Abstract
Background:
A lot of research evidence shows that exosomes play an indelible role in the prognosis of lung cancer, but there are many disputes. Therefore, we conduct a meta-analysis to further demonstrate.
Methods:
A literature retrieval was performed through a search of PubMed, Embase, Web of Science, Cochrane, CKNI, Wanfang, and other databases to locate documents from the literature that satisfied the inclusion criteria. There were four outcome indicators: overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS). Subgroup analysis was conducted according to sample size, country, detection method, analysis method, and pathological type. Stata 14.0 software was used to evaluate the prognostic value of exosomes in lung cancer.
Results:
A total of 2456 patients with lung cancer from 29 studies in 16 articles were included. The expression level of exosomes was closely associated with the OS and DFS of patients, although no statistical difference was observed between exosomes and DSS or PFS. Eighteen studies with 2,110 patients were evaluated to examine the prognostic value of exosomes in lung cancer by exploring the association between exosomes and OS. The results showed that exosomes were strongly associated with worse OS, and the combined hazard ratio (HR) was 2.01 (95% confidence interval [CI]: 1.70–2.39, P = .000). Six studies investigated the association between exosomes and DFS, and showed a pooled HR of 2.48 (95% CI: 1.75–3.53, P = .000).
Conclusion:
Our analysis indicated that the expression level of exosomes was closely associated with the OS and DFS of patients with lung cancer, suggesting that exosomes are associated with poor prognosis of lung cancer. Exosomes may be a new biomarker for the prognosis of lung cancer, although a large number of prospective studies are still needed to support this.
Keywords: exosomes, lung cancer, meta-analysis, prognosis
1. Introduction
Lung cancer is one of the main causes of cancer death in the world, ranking first in terms of morbidity and prevalence.[1] Recently, considerable progress has been achieved in the diagnosis and treatment of lung cancer, although the 5-year survival rate is still very low, at only about 15%.[2,3] Finding biomarkers for more sensitive diagnosis or prognostic monitoring remains an ongoing task and goal for the medical field. At present, blood-based biomarkers are receiving more and more attention in the diagnosis and prognosis of cancer.
Exosomes play an important role in the transmission of substances and information in the occurrence and development of tumors, which can affect their progression.[4,5] Therefore, the bioactive substances carried by exosomes may be used as biomarkers for diagnosis and prognosis of tumors.[6] Exosomes are produced by a number of cell types and are mainly secreted by hematopoietic cells, such as reticulocytes, B lymphocytes, T lymphocytes, and dendritic cells, through a series of processes, such as “endocytosis-fusion-exclusion”. [7–9] Exosomes are also produced by cells of non-hematopoietic origin, like epithelial cells (intestinal epithelial cells), neurons, fibroblasts, and tumor cells.[7–9] Exosomes are membrane-encapsulated extracellular small vesicles, with a diameter of about 30–100 nm.[7] They have a lipid bilayer membrane structure,[10] and their vesicles also contain a large amount of bioactive substances, including proteins, RNA, DNA and lipids.[11,12] Exosomes can be separated from various fluids, including plasma, serum, urine, saliva, nasal secretions, bronchoalveolar lavage, pleural effusion, bile, ascites, semen, breast milk, amniotic fluid, and cerebrospinal fluid.[13–17] They can also be stored very stably at temperatures as low as –20°C.[10] Exosome membranes are mainly composed of lipids, which make them highly stable and easily absorbed by target cells.[18] These characteristics make exosomes useful as biomarkers.
With the development of proteomics and genomics, there is an increasingly profound understanding of the source, structure, and function of exosomes. In recent years, research on exosomes and lung cancer prognosis has become a medical hotspot. Majority of studies have found that exosomes play a vital role in the diagnosis, treatment, and prognosis of lung cancer, although the results remain controversial. This study aimed to assess the association between exosomes and the prognosis of lung cancer through meta-analysis.
2. Materials and methods
2.1. Document retrieval
Studies were identified via an electronic search of PubMed, Embase, Web of Science, Cochrane, CKNI, and Wanfang. Data up to February of 2020 were included. Search terms included: exosome OR exosomes, lung cancer OR lung carcinoma OR lung neoplasm, OS OR overall survival OR mortality OR survival, prognostic value, PFS OR progression-free survival, and DFS OR disease-free survival. The retrieval languages were Chinese and English. All analyses were based on previous published studies, thus no ethical approval and patient consent are required.
2.2. Literature inclusion and exclusion criteria
Inclusion criteria were
-
1.
publicly published studies on the association between exosomes and the prognosis of lung cancer;
-
2.
patients were diagnosed with lung cancer by pathology or cytological examination;
-
3.
outcome indicators of overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS), and
-
4.
ability to extract hazard ratio (HR) and 95% confidence intervals (CIs) from published Chinese and English literature directly or indirectly.
Exclusion criteria were:
-
1.
documents that did not comply with the inclusion criteria;
-
2.
documents that could not provide complete information or data;
-
3.
literature such as reviews, case reports, and animal or cell experiments; and
-
4.
repeated publications.
2.3. Data extraction and quality evaluation
Two researchers independently screened the recovered documents according to the inclusion and exclusion criteria of the literature. If there were disagreements, a third researcher was consulted to provide assistance in resolution. The researchers assessed the quality of the literature that met the requirements and extracted data from them, including author, nationality of the research subjects, publication time, sample size, and detection method. The Newcastle-Ottawa Scale (NOS) score was used to evaluate the quality of the included studies, with 9 points possible. Studies with scores of ≥6 points were included.[19]
2.4. Statistical analyses
Stata 14.0 was used for analysis. Subgroup analysis was performed based on sample size, country, detection method, analysis method, and pathological type to evaluate the potential sources of heterogeneity. Outcome indicators were survival rate, survival curve, and hazard ratio (HR). If the Kaplan-Meier survival curve was provided in the literature, the 5-year survival rate was obtained using Engauge Digtizer 1.4, and the HR was obtained using RevMan 5.3.[20–22] Heterogeneity analyses used the I2 and Q tests. When I2<50% and the Q test P > .1, it was presumed that there was no heterogeneity, and the fixed effect model was used for analysis; otherwise, the random effect model was selected. If heterogeneity was present, sensitivity analysis was used by eliminating a single study in a queue to identify the potential sources of heterogeneity and assess whether they would affect the stability of the overall results. The main source of heterogeneity was analyzed by meta-regression. Funnel charts and Begg and Egger tests were used to detect publication bias. The trim-and-fill method was used to further analyze the impact of publication bias.
3. Results
3.1. Literature screening results
A total of 235 articles were initially screened, and 190 articles remained after 45 duplicates were eliminated. The literature was then screened according to title and abstract, and 153 more studies were excluded. The full text of the remaining 37 studies was read closely, and 21 additional studies were excluded for the following reasons: 14 studies lacked complete data, and 7 studies had no data available. Finally, the remaining 16 articles met the requirements and were included in this study (Fig. 1).[23–38]
Figure 1.
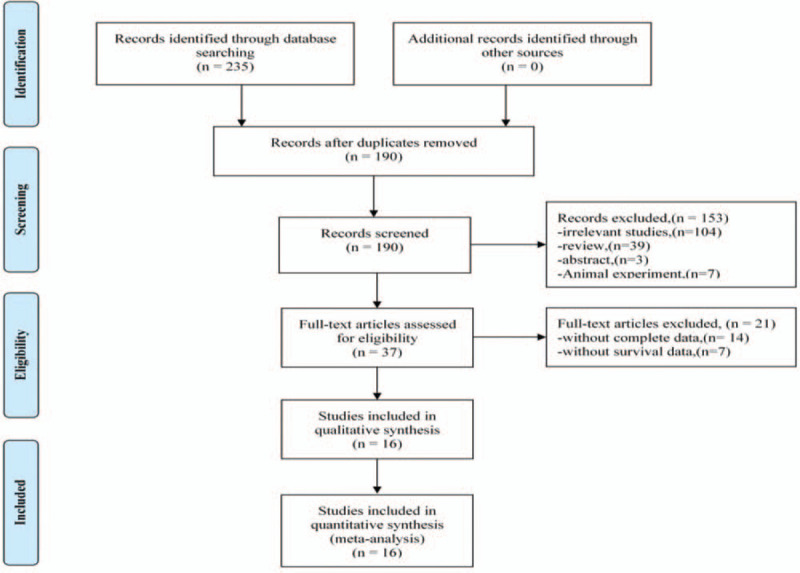
Flow diagram showing study retrieval and selection process.
3.2. Basic characteristics and quality evaluation of literature
Our study included 16 articles that included 29 studies, with 18 studies on OS, six on DFS, two on DSS, and three on PFS. Because of the different types of exosomes mentioned in the articles, it was possible there were multiple studies in the same article, although quality evaluations were conducted to ensure the inclusion criteria were met. Of the 18 studies on exosomes and OS, 15 were from China, and the rest were from Japan and Europe. There were 11 studies with sample sizes >100, and there were 15 that used qRT-PCR as a measure of exosome expression. There were eight studies that used univariate regression analysis; the others used multivariate regression analysis. The pathological types of cases included in the 18 OS studies were: non-small cell lung cancer (NSCLC), adenocarcinoma (ADC), and small cell lung cancer (SCLC). Among them, nine studies were about NSCLC, eight were about ADC, and one was about SCLC. Of the nine studies on NSCLC, Xiong[25] included ADC (42 cases) and SQCC (58 cases); Liu[37] included ADC (115 cases), SQCC (73 cases), large cell carcinoma (LCC) (3 cases), and others (1 case); Dejima[36] included ADC (134 cases), SQCC (53 cases), and others (one case); Paulsen[38] included ADC (198 cases), SQCC (69 cases), and others (nine cases); Xu[28] included ADC (12 cases) and SQCC (31 cases); Zhang[23] included ADC (53 cases), SQCC (46 cases), and others (4 cases); and Kanaoa[34] included ADC (201 cases) and SQCC (72 cases). The specific number of lung cancer subtypes could not be obtained in Yuwen[30] or Nanou.[27] The included documents were evaluated using the NOS score, with a total possible score of 9 points, and scores of ≥6 points indicated high-quality studies, as shown in Table 1.
Table 1.
Basic characteristics of included studies.
| Auuthor | Year | Country | Age | Exosome type | Dysregulation | Sample (n) | Pathological type | Analysis method | Detection method | Oncologic outcomes | NOS | |
| Zhang | 2020 | China | 57.4 | miR-378 | Upregulation | 103 | NSCLC | MVA | qRT-PCR | OS | 8 | |
| Xue | 2020 | China | NA | miR-151a-5p | Upregulation | 6 | ADC | UVA | qRT-PCR | OS | 7 | |
| miR-10b-5p | Upregulation | 6 | ADC | UVA | qRT-PCR | OS | ||||||
| miR-192-5p | Upregulation | 6 | ADC | UVA | qRT-PCR | OS | ||||||
| miR-106b-3p | Upregulation | 6 | ADC | UVA | qRT-PCR | OS | ||||||
| miR-484 | Upregulation | 6 | ADC | UVA | qRT-PCR | OS | ||||||
| Xiong | 2020 | China | NA | miR-214 | Upregulation | 100 | NSCLC | UVA | qRT-PCR | OS | 6 | |
| Shimada | 2020 | Japan | NA | UCHL1 | Upregulation | 72 | SCLC | MVA | qRT-PCR | DFS | 7 | |
| Nanou | 2020 | Netherland | 65 | tDEVs | Upregulation | 137 | NSCLC | UVA | Cell search | OS | 8 | |
| Xu | 2019 | China | 64 | miR-32 | Downregulation | 43 | NSCLC | UVA | qRT-PCR | OS/PFS | 6 | |
| Sun | 2019 | China | NA | miR-423-3p | Upregulation | 155 | ADC | MVA | qRT-PCR | OS | 7 | |
| miR-4270 | Downregulation | 155 | ADC | MVA | qRT-PCR | OS | ||||||
| Yunwen | 2018 | China | NA | miR-425-3p | Upregulation | 170 | NSCLC | UVA | qRT-PCR | PFS | 8 | |
| Li | 2018 | China | NA | FECRS | Downregulation | 56 | SCLC | UVA | qRT-PCR | PFS | 7 | |
| Koh | 2018 | Korea | 66 | Rab27B | Upregulation | 96 | SQCC/ADC | MVA | IHC | DFS/DSS | 6 | |
| Rab27B | Upregulation | 37 | SQCC/ADC | MVA | IHC | DFS/DSS | ||||||
| Xu | 2018 | China | 60 | miR-21 | Upregulation | 437 | ADC | UVA | qRT-PCR | OS | 7 | |
| Kanaoka | 2018 | Japan | NA | miR-451a | Upregulation | 285 | NSCLC | MVA | qRT-PCR | OS/DFS | 9 | |
| Zeng | 2017 | China | 55 | LncRNA | Upregulation | 86 | SCLC | MVA | qRT-PCR | OS | 7 | |
| Dejima | 2017 | Japan | NA | miR-21 | Upregulation | 201 | NSCLC | MVA | qRT-PCR | DFS | 7 | |
| miR-4257 | Upregulation | 201 | NSCLC | MVA | qRT-PCR | DFS | ||||||
| Liu | 2016 | China | 58.5 | miR-23b-3p | Upregulation | 196 | NSCLC | MVA | qRT-PCR | OS | 8 | |
| miR-10b-5p | Upregulation | 196 | NSCLC | MVA | qRT-PCR | OS | ||||||
| miR-21-5p | Upregulation | 196 | NSCLC | MVA | qRT-PCR | OS | ||||||
| Paulsen-S | 2016 | Danmark | 68.6 | Alix | Upregulation | 276 | NSCLC | MVA | Spotbot | OS | 8 |
3.3. Meta-analysis results
There were 18 studies in the OS group, and there was heterogeneity among the studies (I2=67.8%, P = .000), therefore, the random effects model was used. The results showed that the HR between exosomes and the OS of patients with lung cancer was 2.01 (95% CI: 1.70–2.39, P = .000) (Fig. 2). The DFS group contained six studies, and no obvious heterogeneity (I2=30.9%, P = .204) was found among them. Therefore, the fixed effect model was selected. The results indicated that the combined HR between exosomes and DFS of patients with lung cancer was 2.48 (95% CI: 1.75–3.53, P = .000) (Fig. 3). In the OS group, patients with abnormal exosome expression had a 101% higher risk of poor prognosis (HR=2.01, 95% CI: 1.70–2.39, P = .000), and for the DFS group, patients with abnormal exosome expression had a 148% higher risk of poor prognosis (HR=2.48, 95% CI: 1.75–3.53, P = .000). The DSS and PFS groups included two and three studies, respectively. The results showed that there was no statistical significance between exosomes and DSS or PFS (P = .356>0.05, P = .058>.05) (Fig. 4).
Figure 2.
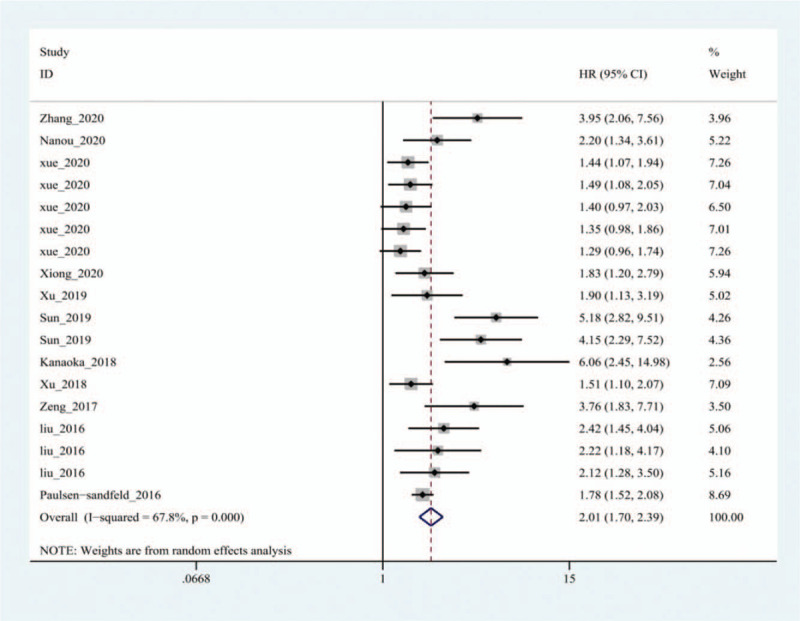
Forest plot of the Hazard ratio for the relationship between exosomes and overall survival of lung cancer patients.
Figure 3.
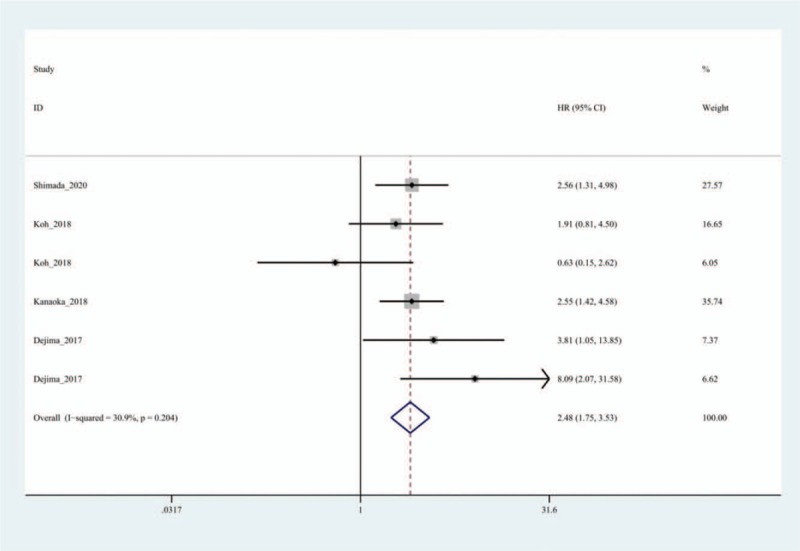
Forest plot of the Hazard ratio for the relationship between exosomes and disease-free survival of lung cancer patients.
Figure 4.
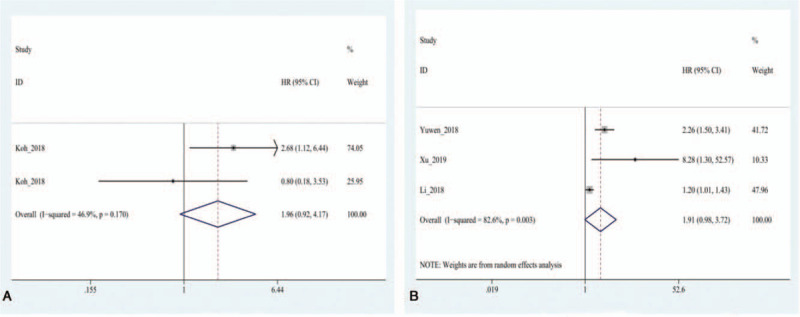
Forest plot of the Hazard ratio for the relationship of exosomes and the prognosis of lung cancer patients. (A) The overall pooled HR for disease-specific survival of lung cancer patients. (B) The overall pooled HR for progression free survival of lung cancer patients.
3.4. Subgroup analysis
To further clarify the source of heterogeneity in the OS group, we conducted a subgroup analysis based on the sample size, country, detection method, analysis method, and pathological type. The results showed that the expression level of exosomes was significantly associated with the OS, as shown in Table 2. After the subgroup analysis, no obvious source of heterogeneity was found, therefore, further meta-regression and sensitivity analyses were required.
Table 2.
Summary of overall and subgroup analyses for exosomes on OS.
| Studies (n) | Combined HR (95%CI) | Weight (%) | I2 | P-value (Q) | |
| overall | 18 | 2.01 (1.70-2.39) | 100.0 | 67.8% | .000 |
| Country | |||||
| China | 15 | 1.98 (1.62–2.42) | 83.53 | 68.7% | .000 |
| Other countries | 3 | 2.43 (1.45–4.08) | 16.47 | 72.5% | .026 |
| Sample size | |||||
| >100 | 11 | 2.48 (1.95–3.15) | 56.41 | 68.1% | .001 |
| <100 | 7 | 1.50 (1.27–1.77) | 43.59 | 30.2% | .198 |
| Detection method | |||||
| qRT-PCR | 15 | 2.08 (1.69–2.55) | 86.09 | 71.2% | .000 |
| Non qRT-PCR | 3 | 1.82 (1.56–2.11) | 13.91 | 0.0% | .424 |
| Analysis method | |||||
| MVA | 10 | 2.90 (2.17–3.86) | 46.88 | 69.1% | .001 |
| UVA | 8 | 1.46 (1.30–1.65) | 53.12 | 0.0% | .877 |
| Pathological type | |||||
| NSCLC | 9 | 2.19 (1.80–2.67) | 45.72 | 38.9% | .109 |
| ADC | 8 | 2.01 (1.70–2.39) | 50.78 | 75.4% | .000 |
| SCLC | 1 | 3.76 (1.83–7.71) | 3.50 | – | – |
3.5. Sensitivity analysis and meta-regression
Sensitivity analysis was used to evaluate the stability of the results by deleting each study in a queue and then recombining the HR for OS. We observed that the heterogeneity of the combined HR for OS was mainly caused by one study[29]; the combined HR after its exclusion was 1.95 (95% CI: 1.68–2.26, P = .008). However, there was no significant change before and after the exclusion, indicating that the combined HR was robust (Fig. 5). The results of the meta-analysis are shown in Table 3, which shows that the analysis method (P = .001) may be the main source of heterogeneity, and other factors, such as sample size (P = .934), country (P = .506), and detection method (P = .63), have no statistical significance.
Figure 5.
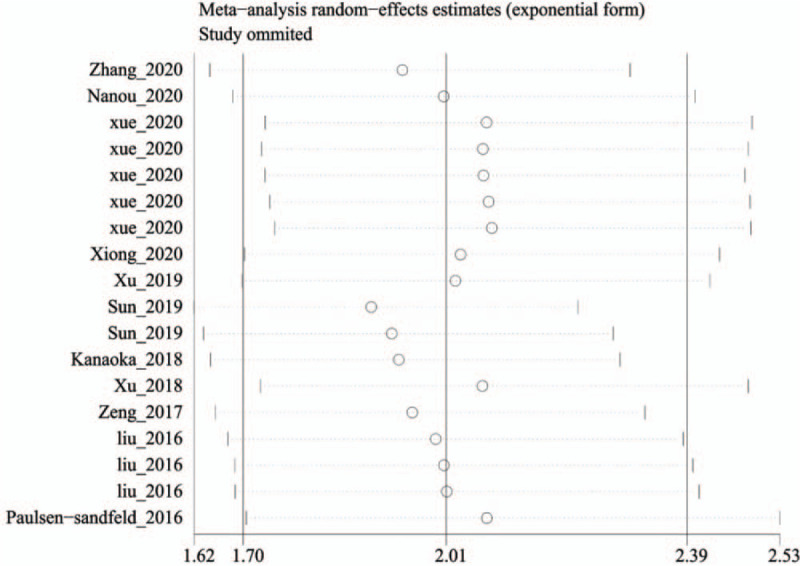
Sensitivity analyses to assess the effect of individual studies on the overall pooled HR for overall survival of lung cancer patients.
Table 3.
Meta-regression analysis of potential source of heterogeneity.
| Heterogeneity factors | OR | SE | t | P-value | 95%CI |
| Sample size | 1.087 | 0.149 | 0.61 | .934 | 0.81–1.46 |
| Country | 0.512 | 0.244 | −1.41 | .506 | 0.18–1.43 |
| Analysis method | 1.988 | 0.316 | 4.32 | .001 | 1.41–2.80 |
| Detection method | 3.338 | 1.562 | 2.58 | .063 | 1.21–9.18 |
3.6. Publication bias
3.6.1. Funnel graph
We detected publication bias using funnel plots in the OS and DFS groups. The funnel plots showed that the OS group was asymmetrical, suggesting that there might be publication bias, while in the DFS group, the funnel plot showed symmetry, which indicated no obvious publication bias, as shown in Figure 6. The bias in the OS group needed to be further verified using the Begg's and Egger's tests.
Figure 6.
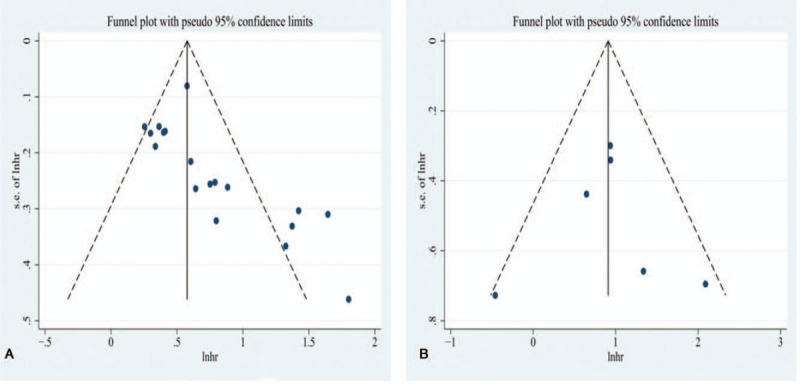
Funnel plots of the relationship between exosomes and the prognosis of lung cancer. (A) Relationship of exosomes and overall survival of lung cancer patients. (B) Relationship of exosomes and disease-free survival of lung cancer patients.
3.6.2. Begg's and Egger's test
The Begg's and Egger's tests were performed to assess the publication bias of the OS group (Fig. 7). These tests suggested that there was a publication bias (P = .000<.05, P = .000<.05). Therefore, we needed to employ the trim and fill method to further evaluate the stability of the combined HR for OS.
Figure 7.
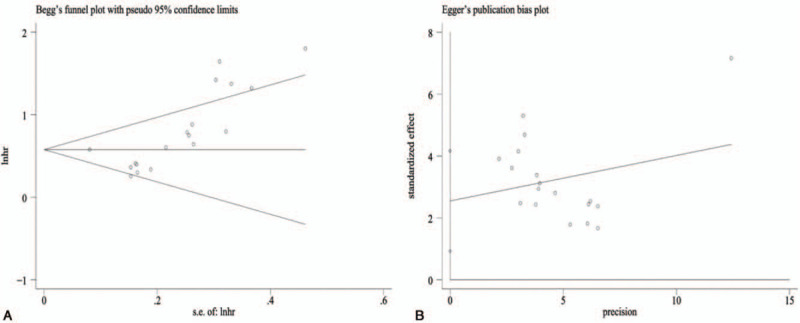
Funnel plots for publication bias of the relationship between exosomes and overall survival of lung cancer patients. (A) Begg's Test. (B) Egger's Test.
3.6.3. Trim and fill method
The final result is shown in Fig. 8. The adjusted funnel plots for OS became symmetrical, and the combined HR (HR = 1.64, 95% CI: 1.35–1.98) for OS only changed marginally after the trim and fill method was applied, indicating the stability and reliability of our analysis.
Figure 8.
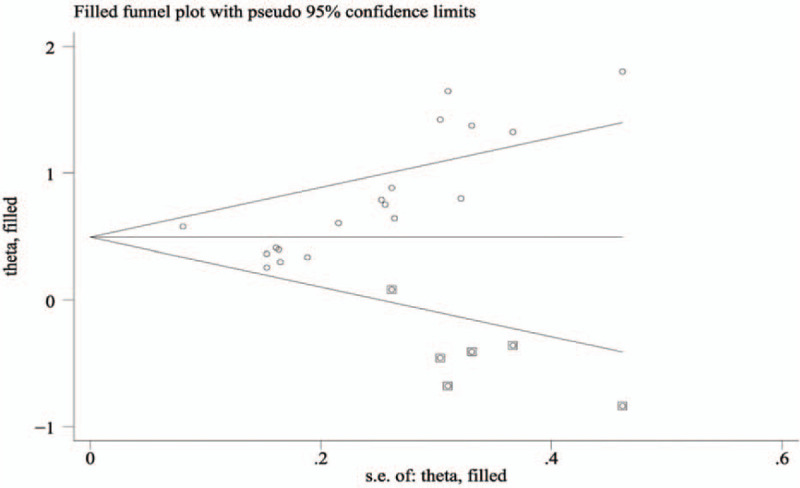
Adjusted Begg's funnel plot from the trim and fill method regarding the publication bias for overall survival.
4. Discussion
It is usually difficult to predict the prognosis of lung cancer. It is well known that tumor size, clinical stage, pathological grading, classification, and other characteristics are the main factors that determine the prognosis of lung cancer, although we still need more and simpler indicators. Like most cancers, lung cancer is inclined to recur, especially in the course of treatment. We hope that these new indicators can reflect their short-term and long-term prognoses to better guide clinical work. A large amount of evidence shows that numerous genetic markers affect the biological behavior of lung cancer, although the inherent nature of gene disorders that lead to cancer recurrence, progression, or metastasis remains elusive.[39] With these factors in mind, it is essential to study the material characteristics associated with the biological behavior of cancer.
Tumor-secreted factors play a guiding role in the local and far-reaching effects of tumors, such as soluble factors and exosome nanovesicles.[40] Exosomes have potent biological functions. They play an important role in mediating tumor immune escape in many cancers, such as gastric cancer, liver cancer, and lung cancer, thereby creating conditions for tumor cells to grow in vivo.[41] They can also cause tumorigenesis by promoting the transformation of cell epithelium and stroma and cell proliferation. Further, they can affect the invasion and metastasis of cancer by regulating cell apoptosis and the formation of the microenvironment before tumor metastasis.[42–47] A majority of studies have shown that exosomes can induce angiogenesis in different tumors by regulating cytokines and growth factor receptors, thus promoting the occurrence and development of tumors.[48–51] Exosomes, as carriers of information, are inextricably associated with the occurrence and development of tumors. They are some of the most important potential biomarkers in cancer, and are worthy of in-depth exploration and research.
Our analysis of the included 16 articles that described 2456 cases showed that the expression level of exosomes was associated with the prognosis of lung cancer. These results suggest that expression of exosomes may play a vital role in the occurrence and progression of lung cancer, and that they may be a potential biological indicator of its prognosis.
Further subgroup analysis of the OS group indicated that the expression level of exosomes was significantly correlated with the OS of patients with lung cancer. There was no significant difference between the expression level of exosomes and the detection methods. At the same time, we found that most studies used qRT-PCR to detect exosomes, and the combined HRs were higher than those of the non-qRT-PCR group (HR = 2.08, 95% CI: 1.69–2.55, HR = 1.82,95% CI: 1.56–2.11). There was no obvious difference between them, therefore, all HRs could be used for the detection of exosomes. The comparison between them needs to be further analyzed, however. Through subgroup analysis, it was found that the effect of abnormal exosome expression on the prognosis of lung cancer remains different. In the multivariate analysis, the combined HRs were significantly higher than those in the univariate analysis (HR = 2.90, 95% CI: 2.17–3.86, HR = 1.46,95% CI: 1.30–1.65). Our results also indicated that the expression level of exosomes was not only associated with the prognosis of lung cancer, but also might be linked to tumor stage, size, and metastasis. However, it is impossible to make a more accurate judgment using the literature alone, and a large amount of detailed research data is needed to further demonstrate these results. We also found regional differences between exosomes and patients’ OS; for example, the combined HRs of studies from China were lower than those from other regions (HR = 1.98,95% CI: 1.62–2.42, HR = 2.43,95% CI: 1.45–4.08). However, due to the small number of studies from other countries, more research is needed to confirm these regional differences. In addition, subgroup analysis indicated that the expression of exosomes was different among the OS of patients with different pathological types of lung cancer (HR = 2.19, 95% CI: 1.80–2.67, HR = 2.01, 95% CI: 1.70–2.39, HR = 3.67, 95% CI: 1.83–7.71). In seven of the nine studies on NSCLC, out of a total of 1198 patients with NSCLC, 754 (63.78%) had ADC, 405 (33.81%) had SQCC, and nine (2.41%) had other cancers. The other two studies included 170 cases and 137 cases of NSCLC. No specific subtypes of lung cancer have been described in the reviewed literature. In addition, none of these seven studies counted the survival data of specific subtypes of lung cancer, which makes it impossible for us to further analyze the association between exosomes and pathological types. In the OS group, there was one study alone on exosomes and SCLC, therefore, whether there is a difference in pathological types needs to be verified by larger samples and multi-center data. Interestingly, many of the included studies are about miRNA, which suggest that miRNA may become a hotspot in exosome research in future studies. Furthermore, we performed meta-regression, which indicated that the analysis methods may be the main source of heterogeneity. Our study used sensitivity analysis to verify the impact of a single study on our analysis results, and the results indicated that our conclusion was relatively stable. At the same time, the included studies had publication bias, although the results of the trim and fill method nonetheless suggested that our analysis results were stable and reliable.
This meta-analysis has the following advantages:
-
1.
strict inclusion criteria were adopted, and the overall NOS score was generally high; and
-
2.
sensitivity analysis, meta-regression, and the trim and fill method were used to further evaluate the robustness of the meta-analysis results.
However, there are still some shortcomings in this study:
-
1.
some studies lacked clinical parameters, such as different subtypes of lung cancer, degree of tumor differentiation, depth of invasion, TNM stage, etc., so that we could not further analyze the association between exosomes and lung cancer from these aspects;
-
2.
because there are too few studies on the PFS and DSS groups, we cannot explain their associations with the expression levels of exosomes.
-
3.
the study only included publications in the Chinese and English literature, which affected the comprehensiveness of the data; and
-
4.
most cases included in the literature came from China, which might have resulted in regional bias.
5. Conclusions
In summary, the expression level of exosomes is closely associated with the poor prognosis of lung cancer. Our results have strong statistical significance, especially for the OS and DFS of patients with lung cancer. In the future, we will look forward to more studies on exosomes and PFS or DSS in lung cancer to more accurately explain the association. Therefore, exosomes can serve as prognostic markers for lung cancer. Our study also has good clinical application prospects. We should combine exosomes with additional lung cancer markers to jointly manage the prognosis of lung cancer.
Author contributions
Conception: Jingying Luo.
Data curation: Hui Xiang, Fan Li, Yuzhui Hu.
Formal analysis: Yunxiao Yuan.
Investigation: Wenting Long.
Project administration: Hui Xiang, Liuyan Hong.
Software: Fan Li, Jingying Luo.
Visualization: Hongying Du.
Writing–original draft: Hui Xiang, Miao Luo.
Writing–review & editing: Hui Xiang, Miao Luo, Jingying Luo.
Footnotes
Abbreviations: ADC = adenocarcinoma, DFS = disease-free survival, DSS = disease-specific survival, HR = hazard ratio, LCC = Large cell carcinoma, NSCLC = non-small cell lung cancer, OS = overall survival, PFS = progression free survival, SCLC = small cell lung cancer.
How to cite this article: Xiang H, Li F, Luo J, Long W, Hong L, Hu Y, Du H, Yuan Y, Luo M. A meta-analysis on the relationship of exosomes and the prognosis of lung cancer. Medicine. 2021;100:15(e25332).
This work was supported by grants from National Natural Science Foundation of China (81860554), Guangxi Natural Science Foundation (2018GXNSFAA138054, 2015GXNSFAA139116), Guilin Scientific Research and Technology Development Project (2016012706-16).
Data availability statement: All data generated or analyzed during this study are included in this article.
The authors have no conflicts of interest to disclose.
Ethics approval and consent to participate: Not applicable.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
ADC = adenocarcinoma, DFS = disease-free survival, DSS = disease-specific survival, IHC = immunochemistry, MVA = multivariate analysis, NA = not available, NOS = Newcastle-Ottawa scale, NSCLC = non small cell lung cancer, OS = overall survival, PFS = progression-free survival, qRT-PCR = quantitative real-time PCR, SCLC = small cell lung cancer, SQCC = squamous cell carcinoma, UVA = univariate analysis.
ADC = adenocarcinoma, DFS = disease-free survival, DSS = disease-specific survival, MVA = multivariate analysis, NSCLC = non small cell lung cancer, OS = overall survival, PFS = progression-free survival, qRT-PCR = quantitative Real-time PCR, SCLC = small cell lung cancer, UVA = univariate analysis.
OR = odds ratio, SE = standard error.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Du Y, Su T, Zhao LJ, et al. Associations of polymorphisms in DNA repair genes and MDR1 gene with chemotherapy response and survival of non-small cell lung cancer. PLoS One 2014;9:e99843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen WL, Kuo KT, Chou TY, et al. The role of cytochrome c oxidase subunit Va in non-small cell lung carcinoma cells: association with migration, invasion and prediction of distant metastasis. BMC Cancer 2012;12:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Frediani JN, Fabbri M. Essential role of miRNAs in orchestrating the biology of the tumor microenvironment. Mol Cancer 2016;15:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang Q, Diamond MP, Al-Hendy A. The emerging role of extracellular vesicle-derived miRNAs: implication in cancer progression and stem cell related diseases. J Clin Epigenet 2016;2: [PMC free article] [PubMed] [Google Scholar]
- [6].Munson P, Shukla A. Exosomes: potential in cancer diagnosis and therapy. Medicines 2015;2:310–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569–79. [DOI] [PubMed] [Google Scholar]
- [8].Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, et al. Exosomes- structure, biogenesis and biological role in non-small-cell lung cancer. Scand J Immunol 2015;81:02–10. [DOI] [PubMed] [Google Scholar]
- [9].Raghu K. The biology and function of exosomes in cancer. J Clin Invest 2016;126:1208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl 2015;9:358–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Akhil S, Justyna F, Katherine M, et al. Exosomes: a role for naturally occurring nanovesicles in cancer growth, diagnosis and treatment. Curr Gene Ther 2015;15:182–92. [DOI] [PubMed] [Google Scholar]
- [12].Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ge R, Tan E, Sharghi-Namini S, et al. Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers. Cancer Microenviron 2012;5:323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rahbarghazi R, Jabbari N, Sani NA, et al. Tumor-derived extracellular vesicles: reliable tools for cancer diagnosis and clinical applications. Cell Commun Signal 2019;17:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles 2013;2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang N, Xie L. Biological characteristics and clincial application of tumor-associated exosomes. China J Cancer Prev Treat 2017;24:653–8. [Google Scholar]
- [19].Stang A. Critical evaluation of the Newcastle—Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [20].Li H, Zhang JB, Chen XL, et al. Different techniques for harvesting grafts for living donor liver transplantation: a systematic review and meta-analysis. World J Gastroenterol 2017;23:3730–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li H, Zhu B, Huang J, et al. Liver hanging maneuver versus conventional approach for open hepatectomy: a meta-analysis. HPB(Oxford) 2019;21:802–9. [DOI] [PubMed] [Google Scholar]
- [22].Lu X, Guo W, Xu W, et al. Prognostic value of the Glasgow prognostic score in colorectal cancer: a meta-analysis of 9,839 patients. Cancer Manag Res 2019;11:229–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Y, Xu H. Serum exosomal miR-378 upregulation is associated with poor prognosis in non-small-cell lung cancer patients. J Clin Lab Anal 2020;e23237-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xue XY, Wang C, Xue ZP, et al. Exosomal miRNA profiling before and after surgery revealed potential diagnostic and prognostic markers for lung adenocarcinoma. Acta Biochim Biophys Sin (Shanghai) 2020;52:281–93. [DOI] [PubMed] [Google Scholar]
- [25].Xiong H, Lu M, Wu F, et al. Exosomal microRNA-214 expression and its prognostic significance in non-small cell lung cancer patients. J King Saud Univ Sci 2020;32:1060–4. [Google Scholar]
- [26].Shimada Y, Kudo Y, Maehara S, et al. Ubiquitin C-terminal hydrolase-L1 has prognostic relevance and is a therapeutic target for high-grade neuroendocrine lung cancers. Cancer Sci 2020;111:610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nanou A, Miller MC, Zeune LL, et al. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br J Cancer 2020;122:801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu S, Li J, Chen L, et al. Plasma miR-32 levels in non-small cell lung cancer patients receiving platinum-based chemotherapy can predict the effectiveness and prognosis of chemotherapy. Medicine (Baltimore) 2019;98:e17335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sun GG, Ding X, Bi N, et al. Molecular predictors of brain metastasis-related microRNAs in lung adenocarcinoma. Plos Genetics 2019;15:e1007888. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [30].Yuwen D, Ma Y, Wang D, et al. Prognostic role of circulating exosomal miR-425-3p for the response of NSCLC to platinum-based chemotherapy. Cancer Epidemiol Biomarkers Prev 2019;28:163–73. [DOI] [PubMed] [Google Scholar]
- [31].Li L, Li W, Chen N, et al. FLI1 Exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res 2019;25:1302–17. [DOI] [PubMed] [Google Scholar]
- [32].Koh HM, Song DH. Prognostic role of Rab27A and Rab27B expression in patients with non-small cell lung carcinoma. Thorac Can 2019;10:143–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xu Z, Liu X, Wang H, et al. Lung adenocarcinoma cell-derived exosomal miR-21 facilitates osteoclastogenesis. Gene 2018;666(undefined):116–22. [DOI] [PubMed] [Google Scholar]
- [34].Kanaoka R, Iinuma H, Dejima H, et al. Usefulness of plasma exosomal microRNA-451a as a noninvasive biomarker for early prediction of recurrence and prognosis of non-small cell lung cancer. Oncol 2018;94:311–23. [DOI] [PubMed] [Google Scholar]
- [35].Zeng L, Yang RX, Wang J, et al. The relationship between the expression of LncRNA EXOC7 and the prognosis of patients with small cell lung cancer. Tumor 2017;37:269–74. [Google Scholar]
- [36].Dejima H, Iinuma H, Kanaoka R, et al. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett 2017;13:1256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017;8:13048–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sandfeld-Paulsen B, Aggerholm-Pedersen N, Boek R, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol 2016;10:1595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hart IR, Saini A. Biology of tumour metastasis. Lancet 1992;339:1453–7. [DOI] [PubMed] [Google Scholar]
- [40].Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell 2016;30:836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion. Nat Rev Immunol 2020;20:209–15. [DOI] [PubMed] [Google Scholar]
- [42].Tang YT, Huang YY, Li JH, et al. Alterations in exosomal miRNA profile upon epithelial-mesenchymal transition in human lung cancer cell lines. BMC Genomics 2018;19:802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yang J, Liu W, Lu X, et al. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget 2015;6:11125–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xiao H, Lässer C, Shelke GV, et al. Mast cell exosomes promote lung adenocarcinoma cell proliferation-role of KIT-stem cell factor signaling. Cell Commun Signal 2014;12:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Al-Nedawi K, Meehan B, Kerbel RS, et al. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A 2009;106:3794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang Y, Yi J, Chen X, et al. The regulation of cancer cell migration by lung cancer cell-derived exosomes through TGF-( and IL-10. Oncol Lett 2016;11:1527–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rahman MA, Barger JF, Lovat F, et al. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget 2016;7:54852–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee HY, Chen CK, Ho CM, et al. EIF3C-enhanced exosome secretion promotes angiogenesis and tumorigenesis of human hepatocellular carcinoma. Oncotarget 2018;9:13193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu Y, Luo F, Wang B, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett 2016;370:125–35. [DOI] [PubMed] [Google Scholar]
- [50].Cui H, Seubert B, Stahl E, et al. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene 2015;34:3640–50. [DOI] [PubMed] [Google Scholar]
- [51].Shrestha S, Hsu SD, Huang WY, et al. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med 2014;3:878–88. [DOI] [PMC free article] [PubMed] [Google Scholar]


