. (2016) Genomics of crop wild relatives: expanding the gene pool for crop improvement. Plant Biotechnol. J. 14, 1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges, R. , Hollricher, K. , Panstruga, R. , Simons, G. , Wolter, M. , Frijters, A. , van Daelen, R. et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell, 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Butt, H. , Eid, A. , Momin, A.A. , Bazin, J. , Crespi, M. , Arold, S.T. and Mahfouz, M.M. (2019) CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 20, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, H. , Boni, R. , Bucher, R. , Kuhn, B. , Buchmann, G. , Sucher, J. , Selter, L.L. et al. (2015) The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathways in transgenic barley. Plant J. 84, 202–215. [DOI] [PubMed] [Google Scholar]
- Chen, F. , Zhang, F.Y. , Xia, X.C. , Dong, Z.D. and Cui, D.Q. (2012) Distribution of puroindoline alleles in bread wheat cultivars of the Yellow and Huai valley of China and discovery of a novel puroindoline a allele without PINA protein. Mol. Breeding, 29, 371–378. [Google Scholar]
- Chen, H. , He, H. , Zhou, F. , Yu, H. and Deng, X.W. (2013) Development of genomics‐based genotyping platforms and their applications in rice breeding. Curr. Opin. Plant Biol. 16, 247–254. [DOI] [PubMed] [Google Scholar]
- Choi, K. (2017) Advances towards Controlling Meiotic Recombination for Plant Breeding. Mol. Cells, 40, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K. and Henderson, I. (2017) Meiotic recombination hotspots – a comparative view. Plant J. 83, 52–61. [DOI] [PubMed] [Google Scholar]
- Choulet, F. , Alberti, A. , Theil, S. , Glover, N. , Barbe, V. , Daron, J. , Pingault, L. et al. (2014) Structural and functional partitioning of bread wheat chromosome 3B. Science, 345, 1249721. [DOI] [PubMed] [Google Scholar]
- Cohen, J. and Desai, N. (2019) With its CRISPR Revolution, China Becomes a World Leader in Genome Editing. Available online at: https://www.sciencemag.org/news/2019/08/its‐crispr‐revolution‐china‐becomes‐world‐leader‐genome‐editing
- Crossa, J. , Pérez‐Rodríguez, P. , Cuevas, J. , Montesinos‐López, O. , Jarquín, D. , de Los Campos, G. , Burgueño, J. et al. (2017) Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. [DOI] [PubMed] [Google Scholar]
- Dakouri, A. , McCallum, B.D. , Walichnowski, A.Z. and Cloutier, S. (2010) Fine‐mapping of the leaf rust Lr34 locus in Triticum aestivum (L.) and characterization of large germplasm collections support the ABC transporter as essential for gene function. Theor. Appl. Genet. 121, 373–384. [DOI] [PubMed] [Google Scholar]
- Doebley, J. (2004) The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59. [DOI] [PubMed] [Google Scholar]
- Doebley, J.F. , Gaut, B.S. and Smith, B.D. (2006) The molecular genetics of crop domestication. Cell 29, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Gustus, C. (1995) teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics, 141, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. and Charpentier, E. (2014) Genome editing. The new frontier of genome engineering with CRISPR‐Cas9. Science 28, 1258096. [DOI] [PubMed] [Google Scholar]
- Ellis, M. , Spielmeyer, W. , Gale, K. , Rebetzke, J. and Richards, A. (2002) "Perfect" markers for the Rht‐B1b and Rht‐D1b dwarfing genes in wheat. Theor. Appl. Genet, 105, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Fauser, F. , Schiml, S. and Puchta, H. (2014) Both CRISPR/Cas‐based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. [DOI] [PubMed] [Google Scholar]
- Ferrie, A.M.R. , Bhowmik, P. , Rajagopalan, N. and Kagale, S. (2020) CRISPR/Cas9‐Mediated Targeted Mutagenesis in Wheat Doubled Haploids. Methods Mol Biol. 2072, 183–198. [DOI] [PubMed] [Google Scholar]
- Filler Hayut, S. , Melamed Bessudo, C. and Levy, A.A. (2017) Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat Commun. 6, 15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary, A. , Nesbitt, T.C. , Grandillo, S. , Knaap, E. , Cong, B. , Liu, J. , Meller, J. et al. (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science, 289, 85–88. [DOI] [PubMed] [Google Scholar]
- Fridman, E. and Zamir, D. (2012) Next‐generation education in crop genetics. Curr. Opin. Plant Biol. 15, 218–223. [DOI] [PubMed] [Google Scholar]
- Gardiner, L.J. , Wingen, L.U. , Bailey, P. , Joynson, R. , Brabbs, T. , Wright, J. , Higgins, J.D. et al. (2019) Analysis of the recombination landscape of hexaploid bread wheat reveals genes controlling recombination and gene conversion frequency. Genome Biol. 20, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli, N.M. , Komor, A.C. , Rees, H.A. , Packer, M.S. , Badran, A.H. , Bryson, D.I. and Liu, D.R. (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliham, M. , Able, J.A. and Roy, S.J. (2017) Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J. 90, 898–917. [DOI] [PubMed] [Google Scholar]
- Halperin, S.O. , Tou, C.J. , Wong, E.B. , Modavi, C. , Schaffer, D.V. and Dueber, J.E. (2018) CRISPR‐guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature, 560, 248–252. [DOI] [PubMed] [Google Scholar]
- Haun, W. , Coffman, A. , Clasen, B.M. , Demorest, Z.L. , Lowy, A. , Ray, E. , Retterath, A. et al. (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. [DOI] [PubMed] [Google Scholar]
- He, X. , Singh, P.K. , Dreisigacker, S. , Singh, S. , Lillemo, M. and Duveiller, E. (2016) Dwarfing genes Rht‐B1b and Rht‐D1b Are associated with both type I FHB susceptibility and low anther extrusion in two bread wheat populations. PLoS ONE, 11, e0162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P. (2003) The genes of the green revolution. Trends Genet. 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Hilton, A.J. , Jenkinson, P. , Hollins, T.W. and Parry, D.W. (1999) Relationship between cultivar height and severity of Fusarium ear blight in wheat. Plant Pathol. 48, 202–208. [Google Scholar]
- Hospital, F. (2005) Selection in backcross programmes. Phil. Trans. R. Soc. B. 360, 1503–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Wang, W. , Ou, S. , Tang, J. , Li, H. , Che, R. , Zhang, Z. et al. (2015) Variation in NRT1.1B contributes to nitrate‐use divergence between rice subspecies. Nat Genet. 47, 834–838. [DOI] [PubMed] [Google Scholar]
- Hu, X. , Wang, C. , Liu, Q. , Fu, Y. and Wang, K. (2016) Targeted mutagenesis in rice using CRISPR‐Cpf1 system. J. Genet. Genomics, 44, 2016–2018. [DOI] [PubMed] [Google Scholar]
- Hussain, J. , Khaliq, T. , Ahmad, A. and Akhtar, J. (2018) Performance of four crop model for simulations of wheat phenology, leaf growth, biomass and yield across planting dates. PLoS ONE, 13, e0197546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W.Z. , Henry, I.M. , Lynagh, P.G. , Comai, L. , Cahoon, E.B. and Weeks, D.P. (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 17, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.C. , Yun, J.Y. , Kim, S.T. , Shin, Y. , Ryu, J. , Choi, M. , Woo, J.W. et al. (2018) Precision genome engineering through adenine base editing in plants. Nat Plants. 4, 427–443. [DOI] [PubMed] [Google Scholar]
- Kelliher, T. , Starr, D. , Su, X. , Tang, G. , Chen, Z. , Carter, J. , Wittich, P.E. et al. (2019) One‐step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 37, 287–292. [DOI] [PubMed] [Google Scholar]
- Kim, Y.A. , Moon, H. and Park, C.J. (2019) CRISPR/Cas9‐targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice, 12, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B.P. , Prew, M.S. , Tsai, S.Q. , Nguyen, N.T. , Topkar, V.V. , Zheng, Z. and Joung, J.K. (2015a) Broadening the targeting range of Staphylococcus aureus CRISPR‐Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, B.P. , Prew, M.S. , Tsai, S.Q. , Topkar, V.V. , Nguyen, N.T. , Zheng, Z. , Gonzales, A.P.W. et al. (2015b) Engineered CRISPR‐Cas9 nucleases with altered PAM specificities. Nature, 523, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmer, J.A. , Singh, R.P. , Garvin, D.F. , Viccars, L. , William, H.M. , Huerta‐Espino, J. , Ogbonnaya, F.C. et al. (2008) Analysis of the Lr34/Yr18 rust resistance region in wheat germplasm. Crop. Sci. 48, 1841–1852. [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Spielmeyer, W. , Singh, R.P. , Huerta‐Espino, J. , McFadden, H. , Bossolini, E. et al. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science, 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Kuang, Y. , Li, S. , Ren, B. , Yan, F. , Spetz, C. , Li, X. , Zhou, X. et al. (2020) Base‐editing‐mediated artificial evolution of OsALS1 in planta to develop novel herbicide‐tolerant rice germplasms. Mol. Plant, 13, 565–572. [DOI] [PubMed] [Google Scholar]
- Lemmon, Z.H. , Reem, N.T. , Dalrymple, J. , Soyk, S. , Swartwood, K.E. , Rodriguez‐Leal, D. , Van Eck, J. et al. (2018) Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants, 4, 766–770. [DOI] [PubMed] [Google Scholar]
- Li, J. , Zhang, X. Su, Y. Zhang, J. Du, W. Guo, X. Li, S. et al. (2018) Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. J. Integr. Plant Biol. 60, 536–540. [DOI] [PubMed] [Google Scholar]
- Li, T. , Yang, X. , Yu, Y. , Si, X. , Zhai, X. , Zhang, H. , Dong, W. et al. (2018) Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Liu, Z.B. , Xing, A. , Moon, B.P. , Koellhoffer, J.P. , Huang, L. , Ward, R.T. et al. (2015) Cas9‐Guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Q. , Zong, Y. , Xue, C. , Wang, S. , Jin, S. , Zhu, Z. , Wang, Y. et al. (2020) Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. [DOI] [PubMed] [Google Scholar]
- Lin, T. , Zhu, G. , Zhang, J. , Xu, X. , Yu, Q. , Zheng, Z. , Zhang, Z. et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226. [DOI] [PubMed] [Google Scholar]
- Lu, Y. and Zhu, J.K. (2017) Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant. 10, 523–525. [DOI] [PubMed] [Google Scholar]
- Lyzenga, W.J. , Harrington, M. , Bekkaoui, D. , Wigness, M. , Hegedus, D.D. and Rozwadowski, K.L. (2019) CRISPR/Cas9 editing of three CRUCIFERIN C homoeologues alters the seed protein profile in Camelina sativa . BMC Plant Biol. 19, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhang, X. , Liu, H. et al. (2020) Highly efficient DNA‐free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants, 6, 773–779. [DOI] [PubMed] [Google Scholar]
- Matus‐Cádiz, M.A. , Pozniak, C.J. and Hucl, P. (2008) Puroindoline allele diversity in Canadian and northern US hard spring wheat varieties differing in kernel hardness. Can. J. Plant Sci. 88, 873–883. [Google Scholar]
- Menz, J. , Modrzejewski, D. , Hartung, F. , Wilhelm, R. and Sprink, T. (2020) Genome edited crops touch the market: a view on the global development and regulatory environment. Front. Plant Sci. 11, 586027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, R.S. , DuVal, A.E. and Jensen, H.R. (2012) Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196, 29–48. [DOI] [PubMed] [Google Scholar]
- Mikami, M. , Toki, S. and Endo, M. (2016) Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 57, 1058–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.W. , Herrera‐Foessel, S. , Lan, C. , Schnippenkoetter, W. , Ayliffe, M. , Huerta‐Espino, J. , Lillemo, M. et al. (2015) A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. [DOI] [PubMed] [Google Scholar]
- Moose, S.P. and Mumm, R.H. (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 147, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morineau, C. , Bellec, Y. , Tellier, F. , Gissot, L. , Kelemen, Z. , Nogué, F. and Faure, J.D. (2017) Selective gene dosage by CRISPR‐Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murovec, J. , Pirc, Ž. and Yang, B. (2017) New variants of CRISPR RNA‐guided genome editing enzymes. Plant Biotechnol. J. 15, 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadolska‐Orczyk, A. , Gasparis, S. and Orczyk, W. (2009) The determinants of grain texture in cereals. J. Appl. Genet. 50, 185–197. [DOI] [PubMed] [Google Scholar]
- Nesbitt, T.C. and Tanksley, S.D. (2002) Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué, F. , Mara, K. , Collonnier, C. and Casacuberta, J.M. (2016) Genome engineering and plant breeding: impact on trait discovery and development. Plant Cell Rep. 35, 1475–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzaki, A. , Ogawa, T. , Koizuka, C. , Kaneko, K. , Inaba, M. , Imamura, J. and Koizuka, N. (2018) CRISPR/Cas9‐mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. [DOI] [PubMed] [Google Scholar]
- Olsen, K.M. and Wendel, J.F. (2013) A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 64, 47–70. [DOI] [PubMed] [Google Scholar]
- Peng, J. , Richards, D.E. , Hartley, N.M. , Murphy, G.P. , Devos, K.M. , Flintham, J.E. , Beales, J. et al. (1999) 'Green revolution' genes encode mutant gibberellin response modulators. Nature, 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Ran, F.A. , Cong, L. , Yan, W.X. , Scott, D.A. , Gootenberg, J.S. , Kriz, A.J. , Zetsche, B. et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520, 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risk, J.M. , Selter, L.L. , Krattinger, S.G. , Viccars, L.A. , Richardson, T.M. , Buesing, G. , Herren, G. et al. (2012) Functional variability of the Lr34 durable resistance gene in transgenic wheat. Plant Biotechnol. J. 10, 477–487. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Leal, D. , Lemmon, Z.H. , Man, J. , Bartlett, M.E. and Lippman, Z.B. (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell, 5, 470–480. [DOI] [PubMed] [Google Scholar]
- Sadhu, M.J. , Bloom, J.S. , Day, L. and Kruglyak, L. (2016) CRISPR‐directed mitotic recombination enables genetic mapping without crosses. Science, 27, 1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno, R. , Vicq, Y. , Uematsu, N. , Luka, M. , Lapierre, C. , Carroll, D. , Bastianelli, G. et al. (2017) Programming sites of meiotic crossovers using Spo11 fusion proteins. Nucleic Acids Res. 45, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. , Hua, Y. , Fu, Y. , Li, J. , Liu, Q. , Jiao, X. , Xin, G. , Wang, J. , Wang, X. , Yan, C. and Wang, K. (2017) Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci China Life Sci. 60, 506–515. [DOI] [PubMed] [Google Scholar]
- Shi, J. and Lai, J. (2015) Patterns of genomic changes with crop domestication and breeding. Curr. Opin. Plant Biol. 24, 47–53. [DOI] [PubMed] [Google Scholar]
- Shimatani, Z. , Ariizumi, T. , Fujikura, U. , Kond, A. , Ezura, H. and Nishida, K. (2019) Targeted base editing with CRISPR‐Deaminase in tomato. Methods Mol. Biol. 1917, 297–307. [DOI] [PubMed] [Google Scholar]
- Simmonds, J. , Scott, P. , Brinton, J. , Mestre, T.C. , Bush, M. , Del Blanco, A. , Dubcovsky, J. et al. (2016) A splice acceptor site mutation in TaGW2‐A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 129, 1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K.J. , Fellers, J.P. , Trick, H.N. , Zhang, Z. , Tai, Y.S. , Gill, B.S. and Faris, J.D. (2006) Molecular characterization of the major wheat domestication gene Q. Genetics, 172, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker, I.M. , Gao, L. , Zetsche, B. , Scott, D.A. , Yan, W.X. and Zhang, F. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science, 351, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, S.J. (2017) Canadian regulatory perspectives on genome engineered crops. GM Crops Food, 8, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.J. , Huang, W. , Shi, M. , Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Srinivasachary, Gosman, N. , Steed, A. , Simmonds, J. , Leverington‐Waite, M. , Wang, Y. et al. (2008) Susceptibility to Fusarium head blight is associated with the Rht‐D1b semi‐dwarfing allele in wheat. Theor. Appl. Genet. 116, 1145–1153. [DOI] [PubMed] [Google Scholar]
- Swinnen, G. , Goossens, A. and Pauwels, L. (2016) Lessons from domestication: Targeting Cis‐regulatory elements for crop improvement. Trends Plant Sci. 21, 506–515. [DOI] [PubMed] [Google Scholar]
- Thygesen, P. (2019) Clarifying the regulation of genome editing in Australia: situation for genetically modified organisms. Transgenic Res. 28, 151–159. [DOI] [PubMed] [Google Scholar]
- Tian, S. , Jiang, L. , Cui, X. , Zhang, J. , Guo, S. , Li, M. , Zhang, H. et al. (2018) Engineering herbicide‐resistant watermelon variety through CRISPR/Cas9‐mediated base‐editing. Plant Cell Rep. 37, 1353–1356. [DOI] [PubMed] [Google Scholar]
- Tsanova, T. , Stefanova, L. , Topalova, L. , Atanasov, A. and Pantchev, I. (2021) DNA‐free gene editing in plants: a brief overview. Biotechnol. Biotechnol. Equip. 35, 131–138. [Google Scholar]
- Uauy, C. , Distelfeld, A. , Fahima, T. , Blechl, A. and Dubcovsky, J. (2006) A NAC gene regulating senescence improves grain protein, zinc and irons content in wheat. Science, 314, 1298–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA (2018). https://www.usda.gov/media/press‐releases/2018/03/28/secretary‐perdue‐issues‐usda‐statement‐plant‐breeding‐innovation
- Veillet, F. , Perrot, L. , Chauvin, L. , Kermarrec, M.P. , Guyon‐Debast, A. , Chauvin, J.E. , Nogué, F. et al. (2019) Transgene‐free genome editing in tomato and potato plants using Agrobacterium‐mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, K.E. (2009) Backcross breeding. Methods Mol. Biol. 526, 161–169. [DOI] [PubMed] [Google Scholar]
- Voss‐Fels, K.P. , Qian, L. , Parra‐Londono, S. , Uptmoor, R. , Frisch, M. , Keeble‐Gagnère, G. , Appels, R. (2017) Linkage drag constrains the roots of modern wheat. Plant Cell Environ. 40, 717–725. [DOI] [PubMed] [Google Scholar]
- Voytas, D.F. (2013) Plant genome engineering with sequence‐specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. [DOI] [PubMed] [Google Scholar]
- Wang, T. , Zhang, H. and Zhu, H. (2019) CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 6, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Simmonds, J. , Pan, Q. , Davidson, D. , He, F. , Battal, A. , Akhunova, A. et al. (2018) Gene editing and mutagenesis reveal inter‐cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 131, 2463–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Weeks, D.P. (2017) Gene editing in polyploid crops: wheat, camelina, canola, potato, cotton, peanut, sugar cane, and citrus. Prog. Mol. Biol. Transl. Sci. 149, 65–80. [DOI] [PubMed] [Google Scholar]
- Wijnker, E. and de Jong, H. (2008) Managing meiotic recombination in plant breeding. Trends Plant Sci. 13(12), 640–646. [DOI] [PubMed] [Google Scholar]
- Wilhelm, E.P. , Turner, A.S. and Laurie, D.A. (2009) Photoperiod insensitive Ppd‐A1a mutations in tetraploid wheat (Triticum durum Desf.). Theor. Appl. Genet. 118, 285–294. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Qin, R. , Li, H. Li, D. , Li, L. , Wei, P. and Yang, J. (2017) Generation of targeted mutant rice using a CRISPR‐Cpf1 system. Plant Biotechnol. J. 15, 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L. , Loukoianov, A. , Blechl, A. , Tranquilli, G. , Ramakrishna, W. , SanMiguel, P. , Bennetzen, J.L. et al. (2004) The wheat VRN2 gene is a flowering repressor down‐regulated by Vernalization. Science, 303, 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q. , Fan, C. , Guo, Z. , Qin, J. , Wu, J. , Li, Q. , Fu, T. et al. (2012) Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele‐specific markers for high oleic and low linolenic acid contents. Theor. Appl. Genet. 125, 715–729. [DOI] [PubMed] [Google Scholar]
- Zetsche, B. , Gootenberg, J. S. , Abudayyeh, O. O. , Slaymaker, I. M. , Makarova, K. S. , Essletzbichler, P. , Volz, S. E. et al. (2015) Cpf1 Is a Single RNA‐Guided Endonuclease of a Class 2 CRISPR‐Cas System. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Rouse, M.N. , Nava, I.C. , Jin, Y. and Anderson, J.A. (2016) Development and verification of wheat germplasm containing both Sr2 and Fhb1. Mol. Breeding, 36, 85. [Google Scholar]
- Zhang, Y. , Li, D. , Zhang, D. , Zhao, X. , Cao, X. , Dong, L. , Liu, J. et al. (2018) Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Massel, K. , Godwin, I.D. and Gao, C. (2018) Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Belcram, H. , Gornicki, P. , Charles, M. , Just, J. , Huneau, C. , Magdelenat, G. et al. (2011) Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA, 108, 18737–18742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Lin, Y. , Chen, S. , Liu, H. , Chen, Z. , Fan, M. , Hu, T. et al. (2019) CRISPR/Cas9‐mediated functional recovery of the recessive rc allele to develop red rice. Plant Biotechnol. J. 17, 2096–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, Y. , Song, Q. , Li, C. , Jin, S. , Zhang, D. , Wang, Y. , Qiu, J.‐L. et al. (2018) Efficient C‐to‐T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953. [DOI] [PubMed] [Google Scholar]
- Zsögön, A. , Cermak, T. , Voytas, D. and Peres, L.E. (2017) Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: case study in tomato. Plant Sci. 256, 120–130. [DOI] [PubMed] [Google Scholar]
- Zsögön, A. , Čermák, T. , Naves, E.R. , Notini, M.M. , Edel, K.H. , Weinl, S. , Freschi, L. et al. (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol. 36, 1211–1216. [DOI] [PubMed] [Google Scholar]


