Abstract
Cell lines are widely used for various research purposes including cancer and drug research. Recently, there have been studies that pointed to discrepancies in the literature and usage of cell lines. That is why we have prepared a comprehensive overview of the most common gynaecological cancer cell lines, their literature, a list of currently available cell lines, and new findings compared with the original studies. A literature review was conducted via MEDLINE, PubMed and ScienceDirect for reviews in the last 5 years to identify research and other studies related to gynaecological cancer cell lines. We present an overview of the current literature with reference to the original studies and pointed to certain inconsistencies in the literature. The adherence to culturing rulesets and the international guidelines helps in minimizing replication failure between institutions. Evidence from the latest research suggests that despite certain drawbacks, variations of cancer cell lines can also be useful in regard to a more diverse genomic landscape.
Keywords: breast neoplasm, cell line, cervix cancer, endometrial neoplasms, gynaecology, in vitro techniques, pathology, tumour cell line
1. INTRODUCTION
There is a variety of methods to study the properties of malignancies. However, many of those methods depend on clinical subjects and/or patients and are in certain regards restricted/limited. A possible to avenue to form a better understanding of the carcinogenesis and behaviour of specific malignancies are cell cultures and cell lines (CLs). 1 , 2 , 3 Areas that commonly utilize CLs are pathology and oncology as well as pre‐clinical areas such as pharmacology. Research on cancer cell lines (CCLs) in these areas holds the potential to lead to translational, clinically applicable results. Several papers have discussed and compared different cell models. This includes CLs, in vivo models, or cell models that are derived from individual patients. It comes as no surprise that all have their drawbacks and advantages. One of the oldest and still most recurring problems in CL culturing remains to be the potential of genetic or epigenetic changes can potentially arise during their growth and use. The latter limits the correlation potential of results based on these with the properties of the primary tissue. 2 , 4 , 5 By this, also their clinical relevance might be questioned. Nevertheless, there are several obvious benefits of CCLs. Figure 1 shows some of these potential applications of CCLs in medicine and translational research.
FIGURE 1.
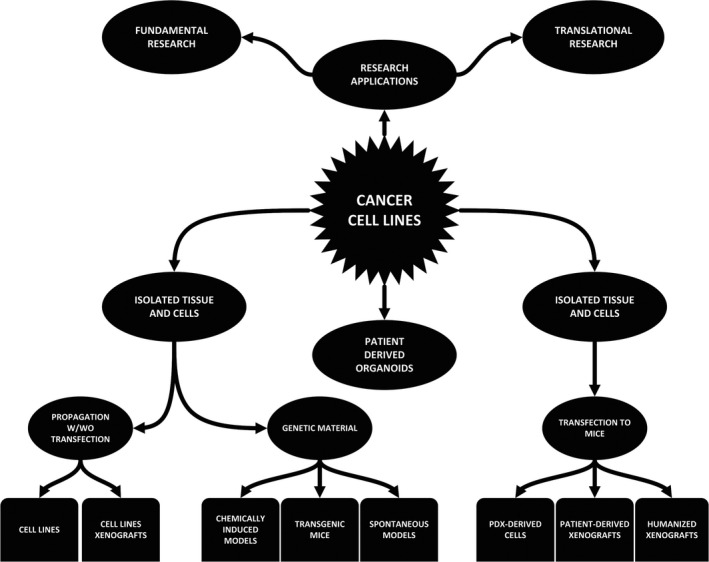
Potential CCL applications in research
The limitations of CCLs, considerations about their relevance for clinical outcomes, and the discrepancies in their characteristics through the decades, were discussed before. For example, a recent paper by Ben‐David et al presented evidence that points to a variety of mutational processes that affect the homogeneity of commonly used CCLs. On a practical level, this translates to the phenotypical characteristics of these CLs and their behaviour (therapy response). 6 , 7 Consequently, when considering the replicability of experiments, one understands that a number of factors can alter the final outcome. Some of these are as follows: (a) the number of population doublings that affect the cell geno‐ and phenotypes; (b) the origin of CCLs could be from the primary source tissue or a metastasis; (c) culture conditions could promote differentiation or even dedifferentiation. Despite this seemingly ‘negative’ prognosis to CL use, multiple mechanisms and rules have been proposed that help either prevent or at least help resolve these and other occurrences. Some of these recommendations and guidelines will be discussed in the later sections of the review.
In comparison with complex culturing techniques (eg transgenic mice, transfection‐based models and xenografts), CLs still provide a combination of a stable culturing setting, control over the experiment, relatively quick results and moderate expenses. 8 , 9 Furthermore, thanks to recent advances in genomic studies and the digitalization of data, specific CL properties can be simply checked via various databanks, including their possible uses and genetic heritage (eg Cellosaurus, ECLA). 10 , 11 , 12 , 13
We present an overview of three types of gynaecological CCLs. Firstly, we present our methodology of search. The next chapter will discuss breast cancer (BC) and its CLs, the following endometrial cancer (EC) and its CLs, the subsequent cervical cancer (CC), and its CLs. Finally, the last chapter will be dedicated to new emerging and potential methods of use for CL research and our own related experiences.
2. METHODS
A literature review was conducted via the biggest medical literature databases (MEDLINE, PubMed, ScienceDirect) to obtain studies related to gynaecological CCLs. The employed search terms in the form of keywords were “breast cancer cell lines”, “endometrial cell lines”, “cervical cancer cell lines”, “Gynaecological cancer cell lines”. Used MeSH identifiers were “Breast Neoplasms”, “cell line”, “cell lines, tumor”, “cell line, transformed”, “uterine cervical neoplasms” and “endometrial neoplasms”. With the help of this search algorithm and specific filters (5 years, human, review), we were able to find relevant new impactful studies on gynaecological CLs (Table 1). We specifically searched for the corresponding originators’ study which was crosschecked via the Cellosaurus database. The search inquiries and presentation of the final number of included studies after exclusion and filtering is presented in Figure 2 and has been prepared in accordance with the PRISMA guidelines for review articles.
TABLE 1.
Overview of the preformed search results
| Search terms | Results |
|---|---|
| ("Breast Neoplasms"[Mesh]) AND "Cell Line, Tumor"[Mesh] with filters (5‐year filter; review, human) | No. 96 |
| ("Cell Line, Tumor"[Mesh]) AND "Endometrial Neoplasms"[Mesh] (5‐year filter; review, human) | No. 3 |
| ("Cell Line"[Mesh]) AND "Uterine Cervical Neoplasms"[Mesh] (5‐year filter; review, human) | No. 7 |
| ("Uterine Cervical Neoplasms"[Mesh]) AND "Breast Neoplasms"[Mesh]) AND "Endometrial Neoplasms"[Mesh]) AND "Cell Line"[Mesh] (5‐year filter; review, human) | No. 1 |
FIGURE 2.
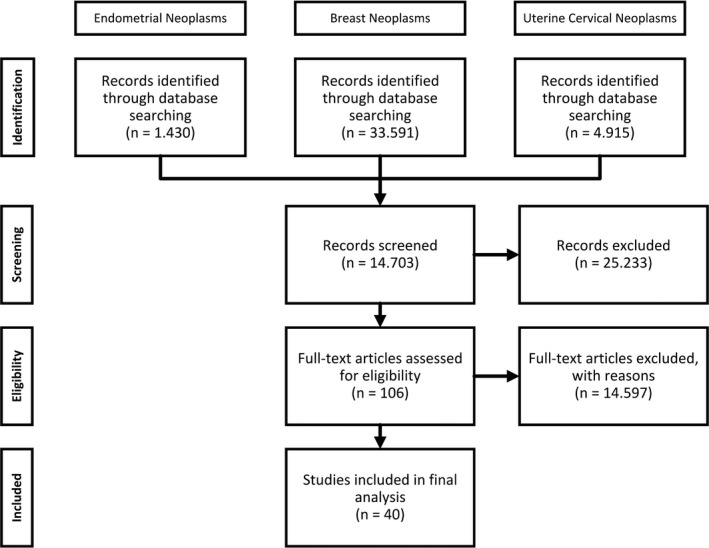
PRISMA diagram of the conducted search inquiries
3. BREAST CANCER AND ITS CELL LINES
3.1. Introduction
The second most common cancer worldwide is breast cancer. This type of cancer is ranked first in incidence in women living either in developed or developing countries (Table 2). It is ranked as the fifth common cause of cancer death (627 000 deaths/per year). 14 A short overview of the most important histological subtypes is shown in Table 3.
TABLE 2.
Epidemiological worldwide cancer statistics for 2018
| Estimated number of incident cases and deaths worldwide, both sexes, all ages (2018) | |||
|---|---|---|---|
| No. | Cancer | Incidence | Mortality |
| 1 | Lung | 2.093.876 | 1.761.007 |
| 2 | Breast | 2.088.849 | 626.679 |
| 3 | Colorectum | 1.849.518 | 880.792 |
| 4 | Prostate | 1.276.106 | 358.989 |
| 5 | Stomach | 1.033.701 | 782.685 |
| 6 | Liver | 841.080 | 781.631 |
| 7 | Oesophagus | 572.034 | 508.585 |
| 8 | Cervix uteri | 569.847 | 311.365 |
| 9 | Thyroid | 567.233 | 41.071 |
| 10 | Bladder | 549.393 | 199.922 |
| Estimated number of incident cases and deaths worldwide, females, all ages (2018) | |||
|---|---|---|---|
| No. | Cancer | Incidence | Mortality |
| 1 | Breast | 2.088.849 | 626.679 |
| 2 | Colorectum | 823.303 | 396.568 |
| 3 | Lung | 725.352 | 576.060 |
| 4 | Cervix uteri | 569.847 | 311.365 |
| 5 | Thyroid | 436.344 | 25.514 |
| 6 | Corpus uteri | 382.069 | 89.929 |
| 7 | Stomach | 349.947 | 269.130 |
| 8 | Ovary | 295.414 | 184.799 |
| 9 | Liver | 244.506 | 233.256 |
| 10 | Non‐Hodgkin lymphoma | 224.877 | 102.755 |
| Estimated age‐standardized incidence and mortality rates (World) in 2018 (Low income, Low‐middle income, females, all ages) | |||
|---|---|---|---|
| No. | Cancer | Incidence | Mortality |
| 1 | Breast | 31.1 | 14.9 |
| 2 | Cervix uteri | 17.5 | 11.5 |
| 3 | Colorectum | 6.3 | 4.1 |
| 4 | Ovary | 5.8 | 4 |
| 5 | Lung | 4.5 | 4.1 |
| 6 | Corpus uteri | 3.8 | 1.3 |
| 7 | Stomach | 3.7 | 3.3 |
| 8 | Liver | 3.4 | 3.3 |
| 9 | Lip, oral cavity | 3.2 | 2.4 |
| 10 | Thyroid | 3.2 | 0.54 |
| Estimated number deaths worldwide, both sexes, all ages (2018) | ||
|---|---|---|
| No. | Cancer | Mortality |
| 1 | Lung | 1.761.007 |
| 2 | Colorectum | 880.792 |
| 3 | Stomach | 782.685 |
| 4 | Liver | 781.631 |
| 5 | Breast | 626.679 |
| 6 | Oesophagus | 508.585 |
| 7 | Pancreas | 432.242 |
| 8 | Prostate | 358.989 |
| 9 | Cervix uteri | 311.365 |
| 10 | Leukaemia | 309.006 |
Data summarized from the Globocan database. 14
TABLE 3.
Breast cancer subtypes
| Non‐invasive histological types | |
|---|---|
| Adenocarcinoma (99.9%) | Ductal carcinoma in situ (DCIS) (80.1%) |
| Lobular carcinoma in situ (LCIS) (15.9%) | |
| Intraductal and lobular in situ carcinoma (3.4%) | |
| Other adenocarcinomas (0.5%) | |
| Other in situ histologies (0.1%) | |
| Invasive histological types a | ||
|---|---|---|
| Carcinoma (99.5%) | Adenocarcinoma (97.7%) | Infiltrating duct carcinoma (72.4%) |
| Lobular carcinoma, NOS (9.9%) | ||
| Mixed NST and special subtypes (9.7%) | ||
| Mucinous adenocarcinoma (1.8%) | ||
| Other adenocarcinomas (1.5%) | ||
| Papillary adenocarcinoma (0.8%) | ||
| Adenocarcinoma NOS (0.6%) | ||
| Tubular adenocarcinoma (0.5%) | ||
| Paget disease (0.3%) | ||
| Inflammatory adenocarcinoma (0.2%) | ||
| Unspecified, carcinoma, NOS (0.9%) | ||
| Other specific carcinoma (0.8%) | ||
| Epidermoid carcinoma (cca. 0.2%) | ||
| Sarcoma and soft tissue tumours (0.1%) | Hemangiosarcomas (cca. 0.1%) | |
| Other sarcomas (cca. 0.1%) | ||
| Other specific types (0.2%) | Phyllodes tumour, malignant (0.2%) | |
| Other (0.0%) | ||
| Unspecified (0.2%) | ||
| Intrinsic subtype | IHC status | Grade | Prevalence |
|---|---|---|---|
| Luminal type A | (ER+, PR+), HER2‐, Ki‐67‐(<15%) | 1/2 | 23.7% |
| Luminal type B |
(ER+, PR+), HER2‐, Ki‐67+(>15%) (ER+, PR‐), HER2‐, Ki‐67+(>15%) |
2/3 |
38.8% 14% |
| HER2 | (ER‐, PR‐), HER2+ | 2/3 | 11.2% |
| Basal | (ER‐, PR‐), HER2‐, basal markers+ | 3 | 12.3% |
| Normal like | (ER+, PR+), HER2‐, Ki‐67‐ (<15%) | 1/2/3 | 7.8% |
3.1.1. Invasive breast cancer
We distinguish multiple different types of invasive breast cancer (IBC). The distinctions are based on the molecular as well as histopathological properties. 15 In a routine diagnostic procedure, multiple different criteria will be gathered and analysed; tumour size and grade (Elston and Ellis); presence of lymphovascular invasion; DCIS and LCIS; assessment of surgical margins; the immunohistochemical profile of hormonal receptors (eg oestrogen – ER (most commonly the alpha receptor), progesterone – PR), the HER2 receptor status and analysis of proliferation index, determined with the Ki‐67 (Mib 1) percentage of nuclear expression, is an additional independent prognostic parameter for DFS and OS in BC patients. 16 Furthermore, proteases and genetic profiling can be of great help in determining the need for additional treatment. These features are not only of great importance for the clinician but also of great importance for the researcher during CL culturing. Namely, for successful culturing, the scientist must know the specific properties to provide and use an adequate environment, culturing methods, characterization methods, etc, to preserve the characteristics of the source tissue. Primary BC can be classified into two categories. The first are the non‐invasive BCs. These are ductal carcinoma in situ (DCIS) and lobular carcinoma in situ (LCIS). These types of cancer are confined within the milk ducts and lobules of the breast and surrounded by an intact basement membrane and myoepithelial cells (Table 3). 16 More common are invasive BC types. The term invasive breast carcinoma (IBC) refers to a large heterogeneous group of malignant epithelial neoplasms of the breast.
Due to treatment purposes, distinct outcomes and responses to therapy, all IBCs are grouped into the following subtypes: (a) ER‐positive, HER2‐negative; (b) ER‐positive, HER2‐positive; (c) ER‐negative, HER2‐positive; (d) ER‐negative, HER2‐negative. 16 The majority of BCs are unifocal and can occur in any quadrant of the breast, with a higher frequency in the upper outer quadrant. A synchronous contralateral tumour is found in approximately 2% of patients. The macroscopically most common features in advanced stages of IBC are skin retraction, nipple inversion, nipple discharge, change of texture or colour of the skin. Otherwise, lesions that are discovered early are mostly asymptomatic and may clinically show themselves on palpation as a lump. About 5.15% of all palpable BCs are not seen on a mammogram but can be identified with ultrasound. MRI is the most sensitive method. 16
The most common is the invasive carcinoma of no special type (NST), which accounts for 40%‐75% of all invasive BC types and is commonly present alongside DCIS. The special morphological patterns include a variety of different patterns. These BCs can be mixed IBC‐NST and special subtypes such as the pleomorphic carcinoma, BC with osteoclast‐like giant cells, carcinoma with choriocarcinomatous features and IBC with melanocytic differentiation, oncocytic pattern, lipid‐rich pattern, glycogen‐rich clear cell pattern and sebaceous pattern. 16 To continue, based on epidemiological data, invasive lobular carcinoma (ILC) comes in second place. 16 It presents itself in the somewhat older female population (57‐65 years) and accounts for 5%‐15% of all invasive BCs. ILC is rather heterogeneous due to its various subtypes that are based on the histological picture. The most common picture (‘classic form’) is comprised of small uniformly looking cancer cells. These cells grow in single file, linear pattern and invade the stroma. 16 Two very rare types of BC are the tubular carcinoma and cribriform carcinoma (1.6% and 0.4%). Despite their rarity, they have a very good prognosis. 16
Mucinous carcinoma presents approximately 2% of all BCs cases. This type is more common in women older than 55 years (median 71 years) and has a good 5‐year disease‐free survival rate as well as a very low local recurrence rate. The metaplastic carcinoma, also known as carcinosarcoma (if the mesenchymal component is malignant), is known for its heterogeneity and different components that arise from neoplastic differentiation. Its characterization can be done based on its morphology that is marked by the presence of squamous cells and/or mesenchymal‐looking elements (cartilage, spindle cells, bone, etc). These present 0.2%‐1% of all BC cases and less frequently metastasize into axillar lymph nodes in comparison with invasive NST carcinomas. Other even rarer types of invasive BCs are micropapillary carcinoma, salivary gland/skin adnexal type tumours, adenoid cystic carcinoma, carcinoma with apocrine differentiation, invasive carcinomas with neuroendocrine differentiation, papillary carcinoma and inflammatory carcinoma. 16
In 2019, the new WHO classification for breast cancer was published. 16 , 17 It includes a number of changes. To briefly summarize the most important changes: (a) carcinoma with medullary features, which was previously a separate entity is now regarded as a tissue infiltrating lymphocyte (TIL)‐rich IBC‐NST; (b) oncocytic, lipid‐rich, glycogen‐rich clear cell, sebaceous, pleomorphic, melanotic, oncocytic and choriocarcinomatous carcinomas, carcinoma with osteoclast‐like giant stromal giant cells, previously separate entities, are now regarded as rare variants of carcinoma NST; (c) inflammatory, bilateral and non‐synchronous breast carcinomas, previously separate entities, are now recognized as distinct clinical presentations rather than special subtypes; (d) lobular carcinoma in situ consists now of classic, pleomorphic and florid types; (e) addition of neuroendocrine neoplasms group, of which true neuroendocrine neoplasms are typed as neuroendocrine tumour (NET), small‐cell neuroendocrine carcinoma, large‐cell neuroendocrine carcinoma; (f) neuroendocrine differentiation was overridden by morphological tumour type (NST, mucinous, solid papillary); (g) well‐differentiated liposarcoma in phyllodes tumours is no longer a histological criterion of malignancy by itself; (h) mucinous cystadenocarcinoma is a new recognized entity; (i) a previously known breast tumour with resemblance to the tall variant of papillary thyroid carcinoma but now categorized and grouped under the name tall cell carcinoma with reversed polarity; (j) periductal stromal tumour is no longer a separate entity but a variant of the phyllodes tumour; (k) mesenchymal tumours, haematolymphoid tumours and genetic tumour syndromes are now covered in dedicated chapters. 16 , 17
3.2. Breast cancer cell lines
The field of BC CLs has been established with the first BC CL from Lasfargues and Ozzello in 1958 called BT‐20. 18 Following an increased interest in this area, researchers have, in the following years, isolated and presented an increasing number of CCLs. We face a growing assortment of different CCLs, whereas the integrity and uniqueness of certain CCLs are doubtful. The most commonly used BC CL is still MCF7, T47D and MDAMB231. 4 , 19 Their properties can be seen in Table 4. We also published some more information on the less known BC CLs in one of our previous studies. 4
TABLE 4.
Properties and names of the most commonly used BC CL
| Name | Properties | Type |
|---|---|---|
| MCF7 | ER+/PR+/HER2‐ | Luminal type A |
| MDAMB231 | ER‐/PR‐/HER2‐ | Triple negative |
| SkBr3 | ER+/PR+/HER2+ | HER2 positive |
| T‐47D | ER+/PR+/HER2‐ | Luminal type A |
| BT‐20 | ER‐/PR‐/HER2‐ | Triple negative |
| Molecular subtypes of cell lines | ||
|---|---|---|
| HER2 |
ER‐negative and HER2‐positive profile Over‐represented genomic profile on the chromosomal region 17q12 Present based on their profile, a bridge between luminal and basal cell lines Heterogeneous with luminal and basal features. |
|
| LUMINAL |
Expression of ER and or PR miRNA specific profile (eg hsa‐miR‐501‐5p, hsa‐miR‐202, hsa‐miR‐760 and hsa‐miR‐62) A subdivision into A (luminal A‐like; ER+, PR+, HER2‐, Ki‐67 low) and B (liminal B‐like; ER+, HER2‐; Ki‐67 high or PR low/‐; luminal B‐like, HER2 + m ER+, Ki‐67 any, PR any) Good differentiation, more intercellular tight junctions. |
|
| TNBC | TNBC basal A |
Common cytokeratins (KRT4/5/6A/6B/13/14/15/16/17) Integrins (ITGA6, ITGB4/6) Specific miRNA Better differentiation |
| TNBC basal B |
Higher expression of genes that promote aggressive behaviour (eg vimentin, moesin, plasminogen‐activating factor) CD44 + and CD24‐ Specific miRNA Worse differentiation |
|
Summarized based on the Cellosaurus database. 12
The first to successfully culture a BC CL was Lasfargues and Ozzello in 1958. The CL was named BT‐20. 18 The tissue was obtained from a 74‐year‐old patient, who had a mammary duct‐cell carcinoma of no special type. The cell collection was performed via sampling of the spillage that occurred during tumour preparation (slicing). 18 Certain characteristics of the CL are as follows: triple‐negative breast cancer (TNBC) with a basal A subtype 20 ; homozygous CDKN2A deletion, homozygous point mutation of TP53 p. Lys132Gln (c.394A>C) and RB1 p. Ile388Ser (c.1163T>G); heterozygous point mutation of RB1 p. Pro515Leu (c.1544C>T) and PIK3CA p. His1047Arg (c.3140A>G) as well as PIK3CA p. Pro539Arg (c.1616C>G). 21
MCF7 is one of the most studied CLs in the world. It was derived from the free‐floating cells of the primary CL 734B. The latter was obtained from a patient with metastatic BC. Specifically, the cells were sampled from a malignant pleural effusion. The patient was a 69‐year‐old Caucasian female (blood type 0, Rh‐positive) named Sister Catherine Frances (Helen Marion). 22 , 23 It has been reported that she underwent surgery for both breasts. At first, a mastectomy was performed to remove a benign tumour in her right breast. Later a radical mastectomy of her left breast was performed due to an adenocarcinoma. This happened 7 and 3 years, respectively, before initiating the primary culture. The CL was first described by Soule HD et al in 1973 and had been, as described by the authors, then maintained for 3 years. 22
The authors described that the primary CL exhibited a typical rapid proliferation that is common for most metastatic tumours in vitro. The primary cells (large, immature, striated) did microscopically not appear to be epithelial in origin. It was concluded that they may have been mesothelial in origin or abnormal fibroblasts, due to morphological characteristics and collagenization of the cultures. 22 Certain characteristics of the CL: ER positivity (first described by Lippman and Horwitz) 24 , 25 ; PR heterogeneity, which is generally attributed to the coexistence of several sublines, each possessing different stages of differentiation as well as dependent on the cell cycle phase and population doubling time (PDT) 26 ; in microarray profiling, the cl genome clusters with the BC luminal type A type; HER2 negativity; additional expression of androgen and glucocorticoid receptors. 23 , 27 However, there have been reports on successful subculturing and generations of MCF‐7 cells overexpressing HER2. This CL and the information it presents was referenced in many studies. 23 , 28
MDAMB231 is another BC CL that was isolated from a pleural effusion in a BC patient. The CL had been together with MDAMB134 (mean chromosome number 43) and MDAMB175 (mean chromosome number 49) first described in 1974 by Cailleau et al. 29 The CL was cultured from a single sample of pleural effusion obtained on 17 October 1973. The patient was a 51‐year‐old Caucasian woman who had had a right radical mastectomy in January 1969 for a poorly differentiated invasive ductal carcinoma. The authors reported that the patient developed a pericardial (June 1973) and left sided pleural effusion (July 1973). The effusions were of metastatic origin due to a BC primum. Subsequently, oophorectomy and systemic treatment were initiated. At first, she was given 5‐FU and prednisone. Later, in September 1973, combined chemotherapy (cyclophosphamide, adriamycin and amethopterin) was started. However, the patient's state deteriorated, and she died on 13 January 1974. 29 The pleural effusion from which the CL was cultured occurred after systemic treatment. Certain characteristics of the CL are as follows: epithelial growth pattern; a near triploid chromosome number (from 60 to 70); ER, PR and E‐cadherin–negative status; heterozygous point mutation for BRAF p. Gly464Val (c.1391G>T) and KRAS p. Gly13Asp (c.38G>A), homozygous point mutation for TP53 p. Arg280Lys (c.839G>A) and CDKN2A gene deletion; basal B subtype. 30 , 31
In the last years, many studies have analysed the field of BC CLs. A large‐scale study was done by Xiaofeng et al, who not only offered a detailed overview of 92 BC CLs and their molecular classification (luminal A, luminal B, HER2‐positive and triple‐negative subtypes divided into basal A and basal B) but also highlighted inconsistencies in studies with regard to primary marker status reports. 31 The authors came to several conclusions. Firstly, as an observation, the genetic and epigenetic categorization between BC CLs and primary tissue is not one to one. Secondly, TNBC CLs can be genetically subdivided into basal A and B groups that have specific properties (eg phenotype, molecular properties). Namely, the TNBC A subtype is supposedly characterized by the expression of basal keratins (KRT4/5/6/13/14/15/16/17). This expression profile shows similarity with the core basal tumours. The TNBC B subtype has a characteristic stem cell profile of CD44+CD24‐ and migration markers such as vimentin. This shows promise for modelling claudin—low and/or metaplastic breast cancers. 31
The study from Holiday et al 2011 was, at that time, one of the rare and most comprehensive studies on this specific subject at that time. Nine years after their study, we can see that although there is now a reasonable number of BC CLs, these still lack some of the rarer histopathological types (eg phyllodes tumours, male BC CLs and inflammatory BC). Another recent study, done by Lima Mota et al, focused on the molecular characterization of BC CLs by clinical immunohistochemical markers. 32 The authors set out to evaluate the hormone receptor and HER2 receptor expression and classify the BC CL based on the molecular subtypes. The used BC CLs were as follows: SKBR3, MCF‐7, MCF‐7/AZ, Hs578T, MDA‐MB‐231, MDA‐MB‐468, BT‐20 and T47D. What is surprising is that their results differ from previously reported CL expression profiles for BT20 CL. This CL has been in a wide variety of studies described as a TNBC BC CL. 12 , 30 , 31 , 33 However, the authors report results of it being HER2 overexpressed. 32 Other BC CLs that have inconsistent expression profiles in various studies are HCC1007, HCC1419, HCC1500, HCC2185, SUM52PE, SUM44PE, EVSA‐T and EVSA‐T. 20 , 31 , 34 , 35 , 36 , 37 The reasons for these inconsistencies can be several. We will give an overview of this topic in the last segment of the paper.
4. CANCER OF THE ENDOMETRIUM AND ITS CELL LINES
4.1. Endometrial cancer
EC presents a very common gynaecological malignancy. It has a yearly incidence rate of 60 000 (United States) new cases and more than 10 000 deaths. 38 , 39 , 40 , 41 On a global scale, there are 382 069 new cases yearly and 89 929 deaths. 38 , 39 , 40 , 41 The incidence of this cancer is very tightly linked to certain epidemiologic factors. According to literature, obesity being one of the most important. 38 , 39 , 40 , 42 Others include the exposure to unopposed oestrogens or tamoxifen, diabetes, nulliparity, early‐onset menarche and late‐onset menopause. In one of our previous publications, we provided an in‐depth overview of EC CLs. 2
We can, based on the latest WHO classification (2020, 5th), divide EC into multiple histological subtypes. The most common is the endometrioid type (up to 80%) followed by mixed cell type (up to 10%), serous (up to 10%); carcinosarcoma (<10%), clear cell (<10%), undifferentiated and dedifferentiated carcinomas (<10%), mixed carcinoma (<10%) and other types (<10%; mesonephric adenocarcinoma, squamous cell carcinoma NOS, mucinous carcinoma, intestinal type and mesonephric‐like adenocarcinoma). 43 , 44 , 45 The endometrioid carcinomas are graded with the use of the FIGO classification system. The grades are determined by the percentage of solid growth patterns (Grade 1: <5%; Grade 2: 6%‐50%; Grade 3: >50% solid growth). Endometrioid types have commonly a positive hormone receptor status (ER, PR) and p53 wild‐type (G1/2) or heterogenous status (G3). Microscopically one can observe tall columnar cells lining back‐to‐back glands without intervening stroma. The growth pattern can also be cribriform. The microscopist has to be careful to not include areas of squamous differentiation when assessing the percentage of solid growth and consequently the grade. Serous carcinomas have a positive hormone receptor expression (ER, PR), are of the abnormal p53 (p53abn) molecular subtype and have a p16‐ and PTEN‐positive status. Histological features include tumour cells (nuclear atypia, mitotic figures) that form papillary structures. The glands have serrated outlines. Serous carcinomas are frequently clinically occult, often invade the lymphovascular space and have therefore a worse prognosis. Clear cell carcinomas are uncommon. They can have an either negative or positive receptor status (ER, PR) p53 heterogenous status, variable PTEN and p16 status (±) and show positive staining for Napsin A, Racemase and HNF1ß. These tumours share many morphologic features with ovarian clear cell carcinomas. The different architectural features include a combination or solely a solid, glandular or papillary architecture. Also, commonly cells with abundant clear cytoplasm can be observed. 45 , 46
Based on The Cancer Genome Atlas (TCGA), endometrial carcinomas are classified into four subgroups. These are as follows: (a) the POLE (DNA polymerase ε) ultramutated group, (b) the hypermutated/microsatellite unstable (MSI) group, (c) the copy number low/microsatellite stable group and d) the copy number high (serous‐like) group. 44 , 47 The routine of identifying and correctly specifying the molecular EC groups can be done by following a system known under the name of Proactive Molecular Risk Classifier for Endometrial Cancer (ProMisE). The first step is identifying specimens with POLE pathogenetic mutations (‘group a’). If the specimen has a non‐pathogenetic mutation or a wild type, then mutation mismatch repair proteins (MMR) are inspected (unstable – ‘group b’). In specimens with intact MMR, immunohistochemical p53 staining is performed. This shows either an aberrant (corresponding with ‘group d’) or wild‐type variant (‘group c’). 48
For a long time, EC was classified into two categories which correlated to its aggressiveness. With the advancement of technology and new studies, the molecular background became much more important due to its implications in prognosis. Therefore, the newest WHO classification (2020, 5th) includes explanations on the new molecular classification system for ECs and how it relates to the traditional histomorphologic evaluation. The characteristics of group 1 are as follows: more common, lower risk, dependent on oestrogen, hormone receptor (mostly) positive (ER, PR), good prognosis, comparatively younger population (between 55 and 65), endometrioid histology (grade 1/2) and with most commonly no specific molecular profile (sometimes mismatch repair deficient—MMRd). The characteristics of group 2 are as follows: no growth dependency on oestrogen, mostly hormone receptor negative, older population of females (over 65), bad prognosis, histologic clear cell or serous carcinoma with respectively no specific molecular profile or abnormal p53 status. Grade 3 endometrioid EC is best considered as a separate category since it can have any of the molecular anomalies and be therefore in any group.
The standard treatment for endometrial cancer consists of primary hysterectomy and bilateral salpingo‐oophorectomy (eg laparoscopic or robotic). The 5‐year overall survival ranges from 74% to 91% in patients without metastatic disease. 49
4.2. Endometrial cancer cell lines
Endometrial CCL that is widely used is Ishikawa, HEC‐1‐A, HEC‐1‐B and KLE. Some other examples of commonly described immortal endometrial CLs include HES and hTERT‐EEC. 2 What is more, reports said that the CLs are commonly contaminated with the MCF‐7 cancer cells and the HeLa CL. 50 Other issues include the misidentification and redundancy of these cell isolates. This was shown by Korch et al in 2012, 50 who presented proof that ECC‐1 isolates were contaminated and genotyped either as Ishikawa cells, MCF‐7 breast cancer cells (or a combination). 2 Furthermore, another issue is the uncertainty regarding their primary type (I or II) due to conflicting reports in the literature. 8 , 51 , 52 Among the studies that tackle the issue of EC CLs some of them show similarities while others differ in certain aspects. 8 , 9 , 50 , 51 , 53 Our findings from the literature were that HEC‐1‐A, HEC‐1‐B and KLE are reported differently. 2 Crucial properties and the originators' information of the most common EC CLs are succinctly summarized in Table 5.
TABLE 5.
Properties of the most common EC CL
| Cell line name | HEC‐1‐A and HEC‐1‐B | Ishikawa | AN3‐CA | KLE |
|---|---|---|---|---|
| First described | Kuramoto H in 1972 | Nishida et al in 1985 | Dawe CJ et al in 1964 | Richardson et al in 1984 |
| Patient | 71‐year‐old woman | 39‐year‐old woman | 55‐year‐old woman | 64 to 68‐year‐old female |
| Tumour | Endometrial adenocarcinoma | Endometrial adenocarcinoma | Uterine neoplasm associated with the clinical syndrome of malignant acanthosis nigricans, obtained from lymph node | Tissue of a colon metastasis from a poorly differentiated G3 endometrial adenocarcinoma |
| Chromosomes |
HEC‐1A – diploid. HEC‐1B – tetraploid |
Diploid chromosomal range | Diploid chromosomal range | ‐ |
| Special remarks |
HER‐1‐A (parent) HEC‐1‐B (child) Heterozygous point mutation KRAS p. Gly12Asp (c.35G > A); homozygous point mutation for TP53 p. Arg248Gln (c.743G > A) and HEC‐1‐B no PTEN mutation |
ER and PR disappear after long‐term culture |
MSI instability; Heterozygous point mutation of MAPK3 p. Pro373Ser (c.1117C > T), heterozygous point mutation of PIK3R1 a and heterozygous point mutation of TP53 p. Gly389Trp (c.1165G > T); homozygous point mutation of PTEN p. Arg130Glnfs*4 (c.389delG); KRAS wild type |
PTEN, KRAS wild type, no mutation Low MSI |
| Literature | 79, 80, 81 | 82, 83 | 84 | 9, 12, 85 |
| Cell line name | HEC‐1‐A | HEC‐1‐B | Ishikawa | AN3‐CA | KLE |
|---|---|---|---|---|---|
| Histopathological properties | Status | Status | Status | Status | Status |
| ER | Low | Low | High | Low | low |
| PR | Low | Low | High | Low | Low |
| PTEN | High | High | Low | Low/Deletion | High |
| Grade | G2 | G2 | G1 | Metastasis | Metastasis |
| Type | II | II | I | I | II |
Summarized from the Cellosaurus databank (Bairoch et al 12 ).
Due to space issues: p. Arg557_Lys561delArgGluIleAspLysinsGln (c.1670_1681delGAGAAATTGACA).
5. CERVICAL CANCER AND ITS CELL LINES
5.1. Cervical cancer
The last cancer type to be discussed in this review is uterine cervix cancer (CC). CC is a common type of cancer and has a relatively high mortality among gynaecological cancers with stark regional differences. The incidence and death toll are on a global scale estimated at approximately 570 000 and 311 000 per year. 54 Interregional differences in mortality can be attributed to countries lacking cervical cancer screening and prevention programmes, since this type of cancer remains the second most common type (17.8 per 100 000 women) as well as the cause of cancer deaths (9.8 per 100 000) among all types of cancer in women that live in lower income countries (Table 2). 14 Furthermore, collectively, most of CC cases (80%‐90%) occur in these countries (eg parts of Africa and Asia). 55 The most important directly linked factor for CC is the human papillomavirus (HPV) infection, 56 which is also the most common sexually transmitted disease. Through research, more than 200 different types have been found, identified and systematically classified into 5 genera (α, β, γ, μ and ν). 57 Based on their oncogenic potential, these are labelled as high risk and low risk. The most studied types are of the α genera, since these have been directly linked to almost all squamous intraepithelial lesions and cancers of the cervix and anus as well as to a subset of penile, vulvar and vaginal cancers. 57 HPV types and their biologic potential can be seen in Table 6. It has been estimated that HPV is responsible for almost a tenth of human malignancies (7%‐8%). It is associated with almost all cases of CCs (96%) and anal cancers (93%). Furthermore, almost two thirds of all vaginal cancers (64%) and oropharyngeal carcinomas (63%) arise due to its oncogenic potential. And lastly, HPV also presents an important factor in the development vulvar cancer (51%) and penile cancers (36%). 57
TABLE 6.
HPV characteristics
| HPV types and their biologic potential | |
|---|---|
| Low‐risk (non‐oncogenic) types | HPV 6, 11, 40, 42, 43, 44, 54, 61, 72, 81 |
| High‐risk (oncogenic or cancer‐associated) types | HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 69, 82 |
| Squamous cell carcinoma | HPV 16 (59%), 18 (13%), 58 (5%), 33 (5%), 45 (4%) |
| Adenocarcinoma | HPV 16 (36%), 18 (37%), 45 (5%), 31 (2%), 33 (2%) |
| HPV‐associated diseases | |
|---|---|
| Disease | HPV type association |
| Cutaneous warts | 1, 2, 3, 4, 27, 57 |
| Anogenital warts | 6, 11, 53 |
| Mucosal cancers | 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 73, 82 |
| Non‐melanoma skin cancers | 1, 5, 8, 9, 17, 20, 23, 38 |
| Bowen disease | 16, 18, 31, 32, 34 |
| Epidermodysplasia verruciformis | 5, 8, 9, 12, 14, 15, 17, 19‐25, 36‐38, 46, 47, 49, 50 |
| Clinical properties | |
|---|---|
| Signs | Irregular or heavy vaginal bleeding; post‐coital bleeding |
| Risk factors | Early onset of sexual activity, multiple sexual partners, a high‐risk sexual partner, history of sexually transmitted infections, history of vulvar or vaginal neoplasia, immunosuppression. |
The most common histologic types of CC are squamous cell carcinoma (64.5%), adenocarcinoma (28.9%) and 6.6% other histology's. 58 , 59 Based on a recent study from 2019, HPV infection may also be implicated in developing some types of breast cancer. 60 The presence of screening programmes for cervical cancer and public health programmes for HPV vaccination and education of the general public led, according to sources, to a significant decrease in the incidence and mortality of cervical cancer over the past 50 years in developed countries (75%). 61 , 62 , 63
It is worth noting that viral load seems to be in an inverse correlation with the malignancy of the lesion. This can be observed predominantly in non‐melanoma skin cancer (NMSC; cutaneous lymphomas, adnexal tumours, Kaposi's sarcomas, Merkel‐cell carcinomas, basal cell carcinomas—BCCs, squamous cell carcinomas—SCCs) and other skin lesions, which is in contrast to the direct carcinogenic effect of genital HPVs. 57 , 64 In such cases, this evidence supports a hit‐and‐run mechanism of carcinogenesis. 64 Treatment depends on the extent of the disease. The more advanced cases may require radical hysterectomy or chemoradiation, or even a combination of both. At the same time, conservative, fertility‐preserving surgical procedures have become the standard of care for women with low‐risk, early‐stage disease. 55 Based on the information from copy number variation (CNV), methylation, mRNA and miRNA profiles, cervical cancer has three distinct molecular subtypes of CC: SCC keratin—high, SCC keratin—low and adenocarcinoma. 65 The differences between these include the following: enriched expression of some genes (eg PIK3CA, ADH7 and SPRR3) in the SCC keratin—high compared with the SCC keratin—low cluster, more frequent CNVs, including common EGFR amplification in SCCs, a high number of aberrations in tumour‐suppressor genes related to TGF‐β pathway in adenocarcinomas including SMAD4 and TGFBR2 deletions, and increased DNA methylation in adenocarcinomas. 65
5.2. Cervical cancer cell lines
The most famous and first CCL in the world is HeLa (Figure 3). The HeLa cells are named after the 30‐year‐old patient Henrietta Lacks who died in 1951 due to an aggressive adenocarcinoma of the cervix. 66 Some of the tissue obtained from a cervical biopsy was supplied to the Tissue Culture Laboratory in the Department of Surgery at The Johns Hopkins Hospital for research purposes. Contrary to their previous results, the cells grew robustly and became the first human CCL immortalized in tissue culture. As is widely known, the naming process consisted of utilizing the initial 2 letters of Henrietta Lacks' first and last names. 66 The cells have been shown to contain human papillomavirus (HPV) 18 DNA, and HPV18‐positive HeLa cells have been linked to changes in microRNA expression. These results were obtained by Gey and colleagues, which published their study in 1952. 67 The most common cervical CCLs are shown in Table 7. What is important to note is the fact that these cells have been in circulation for more than 60 years. Subsequently, they are almost ubiquitous. 68 This has, through the years, also led to many contaminations and has become a crucial problem in CL culturing. This furthermore led to an accumulation of articles and studies that reported their research on contaminated or mutated CLs. The authors Horbach and Halffman were able to identify 32 755 such articles (up to the year 2017). 68 For their search, the authors utilized two methods. One was via the cross‐search of the CLs documented in the eight versions of the International Cell Line Authentication Committees (ICLAC) list of misidentified CLs and by using the WoS database for all articles stating the names of one of the 451 listed CLs. 68 Figure 4 shows, based on the authors published data from 2017, a simplified geographical overview of the areas with the highest percentage of contaminated primary articles as a fraction of the total number of articles on cells per country (primary data set is available at the primary authors publication). 68 The authors utilized the ICLAC list of misidentified cells. The latest version is version 10 and was released 25 March 2020. The register currently lists 552 cell lines. 69 Besides the importance of HPV status, karyotyping, p53 and pRB expression, a recent study compared the secretomes of different cervical CCLs and highlighted the role of cytoplasmic peroxiredoxin‐2 (PRDX2), transforming growth factor‐beta‐induced protein ig‐h3 and NRF2. 70 Their analysis pointed out that the expression of NRF2 indicates that aberrant NRF2‐mediated oxidative stress response (OSR) is a prominent feature of cervical carcinogenesis. 70 Another fascinating study showed that cervical CCLs express markers associated with immunosurveillance. These were as follows: (a) MICA/B and CD95 (involved in tumour cell recognition); (b) CD39, CD73, CTLA‐4 (immune system escape); and (c) NKp30, NKp46, NKG2A and KIR3DL1 (typical markers of NK cells like). The authors concluded that these molecules might allow the CCLs to mimic the immune system. 71
FIGURE 3.
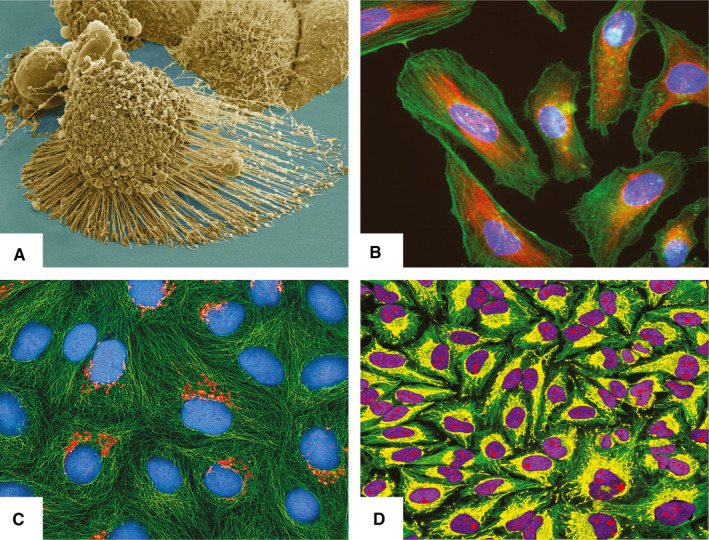
HeLa cell line. (A) Electron micrograph of an apoptotic HeLa cell (source: National Institutes of Health—NIH; https://imagebank.nih.gov/details.cfm?imageid=1463); (B) Immunofluorescence image of HeLa cells grown in tissue culture and stained with antibody to actin in green, vimentin in red and DNA in blue (source: GerryShaw—wikimedia.commons; https://commons.wikimedia.org/wiki/File:HeLa_cells_stained_with_antibody_to_actin_(green)_,_vimentin_(red)_and_DNA_(blue).jpg); (C) Multiphoton fluorescence image of cultured HeLa cells with a fluorescent protein targeted to the Golgi apparatus (orange), microtubules (green) and counterstained for DNA (cyan) (source: NIH; https://commons.wikimedia.org/wiki/File:HeLa‐I.jpg); D) Immunofluorescence of HeLa cells showing microtubules in green, mitochondria in yellow, nucleoli in red and nuclear DNA in purple (source: GerryShaw—wikimedia.commons; https://commons.wikimedia.org/wiki/File:HeLa‐Tubulin‐HSP60‐Fibrillarin‐DNA.jpg). All material is published under the CC license or is in the public domain
TABLE 7.
List of common cervical cancer cell lines
| Cell line | Patient | HPV status | Mutations | Primary tissue | Lit |
|---|---|---|---|---|---|
| C33A | 66Y | Negative |
MSI‐high DT – 1.36 d pseudodiploid p53 +; pRB + |
Cervical squamous cell carcinoma. Part of CCLE and COSMIC. |
Auersperg in 1964 86 |
| OMC‐4 (Osaka Medical College‐4) | 47Y | Negative |
Unknown MSI status DT – 63 h |
Cervical adenocarcinoma. | Yamada T et al in 1987 87 |
| CaSki | 40Y | Pos (HPV16) |
MSI stable no TP53 mutation beta subunit of human chorionic gonadotropin (hCG) |
Human papillomavirus‐related cervical squamous cell carcinoma. Metastatic site: Small intestine. Part of CCLE and COSMIC. |
Pattillo R.A in 1977 88 |
| SiHa | 55Y | Pos (HPV16) |
MSI stable DT – 2.6 d p53 +; pRB + hypertriploid CL |
Cervical squamous cell. Part of CCLE and COSMIC. |
Friedl F. et al in 1970 89 |
| HeLa | 30Y6M | Pos (HPV18) |
MSI stable DT – 1.3‐2 d Four marker chromosomes P53 low, pRB normal |
Endocervical adenocarcinoma. Part of CCLE and COSMIC. |
Gey GO et al in 1952 67 |
| TMCC‐1 | Age unspecified | Pos (HPV18) |
Unknown MSI status DT – 53 h |
Endocervical adenocarcinoma. Metastatic site: Pleural effusion. |
Sakamoto M. et al in 1987 90 |
| ME180 | 66Y | Pos (HPV68) |
MSI stable DT – 1.5 d P53 neg/pos, pRB + Heterozygous point mutation of PIK3CA p. Glu545Lys (c.1633G > A) |
Cervical squamous cell carcinoma Metastatic site: Omentum. Part of CCLE and COSMIC |
Sykes J. A. et al in 1970 91 |
| HT‐3 | 53‐58y a | Negative |
MSI stable hypotriploid to hypertriploid DT – 2.48 d p53 +; pRB + |
Metastatic site: Lymph node. Part of CCLE and COSMIC |
Fogh J. and Trempe G. (1975) 92 |
| C‐4‐I | 41Y | Pos (HPV18) |
MSI stable DT – 2 d |
cervical squamous cell carcinoma Part of CCLE and COSMIC |
Auersperg N in 1962 93 |
| C‐4‐II | 41Y | Pos (HPV18) |
MSI stable DT – 2.4 d |
cervical squamous cell carcinoma | Auersperg N in 1962. 93 |
| MS751 | 47Y |
Pos (HPV18) Pos (HPV45) |
MSI stable DT – 2.4 d hypodiploid human cell line |
epidermoid carcinoma Metastatic site: Lymph node Part of CCLE and COSMIC |
Sykes J. A. et al in 1974 |
| SW756 | 46y | Pos (HPV18) |
MSI stable DT – 1.6 d Expressed genes: HLA A1, A24, B8, B44, Cw2, Cx, DR6Y; Le3; Le4; Le5 |
squamous cell carcinoma | Leibovitz A. in 1974 |
Data summarized after the originators works as well as from the CELLOSAURUS databank.
Abbreviations: DT, doubling time; MSI, microsatellite.
Different indications – Indicated to be from a 58‐year‐old female patient on ATCC and from a 53‐year‐old on the Sloan Kettering tech transfer site.
FIGURE 4.
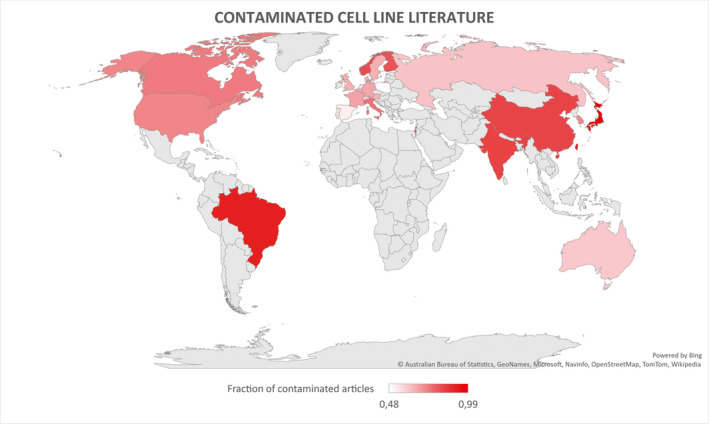
A simplified geographical overview of the percentage of contaminated primary articles as a fraction of the total number of articles on cells per country (Top 5: Japan, Brazil, Taiwan, India, China). Adapted from the dataset from Horbach and Halffman 68 under the CC license
6. CELL LINE CULTURING
The principle of cell culturing was established by Wilhelm Roux. He was an embryologist, who used warm saline to maintain chicken embryos for several days, thereby coming up with the tissue culture principle in 1885. 72 The culturing process is a crucial step, and every mistake in its procedure can lead to a failed experiment (hence cease of cell growth). Changes in the environment can, via alterations in differentiation and gene expression signalling cascades drastically impact the cell morphology, intercellular interactions and cell polarity. Moreover, CCLs mostly stem from invasive high‐grade cancers, which means that genetic changes can occur much more often. These new mutations (de novo) may lead to phenotype changes. Furthermore, even perhaps seemingly small changes in the cell environmental such as changing the growth medium or temperature, different methods of cell culturing (xenografts, the addition of growth factors, 2D or 3D, etc) can lead to epigenetic alterations that affect the expression of genes. 73 Through the years, a number of errors, contaminations and finally also reports with questionable reproducibility, consistency and validity have been exposed. 50 These findings can have, in certain cases, little or, in other instances, dire consequences on the work and results of many. Ben‐David et al recently described the heterogenous nature of commonly used CCLs. The described differences extend from genetic information to phenotype and are proof of the instability and mutational potential of CLs (Figure 5). Consequently, the authors again stressed the need for a system of rules and measures which must be strictly enforced and followed. This system has been published in the international guidelines for the use of CLs in biomedical research. An important suggestion is determining the authenticity of the CL, which can be done via short tandem repeat (STR) profiling. The mechanism is the identification of variants in tetranucleotide microsatellite loci on multiple human chromosomes and comparing those to established databases. The same principle is used in paternity tests or other medico‐legal affairs. As an example: when applying this method in specific time or culturing intervals (eg first, third, fifth passage), the researcher is able to tell when the culture has undergone mutation (different STR profile). If we were all to follow the recommendations, then a lot of the problems would be solved. 74 A succinct review on this topic has been given by Hynds and colleagues. 7 An important point that the authors proposed is the reassessment of the ‘genomic landscape’, which undoubtedly changed a lot during the last years. 7 Some measures for better consistency and reproducibility in cell culturing can be seen in Table 8.
FIGURE 5.
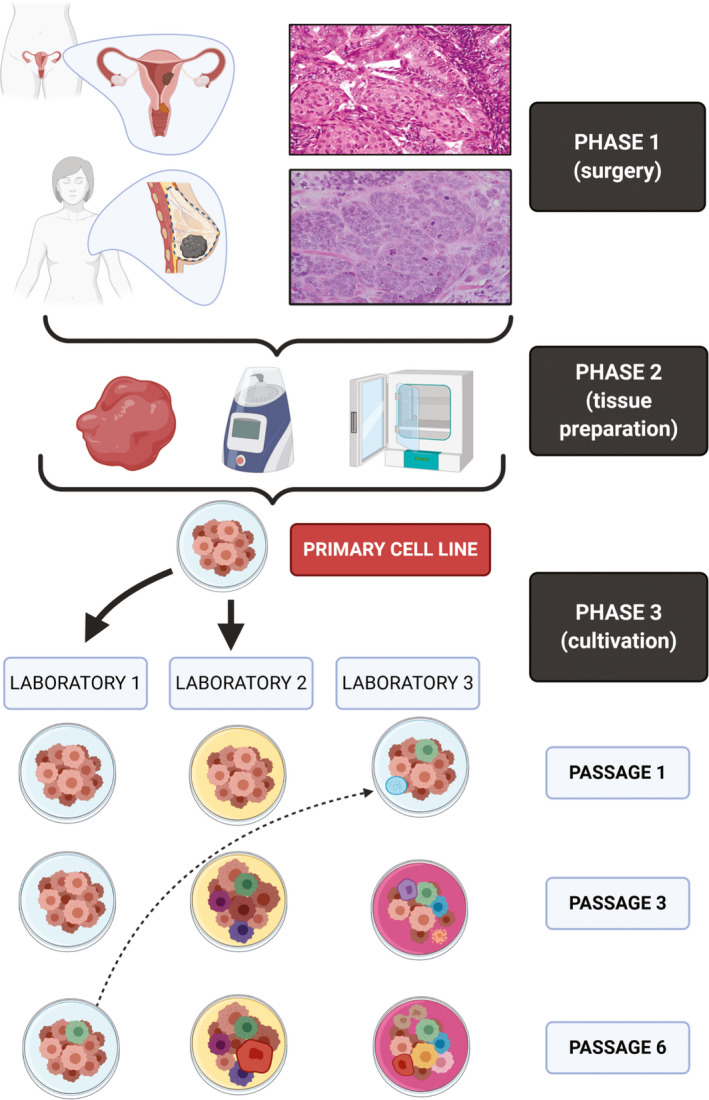
Cell line isolation and changes during cultivation. Phases 1 and 2 encompass the logistics of tissue retrieval and pre‐cultivation procedures (eg homogenization). Phase 3 shows the heterogeneity and susceptibility of CLs to mutate due to various changes. Laboratory 1 and laboratory 2 received commercially bought CLs. The CL from laboratory 1 mutated somewhere between passage 3 and 6. Laboratory 2 used from the beginning a different medium, which led to a variety of genotypical and phenotypical changes. Laboratory 3 borrowed a CL sample (already mutated) from laboratory 1. Due to careless handling, the CL got infected with mycoplasma. The addition of an antibiotic agent again led to a series of genotypical and phenotypical changes in the CL. Source: Histological images were used under the CC license from Wikimedia Commons (https://commons.wikimedia.org/wiki/File:Invasive_Ductal_Carcinoma_40x.jpg; https://commons.wikimedia.org/wiki/File:Endometrioid_endometrial_adenocarcinoma_very_high_mag.jpg). The figure itself was created with BioRender.com
TABLE 8.
Measures for better consistency and reproducibility
| Measures | Explanation | LIT |
|---|---|---|
| CL identification | To avoid misidentification, acquired CLs should come from a reliable source and must be authenticated, bought from a reliable source and banked for future use. Additional STR profiling is also important. | 94 |
| Mycoplasma testing | To avoid contamination good tissue culture practice and frequent testing should be performed to ensure that CLs are clear of contamination. | 95 |
| Use of validated reagents | To avoid a variety of errors only reagents of certified laboratory purity should be used. Decontamination should not be avoided but rather the experiment repeated. | 96 |
| Statistical standards | To avoid misinterpretations and promote transparency, mandatory reporting checklist that catalogued details of statistical information, experimental design and reagents, should be included. | . 7 , 97 , 98 , 99 |
| Profiling | To avoid contaminations with other CLs and possible erroneous results, laboratory's own CLs should be compared to reference CL genomes. | 74 |
| Cryopreservation | To avoid loss of data and ensure replicability, preservation of the primary cultures and early passages with subsequent final comparison and validation of key findings before publication is of grave importance. | 74 |
| Reporting DTs | To promote transparency and replicability, accurate and diligent monitoring as well as reporting of DTs as well as a finite usage number of passages should be standard practice. | 74 |
| Standardized conditions | To ensure interlaboratory replicability, international standardized culture conditions for individual CLs and documentation of the heterogeneity metrics in datasheets should be standard practice. | 74 |
| Naming | To ensure coherent scientific reporting, the international naming guidelines should be used. | 100 |
Abbreviations: CLs, cell lines; DT, doubling time; STR, short tandem repeat.
6.1. Author experience and recommendations
Culturing procedures have drastically changed over the years, especially with the development of new reagents and new materials. If comparing our current procedures with the past, we can now appreciate a much more streamlined process. 2 The core principle, which is the usage of a culturing medium that supports the growth of cells as well as additions in the form of antibiotics that stop the growth of bacteria, is of course universally the same. However, the current procedures are much more uniform and perhaps easier to use as those described by the CL ‘pioneers’ (eg Dawe et al, AN3‐CA). Nevertheless, the protocols can still differ between laboratories. To illustrate, we present a simple workflow protocol (Figure 6) that we used for the isolation of our cell cultures and our characterization. 3 , 75 , 76 Based on this workflow, we successfully cultured a TNBC CL. The naming was done in accordance with the current international guidelines (eg MFUM‐BrTNBC‐1). 3 , 77 Some of our other CLs include a human intestinal epithelial CL (HUIEC) 75 as well as human chondrocytes. 76
FIGURE 6.
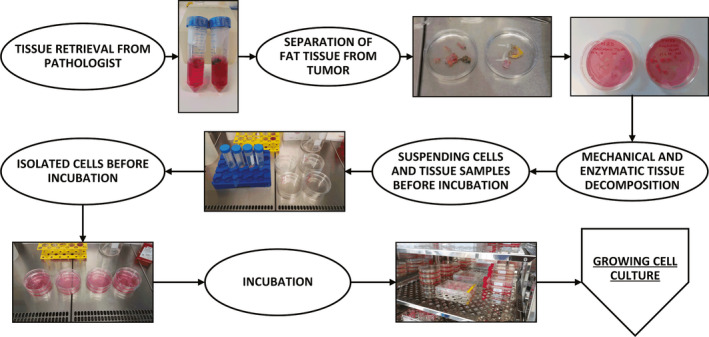
Cell culturing protocol
7. CONCLUSIONS
We presented different types of gynaecological cancers and their CLs as well as discussed aspects of their culturing. There is a growing body of evidence that, despite certain drawbacks, variations within CCLs can also be useful in regard to a more diverse genomic landscape. With more complex characterization methods, researchers would be able to expand their databases, investigate and research new interactions between CCLs as well as possibly discover new genotype/phenotype associations. 6 , 7 A good example is the study published in Nature about the results and genetic data of more than a 1000 CLs (eg NA splicing, DNA methylation, microRNA expression) 11 and the recently established Models in Translational Oncology (MiTO) database, which will also help in the exchange of information on pre‐clinical model data relevant in translational cancer research. 1 CLs remain an important and powerful tool for cancer research, but the genetic changes lead to variation across CL strains. 6 , 7 The adherence to the previously discussed rulesets, and the international guidelines help in minimizing replication failure between institutions. Nevertheless, the already present body of studies that report ‘false’ CLs remains. The authors Horbach and Halffman, therefore, proposed that notifications should be posted alongside previously published articles using misidentified CLs (eg ‘expressions of concern’). This would not be a ‘witch hunt’ (eg retraction hunt) but simply alert the reader that there may be an issue with a paper, when, as the authors nicely said, ‘the full story is not yet clear’. Furthermore, the authors recommended to increase the visibility of utilized CLs in articles by mentioning the employed CLs in easily searchable parts of their article (eg abstract and keywords). 68
CONFLICT OF INTEREST
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
AUTHOR CONTRIBUTIONS
Kristijan Skok: Conceptualization (lead); Formal analysis (equal); Investigation (equal); Methodology (equal); Visualization (equal); Writing‐original draft (lead); Writing‐review & editing (equal). Lidija Gradišnik: Conceptualization (equal); Formal analysis (equal); Investigation (equal); Writing‐review & editing (equal). Uroš Maver: Conceptualization (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Nejc Kozar: Writing‐review & editing (equal). Monika Sobočan: Writing‐review & editing (equal). Iztok Takač: Formal analysis (equal); Investigation (equal); Writing‐review & editing (equal). Darja Arko: Formal analysis (equal); Investigation (equal); Writing‐review & editing (equal). Rajko Kavalar: Conceptualization (equal); Formal analysis (equal); Investigation (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal).
Skok K, Gradišnik L, Maver U, et al. Gynaecological cancers and their cell lines. J Cell Mol Med. 2021;25:3680–3698. 10.1111/jcmm.16397
Funding information
This research was funded by the Slovenian Research Agency (slo. Javna Agencija za Raziskovalno Dejavnost Republike Slovenije), Grant/Award Numbers: P3‐0036, I0‐0029, J3‐9272 and L4‐1843; University Medical Center Maribor, Grant/Award Number: IRP 2018/01‐10, IRP 2018/01‐19.
Contributor Information
Kristijan Skok, Email: kristijan.skok@gmail.com.
Rajko Kavalar, Email: rajko.kavalar@ukc-mb.si.
REFERENCES
- 1. Galuschka C, Proynova R, Roth B, Augustin HG, Müller‐Decker K. Models in translational oncology: a public resource database for preclinical cancer research. Cancer Res. 2017;77(10):2557‐2563. 10.1158/0008-5472.CAN-16-3099 [DOI] [PubMed] [Google Scholar]
- 2. Skok K, Maver U, Gradišnik L, Kozar N, Takač I, Arko D. Endometrial cancer and its cell lines. Mol Biol Rep. 2020;47(2):1399‐1411. 10.1007/s11033-019-05226-3 [DOI] [PubMed] [Google Scholar]
- 3. Skok K, Gradišnik L, Čelešnik H, et al. Isolation and characterization of the first Slovenian human triple‐negative breast cancer cell line. Breast J. 2020;26(2):328‐330. 10.1111/tbj.13695 [DOI] [PubMed] [Google Scholar]
- 4. Skok K, Maver U, Gradišnik L, Sobočan M, Takač I. Humane celične linije raka dojk. Slovenian Medical Journal. 2019;88 (9‐10):427–43. 10.6016/zdravvestn.2842 [DOI] [Google Scholar]
- 5. Lin CY, Erkek S, Tong Y, et al. Active medulloblastoma enhancers reveal subgroup‐specific cellular origins. Nature. 2016;530(7588):57‐62. 10.1038/nature16546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben‐David U, Siranosian B, Ha G, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560(7718):325‐330. 10.1038/s41586-018-0409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hynds RE, Vladimirou E, Janes SM. The secret lives of cancer cell lines. DMM . Dis Model Mech. 2018;11(11):dmm037366. 10.1242/dmm.037366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou X, Wang Z, Zhao Y, Podratz K, Jiang S. Characterization of sixteen endometrial cancer cell lines. Cancer Res. 2007;67(9 Suppl):3870 LP. [Google Scholar]
- 9. Kozak J, Wdowiak P, Maciejewski R, Torres A. A guide for endometrial cancer cell lines functional assays using the measurements of electronic impedance. Cytotechnology. 2018;70(1):339‐350. 10.1007/s10616-017-0149-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in‐depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784‐3788. 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghandi M, Huang FW, Jané‐Valbuena J, et al. Next‐generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569(7757):503‐508. 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bairoch A. The Cellosaurus, a cell‐line knowledge resource. J Biomol Tech. 2018;29(2):25‐38. 10.7171/jbt.18-2902-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dutil J, Chen Z, Monteiro AN, Teer JK, Eschrich SA. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res. 2019;79(7):1263‐1273. 10.1158/0008-5472.CAN-18-2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 15. Bateman AC, Shaw EC. Breast pathology. Surg. 2016;34(1):1‐7. 10.1016/J.MPSUR.2015.10.002 [DOI] [Google Scholar]
- 16. The WHO Classification of Tumours Editorial Board , ed. Breast Tumors, 5th ed. International Agency for Research on Cancer; 2019. [Google Scholar]
- 17. Tan PH, Ellis I, Allison K, et al. The 2019 World Health Organization classification of tumours of the breast. Histopathology. 2020;77(2):181‐185. 10.1111/his.14091 [DOI] [PubMed] [Google Scholar]
- 18. Lasfargues EY, Ozzello L. Cultivation of human breast carcinomas. J Natl Cancer Inst. 1958;21(6):1131‐1147. 10.1093/jnci/21.6.1131 [DOI] [PubMed] [Google Scholar]
- 19. Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13(4):215. 10.1186/bcr2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riaz M, van Jaarsveld MTM, Hollestelle A, et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation‐specific miRNAs. Breast Cancer Res. 2013;15(2):R33. 10.1186/bcr3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wasielewski M, Elstrodt F, Klijn JGM, Berns EMJJ, Schutte M. Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines. Breast Cancer Res Treat. 2006;99(1):97‐101. 10.1007/s10549-006-9186-z [DOI] [PubMed] [Google Scholar]
- 22. Soule HD, Vazquez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma2. JNCI J Natl Cancer Inst. 1973;51(5):1409‐1416. 10.1093/jnci/51.5.1409 [DOI] [PubMed] [Google Scholar]
- 23. Lee AV, Oesterreich S, Davidson NE. MCF‐7 cells—changing the course of breast cancer research and care for 45 years. J Natl Cancer Inst. 2015;107(7):djv073. 10.1093/jnci/djv073 [DOI] [PubMed] [Google Scholar]
- 24. Lippman ME, Bolan G. Oestrogen‐responsive human breast cancer in long term tissue culture. Nature. 1975;256(5518):592‐593. 10.1038/256592a0 [DOI] [PubMed] [Google Scholar]
- 25. Horwitz KB, Costlow ME, McGuire WL. MCF‐7: A human breast cancer cell line with estrogen, androgen, progesterone, and glucocorticoid receptors. Steroids. 1975;26(6):785‐795. 10.1016/0039-128X(75)90110-5 [DOI] [PubMed] [Google Scholar]
- 26. Cassanelli S, Louis J, Seigneurin D. Progesterone receptor heterogeneity in MCF‐7 cell subclones is related to clonal origin and kinetics data. Tumor Biol. 1995;16(4):222‐229. 10.1159/000217939 [DOI] [PubMed] [Google Scholar]
- 27. Benz CC, Scott GK, Sarup JC, et al. Estrogen‐dependent, tamoxifen‐resistant tumorigenic growth of MCF‐7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24(2):85‐95. 10.1007/BF01961241 [DOI] [PubMed] [Google Scholar]
- 28. Vinckevicius A, Chakravarti D. Chromatin immunoprecipitation: advancing analysis of nuclear hormone signaling. J Mol Endocrinol. 2012;49(2):R113‐R123. 10.1530/JME-12-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cailleau R, Young R, Olivé M, Reeves WJ Jr. Breast tumor cell lines from pleural effusions. JNCI J Natl Cancer Inst. 1974;53(3):661‐674. 10.1093/jnci/53.3.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2011;32(1‐2):35‐48. 10.3233/BD-2010-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai X, Cheng H, Bai Z, Li J. Breast cancer cell line classification and its relevance with breast tumor subtyping. J Cancer. 2017;8(16):3131‐3141. 10.7150/jca.18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mota A, Evangelista A, Macedo T, et al. Molecular characterization of breast cancer cell lines by clinical immunohistochemical markers. Oncol Lett. 2017;13(6):4708‐4712. 10.3892/ol.2017.6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grigoriadis A, Mackay A, Noel E, et al. Molecular characterisation of cell line models for triple‐negative breast cancers. BMC Genom. 2012;13:619. 10.1186/1471-2164-13-619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gazdar AF, Kurvari V, Virmani A, et al. Characterization of paired tumor and non‐tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78(6):766‐774. [DOI] [PubMed] [Google Scholar]
- 35. Kao J, Salari K, Bocanegra M, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4(7):e6146. 10.1371/journal.pone.0006146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hollestelle A, Nagel JHA, Smid M, et al. Distinct gene mutation profiles among luminal‐type and basal‐type breast cancer cell lines. Breast Cancer Res Treat. 2010;121(1):53‐64. 10.1007/s10549-009-0460-8 [DOI] [PubMed] [Google Scholar]
- 37. Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515‐527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5‐29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 39. Tran A‐Q, Gehrig P. Recent advances in endometrial cancer. F1000Research. 2017;6:81. 10.12688/f1000research.10020.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 41. Zadnik V, Primic Zakelj M, Lokar K, Jarm K, Ivanus U, Zagar T. Cancer burden in Slovenia with the time trends analysis. Radiol Oncol. 2017;51(1):47‐55. 10.1515/raon-2017-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Society C . Canadian cancer. Statistics (Ber). 2016;2016(2016):1‐142. [Google Scholar]
- 43. Lax SF. Neues in der WHO‐Klassifikation 2014 der Tumoren des Corpus uteri. Pathologe. 2016;37(6):500‐511. 10.1007/s00292-016-0230-4 [DOI] [PubMed] [Google Scholar]
- 44. Talhouk A, McAlpine JN. New classification of endometrial cancers: the development and potential applications of genomic‐based classification in research and clinical care. Gynecol Oncol Res Pract. 2016;3(1):14. 10.1186/s40661-016-0035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matias‐Guiu X, Longacre T, McCluggage WG, et al. Tumors of the uterine corpus: Introduction. In: Kim K‐R, Lax SF, Lazar A, eds. WHO Classification of Tumours: Female Genital Tumours, 5th edn. IARC; 2020:246‐247. [Google Scholar]
- 46. Lax SF. Pathology of endometrial carcinoma. Adv Exp Med Biol. 2017; 943: 75‐96. 10.1007/978-3-319-43139-0_3 [DOI] [PubMed] [Google Scholar]
- 47. Bell DW, Ellenson LH. Molecular genetics of endometrial carcinoma. Annu Rev Pathol Mech Dis. 2019;14(1):339‐367. 10.1146/annurev-pathol-020117-043609 [DOI] [PubMed] [Google Scholar]
- 48. León‐Castillo A, Britton H, McConechy MK, et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol. 2020;250(3):323‐335. 10.1002/path.5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morice P, Leary A, Creutzberg C, Abu‐Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387(10023):1094‐1108. 10.1016/S0140-6736(15)00130-0 [DOI] [PubMed] [Google Scholar]
- 50. Korch C, Spillman MA, Jackson TA, et al. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127(1):241‐248. 10.1016/j.ygyno.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qu W, Zhao Y, Wang X, et al. Culture characters, genetic background, estrogen/progesterone receptor expression, and tumorigenic activities of frequently used sixteen endometrial cancer cell lines. Clin Chim Acta. 2019;489:225‐232. [DOI] [PubMed] [Google Scholar]
- 52. Hevir‐Kene N, Rižner TL. The endometrial cancer cell lines Ishikawa and HEC‐1A, and the control cell line HIEEC, differ in expression of estrogen biosynthetic and metabolic genes, and in androstenedione and estrone‐sulfate metabolism. Chem Biol Interact. 2015;234:309‐319. [DOI] [PubMed] [Google Scholar]
- 53. Van Nyen T, Moiola CP, Colas E, Annibali D, Amant F. Modeling endometrial cancer: past, present, and future. Int J Mol Sci. 2018;19(8):2348. 10.3390/ijms19082348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7‐30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 55. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169‐182. 10.1016/S0140-6736(18)32470-X [DOI] [PubMed] [Google Scholar]
- 56. Cibula D, Pötter R, Planchamp F, et al. The European Society of Gynaecological Oncology/European Society for Radiotherapy and Oncology/European Society of Pathology Guidelines for the Management of Patients with Cervical Cancer. Virchows Arch. 2018;472(6):919‐936. 10.1007/s00428-018-2362-9 [DOI] [PubMed] [Google Scholar]
- 57. Brianti P, De Flammineis E, Mercuri SR. Review of HPV‐related diseases and cancers. New Microbiol. 2017;40(2):80‐85. [PubMed] [Google Scholar]
- 58. Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975‐2017 SEER Cancer Statistics. Natl Cancer Inst. 2017:1975‐2008. [Google Scholar]
- 59. Lax S. Histopathology of cervical precursor lesions and cancer. Acta Dermatovenerologica Alpina, Pannonica Adriat. 2011;20(3):125‐133. [PubMed] [Google Scholar]
- 60. Khodabandehlou N, Mostafaei S, Etemadi A, et al. Human papilloma virus and breast cancer: The role of inflammation and viral expressed proteins. BMC Cancer. 2019;19(1):61. 10.1186/s12885-019-5286-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pesola F, Sasieni P. Impact of screening on cervical cancer incidence in England: a time trend analysis. BMJ Open. 2019;9(1):e026292. 10.1136/bmjopen-2018-026292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Landy R, Pesola F, Castañón A, Sasieni P. Impact of cervical screening on cervical cancer mortality: estimation using stage‐specific results from a nested case–control study. Br J Cancer. 2016;115(9):1140‐1146. 10.1038/bjc.2016.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Quinn M, Babb P, Jones J, Allen E. Effect of screening on incidence of and mortality from cancer of cervix in England: evaluation based on routinely collected statistics. BMJ. 1999;318(7188):904‐908. 10.1136/bmj.318.7188.904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hasche D, Vinzón SE, Rösl F. Cutaneous papillomaviruses and non‐melanoma skin cancer: Causal agents or innocent bystanders? Front Microbiol. 2018;9(MAY):874. 10.3389/fmicb.2018.00874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rosa MN, Evangelista AF, Leal LF, et al. Establishment, molecular and biological characterization of HCB‐514: a novel human cervical cancer cell line. Sci Rep. 2019;9(1):1913. 10.1038/s41598-018-38315-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Greely HT, Cho MK. The Henrietta Lacks legacy grows. EMBO Rep. 2013;14(10):849. 10.1038/embor.2013.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gey GO, Coffmann WD, Kubicek MT. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12(4):264‐265. [Google Scholar]
- 68. Horbach SPJM, Halffman W. The ghosts of HeLa: How cell line misidentification contaminates the scientific literature. PLoS One. 2017;12(10):e0186281. 10.1371/journal.pone.0186281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Capes‐Davis A, Theodosopoulos G, Atkin I, et al. Check your cultures! A list of cross‐contaminated or misidentified cell lines. Int J Cancer. 2010;127(1):1‐8. 10.1002/ijc.25242 [DOI] [PubMed] [Google Scholar]
- 70. Kontostathi G, Zoidakis J, Makridakis M, et al. Cervical cancer cell line secretome highlights the roles of transforming growth factor‐beta‐induced protein ig‐h3, peroxiredoxin‐2, and NRF2 on cervical carcinogenesis. Biomed Res Int. 2017;2017:4180703. 10.1155/2017/4180703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gutiérrez‐Hoya A, Zerecero‐Carreón O, Valle‐Mendiola A, et al. Cervical cancer cells express markers associated with immunosurveillance. J Immunol Res. 2019;2019:1‐10. 10.1155/2019/1242979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kurz H, Sandau K, Christ B. On the bifurcation of blood vessels – Wilhelm Roux’s Doctoral Thesis (Jena 1878) – A seminal work for biophysical modelling in developmental biology. Ann Anat. 1997;179(1):33‐36. 10.1016/S0940-9602(97)80132-X [DOI] [PubMed] [Google Scholar]
- 73. Pickl M, Ries CH. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2009;28(3):461‐468. 10.1038/onc.2008.394 [DOI] [PubMed] [Google Scholar]
- 74. Geraghty RJ, Capes‐Davis A, Davis JM, et al. Guidelines for the use of cell lines in biomedical research. Br J Cancer. 2014;111(6):1021‐1046. 10.1038/bjc.2014.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gradisnik L, Trapecar M, Rupnik MS, Velnar T. HUIEC, Human intestinal epithelial cell line with differentiated properties: process of isolation and characterisation. Wien Klin Wochenschr. 2015;127(S5):204‐209. 10.1007/s00508-015-0771-1 [DOI] [PubMed] [Google Scholar]
- 76. Naranda J, Gradišnik L, Gorenjak M, Vogrin M, Maver U. Isolation and characterization of human articular chondrocytes from surgical waste after total knee arthroplasty (TKA). PeerJ. 2017;5:e3079. 10.7717/peerj.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Skok K, Takač I, Kavalar R, Gradišnik L, Milojević M, Maver U. Simple protocol for effective preparation of breast cancer cell cultures for possible 3D tumor modeling. EPNOE Newletter. 2019;51:7‐8. [Google Scholar]
- 78. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359‐E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 79. Kuramoto H. Studies of the growth and cytogenetic properties of human endometrial adenocarcinoma in culture and its development into an established line. Acta Obstet Gynaecol Jpn. 1972;19(1):47‐58. [PubMed] [Google Scholar]
- 80. Fogh J. Human tumor lines for cancer research. Cancer Invest. 1986;4(2):157‐184. 10.3109/07357908609038260 [DOI] [PubMed] [Google Scholar]
- 81. Wang Y, Yang DA, Cogdell D, et al. Genomic characterization of gene copy‐number aberrations in endometrial carcinoma cell lines derived from endometrioid‐type endometrial adenocarcinoma. Technol Cancer Res Treat. 2010;9(2):179‐189. 10.1177/153303461000900207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nishida M, Kasahara K, Kaneko M, Iwasaki H, Hayashi K. Establishment of a new human endometrial adenocarcinoma cell line, Ishikawa cells, containing estrogen and progesterone receptors. Nihon Sanka Fujinka Gakkai Zasshi. 1985;37(7):1103‐1111. [PubMed] [Google Scholar]
- 83. Nishida M. The Ishikawa cells from birth to the present. Hum Cell. 2002;15(3):104‐117. [DOI] [PubMed] [Google Scholar]
- 84. Dawe CJ, Banfield WG, Morgan WD, Slatick MS, Curth HO. Growth in continuous culture, and in hamsters, of cells from a neoplasm associated with Acanthosis nigricans . JNCI J Natl Cancer Inst. 1964;33(3):441‐456. 10.1093/jnci/33.3.441 [DOI] [PubMed] [Google Scholar]
- 85. Richardson GS, Dickersin G, Atkins L, et al. KLE: A cell line with defective estrogen receptor derived from undifferentiated endometrial cancer. Gynecol Oncol. 1984;17(2):213‐230. 10.1016/0090-8258(84)90080-5 [DOI] [PubMed] [Google Scholar]
- 86. Auersperg N. Long‐term cultivation of hypodiploid human tumor cells. J Natl Cancer Inst. 1964;32:135‐163. [PubMed] [Google Scholar]
- 87. Yamada T, Ueda M, Maeda T, et al. Establishment and characterization of a cell line (OMC‐4) originating from a human adenocarcinoma of the uterine cervix. Nihon Sanka Fujinka Gakkai Zasshi. 1987;39(5):859‐860. [PubMed] [Google Scholar]
- 88. Pattillo RA, Hussa RO, Story MT, Ruckert ACF, Shalaby MR, Mattingly RF. Tumor antigen and human chorionic gonadotropin in CaSki cells: A new epidermoid cervical cancer cell line. Science (80‐ ). 1977;196(4297):1456‐1458. 10.1126/science.867042 [DOI] [PubMed] [Google Scholar]
- 89. Friedl F, Kimura I, Osato T, Ito Y. Studies on a New Human Cell Line (SiHa) derived from carcinoma of uterus. I. Its establishment and morphology. Proc Soc Exp Biol Med. 1970;135(2):543‐545. 10.3181/00379727-135-35091a [DOI] [PubMed] [Google Scholar]
- 90. Sakamoto M, Okabe K, Matayoshi K, Negishi Y, Akiya K. Characterization of a Newly Established Cell Line (TMCC‐1) from human uterine cervical adenocarcinoma. 日本産科婦人科學會雜誌. 1987;39(8):1489. [Google Scholar]
- 91. Sykes JA, Whitescarver J, Jernstrom P, Nolan JF, Byatt P. Some properties of a new epithelial cell line of human origin. JNCI J Natl Cancer Inst. 1970;45(1):107‐122. 10.1093/jnci/45.1.107 [DOI] [PubMed] [Google Scholar]
- 92. Fogh J, Trempe G. New human tumor cell lines. In: Fogh J, ed. Human Tumor Cells in Vitro. US: Springer; 1975:115‐159. 10.1007/978-1-4757-1647-4_5 [DOI] [Google Scholar]
- 93. Auersperg N, Hawryluk AP. Chromosome observations on three epithelial‐cell cultures derived from carcinomas of the human cervix. J Natl Cancer Inst. 1962;28(3):605‐627. 10.1093/jnci/28.3.605 [DOI] [PubMed] [Google Scholar]
- 94. Fusenig NE, Capes‐Davis A, Bianchini F, Sundell S, Lichter P. The need for a worldwide consensus for cell line authentication: Experience implementing a mandatory requirement at the International Journal of Cancer. PLOS Biol. 2017;15(4):e2001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lorsch JR, Collins FS, Lippincott‐Schwartz J. Fixing problems with cell lines. Science. 2014;346(6216):1452‐1453. 10.1126/science.1259110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shah RN, Grzybowski AT, Cornett EM, et al. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol Cell. 2018;72(1):162‐177.e7. 10.1016/j.molcel.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Loken E, Gelman A. Measurement error and the replication crisis. Science. 2017;355(6325):584‐585. 10.1126/science.aal3618 [DOI] [PubMed] [Google Scholar]
- 98. Weissgerber TL, Savic M, Winham SJ, Stanisavljevic D, Garovic VD, Milic NM. Data visualization, bar naked: A free tool for creating interactive graphics. J Biol Chem. 2017;292(50):20592‐20598. 10.1074/jbc.RA117.000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. An update on data reporting standards. Nat Cell Biol. 2014;16(5):385. 10.1038/ncb2964 [DOI] [PubMed] [Google Scholar]
- 100. International Cell Line Authentication Committee . Naming a Cell Line ‐ ver. 1.6; 2015. http://iclac.org/wp‐content/uploads/Naming‐a‐Cell‐Line_v1_6.pdf. Accessed April 14, 2018


