Summary
The translation inhibitor rocaglamide A (RocA) has shown promising antitumor activity because it uniquely clamps eukaryotic initiation factor (eIF) 4A onto polypurine RNA for selective translational repression. As eIF4A has been speculated to be a unique target of RocA, alternative targets have not been investigated. Here, we reveal that DDX3 is another molecular target of RocA. Proximity-specific fluorescence labeling of an O-nitrobenzoxadiazole-conjugated derivative revealed that RocA binds to DDX3. RocA clamps the DDX3 protein onto polypurine RNA in an ATP-independent manner. Analysis of a de novo-assembled transcriptome from the plant Aglaia, a natural source of RocA, uncovered the amino acid critical for RocA binding. Moreover, ribosome profiling showed that because of the dominant-negative effect of RocA, high expression of eIF4A and DDX3 strengthens translational repression in cancer cells. This study indicates that sequence-selective clamping of DDX3 and eIF4A, and subsequent dominant-negative translational repression by RocA determine its tumor toxicity.
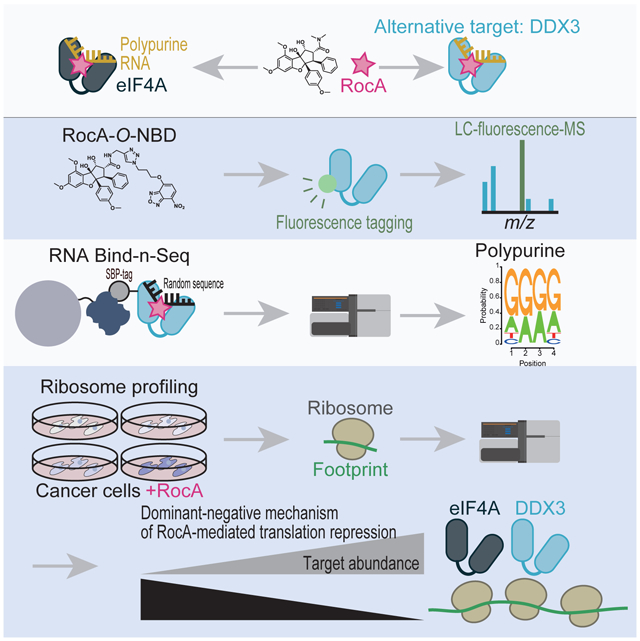
Graphical Abstract
eTOC Blurb
Chen et al. demonstrated the translation inhibitor rocaglamide A (RocA) alternatively targets DDX3, in addition to eIF4A. As RocA converts DDX3 and eIF4A into dominant negative translational repressors, the abundance of those proteins in cells is an indicator of RocA sensitivity.
Introduction
In recent years, great interest in RNA-selective small molecules has arisen due to their therapeutic potential as alternatives for undruggable targets (Mullard, 2017). Rocaglate-family compounds have been identified as candidate small molecules for such applications (Iwasaki et al., 2016; Iwasaki et al., 2019; Chu et al., 2019; Chu et al., 2020). This group of drugs has shown promise in a number of preclinical trials (Bordeleau et al., 2008; Cencic et al., 2009; Santagata et al., 2013; Wolfe et al., 2014; Manier et al., 2017; Chan et al., 2019), and a derivative (zotatifin or eFT226) (Ernst et al., 2020) is currently under clinical trial. Rocaglates target eukaryotic translation initiation factor (eIF) 4A, an ATP-dependent DEAD-box RNA-binding protein, and confer upon eIF4A a strong preference for A- and G-repeat (or polypurine) sequences. In the absence of drugs, eIF4A hydrolyzes ATP and dissociates from RNA; however, rocaglates clamp eIF4A on polypurine RNA even after eIF4A hydrolyzes ATP (Iwasaki et al., 2016; Iwasaki et al., 2019; Chu et al., 2019; Chu et al., 2020). Ultimately, rocaglates block translation of select mRNAs (Wolfe et al., 2014; Rubio et al., 2014; Iwasaki et al., 2016; Iwasaki et al., 2019; Chu et al., 2019; Chu et al., 2020).
The base recognition exhibited by rocaglates is based on binding to the interface between protein and RNA. Structural analysis of the eIF4A•rocaglate•polypurine RNA complex has revealed that rocaglates specifically bind in a cavity formed between human eIF4A1 (at Phe163, Gln195, Asp198, and Ile199) and adjacent purines (A and G) (Iwasaki et al., 2019). Consistent with this observation, yeast genetic screening (Sadlish et al., 2013), mammalian cell mutagenesis (Chu et al., 2016), and RNA sequencing of the plant genus Aglaia (Iwasaki et al., 2019), a natural source of rocaglates, have shown that substitution of Phe163 with Leu in eIF4A leads to rocaglate resistance.
Uniquely, targeting by rocaglates does not simply phenocopy inactivation of eIF4A but rather exhibits gain-of-function activity. As a result of the stable interaction of eIF4A with the polypurine sequence, the rocaglate•eIF4A complex is a roadblock for scanning of the 43S preinitiation complex (PIC) in the 5′ UTR (Iwasaki et al., 2016). Consequently, rocaglate-mediated translational repression is dominant-negative (Iwasaki et al., 2016): excess targets do not counteract, but rather enhance, the effect of the drug. Moreover, rocaglates trap eIF4F complexes, trimers of the cap-binding protein eIF4E, the scaffold protein eIF4G, and eIF4A, on mRNA cap structures containing downstream polypurine motifs (Chu et al., 2020). In addition to exerting cis-effects on directly bound mRNAs, rocaglates sequester eIF4A and eIF4F from the available pool and limit overall protein synthesis (i.e., they exert a “trans bystander effect”) (Chu et al., 2020).
Despite the wealth of evidence that eIF4A is a direct target of rocaglates, whether rocaglates also inhibit additional DEAD-box proteins has not been explored. Moreover, although differential rocaglate sensitivities among diverse cancer cell types have been reported (Santagata et al., 2013), the underlying mechanism remains enigmatic. In this study, we found that the DEAD-box protein DDX3 is an alternative target of rocaglamide A (RocA), a natural derivative of rocaglates. Since RocA converts both eIF4A and DDX3 into dominant-negative repressors, the expression levels of these proteins are associated with the translational repression, thereby the cytotoxicity induced by RocA. Elucidation of the full spectrum of RocA targets will enable prediction of the efficacy of rocaglates in cancer cells.
Results
RocA targets another protein besides eIF4A1 and eIF4A2
Although mutations in the RocA-binding site in eIF4A1 confer complete RocA resistance in vitro (Chu et al., 2016; Iwasaki et al., 2019), we observed a limited impact in cellulo. In an earlier study, we generated a HEK293 cell line in which endogenous eIF4A1 was knocked out by CRISPR-Cas9 and exogenous eIF4A1 with RocA-resistant mutations (Phe163Leu-Ile199Met) was simultaneously expressed [HEK293 SBP-eIF4A1 (Phe163Leu-Ile199Met) eIF4A1em1SINI] (Iwasaki et al., 2019). Mutated eIF4A1 in the cells reversed RocA-mediated cell death, but only partially (Figure S1A and S1B). These contradictory findings suggest the presence of other intracellular RocA target(s) in addition to eIF4A1.
eIF4A2 is an apparent candidate RocA target since it shares approximately 90% sequence identity with its paralog eIF4A1 (Lu et al., 2014). Indeed, eIF4A2 is directly inhibited by RocA. To monitor the effect of RocA, we tested the affinity between proteins and RNAs by fluorescence polarization. As previously observed, RocA imposed polypurine RNA selectivity on eIF4A1 (Iwasaki et al., 2016; Iwasaki et al., 2019; Chu et al., 2019) irrespective of the ATP analog form (Figure S1C, S1D, and Table 1). The recombinant eIF4A2 proteins (Figure S1C) phenocopied the ATP-independent clamping to polypurine RNA mediated by RocA (Figure S1E and Table 1). Similar biochemical effects on eIF4A2 by rocaglates have been reported in earlier studies (Chambers et al., 2013; Chu et al., 2019). Moreover, knockdown of eIF4A2 in the eIF4A1-mutated cells further facilitated the reversal of cell growth inhibition by RocA (Figure S1A and S1B). These data confirm that eIF4A2 is an alternative target of RocA.
Table 1. Summary of the Kd (μM) values between DEAD-box proteins (eIF4A1, eIF4A2, and DDX3X) and RNAs.
The fluorescence polarization of FAM-labeled RNAs was assayed. ND, not determined. See also Figure 3, 4, S1, and S5.
| (AG)10 | ||||
|---|---|---|---|---|
| ADP + Pi | AMP-PNP | |||
| Protein | DMSO | RocA | DMSO | RocA |
| eIF4A1 | ND | 0.25 ± 0.0092 | 1.4 ± 0.38 | 0.15 ± 0.0051 |
| eIF4A2 | ND | 1.3 ± 0.14 | 27 ± 8.4 | 0.14 ± 0.038 |
| DDX3X | ND | 0.79 ± 0.14 | 4.3 ± 0.40 | 0.17 ± 0.018 |
| Pro324Thr | ND | 0.40 ± 0.0079 | ||
| Val328Leu | ND | 0.20 ± 0.0090 | ||
| Gln360Pro | ND | ND | 3.3 ± 0.37 | |
| Gln360Leu | ND | ND | 1.7 ± 0.066 | |
| Arg363Asn | ND | 0.39 ± 0.0087 | ||
Nonetheless, although RocA was expected to be inactive due to the resistance-inducing mutations in eIF4A1 and knockdown of eIF4A2, we still observed substantial suppression of cell growth by RocA (Figure S1A and S1B), suggesting the existence of target(s) other than eIF4A1 and eIF4A2.
Proximity-specific fluorescence tagging by RocA-O-NBD
To determine the unidentified target(s) of RocA, the bifunctional subunit O-nitrobenzoxadiazole (O-NBD) was applied in our study (Figure 1A). O-NBD has been reported to covalently bind to lysine (Yamaguchi et al., 2014). Whereas O-NBD itself is nonfluorescent, its conversion to N-NBD upon lysine conjugation provides “turn-on” fluorescence. Thus, conjugation of O-NBD to another compound enables both proximity labeling close to the compound of interest and target protein identification (Yamaguchi et al., 2014). Here, we conjugated O-NBD to RocA via a dimethylamide group that contacts neither eIF4A1 nor polypurine RNA in the solved structure (Iwasaki et al., 2019) (Figure 1A, S2A, and S2B). The chemical probe, RocA-O-NBD, can be used for three different types of protein labeling: 1) labeling of known targets (eIF4A1 and eIF4A2), 2) labeling of unknown target(s), and 3) labeling of proteins interacting with the direct target proteins (Figure 1B). As unmodified RocA did, RocA-O-NBD blocked translation of a reporter mRNA bearing polypurine motifs in the 5′ UTR in an in vitro translation system with rabbit reticulocyte lysate (RRL) (Figure 1C, right). On the other hand, the effects on a control reporter with CAA repeats in the 5′ UTR were limited (Figure 1C, left). These data indicate that RocA-O-NBD acts as an mRNA-selective translation inhibitor.
Figure 1. RocA-O-NBD allows proximity labeling of eIF4A1.
(A) Chemical structures of RocA and RocA-O-NBD.
(B) Schematic representation of protein types that can be fluorescently labeled by RocA-O-NBD.
(C) In vitro translation (in RRL) from a Renilla luciferase reporter possessing CAA repeats (left) and 7×AGAGAG motifs (right) in the 5′ UTR. The reactions included RocA or RocA-O-NBD.
(D) Fluorescent labeling of recombinant human eIF4A1 WT or Aglaia mutant (Phe163Leu-Ile199Met, Mut) with RocA-O-NBD in the presence of (UC)10 or (AG)10 RNAs. RocA itself was used as a control.
(E) Western blotting of eIF4A (using a pan-eIF4A antibody) to O-NBD-labeled proteins immunopurified with an anti-NBD antibody from the in vitro translation reaction. The same experiment was performed with DMSO or RocA as a control.
(F) The O-NBD-labeled residue in rabbit eIF4A1 from RRL was searched by LC-MS/MS.
(G) The position of the labeled residue (Lys174 in human eIF4A1, red) in the resolved crystal structure of human eIF4A1•RocA•polypurine RNA (PDB ID: 5ZC9) (Iwasaki et al., 2019). Human eIF4A1 N-terminal domain (NTD), green; C-terminal domain (CTD), cyan; RNA, yellow; RocA, salmon; Aglaia substitution sites (Phe163 and Ile199), purple; O-NBD-labeled Lys174, red.
We initially validated the potential of RocA-O-NBD for use in proximity-specific labeling in a purified setup. As an interfacial ligand, RocA targets eIF4A1 only when it is bound to polypurine RNA (Iwasaki et al., 2019). Consistent with this fact, polypurine RNA, but not polypyrimidine RNA, provided fluorescence to recombinant eIF4A1 upon RocA-O-NBD supplementation (Figure 1D). In addition, the labeling efficiency was largely reduced when the recombinant eIF4A1 protein with Aglaia mutations (Phe163Leu-Ile199Met), which does not form the RocA-binding pocket, was used. Thus, RocA-O-NBD tags eIF4A1 in accordance with the specificity of RocA targeting.
Even in crude RRL, RocA-O-NBD maintained its specificity for fluorescence labeling of eIF4A1. The products of an in vitro translation reaction with RocA-O-NBD (Figure 1C) were subjected to immunopurification with an anti-NBD antibody. Western blot analysis showed that the presence of polypurine reporter mRNA in the reaction enabled efficient NBD tagging of eIF4A (Figure 1E). Moreover, liquid chromatography-mass spectrometry with a laser-induced fluorescence detector (LC-fluorescence-MS) (Yamaguchi et al., 2014) identified the fluorescence-tagged peptide from rabbit eIF4A1 (Figure S2C-E) and precisely annotated the labeled residue at Lys166 of rabbit eIF4A1 (Figure 1F). Given the homology to human eIF4A1 (Figure S2E) and the structure of human eIF4A1•RocA•polypurine RNA (Iwasaki et al., 2019), the labeled lysine (Lys174 in human eIF4A1) was located in a reasonable position relative to the dimethylamide group where O-NBD was conjugated (at a distance of ~29 Å) (Figure 1G), since the linker between RocA and O-NBD provides enough distance for reaction (Yamaguchi et al., 2014). LC-fluorescence-MS also detected a rabbit eIF4A2 peptide originating from essentially the same position as rabbit eIF4A1 (Figure S2F-H), indicating again that both eIF4A paralogs are targeted by RocA. All these data demonstrate that RocA-O-NBD may be a molecular tool that can be used to probe the proximal proteins of compounds.
RocA-O-NBD identifies DDX3X as a target of RocA
Using RocA-O-NBD, we surveyed RocA target proteins. We observed that RocA-O-NBD conferred fluorescence to several proteins in RRL (Figure 2A). The fluorescence tagging was enhanced by the addition of polypurine reporter mRNA, as found for purified eIF4A1 (Figure 1D). Subsequent mass spectrometry identified these proteins as eIF3A, eIF4G1, eIF4B, DDX3X, eIF4E, and eIF4H, in addition to the known eIF4A1/2 (Figure 2A). Given that eIF4A1/2 directly interacts with eIF4G1, eIF4B, and eIF4H and forms a complex with eIF4E and eIF3A through parts of the eIF4F complex and the 43S PIC, respectively [reviewed in (Shirokikh and Preiss, 2018)], these proteins were most likely indirectly tagged due to their proximity to eIF4A1/2 (type 3 in Figure 1B). In addition to the candidate concomitantly labeled proteins, we were intrigued by DDX3X since it belongs to the family of DEAD-box RNA-binding proteins (Linder and Jankowsky, 2011) as does eIF4A1/2, although this protein has also been reported to interact with eIF4A in yeast (Gao et al., 2016).
Figure 2. DDX3X is an alternative RocA target.
(A) RocA-O-NBD fluorescent labeling in RRL with or without 7•AGAGAG motif reporter mRNAs. DMSO- or RocA-treated conditions were used as controls. MS-identified proteins are depicted.
(B) Fluorescent labeling of human DDX3X recombinant protein (helicase core) with RocA-O-NBD in the presence of (UC)10 or (AG)10 RNAs. RocA itself was used as a control.
(C) The O-NBD-labeled residue in human DDX3X from the recombinant protein was searched by LC-MS/MS. CAM: carbamidomethylation.
(D) The position of the labeled residue (Lys335 in human DDX3X) in the resolved crystal structure of human DDX3X (PDB ID: 5E7M) (Floor et al., 2016). The NTD of DDX3X was overlaid with the NTD of eIF4A1 (PDB ID: 5ZC9) (Iwasaki et al., 2019). Human eIF4A1 NTD, green; human DDX3X NTD, beige; RocA, salmon; RocA-binding sites in eIF4A1, light gray; homologous residues in DDX3X, dark gray; O-NBD-labeled Lys174 in eIF4A1 and Lys335 in DDX3X, red.
(E) Cell viability assay of naïve HEK293 cells or HEK293 SBP-eIF4A1 (Phe163Leu-Ile199Met) eIF4A1em1sINI cells treated with DMSO or 30 nM RocA. eIF4A2 and/or DDX3X was knocked down by siRNA. The data are presented as the mean ± SD (n = 3).
See also Figure S3.
To address whether DDX3X is a direct target of RocA, we prepared recombinant DDX3X of the functional helicase core region (amino acids 132–607) (Floor et al., 2016) (Figure S5B) and performed RocA-O-NBD labeling. Indeed, the purified protein was fluorescently labeled with RocA-O-NBD in a polypurine RNA-dependent manner (Figure 2B). Moreover, LC-fluorescence-MS analysis identified NBD tagging on Lys335 (Figure 2C, S3A-C). Consistent with the structural similarities of the DEAD-box helicases, the labeled Lys residues were in the same loops of eIF4A1 and DDX3X (Figure 2D).
Consistent with the in vitro assay results, cellular DDX3X was also targeted by RocA. In a cell viability assay, knockdown of DDX3X strengthened RocA resistance coordinately with eIF4A1/2 inhibition (Figure 2E and S3D). Taking the in vitro and in cellulo data together, we conclude that DDX3X is an alternative target of RocA.
RocA clamps DDX3X onto polypurine sequences in an ATP-independent manner
The dependence of the RocA-O-NBD labeling assays on polypurine RNA (Figure 2A and 2B) suggested that clamping of DDX3X by RocA occurs on a specific sequence. Fluorescence polarization assays clearly showed that DDX3X binds to polypurine RNA more tightly in the presence of RocA than in the absence of RocA but does not bind to polypyrimidine RNA (Figure 3A and Table 1). Strikingly, the selective binding evoked by RocA did not require ATP (Figure 3B and Table 1). Thus, as found for eIF4A1/2, RocA converts DDX3X into an ATP-independent and sequence-selective RNA-binding protein.
Figure 3. RocA clamps DDX3X onto polypurine sequences in an ATP-independent manner.
(A and B) Fluorescence polarization assay between FAM-labeled (AG)10 or (UC)10 RNAs and human DDX3X recombinant proteins in the presence of AMP-PNP (A) or ADP + Pi (B).
(C and D) Correlations of tetramer motifs enriched in Bind-n-Seq with eIF4A1 and DDX3X proteins in the presence of RocA and ADP + Pi. The data were normalized to motif frequency in the sequenced input RNA. The motifs containing polypurine tetramers and trimers are highlighted in magenta and pink, respectively. The logos of the enriched motifs (defined as motifs with mean + 2 SD or more) are shown (D).
See also Figure S4.
The sequence bias led us to comprehensively explore the preference of DDX3X induced by RocA. We investigated a full suite of possible sequence combinations and compared the motifs with the motif preferentially bound by eIF4A1. For this purpose, we performed RNA Bind-n-Seq (Lambert et al., 2014; Iwasaki et al., 2016). Briefly, randomized 30-nucleotide (nt) long RNAs bound to SBP-tagged eIF4A1 and DDX3X proteins (Figure S4A) were isolated on streptavidin-conjugated beads and sequenced. In the presence of ADP, the binding between DEAD-box proteins and RNA is weakened. However, RocA allowed a fraction of RNA bound to DDX3X to be recovered, as reported for eIF4A1 (Iwasaki et al., 2016) (Figure S4B). The enriched RNA sequences on DDX3X showed high similarity to those on eIF4A1 (Figure 3C); they were biased toward A- and G-repeat motifs (Figure 3C and 3D). Essentially the same sequence selectivity for DDX3X was observed in AMP-PNP as well (Figure S4C and S4D). Thus, RocA confers both DEAD-box RNA helicases with the same polypurine RNA selectivity.
Gln360 in DDX3X is essential for RocA binding
Analogous to the situation with eIF4A1, these data suggest that RocA anchors to the bimolecular cavity between DDX3X and purine bases. Given the structural alignment of eIF4A1 and DDX3X, however, the phenyl ring C of RocA (Figure 1A) that anchors it to eIF4A1 is likely to be incompatible with DDX3X binding (Figure S5A). Thus, we hypothesize that the binding mode between RocA and DDX3X is not completely the same as that between RocA and eIF4A1.
To understand the important amino acid residues in DDX3X for RocA targeting, we focused on the transcriptome of Aglaia odorata assembled in a previous study (Iwasaki et al., 2019). Considering that plants in the genus Aglaia resist RocA toxicity via amino acid substitutions in their eIF4A proteins (Iwasaki et al., 2019), we hypothesized that the same scenario exists for the alternative RocA target DDX3. Alignment of the assembled Aglaia DDX3 sequence to the DDX3 sequences in other species (Figure 4A) confirmed that the amino acid substitutions in potential RocA-binding sites or their proximal sites were Aglaia-specific (Figure 2D and S5A).
Figure 4. Pro/Leu substitutions for Gln360 in Aglaia DDX3 confer RocA resistance.
(A) Alignments of DDX3 protein sequences from representative eukaryotes with de novo-assembled A. odorata DDX3 proteins.
(B) Fluorescence polarization assay between FAM-labeled (AG)10 RNAs and mutant DDX3X proteins. RocA or DMSO was added in the presence of ADP + Pi.
(C) Fluorescent labeling of WT and mutant DDX3X proteins with RocA-O-NBD in the presence of (AG)10 RNAs. RocA itself was used as a control.
See also Figure S5.
Thus, we biochemically screened the RocA resistance-conferring substitutions. Using ATP-independent polypurine RNA binding as a marker of RocA activity, we monitored affinity changes in the mutant proteins (Figure S5B). Strikingly, substitution of Gln360 with Pro/Leu abolished RocA-induced polypurine RNA clamping (Figure 4B and Table 1). This defect did not result from a disruption in basal RNA-binding ability since the mutant proteins still retained their basal affinities for RNA in the absence of RocA (Figure S5C). Consistent with RocA affinity changes, the mutant proteins were less susceptible to RocA-O-NBD labeling than the wild-type (WT) proteins (Figure 4C), showing the importance of Gln360 for DDX3X targeting by RocA.
To test whether Gln360 is responsible for RocA binding to DDX3X, we measured the affinity of RocA for a DDX3X•AMP-PNP•RNA complex via microscale thermophoresis (Wienken et al., 2010). As expected, mutations in Gln360 perturbed the interaction of the complex with RocA (Table 2).
Table 2. Summary of the Kd (μM) values between the RNA•DEAD-box protein complex and RocA.
Microscale thermophoresis of the RNA•protein complex fluorescently labeled with His tags was performed. ND, not determined.
| Protein | (AG)5A AMP-PNP RocA |
|---|---|
| eIF4A1 | 0.014 ± 0.0023 |
| DDX3X | 0.42 ± 0.31 |
| Gln360Pro | 3.4 ± 1.5 |
| Gln360leu | ND |
In summary, to avoid the deleterious effects of RocA, plants in the genus Aglaia preserve eIF4A and DDX3 with substitutions in their RocA-binding pockets. We note that the corresponding residue in eIF4A1 (Gln195) (Figure S5D) significantly contributes to the RocA binding energy (Iwasaki et al., 2019), and mutation in yeast eIF4A confers RocA resistance (Sadlish et al., 2013), suggesting the key role of the Gln residues in RocA targeting for the both DEAD-box proteins.
RocA•DEAD-box helicase complexes repress translation in a dominant-negative manner
Since RocA converts eIF4A1 into a dominant-negative repressor (Iwasaki et al., 2016), we hypothesized that DDX3X targeted by RocA functions in a similar way. To test this possibility, we added recombinant proteins into an in vitro translation system in RRL. As with eIF4A1, supplementation of DDX3X proteins further enhanced RocA-mediated translational repression of polypurine reporter mRNA (Figure 5A) but not control reporter mRNA with CAA repeats in the 5′ UTR (Figure S6A). The milder translation reduction for DDX3X than for eIF4A1 was consistent with the different affinities of RocA for these proteins (Table 2). Thus, irrespective of the helicase binding partner, the effect of RocA was dominant-negative, and the presence of more target proteins resulted in stronger translational repression (Figure 5B).
Figure 5. The DDX3X•RocA complex inhibits translation in a dominant-negative manner.
(A) In vitro translation from a Renilla luciferase reporter mRNA possessing 7×AGAGAG motifs in the 5′ UTR in RRL with 1 μM RocA. Recombinant eIF4A1 or DDX3X proteins were added to the reaction. Data represents mean ± SD. Three replicates were considered, except 6 replicates at protein concentration “0”.
(B) Schematic representation of RocA-mediated dominant-negative translational repression. Clamped eIF4A1/2 or DDX3X proteins on polypurine motifs in 5′ UTRs confer scanning ribosomes with steric hindrance.
eIF4A1, eIF4A2, and DDX3X abundance predicts RocA sensitivity in cancer cells
RocA has been reported to exert antitumor activity of different magnitudes among different cancer cell types (Santagata et al., 2013) for unknown reasons. The dominant-negative effect of RocA (Figure 5) led us to hypothesize that cancer cell sensitivity to RocA may be correlated with the levels of the RocA targets eIF4A1, eIF4A2, and DDX3X.
To test this hypothesis, we explored the landscape of eIF4A1, eIF4A2, and DDX3X abundance across cancer cell types with published RNA-Seq databases (Barretina et al., 2012; Klijn et al., 2015). Taking the broad range of RocA target levels in cancer cells into account, we chose Hs 936.T, A375, NCI-H1650, and NCI-H520 cells (Figure S6B). The protein levels of the DEAD-box proteins were confirmed by Western blotting (Figure 6A) and quantified (Figure 6B). The order of the DEAD-box protein expression followed: NCI-H520 > NCI-H1650 > A375 > Hs 936.T.
Figure 6. eIF4A1, eIF4A2, and DDX3X levels determine RocA sensitivity in cancer cells.
(A and B) Western blotting of eIF4A1, eIF4A2, and DDX3X from Hs 936.T, A375, NCI-H1650, and NCI-H520 cells (A). β-Actin was used as an internal control. The quantification is shown in (B).
(C and D) Cell viability upon RocA treatment for 48 h in each cell line (C). The IC50 values are shown (D).
(E) Box plots of the changes in translation observed via ribosome profiling of the different cell lines. The cells were treated with 0.3 μM RocA. mRNAs are classified into groups with or without tetramer polypurine motifs in their 5' UTRs.
See also Figure S6.
The abundance of RocA targets accounted for the cytotoxicity of RocA. We tested cell viability upon RocA treatment and observed different half-maximal inhibitory concentration (IC50) values among the cells (Figure 6C and 6D). The IC50 values were negatively correlated with the levels of RocA-targeting DEAD-box proteins: higher protein expression was associated with stronger cytotoxicity (Figure 6B and 6D).
We hypothesized that cytotoxicity is caused by translational repression. To survey the translational impact of RocA in the selected cells, we performed ribosome profiling (Ingolia et al., 2009; Iwasaki and Ingolia, 2017). Cancer cell lines were treated with 0.3 or 3 μM RocA for 30 min, a duration that was chosen because mRNA abundance changes due to drug treatment are limited during this time frame (Iwasaki et al., 2016), and used for ribosome profiling library preparation. As previously reported in HEK293 cells (Iwasaki et al., 2016), the translational repression induced by RocA was mRNA-selective; specifically, it was biased toward mRNAs with polypurine motifs in their 5′ UTRs (Figure 6E, compare polypurine + to polypurine −). Although we observed this trend in all the tested cancer cells, the strength of the trend was different among the cells (Figure 6E). For example, the translational repression of the polypurine 5′ UTR in NCI-520 cells was stronger than that in Hs 936.T cells (Figure 6E) as associated with the abundance of eIF4A1, eIF4A2, and DDX3X (Figure 6A and 6B). Notably, this difference was clearer for the low dose (0.3 μM) of RocA than for the high dose (3 μM), which may have saturated translational repression (Figure S6C).
Overall, the correspondence among stronger translation inhibition by additional target proteins (Figure 5A and Figure 6E), differential target protein (eIF4A1, eIF4A2, and DDX3X) abundance (Figure 6A and 6B), and cell growth repression (Figure 6C and 6D) indicated that RocA functions in a dominant-negative manner that correlates with target protein abundance. These findings enable prediction of the effects of the compound on tumor cells.
Discussion
Earlier works have identified eIF4A1/2 as a target of rocaglates but failed to find DDX3X. This was probably due to the complicated nature of the action of RocA and the target proteins. A biochemical approach with biotinylated RocA and subsequent purification in a mammalian system (Chambers et al., 2013; Chambers et al., 2016) was not performed with polypurine RNA during purification, as we did in this study (Figure 2A). Given that RocA binds in the bimolecular cavity between polypurine RNA and the targeted DEAD-box protein (Iwasaki et al., 2019), the ability to capture the full target proteins may be limited. Moreover, the strong affinity of RocA for the eIF4A1•polypurine RNA complex (Table 2) may have hampered the identification of DDX3X. Similarly, the higher expression of eIF4A (Tif1 and Tif2) than DDX3 (Ded1) in yeast (Figure S6D) may have also masked the readout of genetic screening (Sadlish et al., 2013).
Despite its complexity, an O-NBD approach enabled us to identify DDX3X as an alternative target of RocA. This approach has advantages over affinity purification via classic biotinylated ligands. While biotin derivatives require purification with extensive washes, which may destroy the fragile complexes, O-NBD derivatives do not. This allows target labeling in solution under optimal conditions. Such an approach led to the identification of DDX3X in this study.
Given the conformational conflict between DDX3X and RocA positioned on eIF4A1 (Figure S5A), RocA likely binds to DDX3X at a different angle than it binds to eIF4A1. For example, whereas the substitution of Phe163 with Leu provides RocA resistance to eIF4A1 (Sadlish et al., 2013; Chu et al., 2016; Iwasaki et al., 2019), the same mutation in DDX3X (Val328Leu) does not affect the sensitivity of the protein to RocA (Figure 4B). Thus, the different amino acid residues may contribute to RocA binding in the two DEAD-box proteins, while Gln195 in eIF4A1 and Gln360 in DDX3X are both important (Figure 4).
In addition to clamping to polypurine RNA and engaging in scanning inhibition, rocaglates have been reported to induce a trans bystander effect (Chu et al., 2020); sequestration of eIF4A and complexation of eIF4F with mRNAs reduces the fraction available for active mRNA translation initiation irrespective of the presence of polypurine motifs. Given the role of DDX3X in translation (Lee et al., 2008; Lai et al., 2008; Lai et al., 2010; Soto-Rifo et al., 2012; Ku et al., 2019; Calviello et al., 2020), the trans bystander effect on translation observed in an earlier study could be partially explained by DDX3X sequestration.
Ubiquitously expressed DDX3X has a paralog, DDX3Y, encoded in the Y chromosome. Although the roles of DDX3Y are restricted in male germ cells (Kotov et al., 2017), the high similarity (~90% homology) between the proteins suggests that DDX3Y could also be targeted by RocA.
The cell type-specific expression patterns of DDX3 homologs might also explain the existence of rocaglate resistance-conferring substitutions in only a portion of Aglaia DDX3s (Figure 4). Given that rocaglates are biosynthesized in specific cell types in Aglaia, the DDX3 paralog expressed in the cells may need only to correspond to the compounds present. Indeed, Arabidopsis has 3 DDX3 paralogs (RH11, RH37, and RH52) with differential expression patterns among tissues (shoot apical meristem, guard cells, etc.) (Berardini et al., 2015; Klepikova et al., 2016).
DDX3X mutations are often associated with cancer progression, although both tumor-promoting and suppressing roles have been reported (He et al., 2018). A wide variety of cancer types are caused by overexpression of DDX3X (He et al., 2018), and such cancer cells may be more likely than other cell types to be sensitive to rocaglates because of the unique dominant-negative effects of the compounds.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Shintaro Iwasaki (shintaro.iwasaki@riken.jp).
Material availability
The materials generated in this study will be distributed upon request. There are restrictions to availability due to a Material Transfer Agreement (MTA).
Data and code availability
The results of ribosome profiling (GEO: GSE148636) and RNA-Bind-n-Seq (GEO: GSE150111) obtained in this study have been deposited in the National Center for Biotechnology Information (NCBI) database. All custom scripts are available upon request. Original images used for the figures are deposited in the Mendeley database (http://dx.doi.org/10.17632/syfc38x8md.1).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
T-Rex-293 (HEK293) cells (Thermo Fisher Scientific) and T-REx293 SBP-eIF4A1 (Phe163Leu-Ile199Met) eIF4A1SINI cells (Iwasaki et al., 2019) were cultured in DMEM (1×) + GlutaMAX-I (Thermo Fisher Scientific) supplemented with 10% FBS. Hs 936.T and A375 cells (American Type Culture Collection, ATCC) were grown in high-glucose DMEM (Nacalai Tesque) with 10% FBS, and NCI-H1650 and NCI-H520 cells (ATCC) were grown in RPMI 1640 medium (Thermo Fisher Scientific). All the cells were grown in a humidified incubator at 5% CO2 and 37°C.
The sex of the HEK293 and A375 cells was female, and that of the Hs 936.T, NCI-H1650, and NCI-H520 cells was male.
Bacterial strains
E. coli BL21 Star (DE3) cells (Thermo Fisher Scientific) were transformed with the corresponding plasmids, grown in LB medium with ampicillin at 37°C, and used for recombinant protein expression.
METHOD DETAILS
DNA constructs
pColdI-DDX3X helicase core WT, Pro324Thr, Val328Leu, Gln360Pro, Gln360Leu, and Arg363Asn
A DNA fragment encoding the human DDX3X helicase core (amino acids 132–607) was amplified from HeLa cell cDNA and inserted into pColdI (TaKaRa) downstream of the His tag with In-Fusion HD (TaKaRa). Amino acid substitutions were introduced by site-direct mutagenesis.
pColdI-eIF4A1 WT and Phe163Leu-Ile199Met (with Aglaia mutations)
These plasmids have been reported previously (Iwasaki et al., 2019).
pColdI-eIF4A2
A DNA fragment encoding human EIF4A2 was cloned from HeLa cell cDNA and inserted into pColdI (TaKaRa) downstream of the His tag with In-Fusion HD (TaKaRa).
pColdI-SBP-eIF4A1 and pColdI-SBP-DDX3X helicase core
A DNA fragment encoding SBP-eIF4A1 was PCR-amplified from pcDNA5/FRT/TO-SBP-eIF4A1 (Iwasaki et al., 2016) and inserted into pColdI (TaKaRa) downstream of the His tag with In-Fusion HD (TaKaRa) to construct pColdI-SBP-eIF4A1. A DNA fragment containing the DDX3X helicase core (amino acids 132–607) was PCR-amplified from the WT pColdI-DDX3X helicase core and inserted into the pColdI-SBP backbone using In-Fusion HD (TaKaRa).
Purification of recombinant proteins
E. coli BL21 Star (DE3) cells (Thermo Fisher Scientific) transformed with the pColdI series (described above) were cultivated to an OD600 of 0.4~0.6 at 37°C in 1 l of LB medium with ampicillin. After chilling at 4°C for 30 min, the cells were cultured at 15°C overnight with 1 mM IPTG, collected by centrifugation at 2280 × g for 30 min, flash-frozen in liquid nitrogen, and stored at −80°C.
The pellets were resuspended in lysis buffer (20 mM HEPES-NaOH pH 7.5, 500 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, and 0.5% NP-40) and homogenized by sonication on ice. The lysate was clarified by centrifugation at 10,000 × g for 20 min at 4°C, and then the supernatant was collected and incubated with 3 ml of pre-equilibrated Ni-NTA Agarose beads (Qiagen) for 1 h at 4°C. The beads were washed in a gravity column (Bio-Rad) with 50 ml of high-salt wash buffer (20 mM HEPES-NaOH pH 7.5, 1 M NaCl, 20 mM imidazole, and 10 mM β-mercaptoethanol) and then 25 ml of low-salt wash buffer (20 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 20 mM imidazole, and 10 mM β-mercaptoethanol). The His-tagged proteins were finally eluted into 8 ml of elution buffer (20 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 250 mM imidazole, 10 mM β-mercaptoethanol, and 10% glycerol).
Using an NGC chromatography system (Bio-Rad), the eluted proteins were loaded onto a HiTrap Heparin HP column (1 ml, GE Healthcare) and fractionated via a gradient increase in salt with mixing buffer A (20 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 10% glycerol, and 1 mM DTT) and buffer B (buffer A with 1 M NaCl). The peak fractions of the proteins were collected, and the buffer (NAP-5 or PD-10, GE Healthcare) was exchanged with storage buffer (20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 10% glycerol, and 1 mM DTT). The proteins were then concentrated with a Vivaspin 6 (10 kDa MWCO) (Sartorius) or an Amicon Ultra-4 (10 kDa MWCO) (Millipore) following the manufacturer’s instructions. The concentrated proteins were flash-frozen in liquid nitrogen and then stored at −80°C.
Synthesis of the RocA-O-NBD probe
The RocA-O-NBD probe was synthesized as shown in Figure S2A. According to the methods in a previous report (Iwasaki et al., 2019), RocA propargyl amide derivatives (2, Figure S2A) were synthesized from aglafoline (methyl rocaglate, MedChem Express). All reactions were carried out under an argon atmosphere with dry solvents unless otherwise stated.
To a stirred solution of 3-azido-1-propanol (5.8 μl, 63 μmol) in dry CH2Cl2 (1 ml) was added NBD-F (35 mg, 189 μmol) and N,N-diisopropylethylamine (DIPEA) (43 μl, 251 μmol). After stirring at room temperature for 14 h, the solvent was removed under reduced pressure. The obtained residue was purified by silica gel column chromatography (hexane:EtOAc = 4:1 to 1:1) and gel permeation chromatography (CHCl3) to give an azide O-NBD linker (1) (15.1 mg, 57.2 μmol, 91%).
A solution of tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) in DMSO (95.0 μl, 2.0 mg/ml, 0.36 μmol), an aqueous solution of sodium ascorbate (35.5 μl, 20 mg/ml, 3.6 μmol), an aqueous solution of CuSO4 (28.5 μl, 2.0 mg/ml, 0.36 μmol), DMSO (400 μl), and H2O (200 μl) were added to a 10 ml amber eggplant-shaped flask. After stirring at room temperature for 10 min, a solution of propargyl amide (2) (6.2 mg, 0.012 mmol) and azide O-NBD linker (1) (5.5 mg, 0.021 mmol) in DMSO (400 μl) and H2O (200 μl) was added to the mixture. The resulting mixture was stirred at 40°C for 21 h. The solvent was removed by freeze-drying, and water was added to the residue. The resulting mixture was extracted with CH2Cl2 four times. The combined organic phases were dried over Na2SO4, filtered, and concentrated in vacuo. The residue was purified by silica gel column chromatography (MeOH:EtOAc = 1:19) to yield the RocA-O-NBD probe (3.0 mg, 0.00385 mmol, 32%) as a yellow solid. The following values were obtained:
1H-NMR (400 MHz, CD3OD): δ 8.57 (d, J = 8.3 Hz, 1H), 7.42 (s, 1H), 7.14 (d, J = 9.2 Hz, 2H), 7.03 (m, 3H), 6.90–6.93 (m, 2H), 6.79 (d, J = 8.3 Hz, 1H), 6.63 (d, J = 9.2 Hz, 2H), 6.26 (d, J = 2.3 Hz, 1H), 6.07 (d, J = 2.3 Hz, 1H), 4.82 (d, J = 6.2 Hz, 1H), 4.56 (t, J = 6.7 Hz, 2H), 4.40 (d, J = 15.6 Hz, 1H), 4.25–4.31 (m, 4H), 3.86 (m, 1H), 3.82 (s, 3H), 3.79 (s, 3H), 3.67 (s, 3H), 2.40–2.46 (m, 2H).
13C-NMR (126 MHz, CD3OD): δ 172.6, 165.1, 162.2, 159.9, 159.2, 155.3, 147.0, 146.8, 145.5, 139.2, 136.1, 131.0, 130.3, 130.0, 129.5, 129.2, 128.6, 127.4, 124.6, 113.2, 109.4, 106.9, 102.6, 95.2, 93.1, 89.9, 80.9, 68.7, 56.8, 56.0, 55.4, 52.4, 47.8, 35.9, 30.3.
IR (neat): 3419, 2923, 2853, 1610, 1547, 1453, 1320, 1147 cm−1.
HRMS (ESI): Calculated for C39H37N7NaO11 [M+Na]+, 802.2443: found, 802.2474. [α]D25: −26.07 (c 0.15, MeOH).
Reporter mRNA preparation
Reporter mRNA of Renilla luciferase with CAA repeats or 7×AGAGAG motifs was prepared as previously described (Iwasaki et al., 2016). Briefly, the DNA fragment was PCR-amplified from psiCHECK2-CAA repeats or psiCHECK2-7×AGAGAG motifs (Iwasaki et al., 2016) and used for in vitro transcription with a T7-Scribe Standard RNA IVT Kit (CELLSCRIPT), after which capping and polyadenylation were performed with a ScriptCap m7G Capping System, a ScriptCap 2′-O-Methyltransferase Kit, and an A-Plus Poly(A) Polymerase Tailing Kit (CELLSCRIPT).
In vitro translation in RRL
RRL (nuclease-treated) (Promega) was used to monitor the effects of the RocA and RocA-O-NBD on the translation of Renilla luciferase mRNA with 7× AGAGAG motifs as previously described (Iwasaki et al., 2016). Briefly, a reaction consisting of 5 μl of lysate, 2 μl of H2O, 1 μl of RocA or RocA-O-NBD dissolved in 1% DMSO, 1 μl of 500 nM mRNA reporter, and 1 μl of premix [100 μM Amino Acid Mixture Minus Methionine (Promega), 100 μM Amino Acid Mixture Minus Leucine (Promega), and 1 U/μl ScriptGuard RNase Inhibitor (CELLSCRIPT)] at 30°C for 1 h.
For the assay with recombinant proteins, the reaction was prepared with 1.3 μl of lysate, 4.2 μl of dilution buffer (20 mM HEPES-NaOH pH 7.5, 10 mM NaCl, 25 mM KCl, 1.1 mM MgCl2, and 1 mM DTT), 2.5 μl of purified recombinant proteins dissolved in storage buffer, 0.5 μl of 20 μM RocA dissolved in 2% DMSO, 1 μl of 50 nM mRNA reporter, and 0.5 μl of 2× premix [200 μM Amino Acid Mixture Minus Methionine (Promega), 200 μM Amino Acid Mixture Minus Leucine (Promega), and 2 U/μl ScriptGuard RNase Inhibitor (CELLSCRIPT)] and incubated at 30°C for 30 min. After quenching translation by adding 30 μl of 1× Passive Lysis Buffer (Promega), 10 μl of the mixture was transferred into a 96-well white assay plate (Costar) to react with the reagents of the Renilla-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. Luminescence was detected with a GloMax Navigator (Promega).
RocA-O-NBD labeling assay
RocA (Sigma-Aldrich) and RocA-O-NBD were dissolved in DMSO. In vitro translation in RRL was performed as described above with 3 μM RocA or RocA-O-NBD.
Reaction mixtures (20 μl) were prepared. The mixtures consisted of 10 μM DDX3X core WT, Gln360Pro, or Gln360Leu protein; 50 μM 5′ FAM-labeled RNA oligonucleotides; and 10 μM RocA or RocA-O-NBD in 20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM MgCl2, and 1 mM AMP-PNP. The mixtures were incubated at 37°C for 15 min, and the reaction was stopped at 4°C. For eIF4A1, 10 μM RNA was incubated for 5 min.
The labeled proteins were mixed with loading dye supplemented with 125 mM Tris-HCl (pH 7.5) and run on SDS-PAGE gels. Heat denaturing was avoided because nonspecific labeling occurred. The gels were fixed with 50% methanol and 7% acetic acid. The fluorescent signals on the gels were captured by a PharosFX imager (Bio-Rad). The total protein was stained with Coomassie Brilliant Blue and imaged with an ODYSSEY CLx (LI-COR Biosciences).
Immunopurification of NBD-labeled proteins
A RocA-O-NBD labeling assay was performed in RRL as described above with a 100 μl reaction volume and a 90 min incubation. Unreacted free RocA-O-NBD was removed by a MicroSpin G-25 Column (GE Healthcare) and equilibrated with equilibration buffer (20 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM MgCl2, and 1 mM DTT) containing 0.1% Triton X-100. Five micrograms of an anti-NBD antibody (Acris Antibodies GmbH, BP086) was bound to 30 μl of Dynabeads Protein G (Thermo Fisher Scientific). The beads were incubated with the column eluate at 4°C for 1 h and washed 3 times with equilibration buffer containing 0.1% Triton X-100. The bound proteins were eluted with loading dye.
Mass spectrometric analysis
In-gel digestion of RocA-O-NBD-labeled proteins in RRL
After immunopurification of NBD-labeled proteins from the RocA-O-NBD labeling of RRL described above, the bound proteins were run on SDS-PAGE gels. The fluorescent bands were subjected to in-gel tryptic digestion. The gel slices were diced into 1 mm cubes, washed with Milli-Q water, and destained with 30% acetonitrile (MeCN) in 20 mM Tris-HCl (pH 8.5). The gels were then dehydrated with MeCN and dried by vacuum centrifugation. The proteins were reduced with DTT (Thermo Fisher Scientific) and then alkylated with iodoacetamide (Thermo Fisher Scientific). The gels were washed with Milli-Q water, dehydrated by addition of MeCN, and dried by vacuum centrifugation. To the dried gels, Sequencing Grade Modified trypsin (Promega) were added wtih TG buffer [50 mM Tris-HCl (pH 8.0), 0.01% (w/v) n-decyl-β-D-glucopyranoside (DG)]. After incubating the gels overnight at 37°C, the excess solution and the digests in the gels, which were extracted twice with extraction buffer (TG buffer:MeCN=1:1), were pooled, concentrated to remove MeCN by vacuum centrifugation, and subjected to immunopurification as described below.
In-solution digestion of RocA-O-NBD-labeled human DDX3X recombinant protein
After labeling of human DDX3X recombinant protein with RocA-O-NBD as described above, unreacted free RocA-O-NBD was removed by chloroform-methanol precipitation. The precipitate was dissolved in denaturation buffer [7 M guanidinium chloride (GuHCl), 0.5 M Tris-HCl (pH 8.5)] and incubated for 30 min at 37°C. Following reduction and alkylation of the proteins with DTT and iodoacetamide, respectively, Milli-Q water was added to the protein solution to reduce the final concentration of GuHCl to below 0.7 M. The proteins were enzymatically digested with lysyl endopeptidase (mass spectrometry grade; FUJIFILM Wako Chemical) for 1 h at 37°C and Sequencing Grade Modified Trypsin (Promega) overnight at 37°C. The digests were desalted using a MonoSpin C18 S column (GL Sciences). The eluate was concentrated by vacuum centrifugation with supplementation of DG. The final solution, which contained approximately 0.01% DG, was subjected to immunopurification as described below.
Immunopurification of NBD-labeled peptides
After conjugation of an anti-NBD antibody to Protein G Mag Sepharose beads (GE Healthcare), the beads were mixed with peptide solution and incubated at 4°C overnight with rotation. The beads were washed 3 times with TG buffer and then 3 times with 0.01% (w/v) DG. The bound NBD-labeled peptides were eluted with a 0.15% (v/v) TFA aqueous solution and desalted using a MonoSpin C18 S column (GL Sciences). The eluted peptides were concentrated by vacuum centrifugation with DG supplementation. The final solution contained approximately 0.01% DG.
Mass spectrometry
LC-MS/MS analysis for protein identification was performed mainly using an Advance nanoLC (Bruker-Michrom) and an LTQ linear ion trap (IT) mass spectrometer (Thermo Fisher Scientific) equipped with a C18 capillary column (0.075 mm i.d. × 150 mm, 3 μm, Nikkyo Technos) using a linear gradient (25 min, 5–35% MeCN/0.1% FA) at a flow rate of 300 nl/min. For analyses of NBD-labeled peptides, an LC-fluorescence-MS system was used. This system was composed of an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nanoESI source (Nikkyo Technos) and an UltiMate 3000 nanoLC system (Thermo Fisher Scientific). An LIF727 fluorescence detector fitted with a 6 nl flow cell (GL Sciences) was fitted between the mass spectrometer and the nanoLC system. The parameters of the LIF727 were as follows: photomultiplier tube (PMT) voltage, 700 V; response time, 0.5 s; output range, 1 RFU/FS1V; and bandpass filter, 500–600 nm. The peptides were separated using an Acclaim PepMap 100 C18 analytical column with nanoViper fittings (0.075 mm i.d. × 150 mm, 3 μm, 100 Å, Thermo Fisher Scientific) after trapping on an Acclaim PepMap 100 C18 trap column with nanoViper fittings (0.1 mm i.d. × 20 mm, 5 μm, 100 Å, Thermo Fisher Scientific). For the analytical column, mobile phase A [distilled water containing 0.1% (v/v) FA and 4% (v/v) MeCN] and mobile-phase B [MeCN containing 0.1% (v/v) FA] were used. Mobile phase C [distilled water containing 0.1% (v/v) TFA] and mobile-phase D [MeCN containing 0.1% (v/v) TFA] were used for the trap column. The peptides were eluted with a gradient of mobile phase A and mobile phase B at a flow rate of 250 nl/min [representative gradient: 0–15% B (15 min), 15–35% (35 min), 35–70% (14 min), 70–90% (1 min), 90% (4 min), and 90–0% (1 min)]. Survey scans were performed in an m/z range of 250–1600 in an Orbitrap mass analyzer (resolution: 60,000), and MS/MS spectra were acquired in the linear IT in data-dependent acquisition (DDA) mode. For detailed MS/MS acquisition of the NBD-labeled peptides, collision-induced dissociation (CID) or higher-energy collisional dissociation (HCD) was applied in the Orbitrap (resolution: 15,000).
Data analysis
MS/MS ion searching was performed using the MASCOT server (Matrix Science) or SEQUEST (Thermo Fisher Scientific) via Proteome Discoverer (Thermo Fisher Scientific). The theoretical mass values of NBD-labeled peptides were calculated by Xcalibur software (Thermo Fisher Scientific). The representative parameters for the identification of NBD-labeled peptides were as follows: precursor mass tolerance, 10 ppm; fragment mass tolerance, 0.8 Da; enzyme, trypsin; maximum number of missed cleavage sites, 3; fixed modification, carbamidomethylation of cysteine (+57.021464 Da); dynamic modifications, N-terminal acetylation (+42.010565 Da) and NBD modification of lysine (+163.001791 Da).
Western blot analysis
Anti-pan-eIF4A (Santa Cruz Biotechnology, H-5, sc-377315), anti-eIF4A1 (Cell Signaling Technology, #2490S), anti-eIF4A2 (Abcam, ab31218), anti-β-Actin (LI-COR Biosciences, 926-42212), and anti-DDX3X (Cell Signaling Technology, #8192S) antibodies were used as primary antibodies. IRDye 800CW anti-rabbit IgG (LI-COR Biosciences, 926-32211) and IRDye 800CW anti-mouse IgG (LI-COR Biosciences, 926-32210) were used as secondary antibodies. Images were captured and quantified by an ODYSSEY CLx (LI-COR Biosciences).
siRNA knockdown and cell viability assay
On 24-well plates, 2 × 104 naïve HEK293 cells or HEK293 SBP-eIF4A1 (Phe163Leu-Ile199Met) eIF4A1em1SINI (Iwasaki et al., 2019) were seeded one day before transfection. The cells were transfected with 55 nM DDX3X-specific siRNA (Dharmacon, L-006874-02-0005), eIF4A2-specific siRNA (Dharmacon, L-013758-01-0005), and/or control siRNA (Dharmacon, D-001206-13-05) using the TransIT-X2 Transfection Reagent System (Mirus). After two days of incubation, 4 × 103 cells were seeded in 96-well microplates and incubated for 6 h. Then, siRNA knockdown was repeated again as described above. After 24 h of incubation, the cells were treated with RocA or DMSO for 48 h. Cell viability was measured with a RealTime-Glo MT Cell Viability Assay System (Promega). Luminescence was detected by a GloMax (Promega). The data were normalized to those for the DMSO-treated group and were fitted by Igor Pro8 (WaveMetrics).
Fluorescence polarization assay
A reaction was prepared with 0–10 μM recombinant proteins, 10 nM FAM-labeled RNA ([AG]10 or [UC]10), 1% DMSO or 50 μM RocA, 1 mM AMP-PNP, 20 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 1 mM DTT, and 5% glycerol and incubated at room temperature for 30 min. The reaction mixture was transferred into black 384-well microplates (Corning), and the anisotropy change was measured using an Infinite F-200 PRO (Tecan). For ADP conditions, AMP-PNP was substituted with 1 mM ADP and 1 mM Na2HPO4. The data were fitted to the Hill equation by Igor Pro 8 (WaveMetrics).
Microscale thermophoresis analysis
Microscale thermophoresis analysis was performed at 23°C with a Monolith NT.115 (NanoTemper) unit according to the manufacturer’s protocol. The eIF4A1, DDX3X WT, DDX3X Gln360Pro, and DDX3X Gln360Leu proteins were labeled with a fluorescent dye using a Monolith His-Tag Labeling Kit RED-tris-NTA (NanoTemper) and adjusted to a final concentration of 50 nM. The (AG)5A (Fasmac, Japan) and AMP-PNP solutions were added to the protein solutions at final concentrations of 50 μM and 2.5 mM, respectively. The resultant protein/RNA/AMP-PNP mixtures were titrated with RocA in PBS (−) buffer (Nacalai Tesque) supplemented with 5 mM MgCl2, 1 mM DTT, and 0.05% Tween-20. Each experiment was repeated three times for calculation of the dissociation constants.
RNA Bind-n-Seq
One hundred picomoles of SBP-tagged recombinant protein (DDX3X helicase core or eIF4A1) was incubated with 60 μl of Dynabeads M-270 Streptavidin (Thermo Fisher Scientific), which were pre-equilibrated with equilibration buffer containing 1% Triton X-100, at 4°C for 30 min. The protein-tethered beads were treated with 2 U/μl Micrococcal Nuclease (TaKaRa) in 1× Micrococcal Nuclease Buffer (TaKaRa), 0.5× equilibration buffer and 0.5% Triton X-100 in a total volume of 30 μl at 25°C for 30 min. After incubation, the beads were washed 5 times with 60 μl of equilibration buffer containing 1% Triton X-100, 1 M NaCl, and 5 mM EGTA (pH 7.4) and rinsed twice with the same volume of equilibration buffer containing 0.1% Triton X-100. The rinsed beads were then incubated with a 50 μM concentration of oligonucleotide (5′-CTCTTTCCCTACACGACGCTCTTCCGATCT-N30-ATCGTAGATCGGAAGAGCACACGTCTGAA-3′; the letters in bold represent the DNA sequence, and N represents a random RNA sequence; Gene Design) in 30 μl of equilibration buffer containing 0.1% Triton X-100, 0.33 U/μl SUPERase•In RNase Inhibitor (Thermo Fisher Scientific), 2 mM ADP (Wako Pure Chemical Industries) and 2 mM Na2HPO4 with 3 μM RocA (or 1% DMSO). Following a reaction at 37°C for 30 min, the beads were washed 5 times with equilibration buffer containing 0.1% Triton X-100, 2 mM ADP, and 2 mM Na2HPO4 with 3 μM RocA or 1% DMSO. The bead-tethered protein-RNA complex was eluted using 30 μl of equilibration buffer containing 0.1% Triton X-100, 5 mM D-biotin (Invitrogen), 2 mM ADP, and 2 mM Na2HPO4 with 3 μM RocA or 1% DMSO at 4°C for 30 min. The eluted RNAs were purified using an Oligo Clean & Concentrator Kit (Zymo Research) and converted into a DNA library as described in the ribosome profiling section. RNA Bind-n-Seq was also performed with 2 mM AMP-PNP (instead of 2 mM ADP and 2 mM Na2HPO4) with a 1 μM concentration of oligonucleotide.
Ribosome profiling
Library preparation for ribosome profiling was performed as previously described (McGlincy and Ingolia, 2017; Iwasaki et al., 2019). Cells were treated with RocA (0.3 μM or 3 μM) or DMSO (final concentration 0.1%) for 30 min. Cell lysates containing 5 μg of total RNA were treated with 10 U of RNase I (Epicentre) at 25°C for 45 min. Sucrose cushions were used to collect ribosomes. The RNAs ranging from 26 to 34 nt in length were selected on a 15% urea PAGE gel, dephosphorylated, and linker-ligated. rRNA was depleted with a Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (Illumina). Following reverse transcription, circularization, and PCR amplification, the DNA libraries were prepared and sequenced on a HiSeq 4000 platform (Illumina).
Analysis of deep sequencing data
Ribosome profiling
The ribosome profiling reads were processed as described previously (Iwasaki et al., 2016). The 5' UTR sequences were downloaded from the University of California, Santa Cruz (UCSC) genome browser. The translation change caused by RocA was calculated by DESeq (Anders and Huber, 2010) and then renormalized to the total mitochondrial footprint (used as the internal spike-in).
RNA Bind-n-Seq
Adapter sequences were trimmed from the reads. Given the high diversity of random sequences (430) compared to the sequence depth, any identical sequences in the library most likely originated from PCR duplication. Thus, such reads were removed. The frequency and enrichment of tetramer motifs were calculated as previously described (Iwasaki et al., 2016). The enriched motifs were defined as those with values more than a mean + 2 SD.
All custom R scripts used for data analysis are available upon request.
QUANTIFICATION AND STATISTICAL ANALYSIS
For Figure 1C, 2E, 3A, 3B, 4B, 5A, 6C, 6D, S1B, S1D, S1E, S5C, and S6A, the data are presented as the mean and mean ± SD (n = 3). At protein concentration “0” in Figure 5A, 6 replicates were considered. Fitting curve was drawn by Igor Pro 8 (WaveMetrics) to calculate the IC50 ± SD and Kd ± SD shown in Figure 6C, Table 1, and Table 2, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-NBD antibody | Acris Antibodies GmbH | Cat#BP086; RRID: AB_972399 |
| Mouse monoclonal Anti-pan-eIF4A antibody | Santa Cruz Biotechnology | Cat#sc-377315; RRID: AB_2868449 |
| Rabbit polyclonal anti-eIF4A1 antibody | Cell Signaling Technology | Cat#2490S; RRID: AB_823487 |
| Rabbit polyclonal anti-eIF4A2 antibody | Abcam | Cat#ab31218; RRID: AB_732123 |
| Mouse monoclonal anti-β-actin antibody | LI-COR Biosciences | Cat#926-42212; RRID: AB_2756372 |
| Rabbit monoclonal anti-DDX3X antibody | Cell Signaling Technology | Cat#8192S; RRID: AB_10860416 |
| Goat polyclonal anti-rabbit IgG antibody conjugated with IRDye 800CW | LI-COR Biosciences | Cat#926-32211; RRID: AB_621843 |
| Goat polyclonal anti-mouse IgG antibody conjugated with IRDye800CW | LI-COR Biosciences | Cat#926-32210; RRID: AB_621842 |
| Bacterial and Virus Strains | ||
| E. coli: BL21 Star (DE3) | Thermo Fisher Scientific | Cat#C601003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Rocaglamide A | Sigma-Aldrich | Cat#SML0656 |
| Aglafoline | MedChem Express | Cat#HY-19354 |
| RocA-O-NBD | This study | N/A |
| Adenylyl-imidodiphosphate (AMP-PNP) | Roche | Cat#10102547001 |
| ADP | Wako Pure Chemical Industries | Cat# 019-25091 |
| D-biotin | Invitrogen | Cat#B20656 |
| Trifluoroacetic acid (TFA) | Kanto Chemical Co., Inc. | Cat#40578-1B |
| Distilled water containing 0.1% (v/v) formic acid (FA) | Kanto Chemical Co., Inc. | Cat#01922-64 |
| Acetonitrile (MeCN) containing 0.1% (v/v) FA | Kanto Chemical Co., Inc. | Cat#16245-64 |
| Recombinant protein: His-DDX3X helicase core WT | This study | N/A |
| Recombinant protein: HIs-DDX3X helicase core Pro324Thr | This study | N/A |
| Recombinant protein: His-DDX3X helicase core Val328Leu | This study | N/A |
| Recombinant protein: His-DDX3X helicase core Gln360Pro | This study | N/A |
| Recombinant protein: His-DDX3X helicase core Gln360Leu | This study | N/A |
| Recombinant protein: His-DDX3X helicase core Arg363Asn | This study | N/A |
| Recombinant protein: His-eIF4A1 WT | Iwasaki et al. 2019 | N/A |
| Recombinant protein: His-eIF4A1 Phe163Leu-Ile199Met | Iwasaki et al. 2019 | N/A |
| Recombinant protein: His-eIF4A2 WT | This study | N/A |
| Recombinant protein: His-SBP-eIF4A1 | This study | N/A |
| Recombinant protein: His-SBP-DDX3X helicase core | This study | N/A |
| Sequencing Grade Modified trypsin | Promega | Cat#v5113 |
| Lysyl endopeptidase (Mass Spectrometry Grade) | FUJIFILM Wako Chemical | Cat#125-05061 |
| RNase I | Epicentre | Cat#N6901K |
| Critical Commercial Assays | ||
| DMEM (1×) + GlutaMAX-I | Thermo Fisher Scientific | Cat#10566-016 |
| High Glucose DMEM | Nacalai Tesque | Cat#16971-55 |
| RPMI 1640 Medium | Thermo Fisher Scientific | Cat#A1049101 |
| Ni-NTA Agarose | Qiagen | Cat#30210 |
| HiTrap Heparin HP column 1 ml | GE Healthcare | Cat#17040601 |
| NAP-5 | GE Healthcare | Cat#17085302 |
| PD-10 | GE Healthcare | Cat#17085101 |
| Vivaspin 6, 10 kDa MWCO | GE Healthcare | Cat#28932296 |
| Amicon Ultra-4, 10 kDa MWCO | Millipore | Cat#UFC8010 |
| T7-Scribe Standard RNA IVT Kit | CELLSCRIPT | Cat#C-AS3107 |
| ScriptCap m7G Capping System | CELLSCRIPT | Cat#C-SCCE0625 |
| ScriptCap 2′-O-Methyltransferase Kit | CELLSCRIPT | Cat#C-SCMT0625 |
| A-Plus Poly(A) Polymerase Tailing Kit | CELLSCRIPT | Cat#C-PAP5104H |
| Rabbit Reticulocyte Lysate System, Nuclease Treated | Promega | Cat#L4960 |
| Renilla-Glo Luciferase Assay System | Promega | Cat#E2720 |
| MicroSpin G-25 Column | GE Healthcare | Cat#27532501 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat#10004D |
| Protein G Mag Sepharose beads | GE Healthcare | Cat# 28-9440-08 |
| MonoSpin C18 S | GL Science | Cat#5010-21701 |
| TransIT-X2 Transfection Reagent System | Mirus | Cat#MIR6000 |
| RealTime-Glo MT Cell Viability Assay System | Promega | Cat#G9713 |
| Monolith His-Tag Labeling Kit RED-tris-NTA | NanoTemper | Cat#MO-L018 |
| Dynabeads M-270 Streptavidin | Thermo Fisher Scientific | Cat#DB65305 |
| Oligo Clean & Concentrator Kit | Zymo Research | Cat#D4061 |
| Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) | Illumina | Cat#MRZG12324 |
| Deposited Data | ||
| Ribosome profiling of cancer cell lines with RocA treatment, raw and processed data | This study | GEO: GSE148636 |
| RNA-Bind-n-Seq of eIF4A1 and DDX3X with RocA treatments, raw and processed data | This study | GEO: GSE150111 |
| Aglaia RNA-Seq, raw data | Iwasaki et al. 2019 | SRA: SRR5947159 |
| Human eIF4A1•AMPPNP•RocA•(AG)5 complex structure | Iwasaki et al. 2019 | PDB: 5ZC9 |
| Human DDX3X core•AMPPNP structure | Floor et al. 2016 | PDB: 5E7M |
| Original images used for the figures | This study | Mendeley Data: doi: 10.17632/syfc38x8md.1 |
| Experimental Models: Cell Lines | ||
| T-Rex-293 (HEK) | Thermo Fisher Scientific | Cat#R71007 |
| T-REx293 SBP-eIF4A1 (Phe163Leu-Ile199Met) eIF4A1SINI | Iwasaki et al. 2019 | N/A |
| Hs 936.T | American Type Culture Collection (ATCC) | Cat#CRL-7687 |
| A375 | American Type Culture Collection (ATCC) | Cat#CRL-1619 |
| NCI-H1650 | American Type Culture Collection (ATCC) | Cat#CRL-5883 |
| NCI-H520 | American Type Culture Collection (ATCC) | Cat#HTB-182 |
| Oligonucleotides | ||
| FAM labeled RNA (AG)10: FAM-AGAGAGAGAG AGAGAGAGAG | Hokkaido System Science, Iwasaki et al. 2019 | N/A |
| FAM labeled RNA (UC)10:FAM-UCUCUCUCUCUCUCUCUCUC | Hokkaido System Science, This paper | N/A |
| (AG)5A: AGAGAGAGAGA | Fasmac, This paper | N/A |
| DDX3X-specific siRNA | Dharmacon | Cat# L-006874-02-0005 |
| eIF4A2-specific siRNA | Dharmacon | Cat# L-013758-01-0005 |
| control siRNA | Dharmacon | Cat# D-001206-13-05 |
| CTCTTTCCCTACACGACGCTCTTCCGATCT-N30-ATCGTAGATCGGAAGAGCACACGTCTGAA (letters in bold represent the DNA sequence and N represents random the RNA sequence) | Gene Design | N/A |
| Recombinant DNA | ||
| pColdI-DDX3X helicase core WT | This study | N/A |
| pColdI-DDX3X helicase core Pro324Thr | This study | N/A |
| pColdI-DDX3X helicase core Val328Leu | This study | N/A |
| pColdI-DDX3X helicase core Gln360Pro | This study | N/A |
| pColdI-DDX3X helicase core Gln360Leu | This study | N/A |
| pColdI-DDX3X helicase core Arg363Asn | This study | N/A |
| pColdI-eIF4A1 WT | Iwasaki et al. 2019 | N/A |
| pColdI-eIF4A1 Phe163Leu-Ile199Met | Iwasaki et al. 2019 | N/A |
| pColdI-eIF4A2 WT | This study | N/A |
| pColdI-SBP-eIF4A1 | This study | N/A |
| pColdI-SBP-DDX3X helicase core | This study | N/A |
| psiCHECK2-CAA repeats | Iwasaki et al. 2016 | N/A |
| psiCHECK2-7×AGAGAG motifs | Iwasaki et al. 2016 | N/A |
| Software and Algorithms | ||
| Igor Pro Version: 8.01 | WaveMetrics | https://www.wavemetrics.com/products/igorpro |
| MASCOT server version: 2.6 | Matrix Science | https://www.matrixscience.com/ |
| Proteome Discoverer version: 1.3 | Thermo Fisher Scientific | N/A |
| Xcalibur software version: 2.1 or 4.1 | Thermo Fisher Scientific | N/A |
| Other | ||
| NGC chromatography system | Bio-Rad | N/A |
| GloMax Navigator | Promega | Cat#GM2010 |
| Image Pharos FX | Bio-Rad | N/A |
| ODYSSEY CLx | LI-COR Biosciences | N/A |
| LTQ Orbitrap XL mass spectrometer | Thermo Fisher Scientific | N/A |
| nanoESI source | Nikkyo Technos | N/A |
| UltiMate 3000 nano LC system | Thermo Fisher Scientific | N/A |
| Infinite F-200 PRO | TECAN | N/A |
| Monolith NT.115 | NanoTemper | N/A |
Significance.
Rocaglamide A (RocA) is a small molecule translation inhibitor which attracts great interest as lead anticancer compounds due to their ability to selectively kill tumor. Here, in addition to the known target of eIF4A, we identified an alternative RocA target DDX3. The effect of RocA on DDX3 is mRNA-selective; RocA clamps DDX3 to polypurine RNA in an ATP-independent manner and represses translation from selective mRNAs. As a natural source of RocA, plant Aglaia harbors the critical amino acid substitution at Gln360 of DDX3 for RocA resistance. Because the RocA•target protein (eIF4A or DDX3) complex shows dominant-negative effects, tumor cells overexpressing eIF4A and DDX3 proteins are susceptible to RocA-induced translational repression and cell growth inhibition. Our discovery of the full spectrum of RocA target proteins will allow prediction of its efficacy in cancer cells based on the target abundance.
Highlights.
RocA clamps DDX3 on polypurine sequences in an ATP-independent manner
Gln360 in DDX3 is an critical residue for RocA binding
RocA•DDX3 complex inhibits protein synthesis in a dominant-negative manner
eIF4A and DDX3 abundance correlates with RocA sensitivity in cancer cells
Acknowledgments
We thank all the members of the Iwasaki laboratory for their constructive discussions, technical help, and critical reading of the manuscript. We are also grateful to the Support Unit for Bio-Material Analysis, RIKEN CBS Research Resources Division, for mass spectrometry analysis. S.I. was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) with a Grant-in-Aid for Scientific Research on Innovative Areas in the “Nascent Chain Biology” category (JP17H05679), the Japan Society for the Promotion of Science (JSPS) with a Grant-in-Aid for Young Scientists (A) (JP17H04998), a Challenging Research (Exploratory) (JP19K22406), and the Grant-in-Aid for Transformative Research Areas (B) (JP20H05784), the RIKEN "Cellular Evolution" Pioneering Projects and Aging Project, AMED-CREST, AMED (20gm1410001), and the Takeda Science Foundation. T.I. was supported by MEXT with a Grants-in-Aid for Scientific Research on Innovative Areas in the “Nascent Chain Biology” category (JP15H01548 and JP17H05677), JSPS with a Grant-in-Aid for Scientific Research (B) (JP16H04756), the RIKEN “Dynamic Structural Biology” Pioneering Project and Aging Project, and AMED-CREST, AMED (20gm1410001). M.S. was supported by RIKEN (the “Chemical Probe” Pioneering Project), AMED-CREST, AMED (JP18gm0710004). K.D. was supported by JSPS with a Grant-in-Aid for Scientific Research (C) (JP19K05747) and the Konica Minolta Science and Technology Foundation. M.A. was supported by JSPS with a Grant-in-Aid for Scientific Research (C) (JP17K01956). Y.S. was supported by JSPS with a Grant-in-Aid for JSPS Fellows (JP19J00920). This work was also supported by the Platform Project for Supporting Drug Discovery and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] (JP18am0101082 and JP18am0101113) and Molecular Profiling Committee of a Grant-in-Aid for Scientific Research on Innovative Areas “Advanced Animal Model Support (AdAMS)” (MEXT, JP16H06276). The DNA libraries were sequenced by the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley and were supported by an NIH S10 OD018174 Instrumentation Grant. Computation was supported by Manabu Ishii, Itoshi Nikaido, the Bioinformatics Analysis Environment Service, and supercomputer HOKUSAI SailingShip in ACCC RIKEN. M.C. was an International Program Associate of RIKEN. Y.S. was a recipient of a JSPS Research Fellowship (PD) and a participant of the Special Postdoctoral Researchers Program of RIKEN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, and Garraway LA (2012). The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, and Huala E (2015). The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA, and Pelletier J (2008). Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 118, 2651–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Venkataramanan S, Rogowski KJ, Wyler E, Tejura M, Thai B, Krol J, Filipowicz W, Landthaler M, and Floor SN (2020). DDX3 depletion selectively represses translation of structured mRNAs. BioRxiv doi: 10.1101/589218, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Carrier M, Galicia-Vázquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, Porco JA, and Pelletier J (2009). Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 4, e5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JM, Lindqvist LM, Savage GP, and Rizzacasa MA (2016). Total synthesis of a biotinylated rocaglate: Selective targeting of the translation factors eIF4AI/II. Bioorg Med Chem Lett 26, 262–264. [DOI] [PubMed] [Google Scholar]
- Chambers JM, Lindqvist LM, Webb A, Huang DC, Savage GP, and Rizzacasa MA (2013). Synthesis of biotinylated episilvestrol: Highly selective targeting of the translation factors eIF4AI/II. Org Lett 15, 1406–1409. [DOI] [PubMed] [Google Scholar]
- Chan K, Robert F, Oertlin C, Kapeller-Libermann D, Avizonis D, Gutierrez J, Handly-Santana A, Doubrovin M, Park J, Schoepfer C, Da Silva B, Yao M, Gorton F, Shi J, Thomas CJ, Brown LE, Porco JA, Pollak M, Larsson O, Pelletier J, and Chio IIC (2019). eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat Commun 10, 5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Galicia-Vázquez G, Cencic R, Mills JR, Katigbak A, Porco JA, and Pelletier J (2016). CRISPR-mediated drug-target validation reveals selective pharmacological inhibition of the RNA helicase, eIF4A. Cell Rep 15, 2340–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Zhang W, Cencic R, Devine WG, Beglov D, Henkel T, Brown LE, Vajda S, Porco JA, and Pelletier J (2019). Amidino-rocaglates: a potent class of eIF4A inhibitors. Cell Chem Biol 26, 1586–1593.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Zhang W, Cencic R, O’Connor PBF, Robert F, Devine WG, Selznick A, Henkel T, Merrick WC, Brown LE, Baranov PV, Porco JA, and Pelletier J (2020). Rocaglates induce gain-of-function alterations to eIF4A and eIF4F. Cell Rep 30, 2481–2488.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JT, Thompson PA, Nilewski C, Sprengeler PA, Sperry S, Packard G, Michels T, Xiang A, Tran C, Wegerski CJ, Eam B, Young NP, Fish S, Chen J, Howard H, Staunton J, Molter J, Clarine J, Nevarez A, Chiang GG, Appleman JR, Webster KR, and Reich SH (2020). Design of development candidate eFT226, a first in class inhibitor of eukaryotic initiation factor 4A RNA helicase. J Med Chem 63, 5879–5955. [DOI] [PubMed] [Google Scholar]
- Floor SN, Condon KJ, Sharma D, Jankowsky E, and Doudna JA (2016). Autoinhibitory interdomain interactions and subfamily-specific extensions redefine the catalytic core of the human DEAD-box protein DDX3. J Biol Chem 291, 2412–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, and Jankowsky E (2016). Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. Elife 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang D, Yang Y, Wang X, Zhao X, Zhang P, Zhu H, Xu N, and Liang S (2018). A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (Review). Oncol Rep 39, 883–892. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, and Ingolia NT (2016). Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534, 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, and Ingolia NT (2017). The growing toolbox for protein synthesis studies. Trends Biochem Sci 42, 612–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Iwasaki W, Takahashi M, Sakamoto A, Watanabe C, Shichino Y, Floor SN, Fujiwara K, Mito M, Dodo K, Sodeoka M, Imataka H, Honma T, Fukuzawa K, Ito T, and Ingolia NT (2019). The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol Cell 73, 738–748.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, and Penin AA (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88, 1058–1070. [DOI] [PubMed] [Google Scholar]
- Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, Degenhardt J, Mayba O, Gnad F, Liu J, Pau G, Reeder J, Cao Y, Mukhyala K, Selvaraj SK, Yu M, Zynda GJ, Brauer MJ, Wu TD, Gentleman RC, Manning G, Yauch RL, Bourgon R, Stokoe D, Modrusan Z, Neve RM, de Sauvage FJ, Settleman J, Seshagiri S, and Zhang Z (2015). A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 33, 306–312. [DOI] [PubMed] [Google Scholar]
- Kotov AA, Olenkina OM, Godneeva BK, Adashev VE, and Olenina LV (2017). Progress in understanding the molecular functions of DDX3Y (DBY) in male germ cell development and maintenance. Biosci Trends 11, 46–53. [DOI] [PubMed] [Google Scholar]
- Ku YC, Lai MH, Lo CC, Cheng YC, Qiu JT, Tarn WY, and Lai MC (2019). DDX3 participates in translational control of inflammation induced by infections and injuries. Mol Cell Biol 39, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, and Mann M (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11, 319–324. [DOI] [PubMed] [Google Scholar]
- Lai MC, Chang WC, Shieh SY, and Tarn WY (2010). DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol 30, 5444–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lee YH, and Tarn WY (2008). The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19, 3847–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, and Burge CB (2014). RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell 54, 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, and Reed R (2008). Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36, 4708–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, and Jankowsky E (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12, 505–516. [DOI] [PubMed] [Google Scholar]
- Lu WT, Wilczynska A, Smith E, and Bushell M (2014). The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans 42, 166–172. [DOI] [PubMed] [Google Scholar]
- Manier S, Huynh D, Shen YJ, Zhou J, Yusufzai T, Salem KZ, Ebright RY, Shi J, Park J, Glavey SV, Devine WG, Liu CJ, Leleu X, Quesnel B, Roche-Lestienne C, Snyder JK, Brown LE, Gray N, Bradner J, Whitesell L, Porco JA, and Ghobrial IM (2017). Inhibiting the oncogenic translation program is an effective therapeutic strategy in multiple myeloma. Sci Transl Med 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A (2017). Small molecules against RNA targets attract big backers. Nat Rev Drug Discov 16, 813–815. [DOI] [PubMed] [Google Scholar]
- Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, and Fanidi A (2014). Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 15, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H, Galicia-Vazquez G, Paris CG, Aust T, Bhullar B, Chang L, Helliwell SB, Hoepfner D, Knapp B, Riedl R, Roggo S, Schuierer S, Studer C, Porco JA, Pelletier J, and Movva NR (2013). Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem Biol 8, 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Mendillo ML, Tang YC, Subramanian A, Perley CC, Roche SP, Wong B, Narayan R, Kwon H, Koeva M, Amon A, Golub TR, Porco JAJ, Whitesell L, and Lindquist S (2013). Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341, 1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, and Preiss T (2018). Translation initiation by cap-dependent ribosome recruitment: Recent insights and open questions. Wiley Interdiscip Rev RNA 9, e1473. [DOI] [PubMed] [Google Scholar]
- Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Décimo D, and Ohlmann T (2012). DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 31, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, and Duhr S (2010). Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1, 100. [DOI] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, Schatz JH, Rodrigo CM, Zhao C, Rondou P, de Stanchina E, Teruya-Feldstein J, Kelliher MA, Speleman F, Porco JA, Pelletier J, Rätsch G, and Wendel HG (2014). RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 513, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Asanuma M, Nakanishi S, Saito Y, Okazaki M, Dodo K, and Sodeoka M (2014). Turn-ON fluorescent affinity labeling using a small bifunctional O-nitrobenzoxadiazole unit. Chem Sci 5, 1021–1029. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results of ribosome profiling (GEO: GSE148636) and RNA-Bind-n-Seq (GEO: GSE150111) obtained in this study have been deposited in the National Center for Biotechnology Information (NCBI) database. All custom scripts are available upon request. Original images used for the figures are deposited in the Mendeley database (http://dx.doi.org/10.17632/syfc38x8md.1).








