Abstract
Background:
Human papillomavirus (HPV) oncogenes E6, E7 and shorter isoforms of E6 (E6*) are known carcinogenic factors in head and neck squamous cell carcinoma (HNSCC). Little is known regarding E6* functions.
Methods:
We analyzed RNA-seq data from 68 HNSCC HPV type 16-positive tumors to determine host genes and pathways associated with E6+E7 expression (E6E7) or the percent of full length E6 (E6%FL). Influence scores of E6E7 and E6%FL were used to test for associations with clinical variables.
Results:
For E6E7, we recapitulated all major known affected pathways and revealed additional pathways. E6%FL was found to affect mitochondrial processes, and E6%FL influence score was significantly associated with overall survival and tumor size.
Conclusions:
HPV E6E7 and E6* result in extensive, dose-dependent compensatory effects and dysregulation of key cancer pathways. The switch from E6 to E6* promotes oxidative phosphorylation, larger tumor size and worse prognosis, potentially serving as a prognostic factor for HPV-positive HNSCC.
Keywords: Head and neck cancer, human papillomavirus, E6, E7, E6*, influence score, survival
Introduction
Infection with oncogenic human papillomavirus (HPV) is well established as a causative factor for an increasing subset of head and neck squamous cell carcinomas (HNSCC), especially in the oropharynx where HPV may now account for up to 70% of tumors 1. The HPV ‘early’ proteins E6 and E7 are the main oncogenic proteins leading to HPV-related cancers, repressing immune response and altering the cell cycle program in the host that together allow replication after squamous differentiation and survival despite abnormal mitoses 2. HPV E6 and E7 play multiple roles, some of them synergistic, in the tumorigenesis of cancer. E7 is well known to bind to the retinoblastoma tumor suppressor (pRB) and mark it for degradation 3, which results in E2F-dependent gene transcription and further cell cycle (S phase) activation and proliferation 4. The loss of pRB activity could lead to p53-dependent apoptosis; however, this is avoided by E6 which induces p53 degradation. E6 recruits the unbiquitin ligase, E6AP, which polyunbiquitinates the p53 tumor suppressor protein for proteosome-mediated degradation, leading to the inhibition of p53-dependent apoptosis and enhanced cell cycle activation 5. Besides the p53-dependent apoptosis pathway, E6 and E7 also inhibit alternative apoptosis pathways, including anoikis and the cytokine-activated extrinsic apoptotic pathway 6.
E6 and E7 are involved in several pathways in addition to apoptosis and the cell cycle. They cooperate to inhibit the host immune response 7. It was shown that high-risk HPV E6 interacts with interferon-regulatory factor 3 (IRF-3) and inhibits its activation of downstream factors 8. Similarly, E7 binds with IRF-1 and blocks its function 9. Recent studies showed that E6 and E7 could interact with Wnt signaling components and regulate their signaling transduction through the canonical Wnt/β-catenin pathway 10. Independent of their cooperative functions, E6 and E7 play roles in epigenetic modulation, telomerase activation, DNA damage response, angiogenesis, cell immortalization and differentiation 11–14.
E6 has multiple common transcripts, including the longer, full length transcript and multiple spliced E6* variants, the most common being a 183bp exclusion from positions 226 – 409bp that was only discovered in high-risk HPV subtypes 15,16, suggesting its role in carcinogenesis. While the long E6 isoform is more commonly observed to be transcribed from episomal HPV, the shorter E6* variants are more common in tumors with HPV integrated into the host genome, which are the majority of HNSCC cases 17. E6 splicing is induced by epidermal growth factor (EGF) depletion, and has been observed to correlate with an increase in TP53 and E7 protein levels and a decrease in pRb 15,18. Much less is known regarding the function of E6* compared with E6 protein. Evidence suggests that E6* in HPV18 might counteract full-length E6 function, as it could elevate p53 levels in vivo 19. Recently, E6* was shown to increase reactive oxygen species (ROS) and oxidative stress, which may lead to increased DNA damage 20. In the same study, E6* was observed to decrease superoxide dismutase isoform 2 (SOD2) and glutathione peroxidase (GPX) expression, suggesting a cause for the oxidative stress.
Research on the oncogenic mechanisms of HPV has been performed on a mix of origin cell types (mainly cervical but some oral), HPV types, and stages of HPV infection, and much of it was performed in vitro, sometimes with E6 or E7 transfected in isolation. The extent to which these cumulative findings remain true in oral primary carcinomas infected with HPV16 is not fully known. Although many of the same HPV behaviors and tumor characteristics have been observed in cervical and oropharyngeal HPV-associated cancers 21,22, it remains unknown whether the findings based on cervical cells can be fully extended to the oropharynx, which has a different tumor microenvironment. Most notably, as opposed to cervical carcinomas, oropharynx tumors arise within or in close proximity to a lymph node and lymphoid follicles. Furthermore, although many direct targets of E6 and E7 are known, the extent that these direct effects extend to downstream pathway events or cause compensatory effects is not known. It is also unclear whether most effects of E6 and E7 are dose-dependent or act as on-off switches, although it has been shown that E6 degrades procaspase 8 in a dose-dependent manner 23.
Here, we present a genome-wide study of the relationship between combined E6 and E7 mRNA levels, the percent of E6 mRNA that is full length versus E6*, and host gene expression levels in primary HPV(+) HNSCC cancers. This has allowed us to observe the extensive, downstream effects of the oncogenic HPV proteins on multiple cancer-related pathways. From the set of 18 HPV(+) HNSCC samples collected at the University of Michigan (UM), and 66 HPV(+) HNSCC samples from The Cancer Genome Atlas (TCGA), we selected the total of 68 HPV16(+) for this study (14 UM and 54 TCGA). Through this analysis, we recaptured all of the main pathways known to be affected by E6 and E7, whether originally described in cervical or oropharynx cells, and identified some novel pathways that are potentially related to the disease pathogenesis. In addition, we propose an influence score to assess the overall impact of E6 + E7 (E6E7) and the percent of E6 that is full length (E6%FL) levels on the host transcriptome, and identified that the E6%FL influence score was inversely significantly associated with tumor mutational burden and tumor size, and associated with better overall survival. These associations represent both the direct and indirect effects of E6*, which may involve changes in E7 translation.
Materials and Methods:
Tumor Tissue Acquisition and sample preparation
HNSCC patients at University of Michigan Hospital with untreated oropharynx or oral cavity tumors between 2011 – 2013 were screened for eligibility; those eligible were asked if willing to provide informed consent for collection of tumor tissue and then collected as described previously17,24. Tumor tissue and blood were collected into a cryogenic storage tube and flash frozen in liquid nitrogen by surgical staff until storage at −80C. The flash frozen tissues were embedded in OCT media in vinyl cryomolds on dry ice and stored in −80C until prepared for histology. H&E slides were sectioned from each frozen tumor specimen on a cryostat and assessed by a board-certified pathologist at the University of Michigan for degrees of cellularity and necrosis. Criteria used for inclusion in the study were a minimum of 70% cellularity and less than 10% necrosis; all others were omitted from further study. The first 36 tumors meeting these criteria were selected for RNA sequencing. mRNA library preparation and RNA sequencing were performed as described for these samples in Zhang, et al. 17. Briefly, sequencing with an Illumina HiSeq 2000 using 100 nt paired-end reads was performed by the University of Michigan DNA Sequencing Core Facility resulting in an average of 47 million reads per sample.
UM RNA-seq preprocessing
RNA-seq preprocessing was performed as described in Zhang Y, et al 17, with an exception in the step of differential gene analysis. The raw sequences were aligned to hg19 using Tophat2 v2.0.1125 using default alignment parameters. Quality control was performed using FastQC 26 and RSeQC 27 before and after alignment. Gene expression levels were quantified using HTSeq v0.6.1p1 with the ‘intersection-strict’ option 28. Normalization and calculation of logCPM values (log counts per million reads mapped) were performed using the limma Bioconductor package 29 as described below.
The libraries were also aligned to HPV genomes (downloaded from NCBI) using STAR 30 to allow gapped alignment over splicing junctions. STAR first-pass alignment detected the most abundant splice junctions which were then used in the second-pass alignment. All the quantification and analysis below were based on the second-pass alignment. Samples were classified as HPV(+) if they had more than 500 read pairs aligned to any HPV genome, and HPV subtype was determined as the subtype with the most aligned reads. We identified 14 HPV type 16 tumors, one type 18, one type 33, and two type 35. Only the 14 HPV type 16 samples were used in this study. HPV is a small genome with short oncogenes; therefore, in order to optimize the comparability between University of Michigan (UM) and TCGA data in terms of HPV gene expression and splicing, all UM libraries were trimmed to the first 48 base pairs, which was the read length of all TCGA HNSC RNA-seq data. UM RNA-seq data is available at Gene Expression Omnibus (GEO) #GSE74956.
TCGA RNA-seq data reprocessing
RNA-seq fastq files for the 66 TCGA HPV(+) tumor samples were downloaded from CGHub. The data were re-aligned and analyzed in the same way as UM RNA-seq data described above. The samples contained 55 HPV type 16, 8 HPV type 33 and 3 HPV type 35. One TCGA sample (TCGA-CN-A6V1, Oropharynx, HPV 16 type) was an outlier with extremely low E6 and E7 expression (1.64 CPM), while the average expression of E6 and E7 in all other samples was 112.5 CPM. This sample was excluded from downstream analysis due to its outlier expression and to avoid this sample from having a disproportionately high effect on the results. Only the 54 HPV type16 sequencing data were used in this study.
Calculating E6E7 expression and E6%FL
The CPM (count per million) values for HPV oncogenes E6 and E7 (heretofore denoted as E6E7 expression) and the percent of full length E6 (E6%FL) were calculated as previously described 17. The CPM values were calculated as the number of read pairs aligned to the relevant HPV genome that intersected E6 or E7, divided by the number of million reads in the total library size. E6 alternative splicing results in multiple forms of spliced E6 (referred as E6*). However, the first intron in all forms is missing. To calculate E6%FL, we computed the ratio of average coverage level in the first intron of E6 (approximate full-length level) divided by the average coverage level in the first exon (estimated all E6 mRNA level), referred to as E6%FL. We identified the major donor-acceptor sites of HPV 16 at 227bp and 408bp (for HPV16) by STAR first-pass alignment.
Association analysis for E6E7 expression and E6%FL versus host gene expression
The limma R Bioconductor package was utilized to examine the association between the host gene expression and E6E7 expression or E6%FL, respectively. Read counts of UM and TCGA mRNA-seq data extracted by HTSeq were transformed to logCPM values by the limma function voom and analyzed together. Linear regression was applied to calculate the slope and intercept of E6E7 or E6%FL to each host gene expression using the formula:
where g=1,…,G is a host gene, i=1,...,n is the covariate, α is the intercept, and β is the slope. P-values of the slopes were reported from the limma function lmfit and False Discovery Rate (FDR) was calculated using the Benjamini-Hochberg method. Genes with FDR < 0.05 were called significant.
Backward selection of clinical variables
To identify the optimal set of covariates, we implemented backward selection for high-throughput data 31. A total of nine categorical and continuous variables were analyzed: age, smoking status, clinical T classification (the size and extent of the main tumor, AJCC 7th Edition), tumor stage (based on the TNM combination), nodal status, tumor site, sex, HPV integration, and cohort. We grouped certain values of variables that had an insufficient number of tumors. For example, for clinical T classification we grouped samples as T1 and T2 versus T3 and T4. We analyzed samples located in oropharynx (52 tumors) versus other sites (16 tumors, consisting of 13 oral cavity samples, 1 from larynx and 2 from hypopharynx). Analysis by tumor stage was performed for groups: stage IV (49 samples) versus other stages (19 samples, consisting of stage I – 1 tumor, stage II – 10 tumors, and stage III – 8 tumors). Nodal status was defined as: N0 and N1 (23 tumors: N0 = 16 and N1 = 6) versus N2 and N3 (45 tumors: N2 = 42 and N3 = 3). The backward selection procedure starts with fitting the weighted linear model of voom-transformed read counts using the lmFit function with the empirical Bayes smoothing implemented by the eBayes function for each of the host genes and includes the effects of primary interest (E6E7 or E6%FL), as well as all nine covariates. For E6%FL, the initial statistical model included covariates for all the same variables, except HPV integration status (8 covariates as the start point) because integration of HPV into the host genome is a likely effect of higher expression of E6* 32. Indeed, we observed a significant difference in E6%FL by viral integration status (Wilcoxon rank-sum test p-value = 0.007). In each iteration, the least relevant covariate was dropped if the number of its significantly associated genes (FDR < 0.05) less than or equal to 0.1%, or if multiple covariates had zero significant genes, they were dropped in the following order: age, smoking status, T classification, tumor stage, nodal status, site, sex, HPV integration and cohort; and the resulting reduced model was fit again for all genes. This process was repeated until the least relevant covariate resulted in < 0.1% associated genes. We also analyzed the correlation of log transformed E6E7 or E6%FL with clinical variables.
Gene set enrichment testing
To identify biological pathways and cellular processes affected by E6E7 or E6 %FL levels, gene set enrichment (GSE) testing was performed using RNA-Enrich 33. Gene sets were Gene Ontology (GO) (Biological Process, Cellular Component, and Molecular Function) and KEGG Pathways. To filter out the closely related GO terms for reporting purposes, we used the R package GO.db34 to determine relationships among significant terms. A GO term was filtered if one or more of its parents, children or siblings had a higher rank in the list. An R/Bioconductor package Pathview 35 was utilized to visualize the list of driving genes in the KEGG pathway (FDR < 0.05) as reported by the association analysis described above.
Influence score calculation and association tests between influence score and tumor features
In order to estimate the overall influence of E6E7 or of E6%FL on the affected host genes, we selected the genes that were significantly correlated with E6E7 or E6%FL, respectively, from the association analysis (FDR < 0.05). For positively-associated genes, we ranked the samples by each of those genes’ voom transformed expression values. For the negatively-associated genes, we ranked the samples in descending order. To calculate the sample-wise E6E7 or E6%FL influence, we summed the ranks of these genes in each sample, centered the sum by the mean, and then scaled it by its standard deviation across samples. We defined the resulting values as the “influence scores”. Influence scores were used in place of mRNA levels for testing associations with E6E7 or E6%FL for two reasons: i) protein levels often have poor correlation with their mRNA levels, and we previously observed that HPV oncogene predicted activity scores correlate better than their mRNA levels with known HPV oncoprotein effects 17; ii) this is consistent with previous reports showing transcription factor activity scores based on their target genes provide better estimates of the protein’s activity level than the mRNA level of the protein 36.
We evaluated the association of E6E7 or E6%FL influence scores with tumor features and demographic variables by linear regression. Variables used in this analysis included clinical T classification as continuous (1–3, 4a&b), smoking (never versus smoker), HPV integration (negative versus positive), N classification (N0 or N1 versus N2 or N3), stage (stages I-III versus stage IV), anatomical site (oropharynx versus other), sex, age and cohort (TCGA vs UM). The associations between influence score and tumor/demographic features were analyzed by ANOVA, and p-values from the F-test were reported.
Calculation of tumor mutational burden and survival analysis
To evaluate the relationship between E6E7 or E6%FL expression levels or influence scores with cancer biology and clinical features, we focused on the TCGA cohort due to the availability of the mutational and survival data. The somatic mutation data (MAF file) and clinical data of the TCGA HNSC cohort were downloaded from the Genomic Data Commons (GDC) data portal, and the 54 HPV16(+) patients were extracted. We calculated tumor mutational burden (TMB) as the total number of somatic mutations in each patient divided by the total number of megabases that the whole exome sequencing covered (mut count/Mb). The correlation between TMB and E6E7, E6%FL levels or their influence scores were assessed by the Pearson correlation test. Overall survival of those patients was analyzed for two groups: the high or low levels of E6E7 or E6%FL expression or influence scores using the R packages: survival37 and survminer38. The median score or the optimal score cutoff (searched by the surv_cutpoint function in survminer) was used to define the two groups. Kaplan–Meier estimates of survival were determined, and a P value was calculated using a univariate log-rank test. Cox proportional hazards regression models were used for adjustment of clinical covariate variables (sex, age, clinical stage, tumor site, and smoking status).
Results
Overview of cohort and data
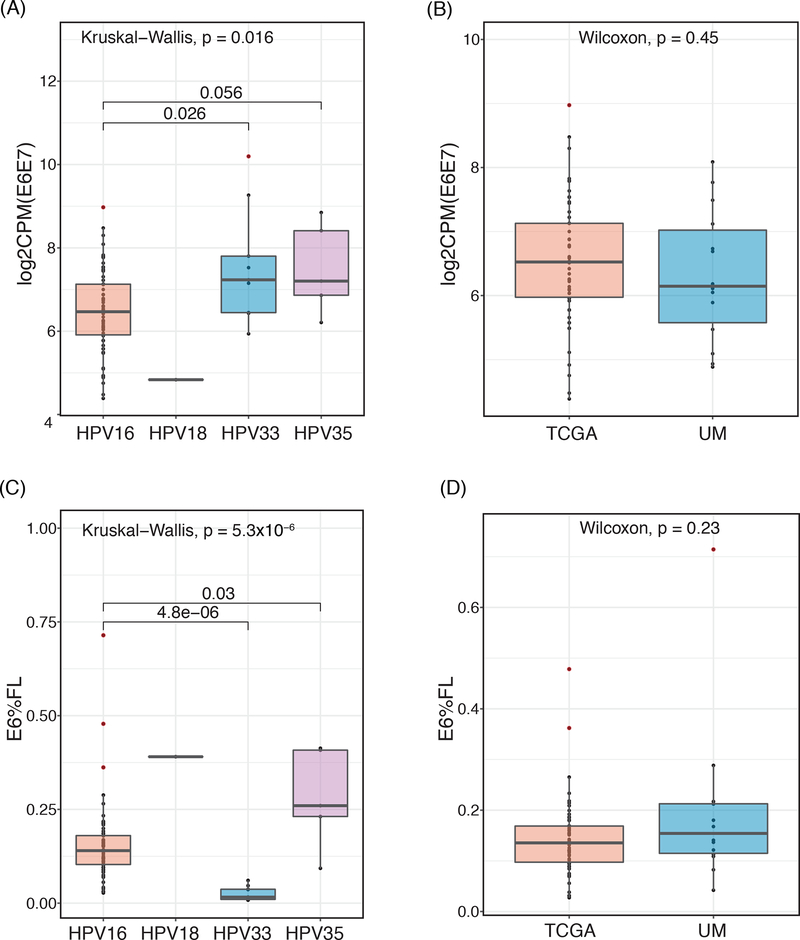
We used previously-published mRNA sequencing results from 18 HPV(+) tumor samples collected at UM and 66 HPV(+) samples from TCGA39. Among these 84 samples, 69 were infected with HPV16 while the other 15 samples contained HPV33 (9), HPV35 (5) and HPV18 (1) (Table 1). The two patient populations showed no differences by age, sex, stage, or smoking status; however, the TCGA population did have a slightly lower proportion of oropharynx tumors (p=0.059) (Table 1). The E6 and E7 proteins of different HPV strains have different behaviors and propensity toward carcinogenesis; consistent with these reports, we found that the expression of E6 and E7 were significantly different among HPV strains in the 83 samples (one TCGA outlier was excluded- see Methods) (Figure 1A), while they were not significantly different between the two cohorts (Figure 1B). In line with the above observations, the percent of E6 transcripts that were of full length (E6%FL) were also significantly different by HPV strains (Figure 1C), but not different by the two cohorts. (Figure 1D). Therefore, to avoid confounding by HPV type, we restricted our analysis to the HPV16 samples, resulting in a total of 68 combined UM and TCGA samples.
Table 1.
Demographic and clinicopathologic characteristics of HPV-positive patients from both UM and TCGA cohort.
| Parameter | HPV(+) UM tumors | HPV(+)TCGA tumors | sum_all_HPV | HPV-16 UM tumors | HPV-16 TCGA tumors | sum_HPV-16 | UM vs TCGA Fisher’s Exact Test p value |
|---|---|---|---|---|---|---|---|
| 18 | 66 | 84 | 14 | 55 | 69 | ||
| Age at diagnosis | Age at diagnosis | ||||||
| Median (std) | 58.5 (7.3) | 57 (9.2) | 58 (7.7) | 57 (9.7) | 0.297 | ||
| Gender | Gender | ||||||
| Male | 17 | 60 | 77 | 13 | 49 | 62 | 1.000 |
| Female | 1 | 6 | 7 | 1 | 6 | 7 | |
| HPV type | HPV type | ||||||
| HPV16 | 14 | 55 | 69 | 14 | 55 | 69 | 0.729 |
| HPV18 | 1 | 0 | 1 | ||||
| HPV33 | 1 | 8 | 9 | ||||
| HPV35 | 2 | 3 | 5 | ||||
| Anatomical Site | Anatomical Site | ||||||
| Oropharynx | 17 | 47 | 64 | 13 | 40 | 53 | 0.059 |
| Oral Cavity | 1 | 16 | 17 | 1 | 12 | 13 | |
| Larynx | 0 | 1 | 1 | 0 | 1 | 1 | |
| Hypopharynx | 0 | 2 | 2 | 0 | 2 | 2 | |
| Tumor Stage | Tumor Stage | ||||||
| I | 0 | 2 | 2 | 0 | 1 | 1 | 0.753 |
| II | 1 | 10 | 11 | 1 | 9 | 10 | |
| III | 2 | 7 | 9 | 2 | 6 | 8 | |
| IV | 15 | 47 | 62 | 11 | 39 | 50 | |
| T stage | T stage | ||||||
| T1 | 1 | 8 | 1 | 5 | 6 | 0.715 | |
| T2 | 7 | 31 | 6 | 28 | 34 | ||
| T3 | 3 | 10 | 13 | 2 | 9 | 11 | |
| T4 | 7 | 16 | 23 | 5 | 12 | 17 | |
| N stage | N stage | ||||||
| N0 | 1 | 18 | 19 | 1 | 15 | 16 | 0.156 |
| N1 | 2 | 6 | 8 | 2 | 4 | 6 | |
| N2 | 11 | 39 | 50 | 10 | 33 | 43 | |
| N3 | 4 | 2 | 6 | 1 | 2 | 3 | |
| Smoking Status | |||||||
| Current | 3 | 13 | 16 | 3 | 10 | 13 | Smoking Status |
| Former | 11 | 30 | 41 | 7 | 26 | 33 | 0.635 |
| Never | 4 | 22 | 26 | 4 | 19 | 23 | |
Figure 1.
(A) Box plot showing the normalized RNA-seq expression level of HPV E6 + E7, denoted as log2CPM(E6E7), in different HPV subtypes across the combined UM and TCGA 84 HPV-positive HNSCC samples. (B) Box plot showing the expression level of HPV E6E7 in each data cohort (UM and TCGA) (p=0.45). (C) Box plot showing the normalized RNA-seq expression level of proportion of HPV E6 that is expressed in full length (E6%FL), in different HPV subtypes across the combined UM and TCGA 84 HPV-positive HNSCC samples. (D) Box plot showing the expression level of E6%FL in each data cohort (UM and TCGA) (p=0.23).
Before studying the association of E6* with host genes, we first studied associations of E6 and E7 with host genes and pathways, many of which are previously known. To identify host genes with E6 and E7 dose-dependent expression levels, we calculated the overall E6 + E7 log normalized read counts (E6E7) and performed multiple linear regression analysis of these E6E7 expression levels with the mRNA-seq results of the 68 samples. E6 and E7 expression levels were combined, due to their extremely high correlation (r = 0.977 for HPV16 UM and r = 0.970 for HPV16 TCGA).
E6 and E7 expression correlates with extensive cell cycle, DNA replication and other cancer pathway gene expression in HNSCC
To test for association between host genes and E6E7 expression, we first investigated known clinical and phenotypic variables that may explain additional heterogeneity in the data. The initial regression model included covariates for patient age, smoking status, clinical T classification, tumor stage, nodal status, sex, anatomical site, HPV integration status, and cohort, and the optimal model with three covariates (cohort, HPV integration and sex) was selected by a backward selection approach (supplementary Table S1) 31. A total of 261 host genes (228 up- and 33 down-regulated) were significantly associated with E6E7 expression (supplementary Table S2; FDR < 0.05). Among the 228 positively-associated genes, 40 were involved in DNA replication; 21 genes were involved in cell cycle checkpoint, whose activation are essential for cell cycle progression; and 42 were involved with DNA repair. The top 6 positively associated genes by p value were: NGFI-A binding protein 1 (NAB1), cell proliferation regulating inhibitor of protein phosphatase 2A (CIP2A), DNA topoisomerase II alpha (TOP2A), timeless circadian regulator (TIMELESS), DLG associated protein 5 (DLGAP5) and cell division cycle 6 (CDC6). These results suggest that higher E6E7 expression in tumors correlates with enhanced DNA replication & repair, and cell cycle progression, indicating an extensive ‘dose response’ to E6E7 by the host cells.
We identified 94 Gene Ontology (GO) terms and 9 KEGG pathways significantly positively associated, and 83 GO terms and 9 KEGG pathways negatively associated with E6E7 expression (FDR<0.05) (Figure 2A and 2B, supplementary Table S3). Consistent with our individual gene results, cell cycle and DNA replication pathways were found to be enriched with positively regulated genes by E6E7 expression (Figure 2B), and cell cycle was the most significant KEGG pathway (OR = 2.33, FDR = 1.04 × 10−21) with 21 out of 88 genes positively correlated at the FDR<0.05 level (supplementary Figure S1, Table S3). Pathways known to be involved in the carcinogenic mechanisms of E6E7 such as regulation of immune response, cell cycle, response to cytokine, Wnt-receptor activity, mismatch repair, nucleotide excision repair, helicase activity, histone binding and histone kinase activity (supplementary Table S3) were also found. In addition, novel pathways and GO terms including oxidative phosphorylation (OXPHOS), ribosome, mitochondrial inner membrane, and cardiac muscle contraction were identified as negatively associated with E6E7 expression (Figure 2, supplementary Table S3). These results confirm the previously reported mechanisms of E6 and E7-related tumorigenesis40, and identify novel pathways that may also contribute to E6 and E7-mediated carcinogenesis.
Figure 2.
Bubble plot of GO terms and KEGG pathways enriched in the genes associated with HPV expression of E6E7 or E6%FL, respectively. (A) Ten of the most enriched GOBP (Gene Ontology in Biological Process domain) terms enriched in host genes positively or negatively associated with E6E7 (or E6%FL) expression. (B) Ten of the most enriched KEGG pathways in host genes positively or negatively associated with E6E7 (or E6%FL) expression. The color of the dots denotes the significant levels (reddish: higher significance; bluish: lower significance), and the size denotes the gene set size.
Percent of E6 that is full length as opposed to E6* (E6%FL) is negatively associated with mitochondrial/oxidative processes, ATP metabolic process, keratinization, and inflammation
In high-risk HPV-related carcinogenesis, E6 mRNA is most often spliced to shorter E6 spliced isoforms (E6*), creating truncated proteins whose functions remain elusive. Previous studies suggest E6* can bind to the full-length E6 and inhibit its function in degrading TP5341. However, it is known that an increasing percentage of E6* predicts a more severe phenotype in cervical cancer 42,43. Thus, we hypothesized that E6* has an alternative carcinogenic mechanism that outweighs any tumor suppressive function it may have, such as allowing higher TP53 activity. In order to investigate a possible relationship between E6 splicing and HNSCC carcinogenesis, we determined the percent of E6 expression that was in the form of the full-length isoform for each sample, which is the reverse of percent of E6* (see Methods for details). We then performed the same linear regression analysis as above, except using E6 percent full-length (E6%FL) instead of E6E7. Similar to the E6E7 analysis, we first investigated known clinical and phenotypic variables that may explain additional heterogeneity in E6%FL (see Methods). Using the backward selection process, we identified the optimal model with the covariates of cohort and sex (supplementary Table S2). A total of 169 differentially expressed genes (156 up- and 13 down-regulated) were significantly associated with E6%FL from this model (supplementary Table S4; FDR < 0.05).
GSE analysis revealed 123 GO terms and 20 KEGG pathways negatively associated and 132 GO terms and 20 KEGG pathways (FDR < 0.05) positively associated with E6%FL (supplementary Table S5). As shown in Figure 2, mitochondrial inner membrane (FDR = 1.2 × 10−43) and oxidative phosphorylation (FDR = 1.86 × 10−26) were the top negative terms, with other oxidoreductase-related terms filling the other top negative terms (supplementary Table S5). These results suggest that mitochondrial/oxidoreductase activity is more activated with a higher rate of spliced E6*, which may lead to heightened oxidative stress and DNA damage. The most highly associated genes related to OXPHOS were NADH:ubiquinone oxidoreductase subunit AB1 (NDUFAB1). However, we did not observe a significant association with either SOD2 or GPX, as was previously observed. Top enriched pathways positively correlated with the E6%FL were chromatin and sequence-specific DNA binding, nuclear division, and microtubule cytoskeleton organization. Also included was stem cell population maintenance, which includes such genes as NOTCH1, SMAD2 & 4, SOX2, STAT3, and FOXO3.
Percent of E6* is positively associated with tumor size and worse survival
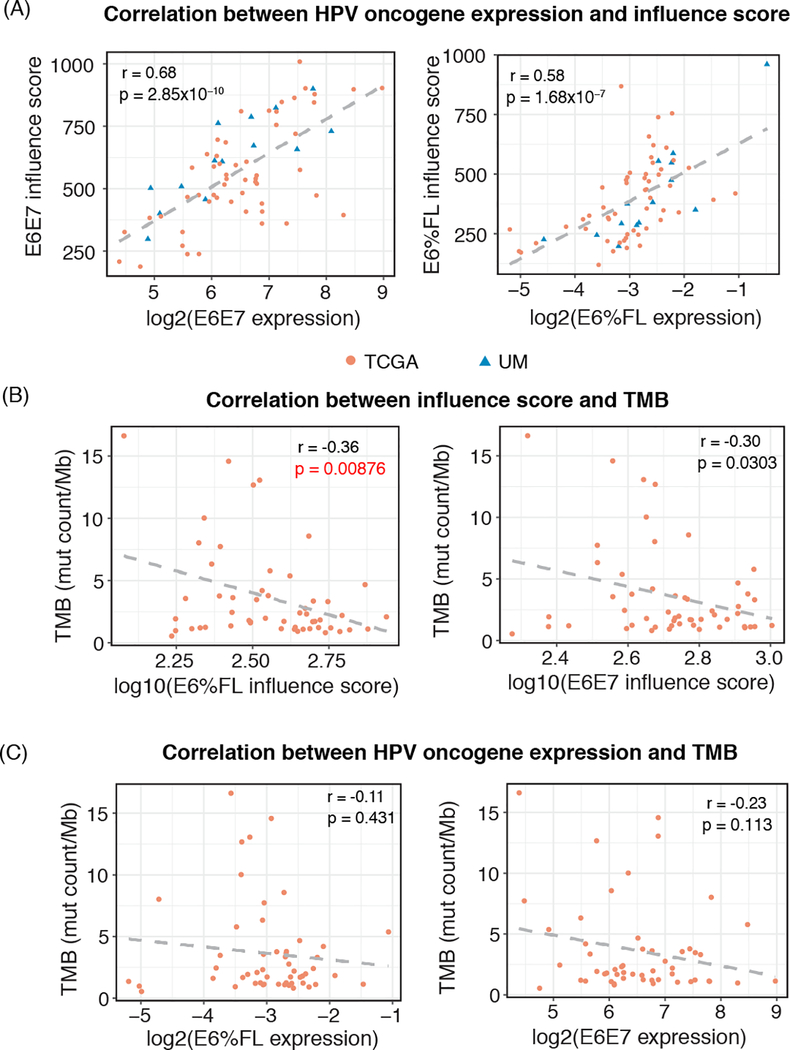
Next, we sought to investigate whether the activity level of E6E7 or E6* has any relationship with tumor features or survival. Since E6E7 or E6* mRNA expression in each patient may not accurately reflect the protein activity levels, respectively 17, we calculated an influence score for each tumor sample, defined by the level of influence E6E7 or E6%FL has on the expression of responsive genes. Genes significantly associated with E6E7 expression or E6%FL (FDR < 0.05) were selected to estimate the influence scores (see Methods for details). As expected, we observed highly significant correlation between the E6E7 influence score and E6E7 mRNA expression (Figure 3A left: Pearson’s r = 0.68, p-value = 2.85×10−10), as well as between E6%FL influence score and E6%FL expression (Figure 3A right: Spearman’s r = 0.58, p-value = 1.68×10−7). We examined the relationship between E6E7 or E6* and tumor mutational burden (TMB). We hypothesized that since E6* was observed to increase oxidative stress and DNA damage in cells, the percent of E6* would be positively associated with TMB (E6%FL would be negatively associated). We also hypothesized that E6E7 may be negatively associated with TMB, since patients with strong E6 and E7 activity may not require as many mutations for carcinogenesis. To test these hypotheses, we calculated the TMB among the 54 TCGA patients (see Methods), and found that TMB had significant negative associations with both influence scores (Figure 3B: E6%FL influence score vs. TMB: Pearson’s r = −0.36, p = 0.00876; E6E7 influence score vs. TMB: Pearson’s r = −0.30, p = 0.0303). As evidence that the influence scores are more relevant to cancer biology than the mRNA levels, no significant association was observed between TMB and either E6%FL or E6E7 expression levels (Figure 3C).
Figure 3.
(A) The correlation between HPV oncogene expression and their corresponding influence scores (left: E6E7 expression level vs. E6E7 influence score; right: E6%FL expression level vs. E6%FL influence score). The color of dots denotes the cohort (red is TCGA and blue is UM cohort). (B) The correlation between the E6E7 or E6%FL influence score and tumor mutational burden (TMB) among the TCGA cohort (left: E6%FL influence score vs. TMB; right: E6E7 influence score vs. TMB; (C) The correlation between E6E7 or E6%FL mRNA expression and TMB among the TCGA cohort (left: E6%FL vs. TMB; and right: E6E7 vs. TMB).
We then carried out multivariable linear regression analysis to model the association of influence score with different tumor variables, specifically with tumor site (Oropharynx (n=52) vs Other (n=16)); tumor T-classification (stage I = 6 samples, stage II = 33 samples, stage III = 11 samples, and stage IV = 17 samples; one sample had undefined T classification); HPV integration status; smoking status; sex; tumor N-classification; cohort and age. The E6%FL influence scores demonstrated a significantly negative association with tumor clinical-T-classification (Figure 4A and supplementary Table S6, p-value = 0.00363), meaning that larger tumors were associated with a higher percent of E6*. Associations resulting in significant unadjusted p-values, but which were not significant after multiple-testing adjustment (Figure 4A), included tumor site (p-value = 0.00374), HPV integration (p-value = 0.017) and smoking status (p-value = 0.0137). On the other hand, the E6E7 influence score had no significant associations after multiple-testing adjustment, and only demonstrated a borderline significant trend with smoking status (p-value = 0.0392) and tumor site (p-value = 0.0509).
Figure 4.
The association between E6%FL influence score and tumor characteristics and clinical features. (A) box plot showing the significant difference in E6%FL influence score by clinical T classification (p=0.00363), tumor anatomical site (p=0.0027), HPV integration status (p=0.0051) and smoking status (p=0.019). (B) the Kaplan-Meier curves showing the significant segregation of overall survival among TCGA patients with HPV16-positive HNSC by E6%FL influence score. Both optimal (left, N = 44 in lower-risk group and N = 10 in higher-risk group at day 0) and median score (right, N = 27 in each sub-group at day 0) cutoffs showed that patients with higher E6%FL influence scores had significantly better survival (Cox proportional hazards: p=0.00027 and 0.024 respectively)
The significant association between E6%FL influence score and tumor features, especially the negative association with tumor size, triggered us to further assess its clinical relevance. We investigated the overall survival (OS) of the 54 TCGA patients stratified by optimal or median cutoff of E6%FL influence scores (Figure 4B). OS was significantly segregated by both cutoffs (optimal cutoff: log-rank test, p-value = 1.00×10−5; median cutoff: log-rank test, p-value = 0.02), and patients with higher E6%FL influence scores, (lower %E6* influence scores), had better survival. In contrast, the mRNA level of E6%FL did not show any association with the patients’ survival (Supplementary Figure S2), again suggesting that the E6%FL influence score rather than the E6%FL level itself is more relative to the clinical characteristics. After control for the clinical variables sex, age, tumor stage, tumor site, and smoking status, the significance remained (Cox hazard regression, p = 0.0198 and interquartile range HR=0.13). In line with the fact that E6* is associated with high-risk HPV(+) cervical cancer, this finding suggests that the E6%FL influence score may also serve as a clinically actionable metric in HNSCC to distinguish patient subtypes, and help guide precision medicine.
Discussion
In this study, we investigated the relationship between HPV oncogene expression and host mRNA expression in HNSCC samples from two cohorts, and used the resulting host gene responses to develop an influence score. This facilitated the identification of the relationship between E6 splicing and tumor size, tumor mutational burden, and overall survival. We also confirmed genes and pathways previously identified to be affected by HPV16 E6 and E7, including genes involved in DNA replication, DNA repair and cell cycle, such as CDK2 and CLSPN (claspin), showing that these responses to E6 and E7 expression display a correlative “dose response.” HPV16 E7 is known to interact with two cyclin/CDK2 complexes and to inhibit the cyclin dependent kinase inhibitors p21 and p27, leading to the activation of CDK2 and disruption of G1/S cell cycle checkpoint 44,45. CLSPN is essential for DNA-replication stress response, and its degradation is associated with DNA damage checkpoint recovery 46. Another top positively-associated gene was RBL1, which may share some functional redundancy with pRB based on the high level of sequence similarity between them, suggesting that pRB degradation may activate a compensatory reaction. In addition to the previously known E6 and E7 transcriptional effects, novel genes and pathways were identified, such as mitochondrial function related biological processes. Notably, some Fanconi Anemia (FA) pathway genes were found to be significantly up-regulated with the increase in E6 and E7 expression, including Fanconi anemia complementation group B (FANCB), Fanconi anemia complementation group C (FANCC), and Fanconi anemia complementation group M (FANCM). This is consistent with the previous finding that patients with FA have much higher risk of HNSCC 47, and HPV(+) HNSCC patients tend to carry more mutations in FA genes 48.
E6 mRNA splicing is a critical step in HPV-induced tumorigenesis. However, the function and effect of truncated E6 (E6*) in the disease remains unclear. Genes that significantly correlate with E6 splicing could provide hints of pathways that E6* affects. We did not see any evidence of downstream effects of E6* inhibiting the degradation of TP53, suggesting this may be a weak effect, does not occur in HNSCCs, or does not affect the expression of downstream genes. The negative correlation observed with E6%FL (i.e. positive correlation with % E6*), which included oxidative phosphorylation (OXPHOS), ATP metabolism, mitochondrial membranes, epithelial differentiation (keratinization), and endoplasmic reticulum, suggests that patients with more E6* rely heavily on OXPHOS for energy production. Interestingly, the Glycolysis/Gluconeogenesis pathway was also found to be negatively associated with percent of E6%FL, although at a lower significance level (FDR = 0.014). This finding appears to contradict the Warburg effect, the tendency of tumors to increase their use of glycolysis for energy production while inhibiting OXPHOS in order to continue growth even in hypoxic conditions. However, an increasing amount of evidence has suggested that OXPHOS is upregulated in some cancers49, and one cause of the increased OXPHOS may be the increased mtDNA content in those cancers, including head and neck, esophageal, thyroid, ovarian, prostate, and colorectal cancers 50. In addition, the cell differentiation process may induce OXPHOS and mitochondrial biogenesis, increasing ROS production51,52, and thus inducing oxidative stress and DNA damage20. Taken together, we hypothesize that HPV16 E6* promotes cancer progression and epithelial differentiation via activating the OXPHOS pathway. The findings also suggest that OXPHOS inhibitors may be an effective treatment for E6*-associated high-risk HNSCC patients. Indeed, some recent studies have highlighted mitochondrial metabolism as a target for anticancer therapy49.
The higher levels of oxidative phosphorylation and mitochondrial genes may explain the larger tumor sizes at diagnosis among tumors that express a higher percent of E6*. By far, the strongest correlations of host genes with E6* involved mitochondrial functions, including PDK3, FH, and numerous subunits of ATP synthase, NADH:ubiquinone oxidoreductase, and ATPase H+ transporters. We also showed that the percent of E6* is positively correlated with tumor mutational burden, suggesting higher E6* may promote carcinogenesis and tumor growth by increasing mutagenesis and/or allowing faster growth via increased energy production. On the other hand, E6* is not associated with E7 at mRNA levels, consistent with the known report that E6* influences translation, not RNA levels of E718, which could help to explain the carcinogenic potential of E6*.
Finally, we found that the percent of E6* is positively associated with worse overall survival. This finding was uncovered using the E6%FL influence scores, and survival analysis adjusting for multiple covariates. Our findings with E6%FL influence scores, which were not found with E6%FL mRNA expression levels, validate the clinical relevance and potential use of our influence score as a prognostic factor. Hong, et al saw a trend in survival based on the ratio of two different shorter isoforms of E6*, E6*I/E6*II53. Our definition of E6* included both of these isoforms, but with E6*I being by far the more prominent one.
Overall, this study demonstrates the dose response effects of E6 and E7 oncogenes on the host transcriptome. This suggests that HPV oncogene expression levels are an important indicator of patient prognosis in HNSCC, consistent with findings in cervical cancer 54. These analyses identified new genes and pathways affected by E6E7 or the splicing ratio of E6, the latter of which has not been well-described. The findings based on the splicing ratio of E6 can guide future studies of the molecular mechanisms underlying E6*-associated carcinogenesis such as mitochondrial metabolism (OXPHOS). The fact that the higher E6%FL influence score was significantly associated with a better overall survival in HPV16-positive patients further supports E6* being associated with high-risk HPV(+) tumors; we are thus optimistic to propose it as a potential prognostic factor for HPV(+) HNSCC patients although a larger cohort is needed for further validation. One of the limitations of this analysis is that we cannot distinguish between the direct influence of E6*, and the indirect influence through increased translation of E7. Further studies are required to investigate the correlation between E6* and E7 at protein level and explore the underlying oncogenic mechanisms of E6*.
Supplementary Material
Acknowledgements
This work was supported by funding from the National Cancer Institute, National Institutes of Health grants [R01CA158286] to MAS and LSR and [P30CA046592], and by the University of Michigan Specialized Programs of Research Excellence (SPORE) Grant [P50CA097248]. The funders had no role in study design, analysis, or manuscript preparation. We would like to acknowledge the support of the University of Michigan Medical School Advanced Genomics Core.
Footnotes
Declarations
The eligible HNSCC patients at University of Michigan Hospital consented to collect tumor tissue. All authors read and approved of the manuscript. The authors declare no competing financial or non-financial interests.
References
- 1.Westra WH. The changing face of head and neck cancer in the 21st century: the impact of HPV on the epidemiology and pathology of oral cancer. Head and neck pathology. 2009;3(1):78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang C, Pauli EK, Biryukov J, et al. The Human Papillomavirus E6 Oncoprotein Targets USP15 and TRIM25 To Suppress RIG-I-Mediated Innate Immune Signaling. Journal of virology. 2018;92(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. The EMBO journal. 1989;8(13):4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vande Pol SB, Klingelhutz AJ. Papillomavirus E6 oncoproteins. Virology. 2013;445(1–2):115–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. [DOI] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nature reviews Cancer. 2010;10(8):550–560. [DOI] [PubMed] [Google Scholar]
- 7.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. Journal of virology. 2001;75(9):4283–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes & development. 1998;12(13):2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park JS, Kim EJ, Kwon HJ, Hwang ES, Namkoong SE, Um SJ. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem. 2000;275(10):6764–6769. [DOI] [PubMed] [Google Scholar]
- 10.Bello JO, Nieva LO, Paredes AC, Gonzalez AM, Zavaleta LR, Lizano M. Regulation of the Wnt/beta-Catenin Signaling Pathway by Human Papillomavirus E6 and E7 Oncoproteins. Viruses. 2015;7(8):4734–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodily JM, Mehta KP, Laimins LA. Human papillomavirus E7 enhances hypoxia-inducible factor 1-mediated transcription by inhibiting binding of histone deacetylases. Cancer research. 2011;71(3):1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duensing S, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer research. 2002;62(23):7075–7082. [PubMed] [Google Scholar]
- 13.Katzenellenbogen RA, Egelkrout EM, Vliet-Gregg P, Gewin LC, Gafken PR, Galloway DA. NFX1–123 and poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16 E6-expressing cells. Journal of virology. 2007;81(8):3786–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfandari J, Shnitman Magal S, Jackman A, Schlegel R, Gonen P, Sherman L. HPV16 E6 oncoprotein inhibits apoptosis induced during serum-calcium differentiation of foreskin human keratinocytes. Virology. 1999;257(2):383–396. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberger S, De-Castro Arce J, Langbein L, Steenbergen RD, Rosl F. Alternative splicing of human papillomavirus type-16 E6/E6* early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc Natl Acad Sci U S A. 2010;107(15):7006–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider-Gadicke A, Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. The EMBO journal. 1986;5(9):2285–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Koneva LA, Virani S, et al. Subtypes of HPV-positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang S, Tao M, McCoy JP Jr., Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. Journal of virology. 2006;80(9):4249–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pim D, Banks L. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene. 1999;18(52):7403–7408. [DOI] [PubMed] [Google Scholar]
- 20.Williams VM, Filippova M, Filippov V, Payne KJ, Duerksen-Hughes P. Human papillomavirus type 16 E6* induces oxidative stress and DNA damage. Journal of virology. 2014;88(12):6751–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Psyrri A, DiMaio D. Human papillomavirus in cervical and head-and-neck cancer. Nat Clin Pract Oncol. 2008;5(1):24–31. [DOI] [PubMed] [Google Scholar]
- 22.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15(22):6758–6762. [DOI] [PubMed] [Google Scholar]
- 23.Filippova M, Johnson MM, Bautista M, et al. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. Journal of virology. 2007;81(8):4116–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koneva LA, Zhang Y, Virani S, et al. HPV Integration in HNSCC Correlates with Survival Outcomes, Immune Response Signatures, and Candidate Drivers. Mol Cancer Res. 2018;16(1):90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S FastQC: a quality control tool for high throughput sequence data. 2010; http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 27.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–2185. [DOI] [PubMed] [Google Scholar]
- 28.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen Y, Nettleton D, Liu H, Tuggle CK. Detecting Differentially Expressed Genes with RNA-seq Data Using Backward Selection to Account for the Effects of Relevant Covariates. J Agric Biol Environ Stat. 2015;20(4):577–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams VM, Filippova M, Soto U, Duerksen-Hughes PJ. HPV-DNA integration and carcinogenesis: putative roles for inflammation and oxidative stress. Future Virol. 2011;6(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C, Patil S, Sartor MA. RNA-Enrich: a cut-off free functional enrichment testing method for RNA-seq with improved detection power. Bioinformatics. 2016;32(7):1100–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M C. GO.db: A set of annotation maps describing the entire Gene Ontology. R package version 370. 2018. [Google Scholar]
- 35.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schacht T, Oswald M, Eils R, Eichmuller SB, Konig R. Estimating the activity of transcription factors by the effect on their target genes. Bioinformatics. 2014;30(17):i401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.T T. A Package for Survival Analysis in S. version 2.38. 2015. [Google Scholar]
- 38.Kosinski AKaM. survminer: Drawing Survival Curves using ‘ggplot2’. R package version 0.4.3. 2018. [Google Scholar]
- 39.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stadler ME, Patel MR, Couch ME, Hayes DN. Molecular biology of head and neck cancer: risks and pathways. Hematol Oncol Clin North Am. 2008;22(6):1099–1124, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene. 1999;18(53):7690–7700. [DOI] [PubMed] [Google Scholar]
- 42.Wanichwatanadecha P, Sirisrimangkorn S, Kaewprag J, Ponglikitmongkol M. Transactivation activity of human papillomavirus type 16 E6*I on aldo-keto reductase genes enhances chemoresistance in cervical cancer cells. The Journal of general virology. 2012;93(Pt 5):1081–1092. [DOI] [PubMed] [Google Scholar]
- 43.Cricca M, Venturoli S, Leo E, Costa S, Musiani M, Zerbini M. Molecular analysis of HPV 16 E6I/E6II spliced mRNAs and correlation with the viral physical state and the grade of the cervical lesion. J Med Virol. 2009;81(7):1276–1282. [DOI] [PubMed] [Google Scholar]
- 44.Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes & development. 1997;11(16):2090–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones DL, Alani RM, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes & development. 1997;11(16):2101–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spardy N, Covella K, Cha E, et al. Human papillomavirus 16 E7 oncoprotein attenuates DNA damage checkpoint control by increasing the proteolytic turnover of claspin. Cancer research. 2009;69(17):7022–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutler DI, Auerbach AD, Satagopan J, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003;129(1):106–112. [DOI] [PubMed] [Google Scholar]
- 48.Qin T, Zhang Y, Zarins KR, et al. Expressed HNSCC variants by HPV-status in a well-characterized Michigan cohort. Scientific reports. 2018;8(1):11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin Cancer Res. 2018;24(11):2482–2490. [DOI] [PubMed] [Google Scholar]
- 50.Reznik E, Miller ML, Senbabaoglu Y, et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraft CS, LeMoine CM, Lyons CN, Michaud D, Mueller CR, Moyes CD. Control of mitochondrial biogenesis during myogenesis. Am J Physiol Cell Physiol. 2006;290(4):C1119–1127. [DOI] [PubMed] [Google Scholar]
- 52.Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26(4):960–968. [DOI] [PubMed] [Google Scholar]
- 53.Hong A, Zhang X, Jones D, et al. E6 viral protein ratio correlates with outcomes in human papillomavirus related oropharyngeal cancer. Cancer Biol Ther. 2016;17(2):181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Boer MA, Jordanova ES, Kenter GG, et al. High human papillomavirus oncogene mRNA expression and not viral DNA load is associated with poor prognosis in cervical cancer patients. Clin Cancer Res. 2007;13(1):132–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.