Abstract
Chromoanagenesis is a genomic event responsible for the formation of complex structural chromosomal rearrangements (CCRs). Germline chromoanagenesis is rare and the majority of reported cases are associated with an affected phenotype. Here, we report a healthy female carrying two de novo CCRs involving chromosomes 4, 19, 21 and X and chromosomes 7 and 11, respectively, with a total of 137 breakpoint junctions (BPJs). We characterized the CCRs using a hybrid-sequencing approach, combining short-read sequencing, nanopore sequencing, and optical mapping. The results were validated using multiple cytogenetic methods, including fluorescence in situ hybridization, spectral karyotyping, and Sanger sequencing. We identified 137 BPJs, which to our knowledge is the highest number of reported breakpoint junctions in germline chromoanagenesis. We also performed a statistical assessment of the positioning of the breakpoints, revealing a significant enrichment of BPJ-affecting genes (96 intragenic BPJs, 26 genes, p < 0.0001), indicating that the CCRs formed during active transcription of these genes. In addition, we find that the DNA fragments are unevenly and non-randomly distributed across the derivative chromosomes indicating a multistep process of scattering and re-joining of DNA fragments. In summary, we report a new maximum number of BPJs (137) in germline chromoanagenesis. We also show that a hybrid sequencing approach is necessary for the correct characterization of complex CCRs. Through in-depth statistical assessment, it was found that the CCRs most likely was formed through an event resembling chromoplexy—a catastrophic event caused by erroneous transcription factor binding.
Electronic supplementary material
The online version of this article (10.1007/s00439-020-02242-3) contains supplementary material, which is available to authorized users.
Background
Complex chromosomal rearrangements (CCRs) are structural variants (SVs) consisting of multiple adjacent breakpoint junctions (BPJs). The vast majority of CCRs are reported in cancers, and most of the current knowledge on CCRs originate from such studies (Collins et al. 2017). However, a growing number of CCRs are reported also in the germline (Collins et al. 2019), and such CCRs are known to cause a variety of disorders, including intellectual disability and dysmorphism (Eisfeldt et al. 2019). CCRs arise through a multitude of events, including breakage-fusion-bridge (BFB) cycles (McClintock 1941), chromoanasynthesis (Liu et al. 2011), chromothripsis (Zhang et al. 2013), and chromoplexy (Baca et al. 2013).
BFB cycles may be initiated through telomeric dysfunction. Chromatids carrying dysfunctional telomeres may undergo rearrangements and fuse with other chromatids. During anaphase, these fused chromatids will be torn apart, resulting in rearranged chromatids that are lacking the telomere. Since the resulting chromatids have lost the telomere, they may fuse and be torn apart the next cell cycle, allowing for continuous cycles of breakage, fusion and bridging (McClintock 1941).
CCRs formed through BFB cycles are, therefore, mainly terminal rearrangements, they include duplications, deletions as well as copy number neutral fragments (Zakov and Bafna 2015). CCRs formed through BFB cycles may also be recognized based on the characteristic, non-random orientation of the DNA fragments involved, and various algorithms are described for such purposes (Kinsella and Bafna 2012).
In contrast to BFB cycles, chromothripsis is a single event of localized scattering of one or a few (< 4) chromosomes; once scattered, the fragments are rapidly joined in a seemingly random fashion (Pellestor 2019). Chromothripsis may involve deletions and copy number neutral fragments (Korbel and Campbell 2013) and may be initiated through numerous events, including the formation of micronuclei, viral insertion, or radiation (Koltsova et al. 2019). Chromothripsis has been observed in cancer (Cortés-Ciriano et al. 2020) as well as in germline (Macera et al. 2015).
Chromoanasynthesis is a catastrophic event, occurring during DNA replication and may be initiated by endogenous factors, including DNA secondary structures, as well as exogenous factors, including radiation, causing the DNA polymerase to replicate the DNA in an aberrant way (Liu et al. 2011). Chromoanasynthesis is characterized by non-clustered breakpoints, copy number states including both deletions and duplications, and templated insertions in the BPJs (Korbel and Campbell 2013; Zepeda-Mendoza and Morton 2019).
Chromoplexy is a recently discovered mechanism of CCR formation (Baca et al. 2013) which usually involves several chromosomes (> 2), and is generally a copy number neutral event (Pellestor 2019). Chromoplexy is believed to occur due to aberrant binding of transcription factors (Haffner et al. 2010). As such, the BPJs will cluster within genes, and may involve multiple co-transcribed genes. Chromoplexy has so far only been observed in cancers (Zhang et al. 2013), and with only a few cases reported.
In aggregate, these events involve different parts of the genetic repair machinery, and occur due to catastrophic errors in the most fundamental activities of the cell, including cell division, DNA replication and transcription.
As such, there is a great value in characterizing CCRs, the molecular characterization of CCRs may provide insights on genetic repair mechanisms and double-stranded breakage (Koltsova et al. 2019), as well as provide details on the structure and function of the genome in general (Pellestor 2019).
However, the correct characterization of CCRs is a difficult task, and commonly involves a large number of experiments, as well as time-consuming and manual analyses. Today, whole-genome sequencing (WGS) is the main method of choice for solving CCRs, and a variety of WGS methods have been applied to solve CCRs, including short-read sequencing (Nazaryan-Petersen et al. 2018), nanopore sequencing (Stancu et al. 2017), linked short-read WGS (Ott et al. 2018), and optical mapping (Chan et al. 2018). Given the novelty of these technologies, and the rarity of the CCR events, the choice of WGS method can be a great challenge. It is clear that each technology comes with a variety of advantages and disadvantages (Eisfeldt et al. 2019) and that the data may be analyzed through a diversity of bioinformatic pipelines (Stancu et al. 2017).
Here, we present a female with no reported health issues except for fertility problems carrying two distinct germline de novo chromosomal rearrangements involving a total of 6 chromosomes and 137 breakpoints, which is, to our knowledge, the largest number of reported BPJs in a germline CCR so far. The rearrangements [(t(X;21;19;4) and t(7;11)] were characterized using Illumina short-read WGS, Oxford nanopore WGS, 10X Genomics Chromium WGS, as well as Bionano optical mapping. We have compared the results of each method, and we find (1) that the methods are complementary, and (2) that a hybrid-sequencing approach is necessary for the complete characterization of these CCRs. Lastly, we provide a detailed description of the two CCRs, showing that they were formed as two separate events and through different cellular mechanisms: while t(7;11) was most likely formed through a replicative mechanism, t(X;4;19;21) was most likely formed through chromoplexy.
Results
Cytogenetic analyses
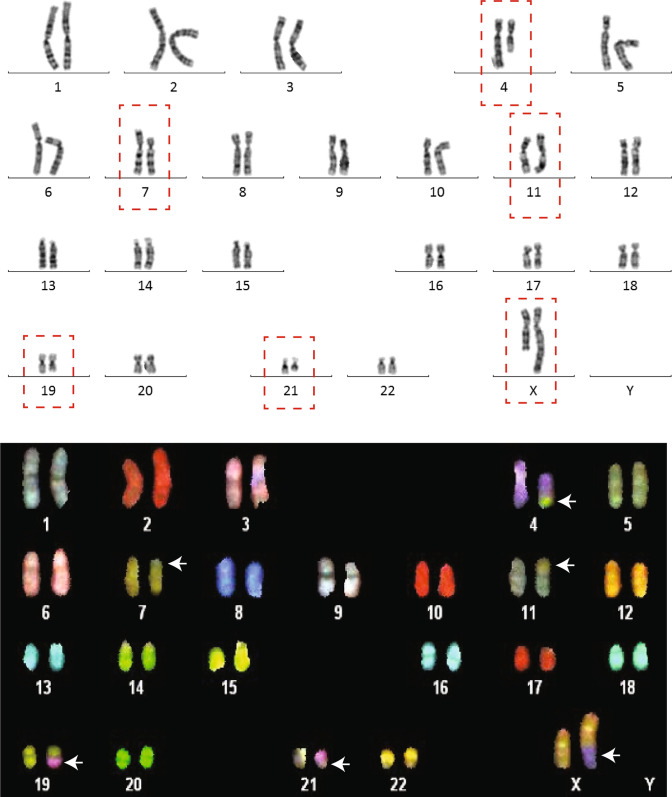
Chromosome analysis revealed two seemingly balanced de novo translocations involving chromosomes 4, 19, 21 and X and chromosomes 7 and 11, respectively, and the initial karyotype was 46,X,t(X;21;19;4)(q26;q21;q13;q21)t(7;11)(p13;p15) (Fig. 1).
Fig. 1.
Chromosome analysis reveals complex translocations involving a total of six chromosomes. The karyotype of the patient was set as 46,X,t(X;21;19;4)(q26;q21;q13;q21)t(7;11)(p13;p15) after initial chromosome analysis (upper figure). Follow-up analysis with spectral karyotyping (SKY) was performed to visualize the chromosomal segments that had been rearranged, especially the chromosome 19 and 21 segments that were not distinguishable from the karyotype. White arrows indicate the different segments (lower figure)
Whole-genome sequencing analyses
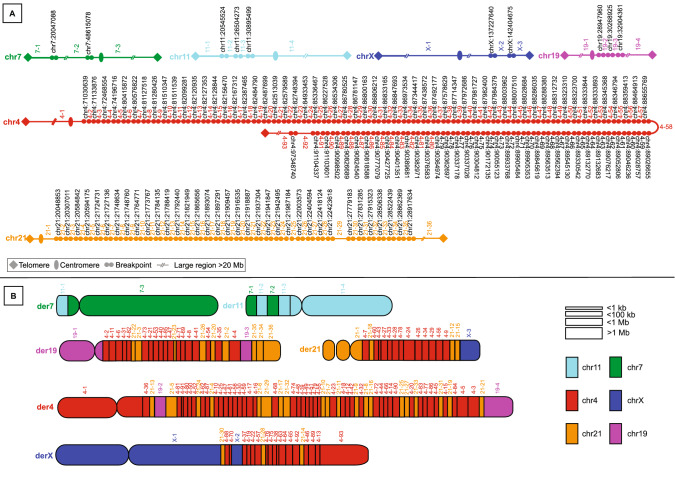
WGS analysis including short-read paired end (PE) WGS, linked-read WGS, nanopore WGS and Bionano optical mapping revealed that the rearrangement was ultra-complex and identified a total of 137 BPJs. The two rearrangements were confirmed to be completely separated and the t(7;11) translocation was far less complex than the t(X;21;19;4) rearrangement, involving 5 and 132 breakpoints, respectively (Figs. 2, 3). The breakpoints were confined to single-chromosome arms on all chromosomes involved and chromosome 4 and chromosome 21 were the most shattered chromosomes with 92 and 35 breakpoints, respectively (Figs. 2, 3). No additional chromosomes except those previously detected by karyotyping were involved in the CCRs.
Fig. 2.
Detailed maps of the t(7;11) and t(X;21;19;4) rearrangements. a The breakpoint positions on chromosome 4, 19, 21, and X. b A diagram detailing the structure of t(X;21;19;4). Each block represents a fragment involved in the CCR, and the coloring illustrates the chromosome of origin of each fragment (turquoise: chromosome 11, green: chromosome 7, purple: chromosome 19, red: chromosome 4, orange: chromosome 21, and dark blue: chromosome X). The exact positioning and orientation of the fragments are presented in Supplementary Table 1
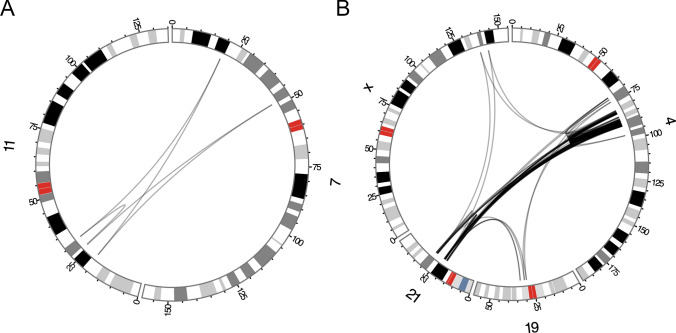
Fig. 3.
Circos plots illustrating the positioning of the BPJs involved in the complex rearrangement. The black arcs indicate the BPJs of a the t(7;11) rearrangement, and b the BPJs of the t(X;21;19;4) rearrangement
Genomic segments involved in the rearrangement, apart from the end segments (i.e., chromosomal segments including the telomere), ranged from 10 s of megabases to less than 100 bp (Supplementary Table 1). Considering the high number of fragments, the rearrangement was remarkably balanced, involving 51 deletions totaling 100 Kbp (Supplementary Table 1). Three of the five breakpoints in the t(7;11) rearrangement were balanced and two breakpoints had deletions of one and two nucleotides, respectively. In the t(X;21;19;4) rearrangement, 84/132 (64%) breakpoints harbored deletions (median size = 80 bp, min = 1 bp, max = 26 Kbp, Supplementary Table 1). No duplications were detected. A total of 23 protein-coding genes were disrupted by one (n = 9) or several (n = 14) breakpoints (Table 1). Two of those genes were associated with human disease (ANO3 associated with Dystonia 24, OMIM 610110, and ABCG2 associated with Gout susceptibility, OMIM 138900) (Table 1). Except for RASGEF1B, all the affected protein-coding genes had a pLI score (Lek et al. 2016) lower than what is expected of genes causing Mendelian disease (0.9) (Table 1).
Table 1.
Genes affected by the complex rearrangements
| Gene | Number of BPJs | Chr | Start | End | Transcriptional strand | MIM phenotype | pLI |
|---|---|---|---|---|---|---|---|
| t(7;11) | |||||||
| ABCA13 | 2 | 7 | 48211057 | 48687091 | + | 8.8e−114 | |
| ANO3 | 2 | 11 | 26210670 | 26684836 | + | 610110 | 1.2e−17 |
| DCDC5 | 2 | 11 | 30885150 | 31014233 | − | 4.7e−8 | |
| LOC101927668 | 2 | 7 | 19958604 | 20180049 | − | N.a. | |
| t(X;21;19;4) | |||||||
| ARHGAP24 | 14 | 4 | 86396284 | 86923823 | + | 3.9e−10 | |
| ABCG2 | 12 | 4 | 89011416 | 89152474 | − | 614490, 138900 | 2.1e−32 |
| AFF1 | 12 | 4 | 87856154 | 88062206 | + | 0.71 | |
| FAM13A | 8 | 4 | 89647105 | 89978346 | − | 1.9e−18 | |
| PRKG2 | 8 | 4 | 82008524 | 82136271 | − | 0.65 | |
| C4orf22 | 6 | 4 | 81256874 | 81884910 | + | 2.1e−9 | |
| CYYR1-AS1 | 6 | 21 | 27765954 | 27941571 | + | N.a. | |
| HERC3 | 6 | 4 | 89513574 | 89629693 | + | 0.07 | |
| MMRN1 | 6 | 4 | 90816052 | 90875780 | + | 2.1e−25 | |
| NCAM2 | 6 | 21 | 22370633 | 22914517 | + | 0.23 | |
| NUDT9 | 6 | 4 | 88343728 | 88380606 | + | 9.2e−6 | |
| CCSER1 | 4 | 4 | 91048684 | 92523370 | + | 0.000789 | |
| HSD17B11 | 4 | 4 | 88257674 | 88312455 | − | 2.1e−7 | |
| LOC101928978 | 4 | 4 | 84889235 | 85220322 | − | N.a. | |
| PTPN13 | 4 | 4 | 87515468 | 87736329 | + | 1.4e−25 | |
| RASGEF1B | 4 | 4 | 82347547 | 82393082 | − | 0.92 | |
| MAPK10 | 3 | 4 | 86933449 | 87374283 | − | 0.28 | |
| CYYR1 | 2 | 21 | 27838528 | 27945723 | − | 0.0004 | |
| DPY19L3 | 2 | 19 | 32896655 | 32976799 | + | 2.9e−7 | |
| GPRIN3 | 2 | 4 | 90165429 | 90229161 | − | 4.1e−13 | |
| HERC6 | 2 | 4 | 89299891 | 89364249 | + | 1.5e−14 | |
| LINC00989 | 2 | 4 | 80413747 | 80497614 | + | N.a. | |
| LOC100420587 | 2 | 19 | 28926295 | 29218601 | − | N.a. | |
| LOC101928942 | 2 | 4 | 82086094 | 82114549 | + | N.a. | |
| MIR548XHG | 2 | 21 | 19933583 | 20132130 | − | N.a. | |
| PRR27 | 2 | 4 | 71019904 | 71032326 | + | 0.0002 | |
Protein-coding genes in bold
BPJ breakpoint junction, N.a. not applicable
A total of 23 fusions of genes (disrupted transcripts positioned next to each other due to the rearrangement) were formed (Supplementary Table 2, Supplementary Table 1). However, these fused genes lack promoter regions, or are fused in incompatible orientations, and are therefore unlikely to be transcribed into a stable RNA.
Next, we analyzed topologically associated domains (TADs) using a publicly available Hi-C dataset (Lajoie et al. 2015), produced from cerebellar astrocytes (Table 2). In total, 28 TADs are affected in this dataset involving all rearranged chromosomes in our patient, 5 by breakpoints from the t(7;11) rearrangement, and 23 by breakpoints from the t(X;21;19;4) rearrangement (Table 2). The breakpoints are unevenly spread across the TADs, and the majority of the breakpoints (156) cluster within the 5 most affected TADs (tad7, tad92, tad90, tad89, tad83). Notably, 127 BPJ were found to bridge across TADs (Supplementary Table 1). Four breakpoints were not located in any TAD and were, therefore, excluded from this analysis.
Table 2.
TADs affected by the complex rearrangement
| TAD | Chromosome | Start | End | Length | BPJ | Inter-TAD BPJ | BP | Protein-coding genes | MIM morbid genes |
|---|---|---|---|---|---|---|---|---|---|
| tad71 | chr4 | 70560001 | 71440000 | 879 | 4 | 4 | 4 | 17 | 2 |
| tad72 | chr4 | 71480001 | 72600000 | 1119 | 2 | 2 | 2 | 8 | 2 |
| tad75 | chr4 | 74040001 | 75080000 | 1039 | 2 | 2 | 2 | 15 | 2 |
| tad82 | chr4 | 79920001 | 81240000 | 1319 | 6 | 6 | 6 | 5 | 3 |
| tad83 | chr4 | 81280001 | 82280000 | 999 | 17 | 16 | 18 | 3 | 0 |
| tad84 | chr4 | 82320001 | 83240000 | 919 | 13 | 12 | 14 | 1 | 0 |
| tad86 | chr4 | 84520001 | 85480000 | 959 | 5 | 4 | 6 | 2 | 0 |
| tad88 | chr4 | 85960001 | 87240000 | 1279 | 16 | 14 | 18 | 2 | 0 |
| tad89 | chr4 | 87280001 | 88200000 | 919 | 22 | 22 | 22 | 6 | 0 |
| tad90 | chr4 | 88240001 | 89480000 | 1239 | 32 | 28 | 36 | 16 | 6 |
| tad91 | chr4 | 89520001 | 90040000 | 519 | 13 | 12 | 14 | 4 | 0 |
| tad92 | chr4 | 90080001 | 91240000 | 1159 | 33 | 28 | 38 | 4 | 1 |
| tad23 | chr7 | 19800001 | 20720000 | 919 | 2 | 2 | 2 | 4 | 0 |
| tad59 | chr7 | 48240001 | 49120000 | 879 | 2 | 2 | 2 | 1 | 0 |
| tad20 | chr11 | 20520001 | 21480000 | 959 | 2 | 2 | 2 | 3 | 1 |
| tad27 | chr11 | 26320001 | 26640000 | 319 | 2 | 2 | 2 | 2 | 1 |
| tad32 | chr11 | 30280001 | 31080000 | 799 | 2 | 2 | 2 | 3 | 0 |
| tad30 | chr19 | 28080001 | 29320000 | 1239 | 2 | 2 | 2 | 0 | 0 |
| tad32 | chr19 | 30080001 | 31200000 | 1119 | 2 | 2 | 2 | 6 | 1 |
| tad35 | chr19 | 32880001 | 32960000 | 79 | 2 | 2 | 2 | 1 | 0 |
| tad6 | chr21 | 19800001 | 20680000 | 879 | 8 | 8 | 8 | 0 | 0 |
| tad7 | chr21 | 20720001 | 22360000 | 1639 | 39 | 36 | 42 | 0 | 0 |
| tad8 | chr21 | 22400001 | 23360000 | 959 | 6 | 6 | 6 | 1 | 0 |
| tad13 | chr21 | 27000001 | 28160000 | 1159 | 6 | 6 | 6 | 5 | 2 |
| tad14 | chr21 | 28200001 | 29080000 | 879 | 6 | 4 | 8 | 2 | 0 |
| tad137 | chrX | 137200001 | 137840000 | 639 | 2 | 2 | 2 | 1 | 0 |
| tad141 | chrX | 141320001 | 142560000 | 1239 | 2 | 2 | 2 | 1 | 0 |
TAD topologically associated domain, BPJ breakpoint junction, BP breakpoint, MIM Mendelian Inheritance in Man
The affected TADs contain a total of 113 genes, of which 21 are known MIM morbid genes (Supplementary Table 1). Eleven of those follow an autosomal-dominant inheritance pattern and span a wide range of phenotypes, including three genes linked to various dental issues (AMTN, ENAM, DSPP) (Nakayama et al. 2015; Dong et al. 2000; Crosby et al. 1995) and APP known to cause Alzheimer disease when duplicated (Adler et al. 1991). Eight genes have a pLI score > 0.9, including one MIM morbid gene (SLC4A4) linked to autosomal recessive disease (Table 2, Supplementary Table 1). The genes located within affected TADs were further analyzed with a PANTHER GO biological process statistical overrepresentation test. This test revealed a total of 32 significantly enriched biological processes (P < 0.05, false discovery rate < 0.05) (Supplementary Table 1).
Analyzing the BPJs on the nucleotide level revealed distinct mutational signatures for the two separate complex rearrangements. The t(7;11) rearrangement showed rare single-nucleotide variants (SNVs), microhomology (4 bp) and a templated insertion in the junctions (Supplementary Fig. 1, Supplementary Table 1), consistent with replicative errors such as fork-stalling and template-switching (FoSTeS)/microhomology-mediated break-induced replication (MMBIR) (Weckselblatt and Rudd 2015) that are typical features of chromoanasynthesis. In contrast, the t(X;21;19;4) rearrangement consistently showed blunt ends, little to no microhomology, non-templated insertions and small deletions in the junctions (Table 2, Supplementary Fig. 1), consistent with non-homologous end-joining (NHEJ) (Weckselblatt and Rudd 2015). Genomic segments as small as 33 bp have previously been shown to sometimes be processed by the repair machinery during reassembly of shattered chromosomes (Slamova et al. 2018), so the large insertions were first manually checked and then checked using BLAT, which generated no specific matches. Next, we used RepeatMasker (Smit et al. 2013) to analyze all insertions larger than 50 bp, revealing that these insertions contain large amounts of simple repeats (29% vs 1.5% in GRCh38 (Smit et al. 2013)). Interestingly, one insertion was found to be a chimera of an SVA and srpRNA (Supplementary Dataset 1).
Out of the 137 breakpoints, 51 (37%) were completely balanced and 45 (33%) had losses of less than 100 nucleotides (Table 3, Supplementary Table 1). Among the total 137 BPJs, 44 (32%) showed microhomology in the junctions, but no more than 4 nucleotides (Table 3, Supplementary Fig. 1, Supplementary Table 1). A total of 53 junctions (39%) had insertions, of which one (1%) seemed templated from nearby sequences.
Table 3.
Breakpoint junction (BPJ) characteristics
| t(7;11) | |||
| By breakpoint | Total number | 5 | 100.0% |
| Balanced | 3 | 60% | |
| <10 nt deletion | 2 | 40% | |
| By junction | Total number | 5 | 100% |
| Microhomology, total | 2 | 40% | |
| < 2 nt | 1 | 20% | |
| 2–10 nt | 1 | 20% | |
| Insertions, total | 1 | 20% | |
| < 2 nt | 0 | 0% | |
| < 20 nt | 0 | 0% | |
| < 100 nt | 1 | 20% | |
| > 100 nt | 0 | 0% | |
| t(X;21;19;4) | |||
| By breakpoint | Total number | 132 | 100% |
| Balanced | 48 | 36% | |
| < 10 nt deletion | 15 | 11% | |
| < 100 nt deletion | 29 | 22% | |
| < 1000 nt deletion | 18 | 13% | |
| < 10,000 nt deletion | 21 | 16% | |
| > 10,000 nt deletion | 2 | 2% | |
| By junction | Total number | 132 | 100.0% |
| Microhomology, total | 39 | 30% | |
| < 2 nt | 18 | 14% | |
| 2–10 nt | 21 | 16% | |
| Insertions, total | 52 | 39% | |
| < 2 nt | 7 | 5% | |
| < 20 nt | 14 | 11% | |
| < 100 nt | 22 | 16% | |
| > 100 nt | 9 | 7% | |
nt nucleotide
Parental origin of both de novo rearrangements could not be determined as no parental samples were available for analysis.
Statistical assessment of the derivative chromosome structure and breakpoint junction characteristics of the t(X;21;19;4) rearrangement
The t(7;11) rearrangement carries features typical to chromoanasynthesis including dispersed interchromosomal translocations, as well as traces of replicative repair mechanisms (Zepeda-Mendoza and Morton 2019). In contrast, an in-depth statistical analysis was required to determine the mechanism of formation of the t(X;21;19;4) CCR. As noted previously, the t(X;21;19;4) CCR BPJ carries signatures consistent with NHEJ, which is found in a diversity of mechanisms of CCR formation, including chromothripsis, chromoplexy (Pellestor 2019), and BFB cycles (Marotta et al. 2013).
Analyzing the distribution of DNA fragments across the derivative chromosomes, we find that the fragments are spread across the derivative chromosomes in a non-random fashion. In particular, we note that derivative chromosome 4 contain 48% (n = 45, p = 10−6) of the fragments originating from chromosome 4, and 56% (n = 20, p = 10−4) of the fragments originating from chromosome 21 (Table 4). Conversely the derivative chromosome 21 is depleted in fragments originating from chromosome 4 (n = 13, p = 0.007), and chromosome 21 (n = 4, p = 0.034). In addition, there is an absence of fragment exchange between some of the derivative chromosome, including chromosome 19 and X. These results indicate that the t(X;21;19;4) CCR was formed either through a non-random process, causing skewed re-assembly of the chromosomes, or that the t(X;21;19;4) CCR was formed through a progressive multistep process, allowing the fragments to be reused in a directed, non-random fashion.
Table 4.
Analysis of the distribution of aberrant fragments in t(X;21;19;4)
| Derivative chromosome | Chromosome of origin Fraction of aberrant fragments |
|||||||
|---|---|---|---|---|---|---|---|---|
| 4 | 19 | 21 | X | |||||
| Fraction | P value | Fraction | P value | Fraction | P value | Fraction | P value | |
| Der(4) | 0.48 | < 0.001 | 0.5 | 0.28 | 0.56 | < 0.001 | 0 | 0.37 |
| Der(19) | 0.19 | 0.12 | 0.5 | 0.28 | 0.25 | 0.5 | 0 | 0.37 |
| Der(21) | 0.14 | 0.01 | 0 | 0.28 | 0.11 | 0.03 | 0.33 | 0.37 |
| Der(X) | 0.18 | 0.08 | 0 | 0.28 | 0.08 | 0.02 | 0.67 | 0.16 |
| Number of fragments in each derivative chromosome | |||
|---|---|---|---|
| Der(4) | Der(19) | Der(21) | Der(X) |
| 67 | 28 | 18 | 29 |
Analyzing the orientation of the fragments, we found no statistically significant patterns, and inverted and non-inverted fragment fusions appear equally likely (Table 5). Such patterns are consistent with a number of mechanisms, including chromoplexy and chromothripsis (Korbel and Campbell 2013), but is inconsistent with BFB cycles, a mechanism known to produce CCRs enriched in head-to-head and tail-to-tail fusions (Kinsella and Bafna 2012).
Table 5.
Orientation of the fused fragments in t(X;21;19;4)
| Derivative chromosome | Tail-to-head | Head-to-tail | Head-to-head | Tail-to-tail | ||||
|---|---|---|---|---|---|---|---|---|
| Count | P value | Count | P value | Count | P value | Count | P value | |
| Der(4) | 19 | 0.29 | 15 | 0.39 | 16 | 0.5 | 16 | 0.5 |
| Der(19) | 3 | 0.06 | 9 | 0.26 | 8 | 0.41 | 8 | 0.41 |
| Der(21) | 4 | 0.45 | 5 | 0.44 | 4 | 0.45 | 4 | 0.45 |
| Der(X) | 6 | 0.45 | 7 | 0.26 | 4 | 0.35 | 4 | 0.35 |
| Overall | 32 | 0.46 | 36 | 0.31 | 32 | 0.46 | 32 | 0.46 |
96 (72%) of the t(X;21;19;4) BPJs affect a total of 26 genes. These genes are spread across chromosomes 4, 19 and 21 (Table 1). The BPJs cluster within these genes; for instance, 14 BPJs were located within ARHGAP24, and 16 genes are affected by 4 or more BPJs. Applying Monte Carlo methods, we find that the t(X;21;19;4) is enriched in intragenic BPJs (p < 0.0001). In addition, although the breakpoints cluster within genes, the genes themselves are not clustered: in total, these genes span roughly 34 Mbp, with 22 Mbp, 4 Mbp, and 8 Mbp distributed on chromosome 4, 19, and 21, respectively.
To investigate the expression pattern of these affected genes, a GTex multigene query (https://gtexportal.org/) was performed on the 15 protein-coding genes affected by at least 3 breakpoints, revealing that all of those genes are co-expressed in testis, but no other tissue (Supplementary Fig. 2).
Comparison of the sequencing technologies
The two rearrangements were characterized using a combination of four technologies: Illumina short-read sequencing, linked short-read sequencing, nanopore sequencing, and optical mapping. The t(7;11) consists of five BPJs, all of which were detected by all four technologies (Supplementary Table 1).
In contrast, none of the technologies were able to detect all of the 132 BPJs in the t(X;21;19;4) rearrangement (Table 6). Nanopore sequencing detected the largest number of the t(X;21;19;4) BPJs (n = 120, 90.9%), and produced the largest number of total calls as well (14,730). Illumina PE sequencing detected the second largest amount of the t(X;21;19;4) BPJs (n = 119, 90.2%), and produced a relatively small number of calls (5084).
Table 6.
The number of detected chromosomal breakpoints in t(X;21;19;4) with the different sequencing technologies
| Illumina PE (TIDDIT) | Nanopore (sniffles) | Optical mapping (bionano access) | Linked reads (Longranger) | |
|---|---|---|---|---|
| Illumina PE (TIDDIT) | 119 (5084) | – | – | – |
| Nanopore (sniffles) | 130 | 120 (14,730) | – | – |
| Optical mapping (bionano access) | 119 | 120 | 19 (12,057) | – |
| 10 × chromium (Longranger) | 130 | 124 | 88 | 87 (1983) |
The table presents the number of gold-standard breakpoints detected for each separate technology, as well as each pairwise combination. The numbers within parenthesis illustrate the total number of calls
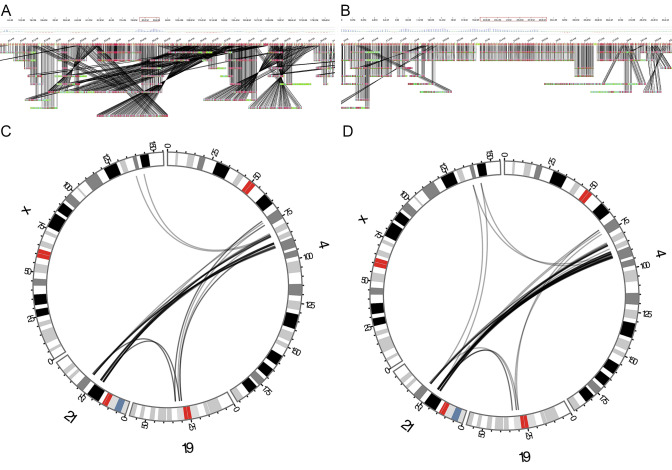
Optical mapping detected the lowest amount of the t(X;21;19;4) BPJs (n = 19, 14.4%), and produced the second largest number of calls (12,057). Analyzing the rearrangement manually in Bionano Access, we do find a larger number of calls corresponding to the BPJs of t(X;21;19;4) (Fig. 4a–d) and comparing all interchromosomal SV Bionano t(X;21;19;4) calls (n = 33) to all confirmed interchromosomal SV BPJs (n = 56), it was found that Bionano produces a significant number of similar calls (Fig. 4c, d). However, only 22 of these calls are located within 100 Kbp of the verified breakpoint positions, indicating low resolution.
Fig. 4.
Manual inspection of the Optical mapping data. a A screenshot of chromosome 4, using Bionano Access. b A screenshot of chromosome 21, using Bionano Access. c A Circos plot presenting all Bionano access translocation calls involving chromosomes 4, 21, 19 and X. d A Circos plot presenting the interchromosomal BPJs involving chromosomes 4, 21, 19 and X
Producing pairwise combinations of each technology, the detection rate is slightly increased. The highest detection rates were obtained by combining Illumina PE with either Nanopore WGS or 10 × Chromium WGS (Fig. 5, Table 6), allowing the automated detection of 130 BPJs (99%). Notably, there is only a small gain in adding a third-sequencing technology (Fig. 5). Inspecting the BPJs not detected by the various pipelines, it was found that the Illumina PE pipeline fails to detect BPJs in highly repetitive regions, as well as BPJs carrying large non-templated insertions (Supplementary Table 1). In contrast, nanopore WGS perform well in repetitive regions, but is limited by the high error rate and relatively low span-coverage. The linked short-read WGS is affected by various sequencing biases, such as GC content, and the sequencing coverage appears noisier than the standard Illumina PE, causing dropouts at the BPJ regions. Optical mapping appears ill-suited for these many and short fragments, and the usefulness of this technology is limited due to its low resolution.
Fig. 5.
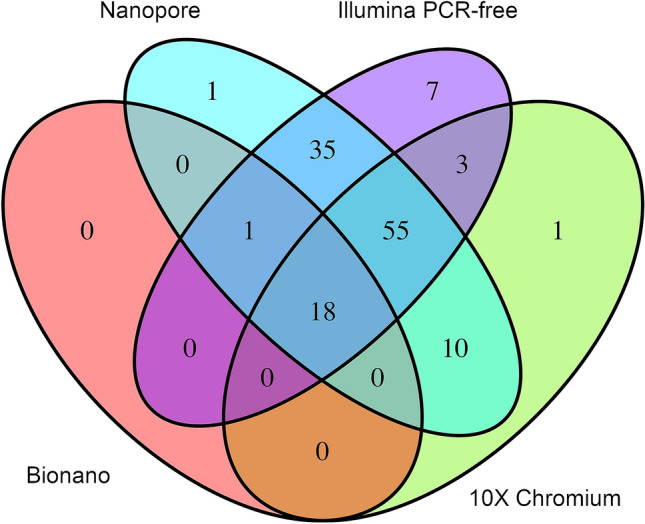
Comparison of the sequencing technologies. A Venn diagram illustrating the number of junctions detected by the four technologies in t(X;21;19;4)
Hybrid de novo assembly of t(X;21;19;4)
We attempted to solve the CCR using a hybrid de novo assembly approach, potentially allowing automated characterization of the CCR. These hybrid assemblies were based on the Wtdgb2 (Ruan and Li 2020) assembly of the nanopore WGS data (Supplementary Fig. 3A), Supernova (Weisenfeld et al. 2017) assembly of the linked short-read WGS data (Supplementary Fig. 3B), and optical maps. Notably, only a few of the BPJs were assembled (Table 7), resulting in low detection rate compared to the mapping assembly approach (Table 6). The highest detection rate is obtained by performing a nanopore–Bionano hybrid assembly. However, with only 71 detected BPJs, the detection rate is clearly lower than most of the mapping-based approaches (Table 6).
Table 7.
De novo assembly statistics summary in t(X;21;19;4)
| Detected BPJ | Calls | N50 (Mbp) | Size (Gbp) | |
|---|---|---|---|---|
| Supernova (10 ×) | 47 | 100,298 | 0.13 | 3.9 |
| Wtdgb2 (nanopore) | 68 | 2870 | 3.9 | 2.6 |
| Quickmerge (10 × + nanopore) | 1 | 6095 | 2 | 4.5 |
| Bionano scaffold 1 (nanopore + Bionano) | 71 | 3941 | 41 | 2.7 |
| Bionano scaffold 2 (Bionano + Quickmerge) | 28 | 118,984 | 45 | 2.8 |
Interestingly, Quickmerge removes the majority of the called BPJs (Table 7); however, it also removes the majority of calls present in the Supernova assembly. Quickmerge is, therefore, better suited for constructing reference genomes, as it removes noise and ambiguities at the cost of true variation.
Despite the low detection rate of these de novo assembly approaches, the resulting contigs were useful for validating the hypothesized derivative chromosomes. Contigs covering multiple aberrant fragments were especially useful, as such contigs allow for validation of the results obtained through mapping assembly (Supplementary Fig. 4).
Quality-wise, the best performance was achieved by combining the nanopore Wtdgb2 assembly with optical maps, yielding a relatively low number of calls (3941) indicating a tolerable amount of misassemblies while maintaining high contiguity and a relatively large assembly size (Supplementary Fig. 3C).
Even higher contiguity is obtained by merging the Supernova-WTDB2 Quickmerge results with optical mapping; however, the large number of calls indicate a large number of misassemblies (Supplementary Fig. 3D).
Discussion
Herein, we utilized four high-throughput genomic technologies to fully resolve the structure of two CCRs that had arisen independently as de novo events in a healthy woman and found Illumina PE WGS and nanopore WGS to be the most promising hybrid-sequencing approach for solving CCRs consisting of a large number of small aberrant fragments. These two technologies offer high detection rate, and are orthogonal, hence variants detected by both methods may be considered validated.
Through a hybrid-sequencing approach and long read de novo assembly, the structure of the two CCRs was fully characterized. Subsequent breakpoint junction analysis enabled the most plausible mechanisms of formation to be determined. The t(7;11) rearrangement was determined to most likely have been formed through a replicative error mechanism, largely due to the templated insertions. Instead, an in-depth statistical analysis was performed to determine the mechanism of formation of t(X;21;19;4). Through these analyses, it was found that the CCR most likely was formed through a progressive multistep process, that the fragments were reinserted in random orientation, and that the breakpoints are widely spread across the genome but cluster within genes. All these signatures are consistent with chromoplexy, and the fact that the rearrangement is largely balanced, and spread across four chromosomes adds to this hypothesis (Pellestor 2019). A growing amount of literature is describing chromoplexy as a catastrophic event occurring during active transcription (Yi and Ju 2018), and as presented here the t(X;21;19;4) CCR, which is highly enriched in intragenic breakpoints would be consistent with this idea.
Our second-best hypothesis would be formation through chromothripsis, however, this hypothesis is likely to be rejected as chromothripsis is defined as one single localized cataclysmic event of breakage, followed by random reintegration of fragments (Korbel and Campbell 2013), which would be the opposite of the formation of t(X;21;19;4). The number of BPJs (132) in t(X;21;19;4) exceed the typical numbers reported in both chromoplexy and germline chromothripsis in the literature. We must note, however, that chromoplexy is a recently described mechanism of CCR formation, and that larger, more complex chromoplexy rearrangements are likely to be reported as they are detected.
In the t(X;21;19;4) CCR, insertions were found in 39% of the BPJs (Table 3) and 24 BPJs harbor repetitive non-templated insertions larger than 50 bp (Supplementary Table 1). In addition, most of the breakpoints (66%) contain deletions (Table 3). These findings provide clues on which cellular repair pathways underly the formation of the CCR. Alternative-NHEJ is a likely candidate, as it is known to produce large insertions and deletions at the BPJs (Seol et al. 2018) and has previously been described in chromoplexy (Zepeda-Mendoza and Morton 2019). In particular, polymerase theta-mediated end-joining (PTMEJ) is known to produce large insertions and these insertions are produced through fill-in synthesis that stabilizes the open ends (Black et al. 2016) and provides the homologous sequences used to join the open ends (Mateos-Gomez et al. 2017). Such DNA synthesis could explain the large random insertions observed at the BPJs of t(X;21;19;4) CCR. Furthermore, polymerase theta is known to include templated insertions at the BPJ (Schimmel et al. 2019) which can be seen at numerous BPJs, including BPJ 4–68 → 21–17 and 21–32 → 4–74 (Supplementary Figure S1).
Analyzing the co-expression pattern of the affected protein-coding genes, it was found that the affected genes are co-expressed in one tissue only, the testis (Figure S2). As t(X;21;19;4) is a germline rearrangement, formed through chromoplexy, these findings could imply that the rearrangement was formed during spermatogenesis. However, since we lack paternal DNA samples, this hypothesis cannot be tested. In particular, we would be interested in confirming whether the t(X;21;19;4) CCR is of paternal or maternal origin, as well as to check whether the CCR was formed before or after meiosis.
Single cell Hi-C studies of the various developmental stages of spermatozoa and ova could be utilized to overcome these issues. Through such analyses, 3D genomic maps could be constructed (https://github.com/lh3/hickit) (Nagano et al. 2017) and these maps could be used to assess the likelihood of a suggested mechanism of formation (Berthelot et al. 2015).
Comparing the four technologies used (Bionano optical mapping, Illumina PE WGS, linked-read WGS, and nanopore sequencing), we would recommend Illumina PE WGS as a first experiment for solving a CCR like the t(X;21;19;4) rearrangement presented here as it is the most cost-efficient approach, offering high detection rate, and a large selection of bioinformatic tools, at a relatively small cost. Follow-up experiments should be chosen based on the biological question, as well as the structure of the CCR.
Except for fertility problems, the proband is unaffected which is surprising given the high number of breakpoints. The reason, however, for the lack of a clinical phenotype is that the CCR largely consists of intragenic breakpoints disrupting no early-onset dominant disease genes (Table 1). In addition, the majority of disrupted genes are unlikely to be haploinsufficient (pLI < 0.9). The fact that 27 TADs containing a total of 113 genes were potentially affected by the CCRs (Table 2, Supplementary Table 1) is remarkable and highlights the importance of continued research into the clinical significance of TAD disruptions. We speculate that the TADs are perturbed so that the change of expression is not consistent with the gene-related disease. The majority of affected TADs and MIM morbid genes are found in the more complex t(X;21;19;4), which is likely to be formed through chromoplexy. As chromoplexy involves regions that are co-expressed and close in the 3D-space of the nucleus (Yi and Ju 2018), the 3D structure of the probands genome may, therefore, be less perturbed than what is shown in these one-dimensional TAD analyses (Table 2, Supplementary Table 1) which could also explain why the patient do not display a phenotype consistent with these disease genes. Alternatively, one could speculate that the rearrangement may cause late-onset disease, explaining why the proband currently do not display any disease phenotype, an example being APP (Supplementary Table 1), which is involved in Alzheimer disease (Adler et al. 1991), a well-known late-onset disease. Whether to test a healthy individual for late-onset disease or not is an ethical dilemma, however, both Hi-C and RNA-seq data would be useful in understanding the functional effect of such a complex rearrangement.
Despite fertility problems, the proband naturally conceived and gave birth to a healthy child carrying only the t(7;11) CCR. The fertility problems observed may be explained by a high number of gametes carrying unbalanced combinations of the two CCRs. Of the 64 gametes possible through alternate or adjacent segregation I (Supplementary Fig. 5), only 4 (6.25%) would carry a balanced set of chromosomes compared to 2/4 (50%) in carriers of reciprocal-balanced translocations (Morel et al. 2004).
Conclusion
In conclusion, we present two de novo complex chromosomal rearrangements involving 6 chromosomes and 137 BPJs in a healthy female. The amount of BPJs are approximately twice as many as have been reported to this day in germline chromothripsis (Collins et al. 2017), and almost sixfold the amount of BPJs that have been reported in a healthy individual (De Pagter et al. 2015). Analysis of the BPJs suggests that a combination of repair machineries have operated in the same cell, and that the distinct rearrangements have occurred through different mechanisms. In this way, we illustrate that present day commercial genomic technologies are suitable for fully characterizing such a rearrangement, however, it is clear that a multi-omics approach is necessary for understanding its full structure and complexity.
The large amount of work and effort put into this single case illustrate how a single a patient can provide significant clues and insights to molecular mechanisms underlying these events; however, we also illustrate that personalized care may require costly and truly personalized analyses.
Methods
Clinical synopsis
The proband is a female who was first referred for genetic investigation at age 3 years because of short stature. The clinical investigation was dropped when the patient started catching up in height and she now reports as an adult as being of normal height compared to other women in her family, and otherwise healthy. She has a history of fertility problems likely due to the chromosomal rearrangement but has a healthy child conceived naturally who only inherited one translocation, the t(7;11).
Cytogenetic analysis
Metaphase slides were prepared from peripheral blood cultures according to standardized protocols. Chromosome analysis was performed according to routine procedures with the GTG-banding technique and an approximate resolution of 550 bands per haploid genome was obtained.
Short-read whole-genome sequencing
Genomic DNA derived from whole blood from the proband was sequenced at National Genomics Infrastructure (NGI), Stockholm, Sweden, using a PCR-free paired-end (PE) protocol; resulting in roughly 35X coverage. Data were processed and analyzed as described previously (Eisfeldt et al. 2019). Briefly, the data were pre-processed using the NGI-piper pipeline (https://github.com/NationalGenomicsInfrastructure/piper) and structural variants were called using the FindSV (https://github.com/J35P312/FindSV) pipeline that combines CNVnator (Abyzov et al. 2011) and TIDDIT (Eisfeldt et al. 2017). Variants of interest were visualized in Integrative Genomics Viewer (IGV) (Thorvaldsdóttir, Robinson, and Mesirov 2013).
Linked-read whole-genome sequencing
Genomic DNA derived from whole blood from the proband was also sequenced using the 10 × Genomics Chromium WGS protocol and data were analyzed and processed as described previously (Eisfeldt et al. 2019). Data were analyzed using 10 × Genomics default pipelines Long Ranger V2.1.2 (https://support.10xgenomics.com/genome-exome/software/downloads/latest).
Optical mapping
Optical mapping was performed on genomic DNA from the proband by running dual enzymes (BspQI, BssSI) on the Bionano Genomics (San Diego, CA, USA) Saphyr platform (https://bionanogenomics.com/support-page/saphyr-system). Analysis was performed as described previously (Eisfeldt et al. 2019). Briefly, the optical maps were analyzed using Bionano-solve (https://bionanogenomics.com/support-page/bionano-solve), aligned to Hg19 reference genome using Bionano RefAligner (version 5649) and output files were converted into VCF files using a custom script (https://github.com/J35P312/smap2vcf). Variants of interest were visualized in Bionano access.
Nanopore whole-genome sequencing
Nanopore WGS was performed at National Genomics Infrastructure (NGI) Uppsala. The sequencing library was prepared using the LSK-109 ligation kit and sequencing was performed on the PromethION platform using the R9.4 flow cell. Bases were called using the Guppy base caller (https://nanoporetech.com) producing 2729,339 reads, with an average length of 15 Kbp, resulting in roughly 13 × coverage.
The resulting nanopore WGS data were aligned to Hg19 using Minimap2 (Li 2018), and variants were called using Sniffles (Sedlazeck et al. 2018), setting the minimum read support parameter to three reads. SV calls shorter than 2 Kbp were considered Indels (insertions/deletions) and were, therefore, removed.
Statistical tests
Statistical tests were performed to describe the t(X;21;19;4) CCR, and to reject implausible mechanisms of formation. Binomial tests were performed to assess the randomness of the distribution of fragments among the derivative chromosomes, and to test for biases regarding the orientation of the aberrant fragments. The binomial tests were performed using the social statistics online binomial test calculator https://www.socscistatistics.com/tests/binomial/.
The tests for biased fragment orientation were run for each derivative chromosome, and each possible orientation (Head–Head, Head–Tail, Tail–Head, and Tail–Tail).
The number of trials were set to the number of fragments within the derivative chromosome being tested, and the probability of the outcome was set to 1/4 (i.e., the number of possible fragment orientations), and the number of observed occasions were set to the orientations as observed.
The randomness of the distribution of fragments was assessed by performing binomial tests for each derivative chromosome. Here, the number of trials were set to the number of fragments involved in t(X;21;19;4), the probability of the outcome was set to 1/4 (i.e., the number of chromosomes involved in t(X;21;19;4)). The number of observed occasions was set to the observed number of fragments within the derivative chromosome being analyzed.
A Monte Carlo test was performed to assess the enrichment of BPJs within genes.
We utilized the RefSeq gene annotation (assembly GCF_000001405.25) to count the number of intragenic t(X;21;19;4) BPJs. A BPJ was considered intragenic if any of its two breakpoints were located within a gene. Next, an equal number of random BPJs were simulated across chromosome 4, 19, 21 and X and the number of intragenic simulated BPJs were counted. The simulated BPJs were selected according to a random uniform distribution, such that all bases across chromosome 4, 19, 21 and X were equally likely to be affected by simulated rearrangements.
This procedure was repeated for 1000 iterations, and a p value was defined as the fraction of simulated CCRs carrying more intragenic BPJs than the observed number of intragenic BPJs in t(X;21;19;4).
Breakpoint PCR and Sanger sequencing
Breakpoint PCR was performed on some BPJs that were too complex for WGS data analysis only. Primers were designed flanking the junctions approximately 500 bp away from the estimated breakpoint with M13 sequences attached. Breakpoint PCR was performed by standard methods using Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific, Waltham, MA, USA). Sanger sequencing of amplicons was performed according to standard protocols with M13 primers. Sequences were aligned using the BLAT tool (Kent 2002) and visualized in CodonCode Aligner (CodonCode Corp., Dedham, MA, USA).
De novoassembly
De novo assembly was performed using Bionano optical mapping, nanopore long-read and linked short-read sequencing data. As mentioned previously, the Bionano optical maps were assembled using Bionano Solve. The nanopore WGS data were assembled using Wtdgb2 (Ruan and Li 2020), and the 10X Genomics Chromium linked reads were assembled using the Supernova assembler (Weisenfeld et al. 2017). The assemblies were aligned to Hg19 using Minimap2 (Li 2018), and SVs were called using Assemblatron (Eisfeldt et al. 2020).
A variety of hybrid assemblies were produced: the Wtdgb2 and Supernova assemblies were merged using the Quickmerge tool (Solares et al. 2018) and all previously mentioned assemblies were merged with Bionano optical maps using the Bionano Solve hybrid-scaffolding tool. The resulting hybrid scaffolds were aligned to Hg19 using Minimap2, and SVs were called using Assemblatron.
Solving the complex chromosomal rearrangements
The BPJs were found by combining the filtered FindSV (Lindstrand et al. 2019) output with the filtered Sniffles (Sedlazeck et al. 2018) output using SVDB (Eisfeldt et al. 2017). Next, the BPJs were manually inspected through IGV (Thorvaldsdóttir et al. 2013), this inspection was necessary to resolve inaccuracies due to repeats or microhomology, as well as to find BPJs not called by any of the pipelines.
The filtered list of BPJs were manually compared the Optical mapping data, using Bionano Access, as well as the linked reads, using IGV and Loupe (https://support.10xgenomics.com). The manual comparison was also necessary due to the low resolution of these technologies, as well as the large amount of false-positive calls produced by the Bionano and Longranger pipelines. BPJs were considered true if they were supported by at least two of the WGS methods (either through calling or manual inspection), or by breakpoint PCR.
Aberrant DNA fragments were defined based on the quality controlled and filtered data. Except for the terminal fragments, each DNA fragment was associated with two BPJs, one at the head and one at the tail, forming chains of fragments, fused together through the BPJs. Lastly, the derivative chromosomes were defined using a custom script. This script “walks” through these chains, starting at a user-defined fragment (such as the first fragment of chromosome 4), and continues to the next fragment through the BPJs (or calls) associated with each fragment. This “walking” is continued until the script finds the terminal fragment in a chain and at that point the script ends and returns the path traveled through the chain of aberrant fragments. This path is a representation of the derivative chromosome, detailing the order and orientation of the fragments involved.
This chain was compared to the various sequencing data, de novo assemblies, as well as the cytogenetic results and was refined until the path was consistent with all signatures in each of the datasets.
Comparison of sequencing technologies
The VCF files produced from each sequencing technology was compared to the confirmed BPJs of the t(X;21;19;4) CCR using SVDB query. The t(X;21;19;4) truth-set was prepared by converting the confirmed BPJ into a bedpe file. In addition, SVDB query was run using the following command:
Svdb –query –query_vcf input.vcf –bedpedb truthset.bedpe –no-var –bnd_distance 100000 –overlap 0.01 > query.vcf
Here, input.vcf represent the VCF produced through the analysis of the various pipelines (TIDDIT, Sniffles, Longranger, Bionano Solve), and truthset.bedpe represent the bedpe file containing the t(X;21;19;4) BPJs.
A call was considered to represent a BPJ if both of its reported breakpoint positions were within 100 Kbp of the confirmed breakpoint. An SV call being close to multiple confirmed BPJs were considered to represent only one BPJ (the closest one). The distance of 100 Kbp was selected based on the size of the aberrant fragments, and to account for the lower resolution of Optical mapping.
Hi-C data and TAD analysis
Hi-C data produced from cerebellar astrocytes were downloaded via the ENCODE experiment ENCSR011GNI website (https://www.encodeproject.org/experiments/ENCSR011GNI/). The analyses were based on the publicly available bed file of TAD regions (Lajoie et al. 2015). TAD regions were coupled with breakpoints using TABIX (Li 2011): in these analyses, the TAD bed file was used as a database that was queried using the coordinates of the BPs. Next, the TADs were searched for protein-coding genes, querying the RefSeq gene annotation (assembly GCF_000001405.25) gff files with the positions of the affected TADs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Breakpoint junction sequences for the 137 junctions. The junction sequences were obtained either through the sequencing data, through breakpoint PCR, or both. Some junctions were only supported by whole-genome sequencing data on one side due to the large inserted sequences, and were either confirmed by breakpoint PCR or through “walking” along the inserted sequence until the sequences met the junction sequence from the other side of the junction (PDF 692 kb)
A GTEx multigene query heatmap. The heatmap illustrate the per tissue expression level of protein-coding genes affected by the t(X;21;19;4) (TIFF 283 kb)
De novo assembly of the t(X;21;19;4). Contiguity plots illustrating contigs larger than 2 Mbp. The chromosomes of the human genome are labeled based on their cytoband patterns, while the contigs are colored based on which chromosome they align. A) Nanopore assembly, B) Linked read assembly, C) Nanopore-Optical map hybrid, D) nanopore-Linked read-Optical map hybrid (TIFF 10229 kb)
A contig consisting of multiple aberrant fragments. The position and origin of the fragments are presented in Supplementary Table 1 (TIFF 387 kb)
Gametes possible through alternate and adjacent I segregation (JPEG 394 kb)
Breakpoints: All breakpoints detected on all of the involved chromosomes, with start and stop coordinates and possibly deleted nucleotides. Segments: All identified segments involved in the rearrangements with start and stop coordinates and segment size, as well as total amount of identified segments originating from each of the chromosomes involved. Junctions: Details of the junctions that were characterized with possible microhomology, insertions or other. Der(4), Der(19), Der(21), Der(X), Der(7) and Der(11): Details of the segments that consisted each derivative chromosome with their size and orientation (XLSX 94 kb)
Acknowledgements
We would like to thank the patient for participating and cooperating throughout the study. Many thanks to Irene White for invaluable help with retrieving the SKY images and Marie Bjurström, Camilla Slyngstadli and Riffat Ajmal for help with lab work. We would also like to thank National Genomics Infrastructure (NGI) Stockholm at Science for Life Laboratory for providing assistance in massive parallel sequencing, as well as UPPMAX for providing computational resources through project sens2017106 and sens2017130.
Author contributions
All the authors took part in the writing of the manuscript. MP performed breakpoint PCR and Sanger sequencing. AP performed nanopore sequencing and extraction of high-weight molecular DNA. JE performed bioinformatic and statistical analyses. AL, LF, and DN performed project planning and experiment design.
Funding
Open access funding provided by Karolinska Institute. AL was supported by the SciLifeLab national sequencing projects grant, the Swedish Research Council [2017-02936, 2019-02078], Stockholm City Council and the Swedish Brain Foundation. MP was supported by grants from Karolinska Institutet funding for doctoral education (KID).
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due the European GDPR law but are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Consent to participate
The local ethical board in Stockholm, Sweden approved the study (ethics permit number KS 2012/222-31/3) and written informed consent was obtained.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jesper Eisfeldt and Maria Pettersson: Equal contribution.
References
- Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and Characterize Typical And Atypical CNVs from family and population genome sequencing. Genome Res. 2011 doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler MJ, Coronel C, Shelton E, Seegmiller JE, Dewji NN. Increased gene expression of alzheimer disease β-amyloid precursor protein in senescent cultured fibroblasts. Proc Natl Acad Sci USA. 1991 doi: 10.1073/pnas.88.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013 doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot C, Muffato M, Abecassis J, Crollius HR. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.02.046. [DOI] [PubMed] [Google Scholar]
- Black SJ, Kashkina E, Kent T, Pomerantz RT. DNA polymerase θ: a unique multifunctional end-joining machine. Genes. 2016 doi: 10.3390/genes7090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Eva KF, Cameron DL, Petersen DC, Lyons RJ, Baldi BF, Papenfuss AT, Thomas DM, Hayes VM. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018 doi: 10.1101/gr.227975.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 2017 doi: 10.1186/s13059-017-1158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Khera AV, Francioli LC, et al. An open resource of structural variation for medical and population genetics. BioRxiv. 2019 doi: 10.1101/578674. [DOI] [Google Scholar]
- Cortés-Ciriano I, Lee JJK, Xi R, Jain D, Jung YL, Yang L, Gordenin D, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020 doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby AH, Scherpbier-Heddema T, Wijmenga C, Altherr MR, Murray JC, Buetow KH, Dixon MJ. Genetic mapping of the dentinogenesis imperfecta type II locus. Am J Hum Genet. 1995;57:832. [PMC free article] [PubMed] [Google Scholar]
- Dong J, Gu TT, Simmons D, MacDougall M. Enamelin maps to human chromosome 4q21 within the autosomal dominant amelogenesis imperfecta locus. Eur J Oral Sci. 2000 doi: 10.1034/j.1600-0722.2000.108005353.x. [DOI] [PubMed] [Google Scholar]
- Eisfeldt J, Vezzi F, Olason P, Nilsson D, Lindstrand A. TIDDIT, an efficient and comprehensive structural variant caller for massive parallel sequencing data. F1000Research. 2017 doi: 10.1268/f1000research.11168.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeldt J, Pettersson M, Vezzi F, Wincent J, Käller M, Gruselius J, Nilsson D, Lundberg ES, Carvalho Claudia MB, Lindstrand A. Comprehensive structural variation genome map of individuals carrying complex chromosomal rearrangements. PLoS Genet. 2019 doi: 10.1371/journal.pgen.1007858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeldt J, Mårtensson G, Ameur A, Nilsson D, Lindstrand A. Discovery of novel sequences in 1,000 Swedish genomes. Mol Biol Evol. 2020 doi: 10.1093/molbev/msz176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010 doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002 doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella M, Bafna V. Combinatorics of the breakage-fusion-bridge mechanism. J Comput Biol. 2012 doi: 10.1089/cmb.2012.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltsova AS, Pendina AA, Efimova OA, Chiryaeva OG, Kuznetzova TV, Baranov VS. On the complexity of mechanisms and consequences of chromothripsis: an update. Front Genet. 2019 doi: 10.3389/fgene.2019.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013 doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Lajoie BR, Dekker J, Kaplan N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods. 2015 doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btq671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrand A, Eisfeldt J, Pettersson M, Carvalho Claudia MB, Kvarnung M, Grigelioniene G, Anderlid BM, et al. From cytogenetics to cytogenomics: whole-genome sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Med. 2019 doi: 10.1186/s13073-019-0675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SCS, Dhar SU, Kołodziejska KE, Dharmadhikari AV, Cooper ML, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011 doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macera MJ, Sobrino A, Levy B, Jobanputra V, Aggarwal V, Mills A, Esteves C, et al. Prenatal diagnosis of chromothripsis, with nine breaks characterized by karyotyping, FISH, microarray and whole-genome sequencing. Prenat Diagn. 2015 doi: 10.1002/pd.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta M, Chen X, Watanabe T, Faber PW, Diede SJ, Tapscott S, Tubbs R, Kondratova A, Stephens R, Tanaka H. Homology-mediated end-capping as a primary step of sister chromatid fusion in the breakage-fusion-bridge cycles. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Gomez PA, Kent T, Deng SK, Mcdevitt S, Kashkina E, Hoang TM, Pomerantz RT, Sfeir A. The helicase domain of Polθ counteracts RPA to promote Alt-NHEJ. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel F, Douet-Guilbert N, Bris MJL, Herry A, Amice V, Amice J, De Braekeleer M. Meiotic segregation of translocations during male gametogenesis. Int J Androl. 2004 doi: 10.1111/j.1365-2605.2004.00490.x. [DOI] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Várnai C, Dudley C, Leung W, Baran Y, Cohen NM, Wingett S, Fraser P, Tanay A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017 doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y, Holcroft J, Ganss B. Enamel hypomineralization and structural defects in amelotin-deficient mice. J Dent Res. 2015 doi: 10.1177/0022034514566214. [DOI] [PubMed] [Google Scholar]
- Nazaryan-Petersen L, Eisfeldt J, Pettersson M, Lundin J, Nilsson D, Wincent J, Lieden A, et al. Replicative and non-replicative mechanisms in the formation of clustered CNVs are indicated by whole genome characterization. PLoS Genet. 2018 doi: 10.1371/journal.pgen.1007780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott A, Schnable JC, Yeh CT, Linjiang W, Liu C, Heng Cheng H, Dalgard CL, Sarkar S, Schnable PS. Linked read technology for assembling large complex and polyploid genomes. BMC Genom. 2018 doi: 10.1186/s12864-018-5040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagter MS, De MJ, Roosmalen V, Baas AF, Renkens I, Duran KJ, Van Binsbergen E, Tavakoli-Yaraki M, et al. Chromothripsis in healthy individuals affects multiple protein-coding genes and can result in severe congenital abnormalities in offspring. Am J Hum Genet. 2015 doi: 10.1016/j.ajhg.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F. Chromoanagenesis: cataclysms behind complex chromosomal rearrangements. Mol Cytogenet. 2019 doi: 10.1186/s13039-019-0415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H. Fast and accurate long-read assembly with Wtdbg2. Nat Methods. 2020 doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, van Schendel R, den Dunnen JT, Tijsterman M. Templated insertions: a smoking gun for polymerase theta-mediated end joining. Trends Genet. 2019 doi: 10.1016/j.tig.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, Von Haeseler A, Schatz MC. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018 doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Shim EY, Lee SE. Microhomology-mediated end joining: good, bad and ugly. Mutat Res Fundam Mol Mech Mutagenes. 2018 doi: 10.1016/j.mrfmmm.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamova Z, Nazaryan-Petersen L, Mehrjouy MM, Drabova J, Hancarova M, Marikova T, Novotna D, et al. Very short DNA segments can be detected and handled by the repair machinery during germline chromothriptic chromosome reassembly. Hum Mutat. 2018 doi: 10.1002/humu.23408. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P (2013) RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org. Accessed 22 Oct 2020
- Solares EA, Chakraborty M, Miller DE, Kalsow S, Hall K, Perera AG, Emerson JJ, Scott Hawley R. Rapid low-cost assembly of the drosophila melanogaster reference genome using low-coverage, long-read sequencing. G3 Genes Genom Genet. 2018 doi: 10.1534/g3.118.200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancu C, Mircea MJ, Roosmalen V, Renkens I, Nieboer MM, Middelkamp S, De Ligt J, Pregno G, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun. 2017 doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013 doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckselblatt B, Rudd MK. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 2015 doi: 10.1016/j.tig.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. Direct determination of diploid genome sequences. Genome Res. 2017 doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Ju Y. Patterns and mechanisms of structural variations in human cancer. Exp Mol Med. 2018 doi: 10.1038/s12276-018-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakov S, Bafna V. Reconstructing breakage fusion bridge architectures using noisy copy numbers. J Comput Biol. 2015 doi: 10.1089/cmb.2014.0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepeda-Mendoza CJ, Morton CC. The iceberg under water: unexplored complexity of chromoanagenesis in congenital disorders. Am J Hum Genet. 2019 doi: 10.1016/j.ajhg.2019.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013 doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Breakpoint junction sequences for the 137 junctions. The junction sequences were obtained either through the sequencing data, through breakpoint PCR, or both. Some junctions were only supported by whole-genome sequencing data on one side due to the large inserted sequences, and were either confirmed by breakpoint PCR or through “walking” along the inserted sequence until the sequences met the junction sequence from the other side of the junction (PDF 692 kb)
A GTEx multigene query heatmap. The heatmap illustrate the per tissue expression level of protein-coding genes affected by the t(X;21;19;4) (TIFF 283 kb)
De novo assembly of the t(X;21;19;4). Contiguity plots illustrating contigs larger than 2 Mbp. The chromosomes of the human genome are labeled based on their cytoband patterns, while the contigs are colored based on which chromosome they align. A) Nanopore assembly, B) Linked read assembly, C) Nanopore-Optical map hybrid, D) nanopore-Linked read-Optical map hybrid (TIFF 10229 kb)
A contig consisting of multiple aberrant fragments. The position and origin of the fragments are presented in Supplementary Table 1 (TIFF 387 kb)
Gametes possible through alternate and adjacent I segregation (JPEG 394 kb)
Breakpoints: All breakpoints detected on all of the involved chromosomes, with start and stop coordinates and possibly deleted nucleotides. Segments: All identified segments involved in the rearrangements with start and stop coordinates and segment size, as well as total amount of identified segments originating from each of the chromosomes involved. Junctions: Details of the junctions that were characterized with possible microhomology, insertions or other. Der(4), Der(19), Der(21), Der(X), Der(7) and Der(11): Details of the segments that consisted each derivative chromosome with their size and orientation (XLSX 94 kb)
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due the European GDPR law but are available from the corresponding author on reasonable request.