Summary
Accumulation of topological stress in the form of DNA supercoiling is inherent to the advance of RNA polymerase II (Pol II) and needs to be resolved by DNA topoisomerases to sustain productive transcriptional elongation. Topoisomerases are therefore considered positive facilitators of transcription. Here, we show that, in contrast to this general assumption, human topoisomerase IIα (TOP2A) activity at promoters represses transcription of immediate early genes such as c-FOS, maintaining them under basal repressed conditions. Thus, TOP2A inhibition creates a particular topological context that results in rapid release from promoter-proximal pausing and transcriptional upregulation, which mimics the typical bursting behavior of these genes in response to physiological stimulus. We therefore describe the control of promoter-proximal pausing by TOP2A as a layer for the regulation of gene expression, which can act as a molecular switch to rapidly activate transcription, possibly by regulating the accumulation of DNA supercoiling at promoter regions.
Keywords: DNA supercoiling, DNA topology, DNA topoisomerases, transcription elongation, promoter-proximal pausing, gene expression
Graphical abstract

Highlights
-
•
Catalytic inhibition of TOP2A results in a global reduction in promoter-proximal pausing
-
•
Catalytic inhibition of TOP2A upregulates transcription of immediate early genes (IEGs)
-
•
IEG upregulation occurs independently of DNA breaks or cellular stress
-
•
IEG upregulation depends on the accumulation of negative supercoiling at promoter regions
Herrero-Ruiz et al. analyze the function of DNA topoisomerase II in transcriptional regulation. The results reveal that TOP2A activity favors promoter-proximal pausing of RNA polymerase II and repression of immediate early genes in a mechanism that is independent of DNA break formation and involves the control of DNA supercoiling at promoters.
Introduction
As originally envisioned by the “twin-supercoiled-domain” model (Liu and Wang, 1987), the advance of RNA polymerase II (Pol II) during transcription is a torque-generating force that results in upstream and downstream regions of over- and underwound DNA and thus positive (+) and negative (−) DNA supercoiling, respectively (Kouzine et al., 2014; Ma and Wang, 2016). This supercoiling of template DNA can cause polymerase stalling (Ma et al., 2013) and therefore needs to be relieved in order to allow productive transcription and gene expression. DNA topoisomerases are the enzymes that relax this topological stress by transiently gating DNA passage, in a controlled cut-and-reseal mechanism that affects either one (type I DNA topoisomerases; mainly TOP1 in eukaryotes) or simultaneously both (type II topoisomerases; TOP2) DNA strands (Pommier et al., 2016). Hence, they are generally considered important facilitators of transcription, especially for long genes in which the load of DNA supercoiling can become particularly burdening (Joshi et al., 2012; King et al., 2013). In this sense, TOP1 and TOP2 have been shown to cooperate to relieve transcription associated (+) and (−) supercoiling in order to maintain appropriate levels of gene expression (Pedersen et al., 2012; Sperling et al., 2011). It is worth noting, however, that DNA supercoiling can also facilitate transcription under some particular circumstances. For example, (−) supercoiling helps duplex melting, the formation of Pol II open complexes, and the recruitment of initiation factors (Parvin and Sharp, 1993; Tabuchi et al., 1993). Furthermore, the accumulation of (+) and (−) supercoiling has been proposed to facilitate the required nucleosome eviction ahead and reloading behind Pol II during transcription elongation (Corless and Gilbert, 2016; Teves and Henikoff, 2014). All this information suggests that the regulation of supercoiling by topoisomerase activity could somehow operate to control transcription and gene expression, although the mechanisms by which this can occur remain largely unknown.
In mammals, TOP1 is essential for efficient transcription elongation, and its activity is induced by direct interactions with the elongating form of Pol II (Baranello et al., 2016; Dujardin et al., 2014; King et al., 2013). In contrast, the functions of TOP2 in transcription seem more complex. Although also involved in facilitating elongation (King et al., 2013), both the α (TOP2A) and β (TOP2B) mammalian TOP2 paralogs are enriched at promoters (Canela et al., 2017; Thakurela et al., 2013; Tiwari et al., 2012), suggesting relevant regulatory functions. Indeed, TOP2A and TOP2B control the expression of neuronal differentiation genes that are essential for proper neural development (Lyu et al., 2006; Thakurela et al., 2013; Tiwari et al., 2012). Accumulating evidence also suggests that TOP2B can be essential for the fast induction of highly regulated genes in response to different types of stimuli, including hormones (Haffner et al., 2010; Ju et al., 2006), growth factors (Bunch et al., 2015), and neuronal activity (Madabhushi et al., 2015). Immediate early genes (IEGs) such as c-FOS, which responds with a transient burst in transcription only a few minutes after cells are stimulated, are a paradigm of this type of transcriptional response (Healy et al., 2013). Interestingly, this has been proposed to operate through the generation of a promoter TOP2-mediated double-strand break (DSB) that would be directly responsible for locally triggering transcription (Bunch et al., 2015; Ju et al., 2006; Madabhushi et al., 2015). As mentioned above, TOP2 enzymes indeed cut duplex DNA as part of their catalytic cycle, remaining covalently linked to the ends of the incised fragment in the so-called cleavage complex (TOP2cc) (Pommier et al., 2016). These intermediates, however, are very rare in cells, unless stabilized with TOP2 poisons such as etoposide (Canela et al., 2017; Gittens et al., 2019; Gothe et al., 2019), and are fully reversible structures that only result in DSB formation upon interference with cellular processes and proteasomal degradation (Canela et al., 2019; Gothe et al., 2019; Sciascia et al., 2020; Zhang et al., 2006). In addition, there is evidence that DSBs, both in promoter and gene bodies, are strong inhibitors of transcription in cis, in a process that facilitates repair of the lesions and protects genome integrity (Caron et al., 2019; Hanawalt and Spivak, 2008; Iacovoni et al., 2010; Pankotai et al., 2012; Shanbhag et al., 2010).
Alternative hypotheses to explain the function of TOP2 and DNA supercoiling in the control of gene expression could therefore be considered. Although in many cases gene regulation implies changes in the recruitment of Pol II to promoters and increased transcription initiation, in mammals, the main regulatory level lies at the entry into productive transcription elongation (Adelman and Lis, 2012). This is exerted through the modulation of promoter-proximal pausing of Pol II, which stops RNA synthesis shortly after initiation and remains primed for a transition into elongation upon P-TEFb-mediated Ser2 phosphorylation of its C-terminal domain (CTD) (Adelman and Lis, 2012; Core and Adelman, 2019; Li and Gilmour, 2011). For this reason, global analyses show that the Pol II signal is concentrated near the transcription-start site (TSS) in a large fraction of genes (Guenther et al., 2007; Kim et al., 2005). Promoter-proximal pausing is controlled by specific complexes such as Negative Elongation Factor (NELF) and DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) sensitivity inducing factor (DSIF) that decrease the elongation efficiency of Pol II (Yamaguchi et al., 2013), as well as by intrinsic features of the template chromatin (Aoi et al., 2020; Jimeno-González et al., 2015; Kwak et al., 2013). Interestingly, promoter-proximal pausing has been recently shown to correlate with TOP2B and spontaneously occurring DSBs at a genome-wide level (Dellino et al., 2019) and interpreted in the context of the TOP2B-mediated DSB model of transcriptional regulation.
Here, we unexpectedly find that the main effect of TOP2 catalytic inhibition in human cells is a quick and global release of Pol II from promoter-proximal pausing that results in a sharp upregulation of immediate early and other highly regulated genes. This is a result of TOP2A repressive functions in transcriptional regulation that are tightly interconnected to TOP1 activity and completely independent of DSB formation. Instead, these functions require the topological integrity of promoter regions, where transcription-associated (−) supercoiling accumulates. Thus, we provide a topological framework for the regulation of promoter-proximal pausing that can explain the typical bursting behavior of IEGs and, in more general terms, advances our understanding of the control of human gene expression by DNA topoisomerases.
Results
TOP2 catalytic inhibitors induce expression of IEGs
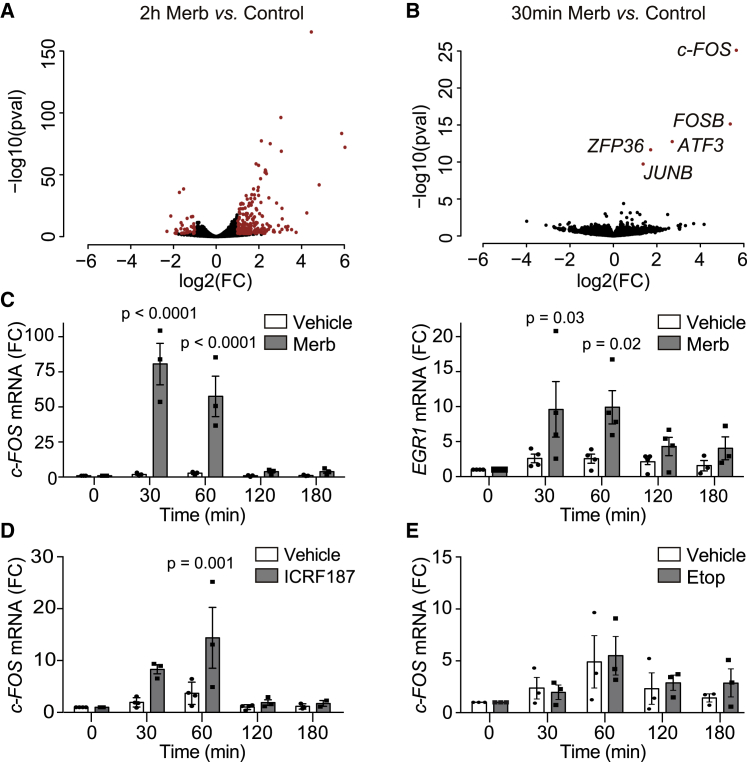
To study a possible function of TOP2 and DNA supercoiling in regulating transcription, changes in gene expression profiles were analyzed in human-telomerase-immortalized retinal pigment epithelial 1 (RPE-1) cells treated with merbarone, a drug that catalytically inhibits TOP2 upstream of DNA cleavage (Fortune and Osheroff, 1998). Cells were previously arrested in G0/G1 by serum starvation in order to avoid possible effects of topoisomerase inhibition on other cellular processes such as replication or chromosome segregation. Upon a 2-h treatment, the comparison between RNA sequencing (RNA-seq) profiles of merbarone-treated and untreated samples revealed that 173 protein-coding genes were differentially expressed, with 148 upregulated and only 25 downregulated upon merbarone treatment (Figure 1A; Data S1), pointing to a mainly repressive role of TOP2 activity on gene expression. Interestingly, most upregulated genes (UP genes) were related with cellular responses to different cellular stimuli (Figure S1A). In fact, there was a clear enrichment of IEGs (Figure S1B), which are genes that, as mentioned above, are highly regulated and activated through protein-synthesis-independent rapid bursts of transcription shortly after different types of stimuli (Bahrami and Drabløs, 2016). A paradigm of this pattern of expression is the c-FOS gene, whose mRNA production peaks at 30–60 min after stimulation and drops after 90 min (Greenberg and Ziff, 1984). Therefore, we analyzed RNA-seq samples taken 30 min after merbarone treatment and found that only 5 genes were upregulated, namely, c-FOS, FOSB, ATF3, ZFP36, and JUNB; and all of them are well-established IEGs (Figure 1B; Data S1). TOP2 activity has been described to be required for the transcription of long genes (Joshi et al., 2012; King et al., 2013). Consistent with this description, UP genes were significantly shorter than the distribution of all human genes (Figure S1C).
Figure 1.
Immediate early genes (IEGs) are upregulated upon TOP2 catalytic inhibition
(A and B) Volcano plots of transcriptional changes upon 2 hours (2h) merbarone (Merb; 200 μM) versus control (untreated) (n = 2) treatment (A) or 30 minutes (30min) Merb (200 μM) versus control (DMSO) treatment (n = 3) (B) measured by RNA-seq on serum-starved RPE-1 cells. The x axis represents fold-induction ratios in a log2 scale, and the y axis represents p values in a log10 scale. Genes with an absolute fold change ≥2 and an adjusted p ≤ 0.05 are shown in red.
(C) c-FOS (left) and EGR1 (right) mRNA levels, as measured by qRT-PCR, at the indicated times following Merb and vehicle (DMSO) treatments (n = 3). Values were normalized to GAPDH mRNA levels and signal under untreated conditions. Individual experimental values and mean ± SEM are shown. Two-way ANOVA test with Bonferroni post-test.
(D) As in (C) for c-FOS expression upon ICRF-187 (200 μM) treatment.
(E) As in (C) for c-FOS expression upon etoposide (Etop; 20 μM) treatment.
In all cases, only the p values indicating statistically significant differences against vehicle treatment are shown.
The changes in expression of c-FOS (used as a representative model hereafter) and EGR1 (another responsive IEG) under TOP2 inhibition were confirmed by quantitative reverse transcription PCR (qRT-PCR), with an outstanding increase in mRNA levels (80- and 10-fold, respectively) 30 min after merbarone treatment and a subsequent decrease at later time points (Figure 1C) that mirrored physiological induction with serum (Figure S1D). As expected, the increased mRNA levels of c-FOS reflected a transcriptional induction, as determined by the analysis of chromatin-associated RNA (Figure S1E). Furthermore, increased c-FOS expression was also observed in the lung carcinoma A549 cell line and in primary mouse embryonic fibroblasts (MEFs) (Figures S1F and S1G), demonstrating conservation among cell types and species, although fold-change inductions were quantitatively diverse due to a strong variability in basal expression (Figure S1H). Importantly, RPE-1 cells treated with ICRF-187, a different TOP2 catalytic inhibitor that targets TOP2 after strand passage and re-ligation, also showed c-FOS upregulation (Figure 1D). In contrast, treatment with the paradigmatic TOP2 poison etoposide resulted in a minor induction of c-FOS expression only at later time points (Figure 1E), similar to that previously reported in other cell types (Bunch et al., 2015; Madabhushi et al., 2015), but negligible compared to the effects of merbarone or serum. We can therefore conclude that treatment with TOP2 catalytic inhibitors results in a rapid and robust induction of highly regulated genes, including a subset of IEGs and c-FOS in particular.
IEG induction is independent of DSBs or stress
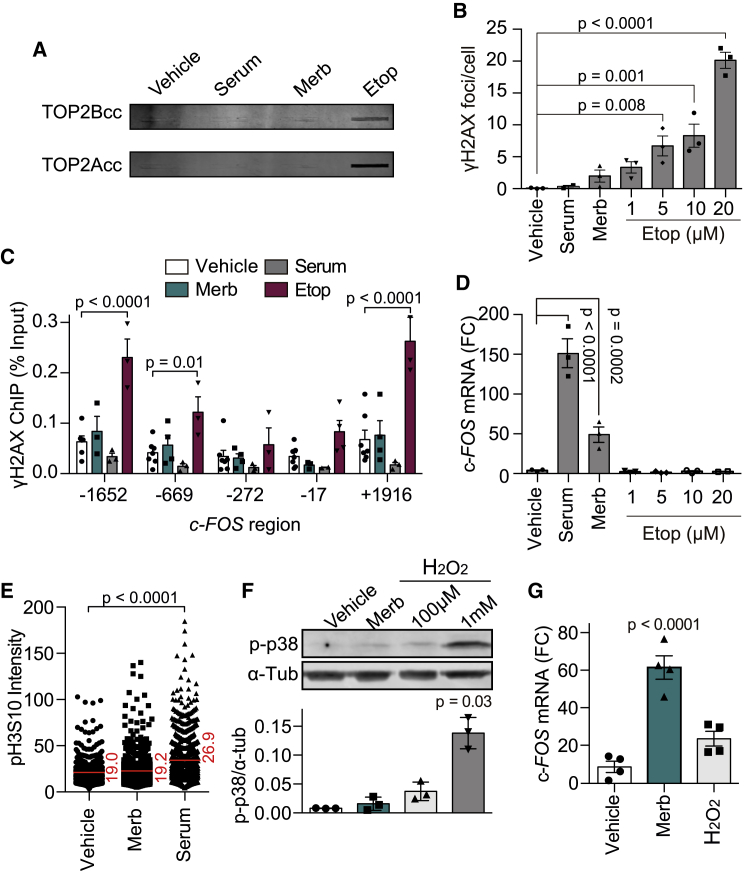
The results described above suggest TOP2 inhibition as a potential molecular mechanism for IEG upregulation, in contrast to current models of TOP2-mediated promoter DSBs (Bunch et al., 2015; Madabhushi et al., 2015). There is, however, some degree of controversy as to whether merbarone can also act as a TOP2 poison in vivo (Pastor et al., 2012). We therefore decided to analyze the accumulation of TOP2ccs and DSBs under conditions of c-FOS upregulation. First, we performed ICE (in vivo complex of enzyme), which measures TOP2ccs by isolating and detecting covalent protein-DNA complexes accumulated in cells (Nitiss et al., 2012; Subramanian et al., 1995; Figure S2A). Clearly, neither merbarone nor serum treatments resulted in a detectable global accumulation of TOP2ccs (Figure 2A). Second, we monitored the generation of DSBs by the accumulation of the γH2AX marker globally, as nuclear foci visualized by immunofluorescence (Kinner et al., 2008), and specifically at the c-FOS locus by using chromatin immunoprecipitation (ChIP). Again, we found that neither merbarone nor serum produced a significant accumulation of γH2AX (Figures 2B, 2C, and S2B), despite concomitantly causing a robust transcriptional upregulation of c-FOS (Figure 2D). In contrast, treatment with etoposide resulted in the expected strong induction of global TOP2ccs and γH2AX signal, which was clearly enriched surrounding the c-FOS gene, without a significant induction of transcription (Figures 2A–2D and S2B). The transcriptional response, therefore, does not correlate with the appearance of TOP2ccs and DSBs. To further confirm that IEG induction was independent of DNA damage, we decided to impair the repair of TOP2-associated DSBs by removing TDP2, a highly specialized DNA-end unblocking enzyme (Cortes Ledesma et al., 2009) whose absence significantly delays repair of this type of DNA lesion (Gómez-Herreros et al., 2013; Schellenberg et al., 2017). As a matter of fact, RPE-1 cells deleted for TDP2 by CRISPR-Cas9 displayed a kinetics of c-FOS induction similar to that of wild-type cells, both with merbarone and serum treatments (Figures S2C and S2D). Altogether, these results disfavor the involvement of stable TOP2ccs or associated DSBs in the mechanism of transcriptional upregulation of IEGs. Interestingly, this finding is not only true for merbarone treatment but also for physiological stimulation with serum.
Figure 2.
IEG induction is independent of DSBs or stress
(A) TOP2A and TOP2B covalently bound to genomic DNA (ICE) under 30 min of vehicle (DMSO), Merb (200 μM), serum (1%), or Etop (20 μM) treatments. A representative image is shown (n = 3).
(B) Quantification of γH2AX foci immunofluorescence upon 30 min of the indicated conditions (40 cells from each experimental condition were manually counted, double-blind) (n = 3). Individual experimental values and mean ± SEM are shown. One-way ANOVA test with Dunnett’s post-test.
(C) γH2AX ChIP at the indicated regions of c-FOS gene in serum-starved RPE-1 cells, as measured by qPCR, at 30 min following vehicle (DMSO), Merb (200 μM), serum (1%), or Etop (50 μM) treatments (n = 3). Individual experimental values and mean ± SEM are shown. Two-way ANOVA test with Dunnett’s post-test.
(D) c-FOS mRNA levels upon 30 min of the indicated conditions (n = 3); details as in Figure 1C. One-way ANOVA test with Dunnett’s post-test.
(E) Quantification of phosphorylated H3S10 (pH3S10) immunofluorescence in serum-starved RPE-1 cells treated for 30 min with vehicle (DMSO), Merb (200 μM), or serum (1%) (n = 3). Individual cellular values and median are shown. Kruskal-Wallis test with Dunn’s post-test.
(F) Representative image (top) and quantification (bottom) of p-p38 MAPK (Thr180/Thr182) western blot upon 30-min incubation with vehicle (DMSO), Merb (200 μM), and H2O2 (100 μM or 1 mM) (n = 3). Individual experimental values and mean ± SEM are shown. One-way ANOVA test with Dunnett’s post-test.
(G) c-FOS mRNA levels upon 30 min of vehicle (DMSO), Merb (200 μM), or H2O2 (100 μM) (n = 3); details as in Figure 1C. One-way ANOVA test with Dunnett’s post-test.
In all cases, only the p values indicating statistically significant differences against vehicle treatment are shown.
Finally, we decided to test whether the transcriptional response to merbarone could be an indirect cause of some type of cellular stress or signaling derived from TOP2 inhibition. There are two main pathways responsible for IEG upregulation in response to stimulus, as follows: the RAS-mitogen-activated protein kinase (MAPK) pathway that is activated by growth factors and mitogens, and the p38-MAPK pathway that is activated by DNA damage or other types of stress (Healy et al., 2013). Merbarone treatment did not significantly trigger these pathways, as determined by increased phosphorylation of histone H3 serine 10 (H3S10) (Figures 2E and S2E) or p38 (Figure 2F) compared to physiological induction with serum or high-stress-inducing hydrogen peroxide concentrations (1 mM). Conversely, mild stress caused by lower hydrogen peroxide exposure (100 μM) did not elicit significant stimulation of c-FOS transcription, in contrast to what was observed in the case of merbarone treatment (Figure 2G). Hence, we conclude that merbarone increases IEG expression directly and not by eliciting a cellular stress response, although we cannot fully rule out the involvement of additional signaling pathways.
TOP2A activity at gene promoters represses transcription of IEGs
To characterize the inhibitory effect of merbarone on TOP2 activity, ICE experiments were carried out following different times of merbarone treatment and a brief (5 min) incubation with a high dose (400 μM) of etoposide in order to “freeze” catalytically engaged enzymes (Figure 3A). In this experimental setup, etoposide-mediated TOP2cc induction is used as an estimate of TOP2 activity, which merbarone is expected to reduce by acting upstream in the catalytic cycle. Strikingly, at times in which IEG upregulation was already evident, merbarone mainly inhibited the TOP2A paralog, whereas TOP2B activity was mildly affected only at later time points (Figure 3B). Consistent with this finding, stable in-pool deletion of TOP2B in RPE-1 cells overexpressing Cas9 (RPE-1 Cas9) (Figure S3A) did not change c-FOS expression (neither its basal levels [Figure 3C] nor its induction upon merbarone or serum treatments [Figure 3D]) compared to cells transfected with a non-targeting guide RNA(gRNA). Importantly, these results not only prove that TOP2B is not a target for the transcriptional response elicited by merbarone but also rule out the previously proposed requirement for TOP2B-mediated DSBs in the regulation of c-FOS expression (Bunch et al., 2015; Madabhushi et al., 2015), at least under our experimental conditions. In order to check the involvement of TOP2A, an essential protein for cell cycle progression, we performed acute TOP2A deletion under conditions of serum starvation by the transfection of an appropriate gRNA in RPE-1 Cas9 cells (Figure S3B). Despite these technical limitations, we achieved substantial levels (>90%) of TOP2A protein depletion (Figure 3E), which were sufficient to cause a significant increase in the basal levels of c-FOS expression compared to cells transfected with a non-targeting gRNA (Figure 3E). Treatment of these control cells with merbarone or serum triggered substantial c-FOS upregulation (Figure 3F), although higher basal c-FOS expression caused by the transient-transfection conditions (Figure S3C) resulted in a lower fold induction. In TOP2A-deleted cells, however, merbarone-mediated c-FOS induction was completely abolished, and interestingly, stimulation with serum was potently enhanced (Figure 3F). These results strongly support inhibition of TOP2A as the direct cause of c-FOS upregulation in response to merbarone treatment, ruling out potential off-target effects of the drug. Furthermore, the de-repression observed in TOP2A−/− cells, both in terms of basal expression levels and upon serum stimulation, suggests that merbarone mainly operates through a reduction in TOP2A activity and not by indirect effects caused by the trapping of the enzyme on chromatin.
Figure 3.
Merb induces IEGs by inhibiting TOP2A
(A and B) Diagram of the experimental design (A) and a representative image (B) of the ICE experiment conducted to measure the inhibitory effect of Merb (200 μM) on TOP2 catalytic activity (n = 3). The 48-h serum-starved (0.1%) RPE-1 cells were subjected to Merb treatment for the indicated time with a final 5-min treatment with either vehicle (DMSO) or Etop (400 μM).
(C) c-FOS mRNA levels in RPE-1 Cas9 cells following transfection with non-targeting (NT) and TOP2B-specific (TOP2B) gRNAs (n = 3), according to the experimental outline illustrated in Figure S3A. Individual values and mean ± SEM are shown, one-sample t test (hypothetic value = 1). A representative western blot image of TOP2B and TOP2A levels in wild-type (WT) and TOP2B−/− RPE-1 cells is shown (right) (n = 3).
(D) c-FOS mRNA levels in cells described in (C) following the indicated times of Merb (left) or serum (right) treatments (n = 3); details as in Figure 1C. Individual experimental values and mean ± SEM are shown. Two-way ANOVA test with Bonferroni post-test.
(E) As in (C) for TOP2A-specific (TOP2A) gRNAs (n = 3). Experimental outline in Figure S3B.
(F) As in (D) for TOP2A-specific (TOP2A) gRNAs (n = 3).
In all cases, only the p values indicating statistically significant differences are shown.
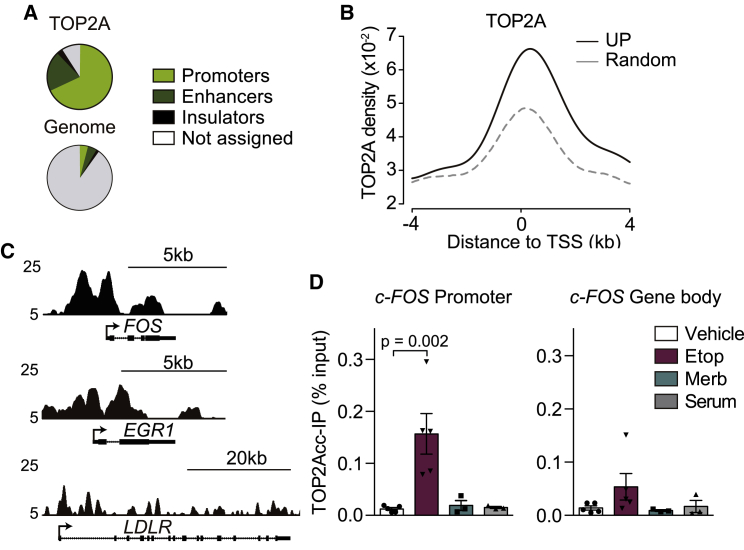
Direct transcriptional roles of TOP2A are somewhat unexpected, as they have been traditionally assigned to TOP2B (Austin et al., 2018). However, TOP2A has been reported to physically interact with Pol II (Mondal and Parvin, 2001) and to accumulate at promoters and nucleosome-free regions (Canela et al., 2017; Thakurela et al., 2013; Yu et al., 2017), similarly to TOP2B. ChIP sequencing (ChIP-seq) in G0/G1-arrested RPE-1 cells confirmed this enrichment at promoters and enhancers under our experimental conditions (Figure 4A). In fact, the distribution of TOP2A strongly correlated with Pol II and marks of active chromatin like H3K4me3 and H3K27ac at these regions (Figure S4A). Interestingly, average TOP2A levels around the TSS were notably higher in the 148 genes upregulated at the mRNA level after 2-h merbarone treatment (UP genes) than in the same number of randomly selected genes (Figure 4B), with similar expression levels (Figure S4B), or also when compared to all human genes (Figure S4C). This result is consistent with a direct involvement of TOP2A in transcriptional repression of these genes under basal conditions. TOP2A profiles in c-FOS and EGR1, as positive examples, and LDLR, an IEG not upregulated upon merbarone treatment (Data S1), illustrate these differences (Figure 4C). To further link TOP2A activity to transcriptional repression, we decided to determine whether TOP2A present at the c-FOS promoter was catalytically active. To do so, we performed TOP2A ICE-IP (Álvarez-Quilón et al., 2020), a technique in which accumulated TOP2ccs were immunoprecipitated from ICE extracts with specific antibodies against TOP2A, and the associated DNA was subsequently amplified by qPCR with pairs of primers in the c-FOS gene (Figures S2A and 4D). Interestingly, etoposide treatment strongly increased the amount of covalently bound TOP2A at the c-FOS promoter (Figure 4D, left), which is indicative of its strong catalytic engagement at this region. Increased signal, although not reaching statistical significance, was also observed in the gene body (Figure 4D, right). TOP2A is, therefore, not only present but also active at c-FOS under basal expression conditions, which is in agreement with a repressive role in the transcriptional regulation of this gene. Furthermore, in contrast to what was observed upon etoposide treatment, ICE-IP signal in c-FOS was not increased in the presence of merbarone or serum (Figure 4D), further confirming that the transcriptional effects observed are independent of TOP2Acc stabilization. We therefore conclude that TOP2A plays a major role in the basal constitutive repression of c-FOS, which is overcome upon inhibition of its catalytic activity, leading to a sharp transcriptional upregulation.
Figure 4.
TOP2A activity at gene promoters represses transcription
(A) Genome-wide distribution of TOP2A peaks compared with the general distribution of human mappable genomic regions.
(B) Average TOP2A ChIP-seq enrichment around the TSS of UP genes and an equal number of randomly selected genes.
(C) Genome browser tracks for TOP2A ChIP-seq at c-FOS, EGR1, and LDLR genes in untreated serum-starved RPE-1 cells. The y axis represents Reads Per Kilobase Million (RPKM).
(D) TOP2A ICE-IP under vehicle (DMSO), Merb (200 μM), serum (1%), or Etop (400 μM) treatment for 5 min, at the indicated regions in serum-starved RPE-1 cells (see Figure S2A) (n = 3). Individual experimental values and mean ± SEM are shown. One-way ANOVA test with Dunnett’s post-test. Only the p values indicating statistically significant differences against vehicle treatment are shown.
TOP2A catalytic inhibition releases Pol II from promoter-proximal pausing
In order to understand the transcriptional effect of TOP2A inhibition in detail, we used ChIP-seq to compare the distribution of Pol II under control conditions and following 30 and 60 min of merbarone treatment. An analysis of differential Pol II gene body occupancy showed a repression of only 4 genes, whereas 48 genes were induced at 60 min of merbarone treatment, with 19 of them (18%) coinciding with the genes upregulated at the mRNA level (UP genes) (Figures S5A and S5B). In fact, Pol II occupancy at the body of UP genes was significantly increased upon merbarone treatment (Figure 5A), confirming that the effect of TOP2A inhibition on gene expression was exerted at the level of transcriptional upregulation. This effect could be clearly observed in c-FOS and EGR1 as representative examples but not in the irresponsive LDLR IEG (Figure 5B). Strikingly however, Pol II occupancy did not increase in the region surrounding the TSS of UP genes, where it was even reduced, although it did not reach statistical significance (Figure 5A). Although this trend was not observed in strongly induced genes, such as c-FOS, in which the Pol II ChIP-seq signal increased throughout the gene (Figure 5B), the ratio of gene-body-bound (at the +1916 region) versus TSS-bound (at the −17 region) polymerase, as determined by ChIP-qPCR, increased significantly upon merbarone treatment (Figure S5C). These transcriptional phenotypes, which can be visualized in the profile of Pol II occupancy (Figure 5C), are consistent with an increased release from promoter-proximal pausing, accompanied in some cases such as c-FOS and EGR1 by a concomitant stimulation of transcription initiation. To quantify the global changes in Pol II distribution, two gene body/promoter ratios were calculated, namely, pause release ratio (PRR) (Chen et al., 2015) and pausing index (PI) (Core et al., 2008; Day et al., 2016), which are positive and negative indicators of pause release, respectively. Merbarone treatment significantly increased PRR (Figure 5C) and decreased PI (Figure S5D) of UP genes, suggesting that TOP2A inhibition leads to gene upregulation through the release of Pol II from the pause site. We then decided to extend our analysis to all (n = 10,471) active genes, as determined by the presence of Pol II at the promoter under control conditions. Again, although more mildly, average Pol II gene occupancy showed a general shift from the TSS to proximal coding regions and significant increases in PRR (Figure 5D) and decreases in PI (Figure S5E) under TOP2A inhibition. Finally, we also evaluated the genome-wide effect of merbarone treatment on promoter-proximal pausing by using an empirical cumulative distribution function (ECDF) of PRR, which resulted in a shift toward pause release (Figure 5E). We therefore conclude that TOP2A inhibition leads to an increase in the release of Pol II from promoter-proximal pausing that can be observed globally, although only a subset of genes becomes significantly upregulated at the mRNA level.
Figure 5.
Merb releases Pol ll from promoter-proximal pausing
(A) Box-plot distribution of Pol II/IgG ChIP-seq reads (RPKM) at the gene body (+0.5 to +2.5 kb; left) and the TSS (−0.5 to +0.5; right) in UP genes following the indicated times of Merb treatment. Wilcoxon rank-sum test.
(B) Genome browser tracks for Pol II ChIP-seq (top) following the indicated times of Merb treatment at c-FOS, EGR1, and LDLR genes. The y axis represents RPKM.
(C) Average Pol II ChIP-seq distribution around the TSS (left) and box-plot distribution of PRR (right) in UP genes following the indicated times of Merb treatment. Wilcoxon rank-sum test.
(D) As in (C) for all (10,471) transcriptionally active genes, determined by Pol II or H3K4me3 ChIP-seq signal at the TSS.
(E) Empirical cumulative distribution function (ECDF) of PRR following the indicated times of Merb treatment.
(F) Correlation between TOP2A levels at the TSS and PI. Genes were stratified in quintiles (Q1 to Q5) regarding their TOP2A levels at the TSS, as measured by TOP2A ChIP-seq counts, and the box-plot distribution of PI in each quintile is represented.
(G) Proximity ligation assay (PLA) quantification and representative image (inset) of TOP2A and NELFE interaction in U2OS cells. The antibody used in each condition is indicated on the y axis (40 cells; n = 3). Individual cells and median are shown. Kruskal Wallis test with Dunn’s post-test.
(H) NELFE ChIP at the indicated regions of the c-FOS gene in serum-starved RPE-1 cells, as measured by qPCR, at 30 min following vehicle (DMSO), Merb (200 μM), or serum (1%) treatment (n = 3). Individual experimental values and mean ± SEM are shown. Two-way ANOVA test with Dunnett’s post-test.
In all cases, only the p values indicating statistically significant differences are shown.
To further link TOP2A to the regulation of promoter-proximal pausing, genes were categorized in quintiles regarding their TOP2A density at promoters, and their average PI and PRR were determined. Interestingly, TOP2A positively correlated with PI (Figure 5F) and negatively with PRR (Figure S5F), indicating a positive association of TOP2A with promoter-proximal pausing. Furthermore, we found that TOP2A was in close proximity to NELF, as determined by a proximity-ligation assay (PLA) against its RNA-binding subunit NELFE (Figure 5G). The specificity of this signal was confirmed by the reduction of PLA foci under NELFE downregulation (Figure S5G). Furthermore, both merbarone and serum treatments caused a mild but significant reduction in TOP2A-NELFE proximity, together with a reduction in NELFE ChIP signal at the c-FOS TSS (Figures 5H and S5H). These results, together with the general pause release and transcriptional upregulation effects observed under merbarone treatment, strongly suggest that TOP2A is an important repressor of transcription that operates by a stimulation of promoter-proximal pausing.
c-FOS expression is affected by DNA supercoiling
TOP2 and TOP1 have been shown to collaborate in the removal of transcription-associated supercoiling (Pedersen et al., 2012; Sperling et al., 2011). We therefore decided to monitor changes in TOP1 activity during c-FOS upregulation by using TOP1 ICE-IP as described above for TOP2A but with a pulse of the TOP1 poison camptothecin (CPT) (Figure S2A). Because merbarone has been reported to partially inhibit TOP1 activity at high concentrations in vitro (Drake et al., 1989), we first checked the effect of merbarone and serum treatments on CPT-induced ICE signal, globally (Figure S6A) and specifically at a control expressed gene such as GAPDH (Figure S6B). Neither treatment caused a significant change in the accumulation of TOP1ccs, suggesting that they do not significantly affect TOP1 cleavage in cells, which validates the use of this technique to monitor TOP1 activity during c-FOS induction. TOP1 had basal activity at the c-FOS promoter and gene body, as shown by a clear CPT-dependent TOP1cc accumulation (Figure S6C). This activity was strongly increased, specially within the gene body (4- and 12-fold at promoter and gene body, respectively), when transcription was induced with serum (Figure 6A), consistent with the reported tight link between TOP1 activity and transcription elongation (Baranello et al., 2016). Upon merbarone treatment, however, despite the dramatic transcriptional stimulation (Figure 1C), TOP1 activity in the c-FOS promoter and gene body remained at basal levels (Figure 6A). Thus, TOP2A inhibition completely bypasses the need of TOP1 for transcription elongation, which is a result that supports a topological nature for merbarone-induced gene upregulation. Conversely, TOP1 inhibition with CPT strongly suppressed c-FOS upregulation upon merbarone treatment (Figure 6B). We can conclude that TOP2A and TOP1, rather than operating redundantly, are negative and positive transcriptional effectors, respectively. The fact that an acute transcriptional response can be achieved without an accompanying increase in topoisomerase activity is remarkable.
Figure 6.
c-FOS expression is regulated by DNA supercoiling
(A) TOP1cc levels, as determined by TOP1 ICE-IP, at the indicated regions of the c-FOS gene (see experimental set up in Figure S2A) following 30-min vehicle (DMSO), Merb (200 μM), or serum (1%) treatments. Under all conditions, TOP1ccs were induced by a 5-min incubation with camptothecin (CPT; 10 μM) (n = 4). Individual experimental values and mean ± SEM are shown. Two-way ANOVA test with Dunnett’s post-test.
(B) c-FOS mRNA levels in RPE-1 cells upon treatment with Merb (200 μM), CPT (10 μM), or their combination, as indicated (n = 3), details as in Figure 1C. Individual experimental values and mean ± SEM are shown. Two-way ANOVA test with Bonferroni post-test.
(C) Biotin-psoralen (bio-psoralen) incorporation, expressed as enrichment after pull-down at the indicated positions of the c-FOS gene and normalized to a non-transcribed intergenic region as an internal control, in serum-starved RPE-1 cells after 10-min treatments with vehicle (DMSO), Merb (200 μM), or serum (1%) (n = 3). Individual experimental values and mean ± SEM are shown; two-way ANOVA test with Bonferroni post-test.
(D) c-FOS mRNA levels in RPE-1 Cas9 cells 2 h after transfection with the indicated gRNA and at different times following induction with Merb (200 μM) (top) or serum (1%) (bottom) (n = 3); other details as in (B).
(E) As in (D) but in RPE-1 Cas9 D10A cells and including 30-min pretreatment with PARP1 inhibitor PJ34 as indicated (PARPi; 10 μM; right).
In all cases, only the p values indicating statistically significant differences are shown.
The involvement of two opposing topoisomerase activities strongly suggests a central role for DNA topology, and likely DNA supercoiling, in the regulation of c-FOS expression. We therefore decided to monitor changes in the incorporation of biotinylated psoralen (bio-psoralen) (Figure S6D), a compound that preferentially intercalates into negatively supercoiled (−) DNA (Bermúdez et al., 2010). Limiting concentrations of bio-psoralen that resulted in a measurable level of transcription-dependent intercalation were selected and experimentally tested in order to minimize the contribution of other variables such as chromatin structure (Figures S6E and S6F). Under these conditions, the stimulation of c-FOS transcription by serum resulted in increased incorporation of bio-psoralen at the promoter and gene body (Figure 6C), consistent with (−) supercoiling accumulation as a result of transcriptional activity (Bermúdez et al., 2010; Kouzine et al., 2013; Naughton et al., 2013). In contrast, merbarone treatment resulted in a specific increase of (−) supercoiling at the promoter that was not observed within the body of the gene (Figure 6C). These results, together with the unchanged TOP1 activity, highlight the particular topological context of merbarone-mediated stimulation of c-FOS transcription.
Based on these results, we considered the possibility that the accumulation of (−) supercoiling at the promoter region could play an active role in the regulation of transcription. In order to tackle this question, we decided to disrupt local promoter supercoiling by the induction of DNA breaks. We individually transfected two different gRNAs in serum-starved RPE-1 Cas9 cells to target a DSB to the c-FOS promoter region and analyzed changes in gene expression with merbarone or serum. Interestingly, in both cases, c-FOS induction was strongly disrupted upon transfection with the targeting gRNAs and not when a non-targeting control was used (Figure 6D). These results further argue against the DSB model of transcriptional stimulation and suggest, instead, that the topological integrity of the promoter region is an essential factor to allow gene expression. One caveat for these experiments, however, is that DSBs are well known in cis repressors of transcription through a canonical γH2AX-dependent DNA damage response (Caron et al., 2019) and could therefore indirectly limit c-FOS expression. We therefore decided to alternatively target a CRISPR-Cas9 D10A nickase that generates single-strand breaks (SSBs) (Chiang et al., 2016), which have not been reported to locally inhibit transcription. Indeed, although transfection with a c-FOS-targeting gRNA in Cas9-expressing cells resulted in a significant increase in the percentage of cells harboring two γH2AX foci, this was not the case in cells expressing the Cas9 D10A variant (Figure S6G). Under these conditions, despite this lack of DDR signaling, we observed a disruption of merbarone-mediated c-FOS induction, which was further enhanced and reached statistical significance when the PARP1 inhibitor PJ34 was used to impair SSB repair (Figure 6E). We conclude that the topological state of the promoter region is important for the transcriptional response of c-FOS. Altogether, our results support the disruption of DNA topology as a key factor driving merbarone-induced transcription upregulation and suggest a tight interdependence between TOP2A and TOP1 activities in order to maintain supercoiling homeostasis and transcriptional control.
Discussion
A model of supercoiling-mediated regulation of promoter-proximal pausing
The results presented here suggest that the removal of (−) supercoiling from gene promoters by TOP2A facilitates promoter-proximal pausing of Pol II and that this is essential to maintain c-FOS and other IEGs under basal repressed conditions. It may seem counterintuitive that a topoisomerase, which relieves torsional stress, acts to negatively regulate transcription. One must bear in mind however that (−) supercoiling is a well-established stimulator of transcription initiation, at least in vitro and in prokaryotic models (Chong et al., 2014; Kim et al., 2019; Parvin and Sharp, 1993; Revyakin et al., 2004; Tabuchi et al., 1993), and has been proposed to operate similarly in yeast and mammalian cells (Baranello et al., 2016; Bermúdez et al., 2010). Thus, (−) supercoiling at promoter regions facilitates transcription initiation by allowing promoter melting and the formation of Pol II open complexes. In this same line, although upstream (−) supercoiling can stall bacterial RNA polymerase (Ma et al., 2013), when encountered downstream, it can favor transcriptional elongation by cancelling the (+) supercoiling generated (Kim et al., 2019). Furthermore, supercoiling has been shown to affect the turnover rate of nucleosomes, binding of chromatin factors, and even the formation of non-B DNA structures with regulatory potential (Dykhuizen et al., 2013; Kaczmarczyk et al., 2020; Kouzine et al., 2004, 2008; Sperling et al., 2011; Teves and Henikoff, 2014), providing different mechanisms by which supercoiling-mediated transcriptional regulation could operate.
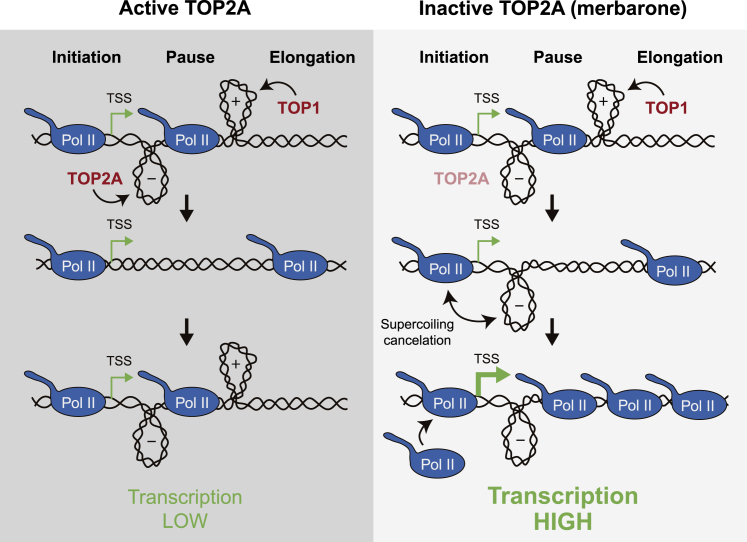
However, the fact that merbarone induces transcription without a concomitant increase in TOP1 activity is more consistent with a direct topological connection. In this context, we propose that by removing (−) supercoiling generated behind advancing Pol II complexes, TOP2A might limit initiation and processivity of subsequent transcriptional cycles, imposing a strong requirement for TOP1-mediated removal of (+) supercoiling ahead of each elongating polymerase (Figure 7). This should be more relevant in the vicinity of the TSS, where TOP1 activity is reduced (Baranello et al., 2016) (Figure 6A), favoring the formation of a (+) topological barrier that reinforces promoter-proximal pausing and maintains transcription under repressed and controlled conditions. Disrupting this balance, either in a regulated manner or by inhibiting TOP2A, would lead to a transcription-dependent accumulation of (−) supercoiling in a topological feedback loop (transcription increases supercoiling and supercoiling increases transcription) that could result in strong transcriptional bursts. In fact, the balance between Top I and gyrase activities in removing (−) and (+) supercoils, respectively, has been shown to regulate transcriptional bursting in bacteria (Chong et al., 2014). It would be extremely relevant to find physiological scenarios in which human cells use this mechanism and control TOP2 activity to regulate transcriptional outbursts.
Figure 7.
A model of supercoiling-mediated regulation of promoter-proximal pausing
Proposed model for transcription regulation through DNA supercoiling. Under basal transcription conditions in the presence of active TOP2A (left), Pol II early elongation generates (+) DNA supercoiling ahead and (−) DNA supercoiling behind. TOP1 removes (+) DNA supercoiling to favor Pol II release from promoter-proximal pausing, and TOP2A removes (−) DNA supercoiling at the promoter, so the topological context is reset for the following initiating polymerase, maintaining transcription under controlled conditions. If TOP2A is inactive (right), (−) DNA supercoiling accumulates at the promoter region and counteracts the (+) DNA supercoiling generated ahead of the next initiating Pol II complex. This supercoiling cancelation allows TOP1-independent advance of Pol II bypassing promotor-proximal pausing. This can be accompanied by increased transcription initiation and/or other modifications at the promoter region that, altogether, increase transcription.
It is worth noting that, mechanistically, under conditions of absent or limited TOP2A-mediated (−) supercoiling removal, multiple Pol II complexes could advance in tandem, taking advantage of (+) and (−) supercoiling cancellation, as recently reported in bacteria (Kim et al., 2019), in “trains” of polymerases in which only the “engine” encounters topological constraints. This way, upregulation of transcription could be achieved by simply adding more “wagons” without additional topological cost. Interestingly, the inherent preference of eukaryotic topoisomerases for removing (+) over (−) supercoils (Fernández et al., 2014; Frøhlich et al., 2007) could extend the supercoiling cancellation effect to favor a cooperative advance of polymerase complexes also during transcription elongation. In any case, the accumulation of (−) supercoiling at promoter regions seems to be a widespread characteristic of eukaryotic gene organization (Bermúdez et al., 2010; Kouzine et al., 2013; Matsumoto and Hirose, 2004; Naughton et al., 2013; Teves and Henikoff, 2014) and could serve additional functions such as, for example, providing an appropriate 3D conformation to favor interaction with regulatory elements (Liu et al., 2001; Racko et al., 2019) or as a means to isolate and maintain the topology of the transcriptional unit (Achar et al., 2020).
It is perhaps difficult to explain that despite a general release from promoter-proximal pausing upon merbarone treatment, similar to that caused by depletion of pausing factors (Chen et al., 2015; Core et al., 2012; Rahl et al., 2010), only a relatively small number of genes is upregulated (Figure 1A). One should bear in mind, however, that pausing of Pol II has been demonstrated to be a necessary step for optimal transcription. In fact, upon depletion of promoter-proximal pausing factors, transcriptional upregulation is not widespread but instead mainly restricted to rapidly inducible genes, like heat-shock or, interestingly, immediate-early genes (Fujita et al., 2009; Gilchrist et al., 2008; Schaukowitch et al., 2014). In this sense, promoter-proximal pausing also indirectly regulates Pol II recruitment to promoters by inhibiting transcription re-initiation (Shao and Zeitlinger, 2017). This might be especially relevant for those highly regulated genes that are characterized by fast and synchronous changes in gene expression under stimulation, in which not only elongation but also initiation has to be rapidly increased. TOP2A inhibition may therefore operate in a similar fashion, with expression levels being significantly altered only in genes, such as IEGs, that are poised for transcription stimulation and lack other repressive mechanisms of regulation. This is in agreement with the increase in promoter occupancy that accompanies c-FOS and EGR1 induction upon merbarone treatment (Figure 5B). The length of the transcriptional unit may also contribute significantly to the effect of TOP2A inhibition on gene expression. Long genes have been reported to be particularly sensitive to the loss of TOP2 activity (Joshi et al., 2012; King et al., 2013). Therefore, the effect of merbarone on stimulating expression might not be observed in long genes that further require TOP2A for efficient transcription elongation. In agreement with this idea, genes upregulated at the mRNA level are significantly shorter than expected (Figure S1C). In summary, we understand that DNA supercoiling is a relevant aspect contributing to transcriptional regulation and may also affect the process at additional levels in addition to promoter-proximal pausing. The relative contribution and interaction between these factors would shape the variable response of individual genes, cell types, and tissues.
TOP2-mediated DSBs and transcription regulation: cause or consequence?
Our model changes the current view of IEG stimulation through the physiological induction of TOP2B-mediated DSBs at gene promoters (Bunch et al., 2015; Madabhushi et al., 2015), an idea that was originally put forward for the regulation of hormone responsive genes (Ju et al., 2006). In contrast, we provide an alternative possibility by which IEG expression is stimulated by (−) DNA supercoiling and actually repressed by the action of TOP2A. Indeed, we present solid evidence demonstrating that neither TOP2A- nor TOP2B-mediated DSBs are necessary for IEG expression. First, we are unable to detect TOP2ccs or DSBs upon merbarone or serum stimulation, neither globally nor specifically at the affected genes, despite a concomitant strong induction of transcription. Second, we show that c-FOS stimulation does not change in cells deleted for the highly TOP2-specialized repair enzyme TDP2. Although alternative TDP2-independent pathways to repair TOP2-induced DSBs exist in cells, they operate at a slower rate and seriously compromise genome integrity (Álvarez-Quilón et al., 2014; Gómez-Herreros et al., 2013) and would therefore inevitably affect the extent and/or kinetics of putative DSB-dependent gene expression. Third, neither TOP2A nor TOP2B deletion reduce c-FOS expression or stimulation with serum, demonstrating the complete dispensability of their activity. As a matter of fact, TOP2A-deleted cells display higher basal c-FOS expression and a more accused serum response, in agreement with its repressive functions. Finally, we show that DSBs targeted to the c-FOS promoter abolish, rather than stimulate, its capacity to respond to merbarone or serum treatments.
Conciliatingly, however, although it is difficult for us to fully explain some of the results that support the TOP2-DSB model, some key observations are actually completely compatible. Thus, a mild (∼2-fold) induction of c-FOS and other IEGs was found at late times of etoposide treatment in neurons (Madabhushi et al., 2015). Indeed, here, we observe a very similar behavior in RPE-1 cells (Figure 1E), but the fact that a much higher and faster induction is observed upon catalytic inhibition (Figures 1C and 1D) strongly suggests the reduction in activity, which also occurs upon etoposide treatment, rather than the generation of TOP2ccs or DSBs, as the molecular trigger of the transcriptional response. Furthermore, the association between transcription and DNA damage is clear and well documented (Gaillard and Aguilera, 2016), and it is plausible that at least part of this can be caused by accidental topoisomerase-mediated DNA breaks (Sun et al., 2020). We therefore favor a scenario in which TOP2-induced DSBs are accidental and a consequence, rather than the cause, of transcriptional upregulation. In support for this scenario, chemical inhibition of transcription elongation dramatically reduces the accumulation of spontaneous DSBs that normally colocalize with TOP2B and Pol II at gene promoters (Dellino et al., 2019).
Conclusions
Our results uncover a layer of transcriptional regulation that operates at the level of promoter-proximal pausing and depends on canonical functions of TOP2A. In addition, our discoveries provide a molecular explanation for the typical bursting behavior of immediate early and potentially other highly regulated genes. Interestingly, this topological control operates at a high hierarchical level and is sufficient to trigger gene expression and overcome other regulatory steps such as signaling cascades, chromatin remodeling, or the recruitment of specific factors. Furthermore, our findings place the functions of TOP2 in transcriptional regulation in a context that, in contrast to previous models based on DSB formation (Ju et al., 2006; Madabhushi et al., 2015), do not entail a risk for genome integrity. In more general terms, this study constitutes a step to understand how TOP2 activity may be used to modulate biological processes in mammalian cells. Future work should be devoted to study the regulation of topoisomerase function and DNA supercoiling in this and other relevant physiological scenarios and how they converge with different aspects of chromatin dynamics to shape genome function, organization, and stability.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rpb1 NTD (D8L4Y) antibody | Cell Signaling Tech. | Cat# 14958; RRID:AB_2687876 |
| Rabbit monoclonal antibody against Topoisomerase II Alpha (Topo IIa)(EP1102Y) | Abcam | Cat# ab52934; RRID:AB_2240762 |
| Anti-Topo IIalpha (F-12) antibody | Santa Cruz Biotech. | Cat# sc-365916; RRID:AB_10842059 |
| TOP2B antibody | Proteintech | Cat# 20549-1-AP; RRID:AB_10700004 |
| Topoisomerase I antibody - ChIP Grade | Abcam | Cat# ab3825; RRID:AB_304095 |
| Topoisomerase I antibody [EPR5375] | Abcam | Cat# ab109374; RRID:AB_10861978 |
| Anti-p38 MAPK, phospho (Thr180 / Tyr182) Antibody, Unconjugated | Cell Signaling Tech. | Cat# 9211; RRID:AB_331641 |
| Anti-α-Tubulin antibody | Sigma-Aldrich | Cat# T9026; RRID:AB_477593 |
| Mouse Anti-Histone H2A.X, phospho (Ser139) Monoclonal antibody, Unconjugated, Clone jbw301 | Millipore | Cat# 05-636; RRID:AB_309864 |
| Rabbit Anti-Histone H3, phospho (Ser10) Mitosismarker Polyclonal antibody, Unconjugated | Millipore | Cat# 06-570; RRID:AB_310177 |
| NELF-E (H-140) antibody | Santa Cruz Biotech. | Cat# sc-32912; RRID:AB_2177858 |
| Bacterial and virus strains | ||
| e. coli: stbl3 competent cells | This paper | N/A |
| e. coli: DH5α competent cells | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| EZ-Link Psoralen-PEG3-Biotin | Thermo Fisher | Cat# 29986 |
| Digitonin | Sigma-Aldrich | Cat# D141 |
| Tn5A Enzyme | CABD | N/A |
| s.p. Cas9 Nuclease 3NLS | Integrated DNA Tech. | Cat# 1074181 |
| Merbarone | Sigma-Aldrich | Cat# M2070 |
| Etoposide | Sigma-Aldrich | Cat# E1383 |
| ICRF-187 | Sigma-Aldrich | Cat# D1446 |
| Camptothecin | Sigma-Aldrich | Cat# C9911 |
| Triptolide | Sigma-Aldrich | Cat#T3652 |
| Critical commercial assays | ||
| DuoLink PLA Kit | Sigma-Aldrich | Cat# DUO92008100RXN |
| RNeasy KIT | QIAGEN | Cat# 74106 |
| TruSeq Stranded mRNA | lllumina | Cat# 20020594 |
| Deposited data | ||
| RNA sequencing Data | This paper | GEO: GSE141800 |
| ChIP-seq data of TOP2A | This paper | GEO: GSE141800 |
| ChIP-seq data of POL2 | This paper | GEO: GSE141800 |
| CTCF ChIP-seq | ENCODE Project Consortium, 2012 | https://www.encodeproject.org/experiments/ENCSR000DVI/ |
| H3K4me3 ChIP-seq | ENCODE Project Consortium, 2012 | https://www.encodeproject.org/experiments/ENCSR000DVK/ |
| H3K27ac ChIP-seq | Sánchez et al., 2018 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2865066 |
| Human reference genome, NCBI build 37, GRCh37/hg19 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/human |
| Unprocessed and uncompressed imaging data (microscopy and blots) | This paper | Mendeley Data at: http://dx.doi.org/10.17632/r69w5y36w2.1 |
| Experimental models: Cell lines | ||
| hTERT-RPE-1 cells | ATCC | CRL-4000 |
| Cas9-overexpressing hTERT RPE-1 | Kindly provided byDr. Durocher Lab | N/A |
| A549 | ATCC | CCL-185 |
| HEK293T | ATCC | CRL-1573 |
| U2OS | ATCC | HTB-96 |
| Mouse Embryonic Fibroblast (MEFs) | This paper | N/A |
| Oligonucleotides | ||
| Primers for qPCR, see Table S1 | This paper | N/A |
| siRNA targeting sequence: NELFE: GGCAUUGCUGGCUCUGAAGUU | Aiyar et al., 2004 | N/A |
| siRNA targeting sequence: LUC: CGUACGCGGAAUACUUCGA | Jimeno-González et al., 2015 | N/A |
| CRISPR-Cas9 tracrRNA | Integrated DNA Tech. | Cat# 1072532 |
| crRNAs targeting sequence: TOP2A:CTCCGCCCAGACACCTACAT | This paper | N/A |
| crRNAs targeting sequence:TOP2B: CTTCGTCCTGATACATATAT | This paper | N/A |
| crRNAs targeting sequence: TDP2:CTTGCTGAGTATCTTCAGAT | This paper | N/A |
| crRNAs targeting sequence: FOS 1#:ACTAGCACTGTTCCTGCGTT | This paper | N/A |
| crRNAs targeting sequence: FOS 2#:CCCTAATTCAGTGCAAAGCG | This paper | N/A |
| crRNAs targeting sequence: Non-targeting | Integrated DNA Tech. | Cat# 1072544 |
| Recombinant DNA | ||
| lentiCas9n(D10A)-Blast plasmid | Sanjana et al., 2014 | Addgene Plasmid #63593 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | N/A |
| R version 3.5.0 | R Core Team, 2018 | https://www.R-project.org/ |
| FASTQC | Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Bowtie version 1.2.0 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| MACS2 version 2.1 | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Felipe Cortés-Ledesma (fcortes@cnio.es).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
Data are available in the main text, supplementary materials and auxiliary files. The accession number for all the sequencing data generated in this manuscript is GEO: GSE141800.
Experimental model and subject details
Cell lines and bacterial strains
hTERT RPE-1 cells (ATCC), a near-diploid human cell line of female origin, were cultured in Dubelcco’s Modified Eagle’s Medium (DMEM) F-12 (Sigma) supplemented with 50 units ml-1 penicillin, 50 units ml-1 streptomycin and 10% Fetal Bovine Serum (FBS) (Sigma) at 37°C in 5% CO2 atmosphere. RPE-1 cells were serum starved by 48h incubation in the same medium, but with reduced 0.1% FBS content. Primary MEFs were isolated at day 13 p.c. and cultured in DMEM with 50 units ml-1 penicillin, 50 units ml-1 streptomycin, 15% FBS and non-essential amino acids at 37°C in 5% CO2 and 3% O2 atmosphere. HEK293T, U2OS and A549 were cultured in DMEM with 50 units ml-1 penicillin, 50 units ml-1 streptomycin and 10% FBS at 37°C in 5% CO2 atmosphere. For the generation of knockout cell lines, Cas9-overexpressing hTERT RPE-1 cell line (kindly provided by Dr. Durocher) was used. Cas9 D10A-overexpressing hTERT RPE-1 cell line was generated using lentiviral particles encoding the Cas9 D10A gene, previously produced by calcium phosphate transfection as describe (Salmon and Trono, 2006). In brief, HEK293T were transfected with a 3:2:1 mixture composed of lentiCas9n(D10A)-Blast plasmid (Addgene # 63593), p8.91 and pVSVG (Packaging plasmids), using 128 μM CaCl and 1xHBS. After 48 hours, the medium was filtered through a 0.45 μm polyvinylidene difluoride (PVDF) filter (SLHV035RS, Millex-HV, Millipore). Then, viral particles were concentrated by centrifugation for 90 minutes at 22000 rpm at 4°C and stored at −80°C. The presence of mycoplasma was routinely checked with MycoAlert PLUS Mycoplasma Detection Kit (Lonza). E.coli strains (DH5α and Stlb3) were grown in Luria-Bertani (LB) media with corresponding antibiotics at 37°C.
Method details
Knock-out cell line generation
Cas9-overexpressing hTERT RPE-1 cell line was transfected with the corresponding gRNAs or non-targeting gRNA as a negative control (Key resources table), using Lipofectamine RNAiMAX (Thermo Fisher, 13778500), following the protocol provided by the manufacturer. Editing efficiency of all gRNAs was validated by in-del analysis of PCR sanger sequencing using TIDE (Brinkman et al., 2014). TOP2B−/− and TOP2A−/− cell lines were generated as a pool. TDP2−/− clones were obtained by limited dilution plating in a 96-well plate.
Antibodies
For ChIP-seq we have used anti-Rpb1-NTD (Cell Signaling, D8L4Y), anti-TOP2A (Abcam, ab52934). For ICE, anti-TOP2B (Proteintech, 20549-1-AP), anti-TOP2A (Santa Cruz, SC-365916), anti-TOP1 (Abcam, ab3825). For ICE-IP, anti-TOP2A (abcam, ab52934), anti-TOP1 antibody (abcam, ab109374). For western blot analysis, anti-TOP2B (Proteintech, 20549-1-AP), anti-TOP2A (Santa Cruz, SC-365916), anti-p-p38 (Cell Signaling, 9211) and anti-tubulin (Sigma, T9026), and as secondary antibodies IRDye 680-labeled anti-mouse (LI-COR Biosciences, 926-68070) and IRDye 800-labeled anti-rabbit (LI-COR BIOSCIENCES, 926-32211).
For immunofluorescence, anti-γH2AX (Millipore, 05-636) and anti-H3S10p (Millipore, 06-570). For PLA, anti-NELF-E (Santa Cruz, sc32912), anti-TOP2A (Santa Cruz, sc-365916), and as secondary antibodies Alexa Fluor 488 anti-mouse (Jackson, 715-545-150) and Alexa Fluor 594 anti-rabbit (Jackson, 111-585-003).
RNA analysis and RNA-seq
Serum-starved RPE-1 cells grown on 60mm plates were treated as required and total RNA was isolated with the RNeasy kit (QIAGEN, 74106), following instructions from the manufacturer. Primers used are described in Table S1. Values were normalized to the expression of GAPDH housekeeping gene. For RNA-seq, total RNA (150ng) cDNA libraries were prepared using TruSeq Stranded mRNA (lllumina). Library size distribution was analyzed with Bioanalyzer DNA high-sensitive chip and Qubit. 1.4pM of each library was sequenced in NextSeq 500 HIGH-Output.
Isolation of chromatin-associated RNA
Isolation of chromatin-associated RNA was performed as previously described with minor modifications (Conrad and Ørom, 2017). Approximately 5x10ˆ6 cells were washed with ice-cold PBS twice and scrapped. The cells were lysed with 400μl ice-cold lysis buffer (10 mM Tris-Hcl pH 7.5, 150 mM NaCl, 0.15% IGEPAL, 20U/μl SUPERSase-IN, 1x proteinase inhibitor Complete (Roche)) and incubated on ice for 5 min. After the incubation, 1 mL of ice-cold sucrose buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 24% Sucrose, 20U/μl SUPERSase-IN, 1x proteinase inhibitor Complete (Roche)) was under-laid and then the nuclei were collected under 16000 g centrifuge at 4C for 10 min. Isolated nuclei were resuspended in 250μl of ice-cold glycerol buffer (20 mM Tris-Hcl l pH 7.5, 75mM NaCl, 0.5 mM EDTA, 20U/μl SUPERSase-IN, 1x proteinase inhibitor Complete (Roche)) followed by 250μl of ice-cold nuclear lysis buffer (10 mM Tris-Hcl pH 7.5, 7.5 mM MgCl2, 0.2 mM EDTA, 300 mM NaCl, 1 M Urea, 1% IGEPAL, 20U/μl SUPERSase-IN, 1x proteinase inhibitor Complete (Roche)). Two minutes incubation was carried out on ice with mixing by max speed vortex for 5 s every minute and then chromatin pellets were precipitated under 13,000 g centrifuge at 4C for 2 min. Chromatin pellets were resuspended in 1ml of Trizol reagent, and RNA isolation was performed following manufacturer’s instructions.
Western blot analysis
For protein extractions, cell pellets were resuspended in RIPA buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40 y 1% sodium deoxycholate) supplemented with protease inhibitors and incubated on ice for 30 min with constant agitation. The lysate was then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was sonicated using a Bioruptor (Diagenode, UCD-200) for 1 cycle of 3 minutes (high power, 30 s on, 30 s off). Protein concentration was determined by Bradford assay (Applied Biochem, A6932). 20 μg of protein was loaded into home-made 10% polyacrylamide gel with SDS or 4%–20% Mini-PROTEAN tris-Glycine Precast Protein Gels (Biorad, 4561096) and electroblotted onto Immobilon-FL Transfer Membranes (Millipore), after 5 minutes methanol activation. Membranes were then blocked in Odyssey Blocking Buffer (LI-COR Biosciences, 927-40000) for 1 hour and then probed with required primary antibodies for 2 hours. Membranes were washed with 0.1% Tween-20 - Odyssey Blocking Buffer and incubated with corresponding secondary antibodies (conc: 1:10.000) for 1 hour and finally, membranes were washed again with 0.1% Tween-20 - Odyssey Blocking Buffer. Membrane were analyzed using Odyssey CLx and ImageStudio Odyssey CLx Software (LI-COR BIOSCIENCES, Lincoln, NE) according to the manufacturer’s protocols.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation was performed as previously described (Jimeno-González et al., 2015). Briefly, serum-starved RPE-1 cells were crosslinked with 1% formaldehyde for 10 minutes at 37°C. Crosslinking reaction was quenched with 125mM glycine for 5 minutes. Cell pellets were resuspended in 2.5 mL lysis buffer A (5 mM Pipes pH 8.0, 85 mM KCl, 0.5% NP40) supplemented with protease inhibitors and incubated for 10 minutes on ice. Chromatin was obtained by centrifugation at 4000 rpm for 5 minutes at 4°C. Nuclear fraction was resuspended in 1 mL of lysis buffer B (50 mM Tris HCl pH 8.1, 1% SDS, 10 mM EDTA, 1 mM PMSF) supplemented with protease inhibitors. Chromatin was sonicated using a Bioruptor (Diagenode, UCD-200), 10 cycles of 30” sonication (high level) and 30” of pause on ice-cold water. 50 μL of sonicated chromatin was reverse-crosslinked using Proteinase K in PK buffer (0.5%SDS, 50mM Tris-Cl, 100mM NaCl, 1mM EDTA) at 65°C overnight. After phenol chloroform extraction, DNA fragmentation was analyzed on 1.2% agarose gel. For each inmunoprecipitation, 20 μg of chromatin and 4 μg of the specific antibody was used in IP buffer (0,1% SDS, 1% TX-100, 2mM EDTA, 20 mM TrisHCl pH8, 150 mM NaCl) at 4°C o/n and then with 40 μL of pre-blocked (1 mg/ml BSA) Dynabeads protein A (ThermoFisher). Beads were sequentially washed with Wash buffer 1 (20 mM Tris HCl pH 8.1, 0.1% SDS, 1% Triton x-100, 2 mM EDTA, 150 mM NaC), Wash buffer 2 (20 mM Tris HCl pH 8.1, 0.1% SDS, 1% Triton x-100, 2 mM EDTA, 500 mM NaCl) Wash buffer 3 (10 mM Tris HCl pH 8.1, 1% NP-40, 1% NaDoc, 1 mM EDTA, 250 mM LiCl), and twice with TE-buffer (10 mM Tris-HCL pH8, 1 mM EDTA pH8). ChIPmentation was carried out as previously described (Schmidl et al., 2015), using Tn5A Enzyme provided by the Proteomic Service of CABD (Centro Andaluz de Biología del Desarrollo). Tagmented DNA was then eluted with 1% SDS in TE at 65°C for 10 minutes and protein was degraded with Proteinase K for 2 hours at 37°C. DNA was purified using QIAGEN PCR purification Kit (QIAGEN, 28106). Libraries were amplified for N-1 cycles (being N the optimum Cq determined by qPCR reaction) using NEBNext High-Fidelity Polymerase (New England Biolabs, M0541). Libraries were purified with Sera-Mag Select Beads (GE Healthcare, 29343052) and sequenced using Illumina NextSeq 500 and single-end configuration.
In vivo Complex of Enzyme (ICE) and ICE-IP
DNA topoisomerase cleavage complexes were analyzed as previously described (Schellenberg et al., 2017). For the induction of cleavage complexes, serum-starved RPE-1 cells were treated as required followed by 400 μM Etoposide (Sigma, E1383), 10 μM Camptothecin (CPT, C9911) (Sigma) or DMSO vehicle (Applichem, A1584) for 5 minutes. Cells were immediately lysed using 1% (w/v) N-Lauroylsarcosine sodium salt (Sigma-Aldrich, L7414) in TE buffer supplemented with protease inhibitors. After homogenization, 0.67 g/ml CsCl (Applichem-Panreac, A1098) was added and lysated were then centrifuged at 57,000 rpm for 20 h at 25°C using 3.3 mL 13 × 33 polyallomer Optiseal tubes (Beckman Coulter) in a TLN100 rotor (Beackman Coulter).
For ICE-IP, 40 μg of ICE material was digested overnight with 0.8 U/μl PstI (NEB, R0140). Samples were incubated at 80°C for 20min to inactivate PstI and then diluted 1/10 in IP buffer (0.1% SDS, 1% TX-100, 2mM EDTA, 20 mM TrisHCl pH8, 150 mM NaCl). Samples were then incubated overnight at 4°C with 4 μg of the required primary antibody and then with 40 μL of pre-blocked (1 mg/ml BSA) Dynabeads protein A (Thermo Fisher). IPs were incubated for 2 hours at RT. Beads were then washed with Wash solution 1 (20 mM Tris HCl pH 8.1, 0.1% SDS, 1% Triton x-100, 2 mM EDTA, 150 mM NaCl), Wash solution 2 (20 mM Tris HCl pH 8.1, 0.1% SDS, 1% Triton x-100, 2 mM EDTA, 500 mM NaCl), Wash solution 3 (10 mM Tris HCl pH 8.1, 1% NP-40, 1% NaDoc, 1 mM EDTA, 250 mM LiCl), and finally with TE. DNA was then eluted with 1% SDS in TE at 65°C for 10 minutes and protein was degraded with Proteinase K for 2 hours at 37°C. Finally, DNA was purified using QIAGEN PCR purification Kit (28106, QIAGEN) and analyzed by qPCR.
Immunofluorescence
Serum-starved RPE-1 cells grown on coverslips were fixed with 4% PFA-PBS for 10 minutes at RT. Immunofluorescence was carried out as previously described (Álvarez-Quilón et al., 2014). In brief, after permeabilization (2 minutes in PBS-0.2% Triton X-100), cells were blocked with 5% BSA-PBS for 30 minutes and incubated with the required primary antibodies in 1% BSA-PBS for 1 hour. Cells were then washed (three times in PBS-0.1% Tween 20) and incubated with the corresponding AlexaFluor-conjugated secondary antibodies (1/1,000 dilution in 1% BSA-PBS) for 30 minutes and washed again. Finally, samples were counterstained with DAPI (Sigma, D9542) and mounted in Vectashield (Vector Labs). γH2AX foci per cell (40 cells per condition and experimental repeat) were manually counted (double-blind). pH3S10 signal was quantified with Metamorph software (100 cells per condition and experimental repeat).
Proximity Ligation Assay (PLA)
U2OS cells were fixed in 4% PFA for 10 min. DuoLink PLA Kit (Sigma-Aldrich, DUO92008100RXN) was used following the protocol from the manufacturer. Foci per cell (40 cells per condition and experimental repeat) were manually counted (double-blind).
Biotin-Psoralen-incorporation assay
Biotinylated-psoralen (bio-psoralen) incorporation was measured as previously described (Naughton et al., 2013) with minor modifications. Briefly, serum-starved RPE-1 cells were treated as required for 10 minutes prior to the addition of 20μM EZ-Link Psoralen-PEG3-Biotin (Thermo, 29986) and 0.01% digitonin (Sigma, D141) for 5 minutes at 37°C in the dark to improve cellular uptake of bio-psoralen (Aw et al., 2016). Bio-psoralen was cross-linked to DNA with 360 nm UV irradiation for 20 minutes on ice. DNA was purified using Proteinase K in PK buffer (0.5%SDS, 50mM Tris-Cl, 100mM NaCl, 1mM EDTA) at 65°C overnight, followed by phenol:chloroform:isoamylalcohol extraction. After RNase treatment and phenol:chloroform:isoamylalcohol extraction, DNA was fragmented by sonication using Bioruptor (Diagenode, UCD-200), 10 cycles of 30” sonication (high level) and 30” of pause on ice-cold water. The Biotinylated-psoralen DNA complex in TE was incubated with avidin conjugated to magnetic beads (Thermo, 6560) for 2 hours at room temperature, and then overnight at 4°C. Beads were washed sequentially for 15 minutes each at room temperature with Wash solution 1 (20 mM Tris pH 8.1, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 0.1% SDS), Wash solution 2 (20 mM Tris pH 8.1, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100 and 0.1% SDS) and Wash solution 3 (10 mM Tris pH 8.1, 0.25 M LiCl, 1 mM EDTA, 1% NP40 and 1% deoxycholate). Beads were then washed twice with TE for 15 minutes. To extract DNA and to release bio-psoralen adducts, samples were treated for 30 mins at 90°C in 50 μL 0.1N KOH (Yeung et al., 1988). Samples were neutralized with 0.5M sodium acetate, pH7, then brought to 500 μL with mQ water and analyzed by qPCR.
High-throughput sequencing analysis
Sequence reads were demultiplexed, quality filtered with FastQC (Andrews, 2010) and mapped to the human genome (hg19) using Bowtie 1.2 (Langmead et al., 2009). We used option “-m 1” to retain those reads that map only once to the genome. Each individual sample contributed with the same number of reads in the ChIP-seq final merged sample. For the computation of ChIP-seq binding sites (peaks), we used MACS2 (Zhang et al., 2008) with option “-q 0.01.” We used the R package DESeq2 (Love et al., 2014) to identify differentially expressed genes from RNA-seq data following authors guidelines. Only genes with associated adjusted p values ≤ 0.05 and absolute fold change ≥ 2 were considered as differentially expressed.For the identification of Pol II differentially bound genes, we followed the same approach than for differentially expressed genes, this time by restricting the gene length to the region stretching 2kb from 500 bp downstream the TSS.
To estimate the level of Pol II recruitment, we used the so-called “pausing index” (PI) and “pause release ratio” (PRR). PI is defined as the ratio of Pol II enrichment within the promoter to that in the gene body, while PRR is an inverse of PI that restricts the gene body to the first 2kb downstream the TSS. For the estimation of both parameters, we based our strategies on Chen et al. (2015) and Day et al. (2016). To calculate PI, the level of Pol II within the promoter was computed as the sum of ChIP-seq reads in 400 bp surrounding the TSS. The level of Pol II within the gene was computed as the average number of reads in 400 bp windows throughout the gene body, from 200 bp downstream the TSS. Finally, the PI was estimated as the ratio of both measures. To calculate PRR, Pol II level within the promoter was estimated as the sum of ChIP-seq reads from 100 bp upstream to 300 bp downstream of the TSS. Gene body Pol II level was computed as the sum of reads within the region stretching from 300 bp downstream of the TSS to 2 kb downstream of the TSS. After normalizing each value by the corresponding window sizes, the PRR was estimated as the level of Pol II within the gene body divided by the level of Pol II within the promoter.
For Gene-filtering, we started from the whole set of protein-coding transcripts associated to Ensembl-annotated genes (GRCh37, release 75), from which we kept only those having a peak of Pol II (see peak calling section) overlapping with the region from 0 to 500 bp downstream of the TSS. If several transcripts of the same gene were found to match this condition, the one whose TSS was closest to the Pol II peak was selected. In order to account for potential false negatives when calling Pol II peaks, some genes with no associated Pol II peak were also selected if the window from 0 to 500 bp downstream of the TSS was found to: 1) have a value of Pol II reads per million (RPM) larger than 0.5, or 2) have a value longer than 0.5 in the difference of H3K4me3 RPM and the corresponding input. Finally, genes that were closer than 1kb of another gene or were smaller than 2kb were excluded. We ended up keeping 9,588 human genes (∼42% of total).
Regulatory regions, namely enhancers, promoters, and insulators, were defined as follows. For promoters, the whole set of transcripts associated to Ensembl-annotated genes were considered. Then, promoters were defined as ± 1 kb from the TSSs. Enhancers were defined as H3K27ac peaks not overlapping with a promoter, and insulators as CTCF peaks not overlapping with promoters and enhancers.
ChIP-seq averaged reads around TSSs were computed using the R package bamsignals (Mammana and Helmuth, 2019) and smoothed using a Gaussian smoothing kernel with the R function ksmooth, respectively. To generate the profile of TOP2A ChIP-seq signal around randomized genes, the 148 upregulated genes upon 2h of merbarone treatment were first considered. Then, 50 sets of 148 genes were randomly selected from the human genome and TOP2A ChIP-seq reads were counted around the TSSs. Finally, the median of such read counts was smoothed and plotted.
Publicly available sequencing data used in this study include ChIP-seq of several proteins and post-translational modifications: CTCF (ENCSR000DVI), H3K4me3 (ENCSR000DVK) and H3K27ac (Sanchez et al., 2018). CTCF and H3K4me3 BAM files (hg19) were batch-downloaded from ENCODE. H3K27ac and H3K27me3 raw sequencing files were processed as described above (Langmead et al., 2009).
The R package TopGO (Alexa and Rahnenfuhrer, 2019) was used to calculate the significance of Gene Ontology (GO) terms associated to differentially expressed genes after merbarone treatment. We computed enrichments using the Fisher’s exact test and the default algorithm (weight01), which is a hybrid between the ‘elim’ and the ‘weight’ algorithms described (Alexa et al., 2006). To perform hypergeometric-based tests, there is a need for defining a ‘gene universe’ (which can be conceptualized as the number of balls in an urn) and a set of ‘interesting genes’ from that universe. To define the gene universe, we started from expressed genes, which were defined as genes for which the sum of RNA-seq reads (combining the replicates) overlapping exons was larger than 10. Then, the gene universe was defined as expressed genes mapping to at least one GO term and the set of interesting genes as differentially expressed genes upon merbarone treatment.
Quantification and statistical analysis
Statistical analyses were carried out using GraphPad prism 8 software. Test methods are described in figure legends. All p values were two-tailed otherwise indicated, and p < 0.05 was considered significant.
Acknowledgments
We thank the Genomics Core facilities at CABIMER and the EMBL (GeneCore) for the generation of the high-thoughput sequencing data and O. Fernández-Capetillo for comments. Computational analyses were run on the High Perfomance Computing cluster provided by the Centro Informático Científico de Andalucía (CICA). This work was funded with grants from the Spanish and Andalusian governments (SAF2017-89619-R, CVI-7948, and European Regional Development Fund) and the European Research Council (ERC-CoG-2014-647359), and with individual fellowships for A.H.-R. (Contratos para la Formación de Doctores, BES-2015-071672, and Ministerio de Economía y Competitividad); S.J.-G. (Ramón y Cajal, RYC-2015-17246, and Ministerio de Economía y Competitividad); J.T.-B. (Formación Profesorado Universitario, FPU15/03656, and Ministerio de Educación, Cultura y Deporte); and G.M.-Z. (AECC Postdoctoral Fellowships). CABIMER is supported by the Andalusian Government.
Author contributions
A.H.-R., S.J.-G., and F.C.-L. conceived and designed the project. A.H.-R., P.M.M.-G., S.J.-G., J.T.-B., J.A.L., and G.M.-Z. performed the experiments and analyzed the results. A.H.-R., S.J.-G., and F.C.-L. wrote the manuscript. All authors read, discussed, and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108977.
Contributor Information
Silvia Jimeno-González, Email: silvia.jimeno@cabimer.es.
Felipe Cortés-Ledesma, Email: fcortes@cnio.es.
Supplemental information
Differential gene expression values for each gen (adjusted p value and fold change) in hTERT-RPE-1 cells upon 30 min (versus DMSO treatment) and 2 h (versus untreated) treatment with 200 μM Merb. List of all differentially expressed (DE), as well as UP and DOWN regulated upon 2 h treatment. List of all genes displaying differential Pol II binding (ChIP-seq) in hTERT-RPE-1 cells treated for 1 h (versus untreated) with 200 μM Merb.
References
- Achar Y.J., Adhil M., Choudhary R., Gilbert N., Foiani M. Negative supercoil at gene boundaries modulates gene topology. Nature. 2020;577:701–705. doi: 10.1038/s41586-020-1934-4. [DOI] [PubMed] [Google Scholar]
- Adelman K., Lis J.T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar S.E., Sun J.L., Blair A.L., Moskaluk C.A., Lu Y.Z., Ye Q.N., Yamaguchi Y., Mukherjee A., Ren D.M., Handa H. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexa A., Rahnenfuhrer J. Bioconductor; 2019. TopGO: Enrichment Analysis for Gene Ontology. [Google Scholar]
- Alexa A., Rahnenführer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Álvarez-Quilón A., Serrano-Benítez A., Lieberman J.A., Quintero C., Sánchez-Gutiérrez D., Escudero L.M., Cortés-Ledesma F. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat. Commun. 2014;5:3347. doi: 10.1038/ncomms4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Quilón A., Terrón-Bautista J., Delgado-Sainz I., Serrano-Benítez A., Romero-Granados R., Martínez-García P.M., Jimeno-González S., Bernal-Lozano C., Quintero C., García-Quintanilla L., Cortés-Ledesma F. Endogenous topoisomerase II-mediated DNA breaks drive thymic cancer predisposition linked to ATM deficiency. Nat. Commun. 2020;11:910. doi: 10.1038/s41467-020-14638-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc FastQC: a quality control tool for high throughput sequence data.
- Aoi Y., Smith E.R., Shah A.P., Rendleman E.J., Marshall S.A., Woodfin A.R., Chen F.X., Shiekhattar R., Shilatifard A. NELF Regulates a Promoter-Proximal Step Distinct from RNA Pol II Pause-Release. Mol. Cell. 2020;78:261–274.e5. doi: 10.1016/j.molcel.2020.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin C.A., Lee K.C., Swan R.L., Khazeem M.M., Manville C.M., Cridland P., Treumann A., Porter A., Morris N.J., Cowell I.G. TOP2B: The First Thirty Years. Int. J. Mol. Sci. 2018;19:2765. doi: 10.3390/ijms19092765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw J.G., Shen Y., Wilm A., Sun M., Lim X.N., Boon K.L., Tapsin S., Chan Y.S., Tan C.P., Sim A.Y. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell. 2016;62:603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- Bahrami S., Drabløs F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016;62:37–49. doi: 10.1016/j.jbior.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H. RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermúdez I., García-Martínez J., Pérez-Ortín J.E., Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H., Lawney B.P., Lin Y.F., Asaithamby A., Murshid A., Wang Y.E., Chen B.P., Calderwood S.K. Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 2015;6:10191. doi: 10.1038/ncomms10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A., Maman Y., Jung S., Wong N., Callen E., Day A., Kieffer-Kwon K.R., Pekowska A., Zhang H., Rao S.S.P. Genome Organization Drives Chromosome Fragility. Cell. 2017;170:507–521.e18. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canela A., Maman Y., Huang S.N., Wutz G., Tang W., Zagnoli-Vieira G., Callen E., Wong N., Day A., Peters J.M. Topoisomerase II-Induced Chromosome Breakage and Translocation Is Determined by Chromosome Architecture and Transcriptional Activity. Mol. Cell. 2019;75:252–266.e8. doi: 10.1016/j.molcel.2019.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P., van der Linden J., van Attikum H. Bon voyage: A transcriptional journey around DNA breaks. DNA Repair (Amst.) 2019;82:102686. doi: 10.1016/j.dnarep.2019.102686. [DOI] [PubMed] [Google Scholar]
- Chen F.X., Woodfin A.R., Gardini A., Rickels R.A., Marshall S.A., Smith E.R., Shiekhattar R., Shilatifard A. PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell. 2015;162:1003–1015. doi: 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang T.W., le Sage C., Larrieu D., Demir M., Jackson S.P. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci. Rep. 2016;6:24356. doi: 10.1038/srep24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S., Chen C., Ge H., Xie X.S. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T., Ørom U.A. Cellular Fractionation and Isolation of Chromatin-Associated RNA. Methods Mol. Biol. 2017;1468:1–9. doi: 10.1007/978-1-4939-4035-6_1. [DOI] [PubMed] [Google Scholar]
- Core L., Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Waterfall J.J., Gilchrist D.A., Fargo D.C., Kwak H., Adelman K., Lis J.T. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–1035. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless S., Gilbert N. Effects of DNA supercoiling on chromatin architecture. Biophys. Rev. 2016;8:51–64. doi: 10.1007/s12551-016-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes Ledesma F., El Khamisy S.F., Zuma M.C., Osborn K., Caldecott K.W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- Day D.S., Zhang B., Stevens S.M., Ferrari F., Larschan E.N., Park P.J., Pu W.T. Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types. Genome Biol. 2016;17:120. doi: 10.1186/s13059-016-0984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino G.I., Palluzzi F., Chiariello A.M., Piccioni R., Bianco S., Furia L., De Conti G., Bouwman B.A.M., Melloni G., Guido D. Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations. Nat. Genet. 2019;51:1011–1023. doi: 10.1038/s41588-019-0421-z. [DOI] [PubMed] [Google Scholar]
- Drake F.H., Hofmann G.A., Mong S.M., Bartus J.O., Hertzberg R.P., Johnson R.K., Mattern M.R., Mirabelli C.K. In vitro and intracellular inhibition of topoisomerase II by the antitumor agent merbarone. Cancer Res. 1989;49:2578–2583. [PubMed] [Google Scholar]
- Dujardin G., Lafaille C., de la Mata M., Marasco L.E., Muñoz M.J., Le Jossic-Corcos C., Corcos L., Kornblihtt A.R. How slow RNA polymerase II elongation favors alternative exon skipping. Mol. Cell. 2014;54:683–690. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- Dykhuizen E.C., Hargreaves D.C., Miller E.L., Cui K., Korshunov A., Kool M., Pfister S., Cho Y.J., Zhao K., Crabtree G.R. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández X., Díaz-Ingelmo O., Martínez-García B., Roca J. Chromatin regulates DNA torsional energy via topoisomerase II-mediated relaxation of positive supercoils. EMBO J. 2014;33:1492–1501. doi: 10.15252/embj.201488091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune J.M., Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIalpha by blocking DNA cleavage. J. Biol. Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- Frøhlich R.F., Veigaard C., Andersen F.F., McClendon A.K., Gentry A.C., Andersen A.H., Osheroff N., Stevnsner T., Knudsen B.R. Tryptophane-205 of human topoisomerase I is essential for camptothecin inhibition of negative but not positive supercoil removal. Nucleic Acids Res. 2007;35:6170–6180. doi: 10.1093/nar/gkm669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Piuz I., Schlegel W. Negative elongation factor NELF controls transcription of immediate early genes in a stimulus-specific manner. Exp. Cell Res. 2009;315:274–284. doi: 10.1016/j.yexcr.2008.10.032. [DOI] [PubMed] [Google Scholar]
- Gaillard H., Aguilera A. Transcription as a Threat to Genome Integrity. Annu. Rev. Biochem. 2016;85:291–317. doi: 10.1146/annurev-biochem-060815-014908. [DOI] [PubMed] [Google Scholar]
- Gilchrist D.A., Nechaev S., Lee C., Ghosh S.K., Collins J.B., Li L., Gilmour D.S., Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittens W.H., Johnson D.J., Allison R.M., Cooper T.J., Thomas H., Neale M.J. A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat. Commun. 2019;10:4846. doi: 10.1038/s41467-019-12802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Herreros F., Romero-Granados R., Zeng Z., Alvarez-Quilón A., Quintero C., Ju L., Umans L., Vermeire L., Huylebroeck D., Caldecott K.W., Cortés-Ledesma F. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013;9:e1003226. doi: 10.1371/journal.pgen.1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothe H.J., Bouwman B.A.M., Gusmao E.G., Piccinno R., Petrosino G., Sayols S., Drechsel O., Minneker V., Josipovic N., Mizi A. Spatial Chromosome Folding and Active Transcription Drive DNA Fragility and Formation of Oncogenic MLL Translocations. Mol. Cell. 2019;75:267–283.e12. doi: 10.1016/j.molcel.2019.05.015. [DOI] [PubMed] [Google Scholar]
- Greenberg M.E., Ziff E.B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner M.C., Aryee M.J., Toubaji A., Esopi D.M., Albadine R., Gurel B., Isaacs W.B., Bova G.S., Liu W., Xu J. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- Healy S., Khan P., Davie J.R. Immediate early response genes and cell transformation. Pharmacol. Ther. 2013;137:64–77. doi: 10.1016/j.pharmthera.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno-González S., Ceballos-Chávez M., Reyes J.C. A positioned +1 nucleosome enhances promoter-proximal pausing. Nucleic Acids Res. 2015;43:3068–3078. doi: 10.1093/nar/gkv149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R.S., Piña B., Roca J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res. 2012;40:7907–7915. doi: 10.1093/nar/gks626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B.G., Lunyak V.V., Perissi V., Garcia-Bassets I., Rose D.W., Glass C.K., Rosenfeld M.G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kaczmarczyk A., Meng H., Ordu O., Noort J.V., Dekker N.H. Chromatin fibers stabilize nucleosomes under torsional stress. Nat. Commun. 2020;11:126. doi: 10.1038/s41467-019-13891-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Beltran B., Irnov I., Jacobs-Wagner C. Long-Distance Cooperative and Antagonistic RNA Polymerase Dynamics via DNA Supercoiling. Cell. 2019;179:106–119.e16. doi: 10.1016/j.cell.2019.08.033. [DOI] [PubMed] [Google Scholar]
- King I.F., Yandava C.N., Mabb A.M., Hsiao J.S., Huang H.S., Pearson B.L., Calabrese J.M., Starmer J., Parker J.S., Magnuson T. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner A., Wu W., Staudt C., Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F., Liu J., Sanford S., Chung H.J., Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat. Struct. Mol. Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F., Levens D., Baranello L. DNA topology and transcription. Nucleus. 2014;5:195–202. doi: 10.4161/nucl.28909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., Fuda N.J., Core L.J., Lis J.T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Gilmour D.S. Promoter proximal pausing and the control of gene expression. Curr. Opin. Genet. Dev. 2011;21:231–235. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Bondarenko V., Ninfa A., Studitsky V.M. DNA supercoiling allows enhancer action over a large distance. Proc. Natl. Acad. Sci. USA. 2001;98:14883–14888. doi: 10.1073/pnas.261477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Y.L., Lin C.P., Azarova A.M., Cai L., Wang J.C., Liu L.F. Role of topoisomerase IIbeta in the expression of developmentally regulated genes. Mol. Cell. Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wang M.D. DNA supercoiling during transcription. Biophys. Rev. 2016;8:75–87. doi: 10.1007/s12551-016-0215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bai L., Wang M.D. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R., Gao F., Pfenning A.R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T.X., Yamakawa H., Pao P.C. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammana A., Helmuth J. Bioconductor; 2019. Bamsignals: Extract read count signals from bam files. R packag. version 1.16.0. [Google Scholar]
- Matsumoto K., Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J. Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- Mondal N., Parvin J.D. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- Naughton C., Avlonitis N., Corless S., Prendergast J.G., Mati I.K., Eijk P.P., Cockroft S.L., Bradley M., Ylstra B., Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat. Struct. Mol. Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J.L., Soans E., Rogojina A., Seth A., Mishina M. Topoisomerase assays. Curr. Protoc. Pharmacol. 2012;Chapter 3:Unit 3.3. doi: 10.1002/0471141755.ph0303s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T., Bonhomme C., Chen D., Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- Parvin J.D., Sharp P.A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Pastor N., Domínguez I., Orta M.L., Campanella C., Mateos S., Cortés F. The DNA topoisomerase II catalytic inhibitor merbarone is genotoxic and induces endoreduplication. Mutat. Res. 2012;738–739:45–51. doi: 10.1016/j.mrfmmm.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Pedersen J.M., Fredsoe J., Roedgaard M., Andreasen L., Mundbjerg K., Kruhøffer M., Brinch M., Schierup M.H., Bjergbaek L., Andersen A.H. DNA Topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS Genet. 2012;8:e1003128. doi: 10.1371/journal.pgen.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Sun Y., Huang S.N., Nitiss J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2018. [Google Scholar]
- Racko D., Benedetti F., Dorier J., Stasiak A. Are TADs supercoiled? Nucleic Acids Res. 2019;47:521–532. doi: 10.1093/nar/gky1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A., Ebright R.H., Strick T.R. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc. Natl. Acad. Sci. USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon P., Trono D. Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. 2006;Chapter 4:Unit 4.21. doi: 10.1002/0471142301.ns0421s37. [DOI] [PubMed] [Google Scholar]
- Sanchez G.J., Richmond P.A., Bunker E.N., Karman S.S., Azofeifa J., Garnett A.T., Xu Q., Wheeler G.E., Toomey C.M., Zhang Q. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res. 2018;46:1756–1776. doi: 10.1093/nar/gkx1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaukowitch K., Joo J.Y., Liu X., Watts J.K., Martinez C., Kim T.K. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg M.J., Lieberman J.A., Herrero-Ruiz A., Butler L.R., Williams J.G., Muñoz-Cabello A.M., Mueller G.A., London R.E., Cortés-Ledesma F., Williams R.S. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science. 2017;357:1412–1416. doi: 10.1126/science.aam6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C., Rendeiro A.F., Sheffield N.C., Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciascia N., Wu W., Zong D., Sun Y., Wong N., John S., Wangsa D., Ried T., Bunting S.F., Pommier Y., Nussenzweig A. Suppressing proteasome mediated processing of topoisomerase II DNA-protein complexes preserves genome integrity. eLife. 2020;9:e53447. doi: 10.7554/eLife.53447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017;49:1045–1051. doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- Sperling A.S., Jeong K.S., Kitada T., Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. USA. 2011;108:12693–12698. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian D., Kraut E., Staubus A., Young D.C., Muller M.T. Analysis of topoisomerase I/DNA complexes in patients administered topotecan. Cancer Res. 1995;55:2097–2103. [PubMed] [Google Scholar]
- Sun Y., Saha S., Wang W., Saha L.K., Huang S.N., Pommier Y. Excision repair of topoisomerase DNA-protein crosslinks (TOP-DPC) DNA Repair (Amst.) 2020;89:102837. doi: 10.1016/j.dnarep.2020.102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi H., Handa H., Hirose S. Underwinding of DNA on binding of yeast TFIID to the TATA element. Biochem. Biophys. Res. Commun. 1993;192:1432–1438. doi: 10.1006/bbrc.1993.1576. [DOI] [PubMed] [Google Scholar]
- Teves S.S., Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurela S., Garding A., Jung J., Schübeler D., Burger L., Tiwari V.K. Gene regulation and priming by topoisomerase IIα in embryonic stem cells. Nat. Commun. 2013;4:2478. doi: 10.1038/ncomms3478. [DOI] [PubMed] [Google Scholar]
- Tiwari V.K., Burger L., Nikoletopoulou V., Deogracias R., Thakurela S., Wirbelauer C., Kaut J., Terranova R., Hoerner L., Mielke C. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. Proc. Natl. Acad. Sci. USA. 2012;109:E934–E943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Shibata H., Handa H. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 2013;1829:98–104. doi: 10.1016/j.bbagrm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Yeung A.T., Dinehart W.J., Jones B.K. Alkali reversal of psoralen cross-link for the targeted delivery of psoralen monoadduct lesion. Biochemistry. 1988;27:6332–6338. doi: 10.1021/bi00417a020. [DOI] [PubMed] [Google Scholar]
- Yu X., Davenport J.W., Urtishak K.A., Carillo M.L., Gosai S.J., Kolaris C.P., Byl J.A.W., Rappaport E.F., Osheroff N., Gregory B.D., Felix C.A. Genome-wide TOP2A DNA cleavage is biased toward translocated and highly transcribed loci. Genome Res. 2017;27:1238–1249. doi: 10.1101/gr.211615.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Lyu Y.L., Lin C.P., Zhou N., Azarova A.M., Wood L.M., Liu L.F. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 2006;281:35997–36003. doi: 10.1074/jbc.M604149200. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential gene expression values for each gen (adjusted p value and fold change) in hTERT-RPE-1 cells upon 30 min (versus DMSO treatment) and 2 h (versus untreated) treatment with 200 μM Merb. List of all differentially expressed (DE), as well as UP and DOWN regulated upon 2 h treatment. List of all genes displaying differential Pol II binding (ChIP-seq) in hTERT-RPE-1 cells treated for 1 h (versus untreated) with 200 μM Merb.
Data Availability Statement
Data are available in the main text, supplementary materials and auxiliary files. The accession number for all the sequencing data generated in this manuscript is GEO: GSE141800.